Abstract
The emergence of the threat of radiological terrorism and other radiological incidents has led to the need for development of fast, accurate and noninvasive methods for detection of radiation exposure. The purpose of this study was to extend radiation metabolomic biomarker discovery to humans, as previous studies have focused on mice. Urine was collected from patients undergoing total body irradiation at Memorial Sloan-Kettering Cancer Center prior to hematopoietic stem cell transplantation at 4–6 h postirradiation (a single dose of 1.25 Gy) and 24 h (three fractions of 1.25 Gy each). Global metabolomic profiling was obtained through analysis with ultra performance liquid chromatography coupled to time-of-flight mass spectrometry (TOFMS). Prior to further analyses, each sample was normalized to its respective creatinine level. Statistical analysis was conducted by the nonparametric Kolmogorov-Smirnov test and the Fisher’s exact test and markers were validated against pure standards. Seven markers showed distinct differences between pre- and post-exposure samples. Of those, trimethyl-l-lysine and the carnitine conjugates acetylcarnitine, decanoylcarnitine and octanoylcarnitine play an important role in the transportation of fatty acids across mitochondria for subsequent fatty acid β-oxidation. The remaining metabolites, hypoxanthine, xanthine and uric acid are the final products of the purine catabolism pathway, and high levels of excretion have been associated with increased oxidative stress and radiation induced DNA damage. Further analysis revealed sex differences in the patterns of excretion of the markers, demonstrating that generation of a sex-specific metabolomic signature will be informative and can provide a quick and reliable assessment of individuals in a radiological scenario. This is the first radiation metabolomics study in human urine laying the foundation for the use of metabolomics in biodosimetry and providing confidence in biomarker identification based on the overlap between animal models and humans.
INTRODUCTION
Radiological incidents, accidental exposures and terrorism have been on the forefront of the news captivating the interest of the public in the past few years. In addition to these events, the potential of a long-duration trip to Mars by astronauts and their unavoidable exposure to high levels of cosmic radiation (1), has made pertinent the development of new methods of rapid and efficient biological dosimetry, not only to be utilized for counter-measures purposes, but also to aid in estimating future risks of radiation-induced malignancy. Current methods to identify exposed individuals rely on the acute radiation syndrome (ARS) treatment guidelines (2) [which is often complicated with combined injuries such as burns and infections (3, 4)], on lymphocyte depletion and the total number of dicentric chromosomes (5). Electron spin resonance dosimetry of teeth has also been utilized providing results comparable to cytogenetics (6). Critical treatment of radiation-exposed individuals may sometimes be delayed due to lack of clinicians’ sufficiently trained to identify radiation victims and the complication of psychological trauma that can manifest in psychosomatic symptoms similar to ARS (7), leading to false identification of victims. The use of hematopoietic stem cell transplantation (HSCT) for treatment of the radiation effects has been utilized in the past, however allogeneic stem cells have been used with limited success and are currently mainly limited to treatment of leukemic patients (8). Banking of autologous peripheral blood stem cells, at least for radiation workers, has been proposed, however ethical and scientific considerations have limited the feasibility (8). Biological dosimetry is further complicated by at risk populations, whether that is based on age, sex or genetic background. The current reference for dose limits and risk determination is based on the “Reference Man” by the International Commission on Radiological Protection (ICRP) (9), even though females, children, immunocompromised individuals and individuals with specific genetic mutations (i.e., ataxia telangiectasia mutated, Nijmegen breakage syndrome, Bloom Syndrome) have an increased risk for radiation-induced cancers and in some cases morbidity compared to this ICRP reference standard (9–12).
Biological dosimetry efforts have primarily concentrated on animal and tissue culture models providing significant understanding of radiation estimates and cancer risks. Human studies have mainly focused on the atomic bomb survivors, the Chernobyl survivors and to a lesser extend exposed individuals of the Fukushima accident. The identification of new markers of radiation exposure, distinct from the classical and laborious cytogenetic methods, are clearly a priority. These markers may be utilized as a first assessment of exposed individuals, which can be followed by cytogenetic analysis for refinement of the dose and lifetime cancer risk assessment. New methods focusing on rapid identification of biological measurements of pre-selected markers in human samples have provided confidence that fast and reliable identification of exposed individuals is possible. Identification has concentrated on gene expression with high levels of classification accuracy (13), miRNA levels (14) and γ-H2AX foci (15). The evolution of other omics technologies, i.e., proteomics and metabolomics, has additionally provided a number of radiation related markers in animal models (4, 16–20).
Metabolomics refers to the identification and quantification of the total small molecule content (<1 kDa) in tissues, cells and easily accessible biofluids such as urine, blood and saliva. A comprehensive review by Coy et al. (21) on radiation metabolomics provides an insight in the potential for biological dosimetry. Although the majority of the studies have concentrated on biofluids from animal models (nonhuman primates, rats and mice) and cell cultures (22), the overlap of radiation markers in different species has provided confidence of the use of animal models in radiation research. In this report, we utilized the power of metabolomics to identify biomarkers in the urine of total body irradiated humans (1.25 Gy) undergoing hematopoietic stem cell transplantation (HSCT) as part of their cancer treatment. The study revealed important perturbations in pathways that have been previously identified in other species. Additionally, important differences in biomarker excretion levels were found to be gender dependent. Separate biomarker signatures are therefore needed for males and females to be utilized for the rapid identification of radiation exposure victims.
MATERIALS AND METHODS
Subjects
Patients at Memorial Sloan-Kettering Cancer Center undergoing total body irradiation (TBI) prior to HSCT were recruited for prospective participation in this study (no. 07–158), which was approved by the institutional review board. After giving informed consent for participation, subjects underwent biospecimen (urine) collection at three time points during fractionated TBI. The first time point was the day of the first fraction of total body irradiation, prior to treatment. The second time point was 4–6 h after irradiation with 1.25 Gy. The third time point was 24 h after the first specimen was collected, after a total of 3 fractions of 1.25 Gy were delivered with a 4–6 h interfraction interval (3.75 Gy total). As part of inpatient clinical management, combined blood count (CBC) with differential and metabolic function was assessed by routine blood sampling every 24 h. TBI was performed using a linear accelerator producing 15 MV photons, with patients standing upright using parallel-opposed anteroposterior beams, with beam compensators and lung shielding. Dose rate was ~0.1 Gy/min. Partial lung shielding and supplemental electron fields were used to limit total lung dose to 8 Gy. A total of 28 patients (17 male and 11 female), diagnosed with diseases including acute myelogenous leukemia, chronic myelogenous leukemia, acute lymphocytic leukemia, non-Hodgkin’s lymphoma, myelodysplastic syndrome and essential thrombocytosis participated in the study (Supplementary Table S1; http://dx.doi.org/10.1667/RR13567.1.S1). All patients were in remission and none had chemotherapy treatment for at least one month prior to the start of the radiation regimen.
Chemicals
All chemicals were of the highest purity available and reagents used were of LC-MS grade. Debrisoquine sulfate, 4-nitrobenzoic acid (4-NBA), creatinine, uric acid, hypoxanthine, xanthine and Nε,Nε,Nε-Trimethyl-l-lysine hydrochloride (Trimethyl-l-lysine or TML) were obtained from Sigma-Aldrich (St. Louis, MO). (±)-Decanoylcarnitine chloride (Decanoylcarnitine) and (±)-Octanoylcarnitine chloride (Octanoylcarnitine) were obtained from Tocris Bioscience (Ellisville, MO). O-Acetyl-l-carnitine hydrochloride (Acetylcarnitine) was obtained from Acros Organics (Fair Lawn, NJ). Creatinine-d3 was obtained from CDN Isotopes (Quebec, Canada).
Sample Preparation and Urine Profiling
The urine samples were prepared and analyzed as previously described (4), although the urine samples were diluted 1:1 in 50:50 acetonitrile:water with internal standards (4 µM debrisoquine [M+H]+ = 176.1188 and 30 µM 4-NBA [M–H]− = 166.0141). Samples were analyzed using an ultra performance liquid chromatography (UPLC) coupled to a Waters Premier® Time-of-Flight mass spectrometer (TOFMS) (Waters, Milford, MA), operating on positive (ESI+) and negative (ESI−) electrospray ionization modes. A Waters Acquity UPLC BEH C18 1.7 µm, 2.1 × 50 mm column was used at 40°C. Liquid chromatography and mass spectrometry conditions were set as previously described (4). Accurate mass was obtained with intermittent injections of sulfadimethoxine ([M+H]+ = 311.0814 and [M–H]− = 309.0658) in 50:50 acetonitrile:water at a concentration of 300 pg/ml introduced as Lock-Spray®.
Data Processing and Statistical Analysis
Chromatographic data were deconvoluted and peak aligned through MarkerLynx software (Waters, Milford, MA). Urine data from each patient were normalized to their respective creatinine ([M+H]+ = 114.0667, retention time 0.32 min) to control for glomerular filtration rate changes. Statistical analysis was conducted by in-house developed software written in Python and the R statistical computing language. When comparing data from pre-exposure vs. post-exposure TBI samples, ions with nonzero abundance values in at least 75% of samples in both groups were first identified (referred to as complete-presence ions). Data from these ions were log transformed and analyzed for statistical significance by the nonparametric Kolmogorov-Smirnov test (P < 0.05). Statistical significance for ions with nonzero abundance values in at least 75% of the samples in only one group (partial-presence ions) was analyzed categorically for presence status (i.e., nonzero abundance) by Fisher’s exact test (P < 0.05). A total of 50 and 134 ions were found to be statistically significant in ESI− and ESI+, respectively. The log-transformed data for statistically significant complete-presence ions was then utilized for principal component analysis (PCA) by singular value decomposition after zero centering and unit variance scaling.
Putative Identification and Metabolic Pathway Analysis
Both statistically significant complete-presence and partial-presence ions were putatively identified utilizing the Human Metabolome Database (HMDB) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Ions were identified by elucidating their neutral mass from the list of possible adducts, which included adducts formed by the addition of H or Na for positive mode and the removal of H for negative mode. These neutral masses were then compared to the monoisotopic mass of small molecules in the HMDB and KEGG compound databases, from which putative metabolites were identified (with a mass error of 20 ppm or less). KEGG annotated pathways associated with these putative metabolites were also identified and aggregated, from which a histogram of top pathways by number of putative metabolite hits was generated.
Validation of Putative Metabolites and Quantification
Verification of the identity of the putative metabolites was performed through tandem mass spectrometry (MS/MS) on the UPLC-TOFMS. Chemicals were diluted in 50:50 acetonitrile:water and fragmented with ramping collision energy between 5 and 70 eV. Fragmentation patterns of the pure chemicals were compared to the fragmentation pattern of putative metabolites in urine samples. Quantification of the validated metabolites was performed by UPLC coupled to the tandem quadrupole mass spectrometer Xevo-TQ (Waters). Two different columns were utilized to account for different chromatographic conditions, a C18 column, as previously mentioned and a Waters Acquity UPLC BEH amide column, 1.7 µm, 2.1 × 150 mm. When the C18 column was used, gradient elution was performed with mobile phase A (0.1% formic acid in water) and B (0.1% formic acid in acetonitrile) at 0.5 mL/min flow rate and at 40°C. The initial composition (2% B) was maintained for 0.5 min, B was increased to 98% over 5.5 min. It was then held for 2 min and returned to initial conditions over 0.5 min followed by 1 min equilibration yielding a total run time of 9.5 min. With the amide column, gradient elution was performed with mobile phase A (95:5 acetonitrile:water, 10 mM ammonium formate pH = 5) and B (50:50 acetonitrile:water, 10 mM ammonium formate pH = 5) at 0.5 mL/min flow rate and at 40°C. The initial composition (0.1% B) was maintained for 0.5 min, B was increased to 58% over 5.5 min, held for 2 min, increased again to 100% and held for 2 min. Return to initial condition was achieved over 1 min followed by 2 min equilibration yielding a total run time of 16 min. Pure chemicals and urine samples were diluted (1:1, 1:5 or 1:100) with either 75–80 % acetonitrile (amide column) or 50 % acetonitrile (C18 column). Two selected reaction monitoring (SRM) transitions were monitored for each internal standards and putative metabolites. Internal standards included debrisoquine (ESI+, quantification transition m/z 176→134), 4-NBA (ESI−, m/z 166→122) and creatinine-d3 (ESI+, m/z 117→47). Hypoxanthine (ESI+, m/z 137→119, internal standards debrisoquine), trimethyl-l-lysine (TML) (ESI+, m/z 189→84, internal standards debrisoquine), xanthine (ESI−, m/z 151→108, internal standards 4-NBA), uric acid (ESI−, m/z 167→124, internal standards 4-NBA) and creatinine (ESI+, m/z 114→44, internal standards creatinine-d3) were analyzed on the amide column. Decanoylcarnitine (ESI+, m/z 316→85, internal standards debrisoquine), octanoylcarnitine (ESI+, m/z 288→85, internal standards debrisoquine), and acetylcarnitine (ESI+, m/z 204→85, internal standards debrisoquine) were analyzed on the C18 column. All standards and urine samples were run in duplicate. Concentrations of the metabolites were calculated with TargetLynx software (Waters) and each metabolite in every sample was normalized to its respective quantified creatinine.
Statistical Analysis of Quantified Data
Whereas a robust nonparametric Kolmogorov-Smirnov (K-S) test was utilized in the discovery phase, a parametric Welch’s t test was used instead in the validation phase due to its greater statistical power. Statistical testing was performed after filtering for outliers utilizing a 1.5 interquartile range (IQR), as well as testing for normality by the Anderson-Darling test at the 10% significance level. Markers that did not pass the Anderson-Darling test for normality were instead analyzed with the K-S test for verification of statistical significance. Markers with a P < 0.05 were considered statistically significant.
RESULTS
Patients who participated in the study had a median age of 44 years and 44% were women. All patients underwent HSCT for leukemia or lymphoma in remission, with the exception of three patients who underwent HSCT for aplastic anemia, myelodysplastic syndrome and essential thrombocytosis, respectively. No subjects had received genotoxic chemotherapy within 4 weeks of starting TBI. Two subjects had received radiotherapy prior to TBI, but had completed this at least 2 weeks prior to starting TBI (Supplementary Table S1; http://dx.doi.org/RR13567.1.S1). Hematopoietic and metabolic parameters were monitored before TBI and 24 h after TBI (Table 1). Consistent with the known effects of TBI, all hematopoietic elements except neutrophil count decreased significantly after TBI. The effect was most pronounced in the most radiosensitive blood elements, lymphocytes and eosinophils. No significant change in blood metabolic indices was noted, except for the expected decrease in blood urea nitrogen and increase in creatinine. Increased creatinine excretion was also identified in the urine at 24 h after TBI, whereas creatinine excretion did not significantly change at the 6 h time point (Supplementary Fig. S1; http://dx.doi.org/RR13567.1.S2). Given the different levels of creatinine in the urine and that the 24 h time point is a result of three fractions of 1.25 Gy (Supplementary Fig. S1; http://dx.doi.org/RR13567.1.S2), biomarker analysis concentrated on the 6 h time point after each sample was normalized to its respective creatinine level.
TABLE 1.
Human Values
| Pre-TBI | Post-TBI | ||||||
|---|---|---|---|---|---|---|---|
| Group mean (n = 24) |
Group standard deviation (n = 24) |
Group mean (n = 24) |
Group standard deviation (n = 24) |
Change after TBI |
Pre-TBI/ Post-TBI |
P value for dependent t test |
|
| WBC (k/mcl) | 4.8 | 4.11 | 4.24 | 3.44 | ↓ | 1.14* | 0.0153 |
| RBC (M/mcl) | 3.4 | 0.64 | 3.26 | 0.66 | ↓ | 1.05* | 0.0003 |
| Hg (g/dl) | 10.6 | 1.59 | 10.13 | 1.60 | ↓ | 1.05* | 0.0001 |
| Hct (%) | 31.5 | 5.00 | 30.00 | 4.92 | ↓ | 1.05* | 0.0002 |
| Plt (10k/mcl) | 18.4 | 16.95 | 16.71 | 14.38 | ↓ | 1.10* | 0.0218 |
| ANC (k/mcl) | 3.3 | 3.74 | 3.37 | 3.24 | 0.98 | 0.8216 | |
| ALC (k/mcl) | 0.9 | 0.48 | 0.39 | 0.24 | ↓↓ | 2.31* | 0.0000 |
| AMC (k/mcl) | 0.4 | 0.45 | 0.31 | 0.29 | ↓ | 1.15* | 0.0346 |
| AEC (k/mcl) | 0.1 | 0.14 | 0.06 | 0.04 | ↓↓ | 2.34* | 0.0060 |
| ABC (k/mcl) | 0.0 | 0.10 | 0.01 | 0.07 | ↓ | 1.89* | 0.0121 |
| Na | 140.5 | 2.21 | 140.43 | 2.02 | 1.00 | 0.8481 | |
| K | 3.8 | 0.34 | 3.65 | 0.35 | 1.03 | 0.1078 | |
| Cl | 106.2 | 2.48 | 106.57 | 2.73 | 1.00 | 0.3458 | |
| CO2 | 26.3 | 2.85 | 25.83 | 1.97 | 1.02 | 0.2830 | |
| BUN | 7.9 | 4.20 | 6.30 | 2.87 | ↓ | 1.26* | 0.0214 |
| Cr | 1.1 | 0.00 | 1.19 | 0.00 | ↑ | 0.91* | 0.0040 |
| Ca | 12.2 | 0.01 | 11.63 | 0.03 | ↓ | 1.05 | 0.2034 |
| Glu | 136.9 | 0.62 | 139.70 | 0.54 | 0.98 | 0.3602 | |
| Mg | 3.6 | 0.09 | 2.44 | 0.00 | ↓ | 1.48 | 0.3182 |
| Ph | 5.7 | 0.02 | 5.92 | 0.02 | 0.96 | 0.2447 | |
Signifies statistical significance with a P value of <0.05.
Notes. Hematopoietic and metabolic parameters were monitored pre-exposure and 24 h after total body irradiation in blood. WBC, white blood cell count; RBC, red blood cell count; Hg, hemoglobin; Hct, hematocrit; Plt, platelet count; ANC, absolute neutrophil count; ALC, absolute lymphocyte count; AMC, absolute monocyte count; AEC, absolute eosinophil count; ABC, absolute basophil count; Na, sodium; K, potassium; Cl, chloride; CO2, carbon dioxide; BUN, blood urea nitrogen; Cr, creatinine; Ca, calcium; Glu, glucose; Mg, magnesium; Ph, phosphorous.
Statistical Analysis and Marker Validation
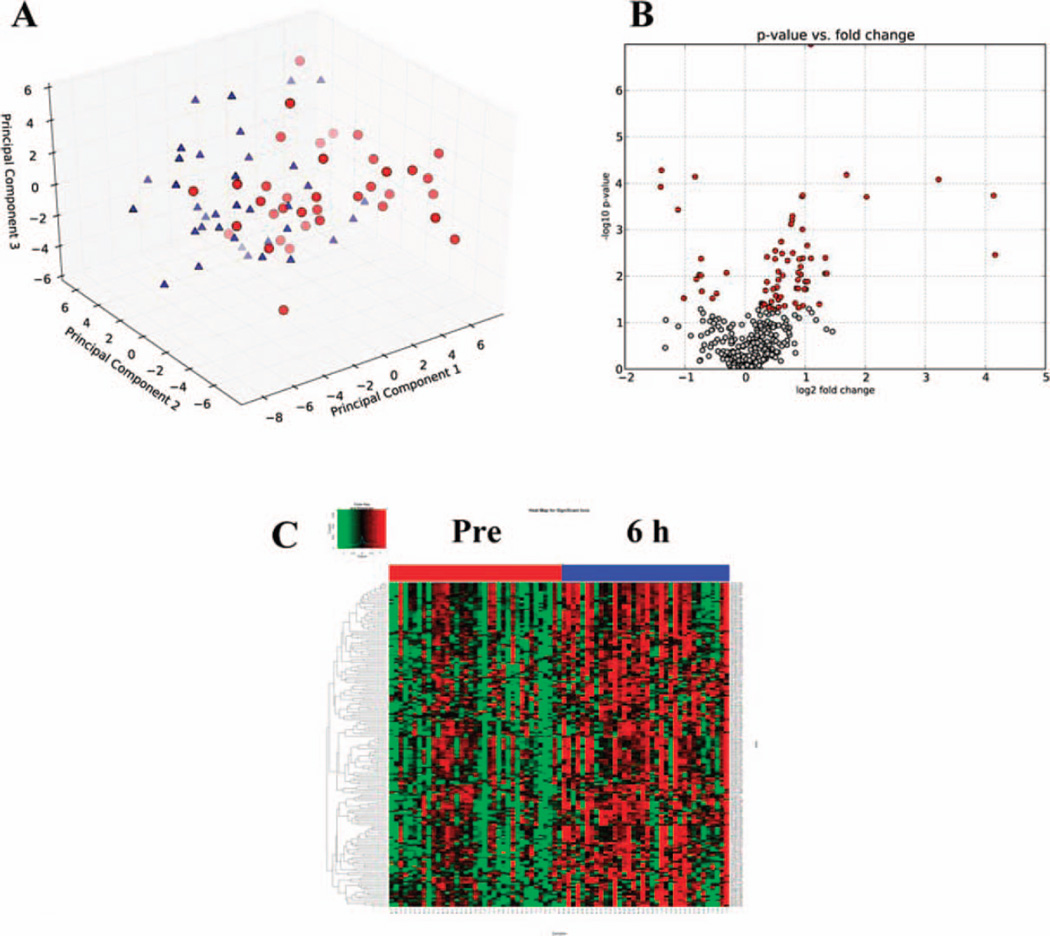
After extraction of abundance values from the raw chromatographic data with MarkerLynx and normalization to creatinine levels for each sample, markers were identified through univariate data analysis with the use of in-house software (23). Through the nonparametric K-S statistical hypothesis test (P < 0.05), a total of 176 ESI− ions and 74 ESI+ ions were found to be statistically significant. The statistically significant ESI+ ions were utilized to construct a PCA scores plot (Fig. 1A), depicting two clusters based on their differential metabolomic profiles. Additionally, a volcano plot was constructed comparing the log of the significance on the y-axis (P value) versus the log of the fold change on the x-axis (Fig. 1B). The volcano plot revealed that metabolic profiles shifted towards an increased excretion pattern (right side of the volcano plot demonstrated by red points) after exposure to 1.25 Gy. Additionally, construction of a heatmap based on hierarchical clustering revealed the individual responses of each patient per treatment/time point for each statistically significant ion (Fig. 1C). Mapping of the putative identities of the statistically significant ions was conducted through the KEGG database to identify possible metabolic pathway perturbations (Supplementary Fig. S2; http://dx.doi.org/RR13567.1.S2) and clues into the effects of radiation in metabolism. Similar statistical analysis was conducted on the ESI− ions identified through the Kolmogorov-Smirnov test for a total of 176 ions (data not shown). The Fisher’s exact test identified 134 ESI+ ions and 50 ESI− with a P < 0.05. Although the primary analysis of this study was concentrated on the 6 h time point, comparable analysis of the 24 h point revealed a more complex metabolic profile, as depicted in Supplementary Fig. S3 (http://dx.doi.org/RR13567.1.S2). The PCA plot showed a less distinct separation of the two groups (pre-exposure and 24 h post-exposure), while the volcano plot and the heatmap revealed distinct differences. However, given that normalization to increased creatinine levels was not a reliable method for this time point, the biomarker levels could produce spurious results, therefore further analysis was not pursued at this point.
FIG. 1.
Principal component analysis (PCA) scores plot with first three components of pre-exposure (red circles) vs. 6 h post-exposure (blue triangles) after total body irradiation (TBI) urine data ESI+ (panel A). Data utilized for the PCA included only statistically significant ions (pre-exposure vs. 6 h post-exposure) through the Kolmogorov-Smirnov test (>75% presence in both groups with P < 0.05). A volcano plot (panel B) and a heatmap (panel C) demonstrate the differences in metabolic profiles between the pre- and post-exposure (6 h) groups. In panel B, the x-axis plots the log2 of the fold change between the two conditions, whereas the y-axis plots the negative log10 of the P value. Red circles signify the statistically significant ions (P < 0.05), showing a more pronounced increased excretion post TBI (right of the plot). A heatmap of the statistically significant ions, as identified through the Kolmogorov-Smirnov test, illustrates the differences in levels of metabolite excretion in the urine after exposure to 1.25 Gy.
For the pre-exposure and 6 h post-exposure comparison, 23 ions were selected for validation based on their biological significance through putative identification from online databases. Validation through tandem mass spectrometry (MS/MS) was successful for seven markers, as listed in Table 2. All ppm errors between predicted and found were below 10. Four markers were either a precursor or a variant of carnitine. In particular, ions [M+Na]+ = 310.2017 with retention time (ret) 5.11 min, [M+H]+ = 189.1586 ret 0.26 min, [M+H]+ = 204.1243 ret 0.36 min and [M+Na]+ = 338.2332 ret 5.43 min were validated as octanoylcarnitine, TML, acetylcarnitine, and decanoylcarnitine, respectively. Ions [M+H]− = 137.0457 ret 0.35 min, [M-H]− = 167.0202 ret 0.34 min, and [M-H]− = 151.0246 ret 0.34 min were validated as hypoxanthine, uric acid and xanthine, respectively. The remaining sixteen statistically significant ions (Supplementary Table S2; http://dx.doi.org/RR13567.1.S1) were not validated and their identity remains unknown at this point. In some cases, some of the most interesting putative markers do not have a chemical that is commercially available and therefore it could not be easily obtained for MS/MS.
TABLE 2.
Identification of Urinary Biomarkers at ~6 h after Irradiation with 1.25 Gy
| Marker no. |
Retention time (min) |
ESI mode | Mass (m/z) found |
Predicted | ppm error | Empirical formula |
Identity | K-S P value |
|---|---|---|---|---|---|---|---|---|
| 1 | 5.11 | positive | 310.2017 | 310.1993 | 7.74 | C15H29NO4 | l-Octanoylcarnitine + Na | 0.0003 |
| 2 | 0.35 | positive | 137.0457 | 137.0463 | 4.38 | C5H4N4O | Hypoxanthine | 0.0006 |
| 3 | 0.26 | positive | 189.1586 | 189.1602 | 8.46 | C9H20N2O2 | N6,N6,N6-Trimethyl-l-lysine | 0.008 |
| 4 | 0.36 | positive | 204.1243 | 204.1235 | 3.92 | C9H17NO4 | Acetyl-dl-carnitine | 0.01 |
| 5 | 5.43 | positive | 338.2332 | 338.2306 | 7.69 | C17H33NO4 | Decanoylcarnitine + Na | 0.01 |
| 6 | 0.34 | negative | 167.0202 | 167.0205 | 1.80 | C5H4N4O3 | Uric acid | 0.001 |
| 7 | 0.34 | negative | 151.0246 | 151.0256 | 6.62 | C5H4N4O2 | Xanthine | 0.02 |
Notes. Markers that exhibited statistical significance through the Kolmogorov-Smirnov test when pre-exposure vs. 6 h post-exposure data were compared. Pathways affected include energy metabolism with implications for fatty acid oxidation, and purine catabolism potentially affecting the purine salvage pathway.
Quantification of Validated Markers
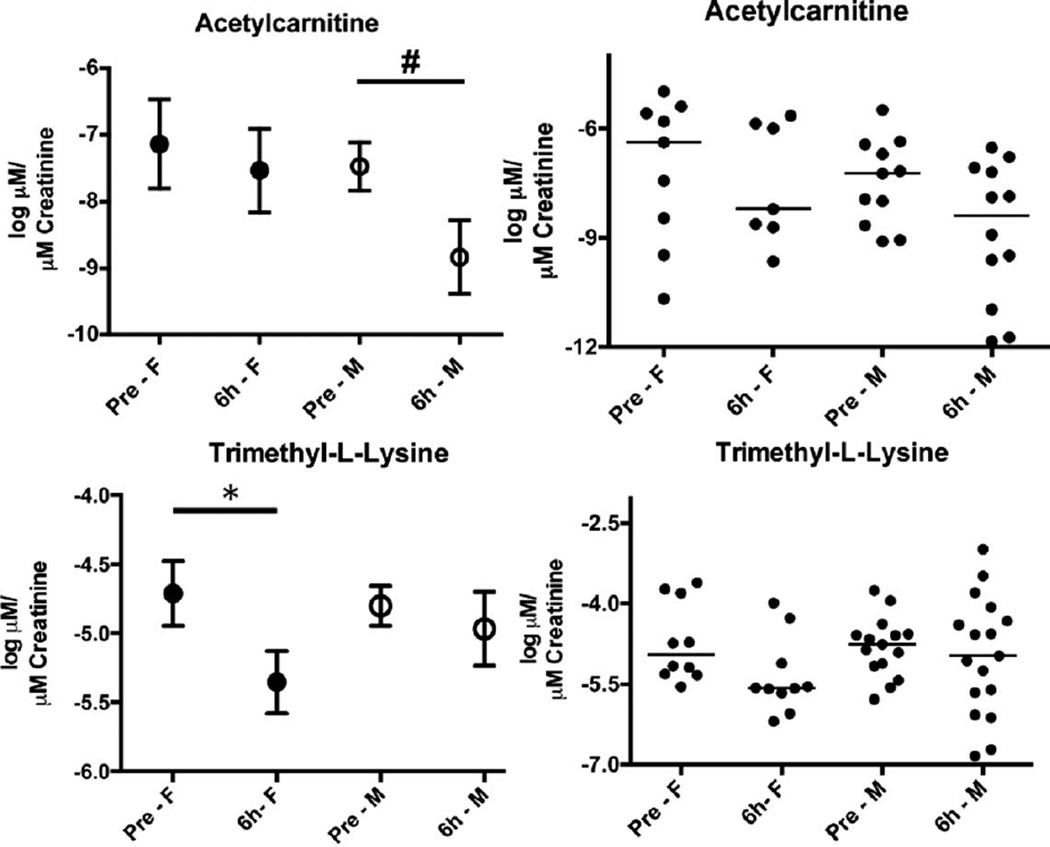
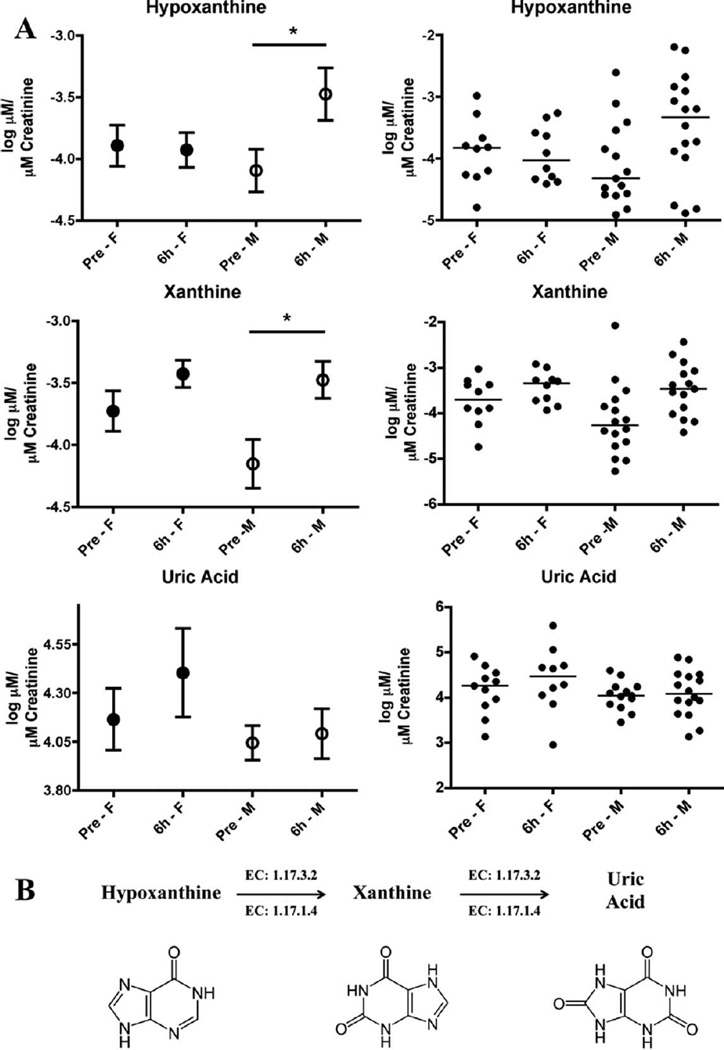
Quantification of the validated markers was performed on a UPLC coupled to a Xevo TQ mass spectrometer. Two different columns were utilized to account for the chemical properties of the markers tested. Concentrations (µM) were calculated from a standard curve for each chemical. Octanoylcarnitine (0.01–0.25 µM, R2 ≥ 0.99), decanoylcarnitine (0.005–0.1 µM, R2 ≥ 0.99) and acetylcarnitine (0.25–5 µM, R2 ≥ 0.99) were quantified on a C18 column with internal standards 4-NBA (30 µM), debrisoquine (4 µM) and creatinine-d3 (5 µM). The more polar compounds that are not well retained and resolved with reverse phase chromatography (C18 column) were quantified by hydrophilic interaction liquid chromatography (HILIC) on an amide column. Those included hypoxanthine (0.25–10 µM, R2 ≥ 0.99), TML (0.1–1 µM, R2 ≥ 0.99), xanthine (5–50 µM, R2 ≥ 0.98), uric acid (25–150 µM, R2 ≥ 0.97), creatinine (0.25–10 µM, R2 ≥ 0.99) and the internal standards 4-NBA (30 µM), debrisoquine (4 µM) and creatinine-d3 (5 µM). Log normalized data were assessed for differences in radiation responses specific for each sex. Figure 2 shows a decrease in acetylcarnitine levels in males at 6 h post-exposure (although not statistically significant by standard definition as the P value was 0.0532) compared to females whose levels remain unaltered. The opposite was observed with TML as the levels were decreased in females post TBI (P = 0.0232) and the urinary levels remained unaltered in the male population. Despite the initial determination of statistical significance of decanoylcarnitine and octanoylcarnitine, the more sensitive quantification method showed only trends of increased excretion for decanoylcarnitine and decreased excretion of octanoylcarnitine for both male and female after radiation exposure (Supplementary Fig. S4; http://dx.doi.org/RR13567.1.S2). The final three validated markers are part of the purine metabolism, mapping to the final steps as shown in Fig. 3B. Hypoxanthine and xanthine showed increased levels in males postirradiation (P values 0.0318 and 0.011, respectively), while only xanthine showed an increase in females, although not statistically significant (Fig. 3A). Finally, uric acid showed a trend of increased excretion only in females after irradiation. One patient [(no. 086) Supplementary Table S1; http://dx.doi.org/RR13567.1.S1] that had received prior radiation exposure was removed from the analysis as the levels of each metabolite were determined to be outliers, whereas patient no. 092 was included in the study, as prior irradiation did not have a significant effect.
FIG. 2.
Quantification of acetylcarnitine and trimethyl-l-lysine (TML). TML is a precursor for biosynthesis of L-carnitine, whereas acetylcarnitine is associated with energy metabolism and neuroprotection. Results on the left hand panels are presented as mean of log (µM of metabolite/µM excreted creatinine) ± SEM, and plotted on the right panels is the distribution in the population with lines signifying the median. The pre-exposure and 6 h post-exposure groups were further analyzed according to sex, demonstrating its importance in different patterns of metabolite excretion. The symbol # represents a borderline P value of 0.0532, as determined through a Welch’s t test. The asterisk * represents a P value of <0.05, as analyzed with the Kolmogorov-Smirnov test. The choice of using either the Welch’s t test or the Kolmogorov-Smirnov test was determined by whether the assumption of statistical normality held by the Anderson-Darling test at the 10% significance level.
FIG. 3.
Quantification of hypoxanthine, xanthine and uric acid. All three metabolites are part of the purine catabolism, as indicated in the enzymatic pathway (xanthine oxidase: 1.17.3.2, xanthine dehydrogenase: 1.17.1.4). Uric acid is the last step of the pathway in humans. Results on the left hand panels are presented as mean of log (µM of metabolite/µM excreted creatinine) ± SEM, and plotted on the right panels is the distribution in the population with lines signifying the median. The pre-exposure and 6 h post-exposure groups were further analyzed based on sex, revealing that the purine catabolism is highly responsive in the adult male population, whereas adult females only exhibit trends of upregulated excretion. The assumption of statistical normality was confirmed for all features by the Anderson-Darling test at the 10% significance level. Asterisks represent a P value of <0.05, according to the t test with Welch’s correction.
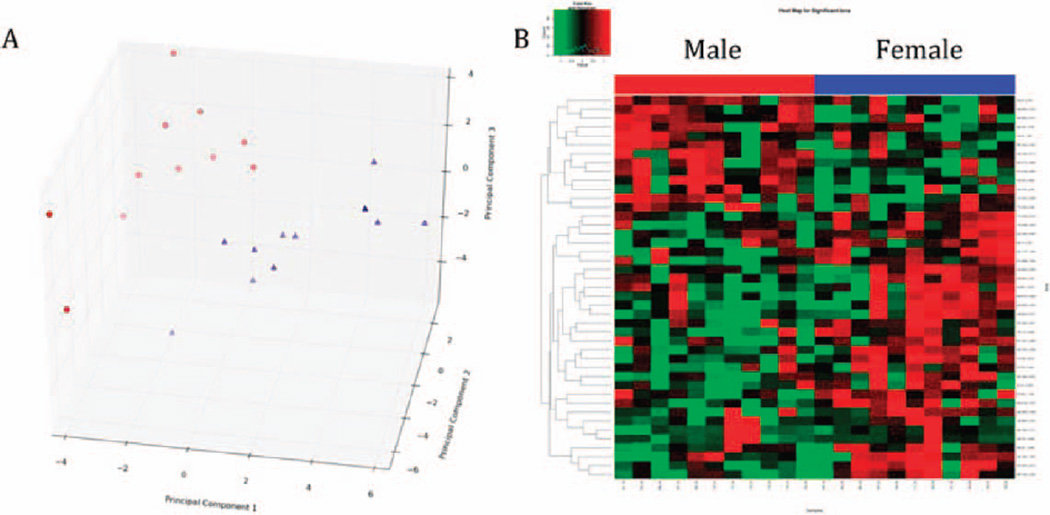
Grouping of the males and females to pre-and post-exposure (6 h) showed decreased levels of TML, acetylcarnitine and octanoylcarnitine, whereas decanoylcarnitine, hypoxanthine, xanthine and uric acid were increased after exposure to IR (Supplementary Figs. S5 and S6; http://dx.doi.org/RR13567.1.S2). Differences in overall metabolic profiles between males and females were observed even before irradiation (data not shown). However, overall metabolic profiles were distinct even after radiation exposure with removal of the pre-exposure statistically significant ions prior to construction of a PCA scores plot and a heatmap (Fig. 4).
FIG. 4.
Male and female metabolic differences are evident after exposure to TBI. Prior to analysis, a list of metabolites that was inherent in sex differences prior to irradiation was removed. Post-exposure statistical analysis showed distinct clustering of the two groups in a PCA score plot (panel A), whereas the differences were more evident as demonstrated on a heatmap (panel B).
DISCUSSION
The threat of radiological sabotage (24) and accidental exposures has increased in the past decades. Nuclear plant accidents, radiological dispersal devices and poisoning of individuals with radioactive materials have led to the need to develop rapid and reliable methods of biological dosimetry and identification of exposed individuals. While cytogenetics remains the gold standard for biological dosimetry and refinement of dose and potential future cancer risk, metabolomics can provide a first assessment (triage) of exposed individuals. Metabolomics has been used with success in the past to identify biomarkers of radiation exposure in easily accessible biofluids, i.e., urine, in animal models such as mice (4, 19, 20, 25), rats (17, 26) and nonhuman primates (16). In this study we extended the metabolomics approach to urine from humans exposed to TBI that was used as a precursor to HSCT. This is the first radiation metabolomics study in urine from humans, laying the foundation for the generation of a radiation signature. Despite the inherent noise in the data, markers were dissected and shown that common pathways are affected and markers overlap between animal models and humans. In addition, this is the first study in human urinary radiation metabolomics to examine the presence of sex differences and determine that separate radiation signatures will need to be developed for utilization of metabolomics as an efficient and rapid triage method.
Radiation metabolomics possesses challenges, as various factors can alter the noise in the matrix or the metabolic signature. Given the nature of the matrix of the samples and the population utilized, noise in this dataset was higher than in previous animal studies. Unlike animal studies, the differences in age, diet, ethnic background, race, genetics and underlying disease status may all play a significant role in alterations of the metabolic profile between individuals. This can easily be assessed by the presence of lower abundance ions in the deconvoluted data and the presence or absence of markers between individuals. Given this inherent issue of this particular dataset, multivariate data analysis failed to provide clear differences between the pre- and post-exposure groups. A more basic univariate approach was adopted, which led to clear identification of radiation biomarkers that are not only injury but also sex specific.
As in previous animal studies, urinary creatinine levels were utilized for normalization purposes to account for differences in glomerular filtration rates. No statistical difference was observed in the levels of creatinine between pre-TBI and after the first radiation fraction, however creatinine was significantly increased after three radiation fractions at 24 h (Supplementary Fig. S1; http://dx.doi.org/RR13567.1.S2). For this reason, investigation of the 24 h time point did not proceed further for this study and all marker identification and quantification focused on the 6 h time point (1.25 Gy). When creatinine levels were assessed according to separation by male or female at 6 h, creatinine showed no statistically significant differences in levels of excretion (Supplementary Fig. S1; http://dx.doi.org/RR13567.1.S2). Creatinine levels, however, have been shown to be higher at rest in males due to higher muscle mass (27), as was observed is our study, although not statistically significant in this population. Creatinine levels were therefore used for normalization purposes at 6 h after TBI. The statistically significant difference observed at 24 h postirradiation could be attributed to either interindividual variability in the hospital setting and variability within each hour, the possibility of oligouria and affected glomerular filtration rates (28). The effect of the fractionated dose and increased total radiation levels may also be contributing factors. It is possible that increased doses and more importantly fractionated dosing could affect the levels of creatinine excretion in a progressively increasing manner. Regardless, different normalization methods such as urinary gravity, freeze drying and reconstitution in the same volume, osmolarity, urine volume, excretion rate, total metabolites or a 24 h urine collection (28, 29), will need to be utilized in future studies.
A number of metabolites identified in the urine are precursors and part of the fatty acid β-oxidation pathway, an important constituent of energy metabolism. TML, a precursor of endogenous carnitine production, and carnitine conjugate levels were found to be altered in the urine of TBI individuals. Carnitine and its derivatives are important in the transportation of fatty acids across the mitochondrial membrane by transformation into long-chain acylcarnitines and conversion of branched-chain amino acids into energy (30). Most of the carnitine is found free in the plasma of humans and the levels are directly correlating with dietary intake (31), however during fasting carnitine uptake into cells is increased resulting in increased β-oxidation (32). Endogenous production of carnitine is based on conversion of the amino acid lysine to TML through methylation of the ε-amino group and subsequent release of TML upon protein degradation (33, 34). TML is further oxidized to γ-butyrobetaine, which is hydroxylated in the liver and kidney to form carnitine (33). Inside the mitochondria, the carnitine conjugate acetylcarnitine regulates the levels of acetyl-CoA (35), which is essential for the balance between fatty acid and carbohydrate metabolism (30). Acetylcarnitine, found to be retained intracellularly by monitoring the urinary levels (Supplementary Fig. S5; http://dx.doi.org/ RR13567.1.S2), has been shown to be cytoprotective by decreasing oxidative stress, whereas carnitine and its esters have no direct antioxidant activity (36). Kocer et al. showed that irradiation of rats and supplementation with l-carnitine led to decreased malondialdehyde levels and increased superoxide dismutase and glutathione peroxidase activities (37). Carnitine and acetylcarnitine have a general role in maintenance of cell viability (36) and restoring activities and protecting membrane permeability (38). Decreased urinary levels of TML, acetylcarnitine and octanoylcarnitine in the urine post TBI (Supplementary Fig. S5; http://dx.doi.org/RR13567.1.S1) are suggestive of increased fatty acid β-oxidation intracellularly. However, the presence of other carnitine conjugates suggests that the process may be incomplete since carnitine levels are altered from the canonical pathway. In fact, the differences in ratios between octanoylcarnitine (C8), decanoylcarnitine (C10) and acetylcarnitine (C2) have been utilized to delineate the different variants of medium-chain acyl-CoA dehydrogenase (MCAD) deficiency of fatty acid β-oxidation (39). When looking at sex differences, a general decrease of excretion is observed after irradiation (Fig. 2), although the female versus the male response is quite different. In females carnitine production in the body appears to be affected, leading to the possibility of utilizing higher levels of the free plasma carnitine. However, males exhibit lower acetylcarnitine loss, suggesting that β-oxidation is active by eliminating toxic acetyl-CoA and recycling it outside of the mitochondria. Overall, energy metabolism appears to be impacted due to radiation, which is reflected in the urine.
Another set of markers that can signify increased DNA damage and oxidative stress belong to the purine catabolism pathway. Hypoxanthine, xanthine and uric acid were prevalent in the urine of irradiated individuals as early as 6 h post-exposure and as low of a dose as 1.25 Gy. As shown in Fig. 3B, these three molecules constitute the last steps of the purine catabolism, with xanthine oxidoreductase (XOR) catalyzing the conversions. XOR exists in the xanthine dehydrogenase (XDH) (E.C. 1.17.1.4) form under normal conditions, with conversion to xanthine oxidase (XO) (E.C. 1.17.3.2) suggested to occur in damaged cells (40, 41). Hypoxanthine can be produced not only from inosine, but also from deamination of adenine, whereas xanthine can also be produced from deamination of guanine. Uric acid, the last step of the pathway, has been identified in previous studies as a radiation marker in urine of mice (4) and nonhuman primates (16) and is considered a natural free radical scavenger (42). Xanthine was found in increased levels in mice and nonhuman primates as well (16, 20), whereas hypoxanthine was only identified in nonhuman primates (16). Although in mammals, during high oxidative stress uric acid is converted to and excreted in the form of allantoin (4), the gene expression of the enzyme responsible for the conversion is silenced by mutations in humans (43), unlike previously identified in mice (4). While ionizing radiation can lead to the conversion of XDH to XO (41) and is the primary source of ROS generation intracellularly, XO is also a major source of free radicals. In addition, Ca2+ disturbances have been known to lead to conversions of XDH to XO (41), however serum Ca2+ in our study remained unaffected after TBI (Table 1). Therefore, IR is most likely the driving force of the pathway, leading to secondary generation of ROS and perpetuation of the oxidative phenotype.
The results of this study also clearly demonstrated the existence of significant differences between females and males in this pathway. Results for males are consistent with previous studies in other species (16, 20) (a summary of identified urinary biomarkers is shown in Table 3) and also with human metabolomics studies of skin from males (44). The attenuated response in females could be attributed to the effect of female hormones, such as estrogen, which has been shown to protect against radiation cataractogenesis (45). Studies on radiation metabolomics of urine in the literature have focused primarily on male subjects, to avoid the complication of estrous cycle effects. The study of Johnson et al. on nonhuman primates (16) did not investigate the differences between the two sexes, although both male and female subjects were utilized. However, the investigators did identify the same pathway (purine catabolism), signifying the commonality of radiation responses between closely related species (Table 3).
TABLE 3.
Comparison of Radiation Markers in Urine of Different Species
| Markers | Humans | Nonhuman primates (16) | Rats (17, 26) | Mice (4, 19, 20) |
|---|---|---|---|---|
| Trimethyl-l-lysine | √ | |||
| Decanoylcarnitine | √ | |||
| Octanoylcarnitine | √ | |||
| Acetylcarnitine | √ | |||
| Hypoxanthine | √ | √ | ||
| Xanthine | √ | √ | √ | |
| Uric acid | √ | √ | ||
| Nicotinate | √* | √ | ||
| Allantoin | √ | |||
| Thymidine | √ | √ | ||
| Xanthosine | √ | |||
| N-hexanoylglycine | √ | √ | ||
| Glyoxylate | √ | |||
| Threonate | √ | |||
| Thymine | √ | |||
| Uracil | √ | |||
| p-cresol | √ | |||
| Citrate | √ | |||
| 2′-Deoxyuridine | √ | |||
| 2′-Deoxyxanthosine | √ | |||
| N(1)-acetylspermidine | √ | |||
| Taurine | √ | √ | √ | |
| N-acetyltaurine | √ | √ | ||
| Isethionic acid | √* | √ | √ | |
| Creatine | √ | |||
| Tyrosol sulfate | √ | |||
| 3-Hydroxytyrosol sulfate | √ | |||
| Tyramine sulfate | √ | |||
| N-acetylserotonin sulfate | √ | |||
| Adipic acid | √ | √ | ||
| Creatinine | √ |
Signifies putative identity.
It is important to note that the samples that were used in these studies originated from cancer patients. However, each patient served as their own control in this study to eliminate disease effects. Additionally, the criteria that were utilized for inclusion in this study were strict to eliminate confounding factors, such as recent chemotherapy. A number of the markers and pathways that were identified have been associated with radiation exposure in other species in studies from our laboratory and other investigators. This, however, is the first metabolomic study in urine from humans exposed to ionizing radiation and the first metabolomic study to identify sex differences in radiation signatures from urine. This study provides the basis for the utilization of metabolomics to identify radiation biomarkers in urine from humans and for future rapid biodosimetry and identification of exposed individuals.
Supplementary Material
Supplementary Fig. S1. Quantified log µM of creatinine for pre-exposure and 6 h post-exposure with 1.25 Gy, males vs. females. Results represent mean of log µM creatinine ± SEM. No statistically significant differences were identified within any of the group analyses. However, creatinine was significantly increased at 24 h after TBI (P < 0.01), making results based on normalization to this metabolite unreliable.
Supplementary Fig. S2. Ions were mapped to KEGG metabolic pathways based on their putative metabolite matches and their corresponding pathways. Although further refined identity determinations of the putative metabolites excluded the majority of the candidates, an overall putative mapping can nonetheless reveal an overall perturbation of the metabolism due to radiation exposure.
Supplementary Fig. S3. PCA scores plot of the pre-exposure vs. 24 h post-exposure (3.75 Gy) results. Although the groups are still distinguishable, overall clear clustering is less evident at this time point. A large number of ions are statistically significant, as indicated on the volcano plot (panel B), whereas distinguished overall metabolic profiles are more prominent in the heatmap (panel C).
Supplementary Fig. S4. Quantification of decanoylcarnitine and octanoylcarnitine revealed no statistically significant differences (Kolmogorov-Smirnov test). However, trends of increased decanoylcarnitine excretion and decreased octanoylcarnitine excretion following exposure to ionizing radiation are evident in both males and females. Results on the left hand panels are presented as mean of log (µM of metabolite/µM excreted creatinine) ± SEM and on the right panels lines signify the median.
Supplementary Fig. S5. Quantification of energy metabolism metabolites, acetylcarnitine, decanoylcarnitine, octanoylcarnitine and trimethyl-l-lysine. Male and female have been grouped under each time point. A general trend of decreased excretion is observed after exposure to 1.25 Gy of ionizing radiation, whereas decanoylcarnitine levels are higher in the exposed group. Results are presented as mean of log (µM of metabolite/µM excreted creatinine) ± SEM. TML is the only metabolite exhibiting statistically significant differences with a P value of <0.05 (Kolmogorov-Smirnov test).
Supplementary Fig. S6. Quantification of three purine catabolism metabolites, hypoxanthine, xanthine and uric acid. Males and females have been grouped together under each time point (pre-exposure vs. 6 h post-exposure). Results are presented as mean of log (µM of metabolite/µM excreted creatinine) ± SEM. Increased excretion of these metabolites is observed after exposure to ionizing radiation, with xanthine being statistically significant with a P value of <0.05 (Kolmogorov-Smirnov test).
ACKNOWLEDGMENTS
This work was funded by the National Institutes of Health (National Institute of Allergy and Infectious Diseases) grant U19AI067773 (P.I. David J. Brenner, performed as part of the Columbia University Center for Medical Countermeasures against Radiation) and 1R01AI101798 (P.I. Albert J. Fornace, Jr.). The authors would like to thank the Lombardi Comprehensive Cancer Proteomics & Metabolomics Shared Resource (PMSR) for data acquisition. The project described above was also supported by Award Number P30 CA051008 (P.I. Louis Weiner) from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
REFERENCES
- 1.Zeitlin C, Hassler DM, Cucinotta FA, Ehresmann B, Wimmer-Schweingruber RF, Brinza DE, et al. Measurements of energetic particle radiation in transit to Mars on the Mars Science Laboratory. Science. 2013;340:1080–1084. doi: 10.1126/science.1235989. [DOI] [PubMed] [Google Scholar]
- 2.Dicarlo AL, Maher C, Hick JL, Hanfling D, Dainiak N, Chao N, et al. Radiation injury after a nuclear detonation: medical consequences and the need for scarce resources allocation. Disaster Med Public Health Prep. 2011;5(Suppl 1):S32–S44. doi: 10.1001/dmp.2011.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dicarlo AL, Ramakrishnan N, Hatchett RJ. Radiation combined injury: overview of NIAID research. Health Phys. 2010;98:863–867. doi: 10.1097/HP.0b013e3181a6ee32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laiakis EC, Hyduke DR, Fornace AJ. Comparison of mouse urinary metabolic profiles after exposure to the inflammatory stressors gamma radiation and lipopolysaccharide. Radiat Res. 2012;177:187–199. doi: 10.1667/rr2771.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waselenko JK, Macvittie TJ, Blakely WF, Pesik N, Wiley AL, Dickerson WE, et al. Medical management of the acute radiation syndrome: recommendations of the Strategic National Stockpile Radiation Working Group. Ann Intern Med. 2004;140:1037–1051. doi: 10.7326/0003-4819-140-12-200406150-00015. [DOI] [PubMed] [Google Scholar]
- 6.Rossi AM, Wafcheck CC, De Jesus EF, Pelegrini F. Electron spin resonance dosimetry of teeth of Goiania radiation accident victims. Appl Radiat Isot. 2000;52:1297–1303. doi: 10.1016/s0969-8043(00)00087-7. [DOI] [PubMed] [Google Scholar]
- 7.Mccurley MC, Miller CW, Tucker FE, Guinn A, Donnelly E, Ansari A, et al. Educating medical staff about responding to a radiological or nuclear emergency. Health Phys. 2009;96:S50–S54. doi: 10.1097/01.HP.0000339001.77899.15. [DOI] [PubMed] [Google Scholar]
- 8.Asano S. Current status of hematopoietic stem cell transplantation for acute radiation syndromes. Int J Hematol. 2012;95:227–231. doi: 10.1007/s12185-012-1027-8. [DOI] [PubMed] [Google Scholar]
- 9.Wilkinson D. Dealing with at-risk populations in radiological/nuclear emergencies. Radiat Prot Dosimetry. 2009;134:136–142. doi: 10.1093/rpd/ncp070. [DOI] [PubMed] [Google Scholar]
- 10.Agency HP. Human Radiosensitivity: Report of the Independent Advisory Group on Ionising Radiation. 2013 [Google Scholar]
- 11.Ncrp. Radiation Protection Guidance for Activities in Low-Earth Orbit. 2000 doi: 10.1016/s0273-1177(02)00160-6. [DOI] [PubMed] [Google Scholar]
- 12.Roberts CJ, Morgan GR, Danford N. Effect of hormones on the variation of radiosensitivity in females as measured by induction of chromosomal aberrations. Environ Health Perspect. 1997;105(Suppl 6):1467–1471. doi: 10.1289/ehp.97105s61467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paul S, Barker CA, Turner HC, Mclane A, Wolden SL, Amundson SA. Prediction of in vivo radiation dose status in radiotherapy patients using ex vivo and in vivo gene expression signatures. Radiat Res. 2011;175:257–265. doi: 10.1667/RR2420.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Templin T, Paul S, Amundson SA, Young EF, Barker CA, Wolden SL, et al. Radiation-induced micro-RNA expression changes in peripheral blood cells of radiotherapy patients. Int J Radiat Oncol Biol Phys. 2011;80:549–557. doi: 10.1016/j.ijrobp.2010.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turner HC, Brenner DJ, Chen Y, Bertucci A, Zhang J, Wang H, et al. Adapting the gamma-H2AX assay for automated processing in human lymphocytes. 1. Technological aspects. Radiat Res. 2011;175:282–290. doi: 10.1667/RR2125.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson CH, Patterson AD, Krausz KW, Kalinich JF, Tyburski JB, Kang DW, et al. Radiation metabolomics. 5. Identification of urinary biomarkers of ionizing radiation exposure in nonhuman primates by mass spectrometry-based metabolomics. Radiat Res. 2012;178:328–340. doi: 10.1667/rr2950.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lanz C, Patterson AD, Slavik J, Krausz KW, Ledermann M, Gonzalez FJ, et al. Radiation metabolomics. 3. Biomarker discovery in the urine of gamma-irradiated rats using a simplified metabolomics protocol of gas chromatography-mass spectrometry combined with random forests machine learning algorithm. Radiat Res. 2009;172:198–212. doi: 10.1667/RR1796.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma M, Halligan BD, Wakim BT, Savin VJ, Cohen EP, Moulder JE. The urine proteome for radiation biodosimetry: effect of total body vs. local kidney irradiation. Health Phys. 2010;98:186–195. doi: 10.1097/HP.0b013e3181b17cbd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tyburski JB, Patterson AD, Krausz KW, Slavik J, Fornace AJJ, Gonzalez FJ, et al. Radiation metabolomics. 1. Identification of minimally invasive urine biomarkers for gamma-radiation exposure in mice. Radiat Res. 2008;170:1–14. doi: 10.1667/RR1265.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tyburski JB, Patterson AD, Krausz KW, Slavik J, Fornace AJJ, Gonzalez FJ, et al. Radiation metabolomics. 2. Dose- and time-dependent urinary excretion of deaminated purines and pyrimidines after sublethal gamma-radiation exposure in mice. Radiat Res. 2009;172:42–57. doi: 10.1667/RR1703.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coy SL, Cheema AK, Tyburski JB, Laiakis EC, Collins SP, Fornace AJ. Radiation metabolomics and its potential in biodosimetry. Int J Radiat Biol. 2011;87:802–823. doi: 10.3109/09553002.2011.556177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patterson AD, Li H, Eichler GS, Krausz KW, Weinstein JN, Fornace AJJ, et al. UPLC-ESI-TOFMS-based metabolomics and gene expression dynamics inspector self-organizing metabolomic maps as tools for understanding the cellular response to ionizing radiation. Anal Chem. 2008;80:665–674. doi: 10.1021/ac701807v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mak TD, Laiakis EC, Goudarzi M, Fornace AJJ. MetaboLyzer: A Novel Statistical Workflow for Analyzing Postprocessed LC-MS Metabolomics Data. Anal Chem. 2014;86:506–513. doi: 10.1021/ac402477z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirkham L, Kuperman AJ. Protecting U.S. Nuclear Facilities From Terrorist Attack: Re-assessing the Current “Design Basis Threat” Approach. 2013 ( http://blogs.utexas.edu/nppp/files/2013/08/NPPP-working-paper-1-2013-Aug-15.pdf) [Google Scholar]
- 25.Chen C, Brenner DJ, Brown TR. Identification of urinary biomarkers from X-irradiated mice using NMR spectroscopy. Radiat Res. 2011;175:622–630. doi: 10.1667/RR2388.1. [DOI] [PubMed] [Google Scholar]
- 26.Johnson CH, Patterson AD, Krausz KW, Lanz C, Kang DW, Luecke H, et al. Radiation metabolomics. 4. UPLC-ESI-QTOFMS-Based metabolomics for urinary biomarker discovery in gamma-irradiated rats. Radiat Res. 2011;175:473–484. doi: 10.1667/RR2437.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slupsky CM, Rankin KN, Wagner J, Fu H, Chang D, Weljie AM, et al. Investigations of the effects of gender, diurnal variation, and age in human urinary metabolomic profiles. Anal Chem. 2007;79:6995–7004. doi: 10.1021/ac0708588. [DOI] [PubMed] [Google Scholar]
- 28.Waikar SS, Sabbisetti VS, Bonventre JV. Normalization of urinary biomarkers to creatinine during changes in glomerular filtration rate. Kidney Int. 2010;78:486–494. doi: 10.1038/ki.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warrack BM, Hnatyshyn S, Ott KH, Reily MD, Sanders M, Zhang H, et al. Normalization strategies for metabonomic analysis of urine samples. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:547–552. doi: 10.1016/j.jchromb.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 30.Malaguarnera M. Carnitine derivatives: clinical usefulness. Curr Opin Gastroenterol. 2012;28:166–176. doi: 10.1097/MOG.0b013e3283505a3b. [DOI] [PubMed] [Google Scholar]
- 31.Steiber A, Kerner J, Hoppel CL. Carnitine: a nutritional, biosynthetic, and functional perspective. Mol Aspects Med. 2004;25:455–473. doi: 10.1016/j.mam.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 32.Krug S, Kastenmuller G, Stuckler F, Rist MJ, Skurk T, Sailer M, et al. The dynamic range of the human metabolome revealed by challenges. FASEB J. 2012;26:2607–2619. doi: 10.1096/fj.11-198093. [DOI] [PubMed] [Google Scholar]
- 33.Hirche F, Fischer M, Keller J, Eder K. Determination of carnitine, its short chain acyl esters and metabolic precursors trimethyllysine and gamma-butyrobetaine by quasi-solid phase extraction and MS/MS detection. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:2158–2162. doi: 10.1016/j.jchromb.2009.05.048. [DOI] [PubMed] [Google Scholar]
- 34.Strijbis K, Vaz FM, Distel B. Enzymology of the carnitine biosynthesis pathway. IUBMB Life. 2010;62:357–362. doi: 10.1002/iub.323. [DOI] [PubMed] [Google Scholar]
- 35.Khan HA, Alhomida AS. A review of the logistic role of L-carnitine in the management of radiation toxicity and radiotherapy side effects. J Appl Toxicol. 2011;31:707–713. doi: 10.1002/jat.1716. [DOI] [PubMed] [Google Scholar]
- 36.Marcovina SM, Sirtori C, Peracino A, Gheorghiade M, Borum P, Remuzzi G, et al. Translating the basic knowledge of mitochondrial functions to metabolic therapy: role of L-carnitine. Transl Res. 2013;161:73–84. doi: 10.1016/j.trsl.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kocer I, Taysi S, Ertekin MV, Karslioglu I, Gepdiremen A, Sezen O, et al. The effect of L-carnitine in the prevention of ionizing radiation-induced cataracts: a rat model. Graefes Arch Clin Exp Ophthalmol. 2007;245:588–594. doi: 10.1007/s00417-005-0097-1. [DOI] [PubMed] [Google Scholar]
- 38.Mansour HH. Protective role of carnitine ester against radiation-induced oxidative stress in rats. Pharmacol Res. 2006;54:165–171. doi: 10.1016/j.phrs.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 39.Okun JG, Kolker S, Schulze A, Kohlmuller D, Olgemoller K, Lindner M, et al. A method for quantitative acylcarnitine profiling in human skin fibroblasts using unlabelled palmitic acid: diagnosis of fatty acid oxidation disorders and differentiation between biochemical phenotypes of MCAD deficiency. Biochim Biophys Acta. 2002;1584:91–98. doi: 10.1016/s1388-1981(02)00296-2. [DOI] [PubMed] [Google Scholar]
- 40.Ichida K, Amaya Y, Okamoto K, Nishino T. Mutations associated with functional disorder of xanthine oxidoreductase and hereditary xanthinuria in humans. Int J Mol Sci. 2012;13:15475–15495. doi: 10.3390/ijms131115475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Srivastava M, Kale RK. Effect of radiation on the xanthine oxidoreductase system in the liver of mice. Radiat Res. 1999;152:257–264. [PubMed] [Google Scholar]
- 42.Vorbach C, Harrison R, Capecchi MR. Xanthine oxidoreductase is central to the evolution and function of the innate immune system. Trends Immunol. 2003;24:512–517. doi: 10.1016/s1471-4906(03)00237-0. [DOI] [PubMed] [Google Scholar]
- 43.Causse E, Pradelles A, Dirat B, Negre-Salvayre A, Salvayre R, Couderc F. Simultaneous determination of allantoin, hypoxanthine, xanthine, and uric acid in serum/plasma by CE. Electrophoresis. 2007;28:381–387. doi: 10.1002/elps.200600205. [DOI] [PubMed] [Google Scholar]
- 44.Hu ZP, Kim YM, Sowa MB, Robinson RJ, Gao X, Metz TO, et al. Metabolomic response of human skin tissue to low dose ionizing radiation. Mol Biosyst. 2012;8:1979–1986. doi: 10.1039/c2mb25061f. [DOI] [PubMed] [Google Scholar]
- 45.Dynlacht JR, Valluri S, Lopez J, Greer F, Desrosiers C, Caperell-Grant A, et al. Estrogen protects against radiation-induced cataractogenesis. Radiat Res. 2008;170:758–764. doi: 10.1667/RR1416.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. S1. Quantified log µM of creatinine for pre-exposure and 6 h post-exposure with 1.25 Gy, males vs. females. Results represent mean of log µM creatinine ± SEM. No statistically significant differences were identified within any of the group analyses. However, creatinine was significantly increased at 24 h after TBI (P < 0.01), making results based on normalization to this metabolite unreliable.
Supplementary Fig. S2. Ions were mapped to KEGG metabolic pathways based on their putative metabolite matches and their corresponding pathways. Although further refined identity determinations of the putative metabolites excluded the majority of the candidates, an overall putative mapping can nonetheless reveal an overall perturbation of the metabolism due to radiation exposure.
Supplementary Fig. S3. PCA scores plot of the pre-exposure vs. 24 h post-exposure (3.75 Gy) results. Although the groups are still distinguishable, overall clear clustering is less evident at this time point. A large number of ions are statistically significant, as indicated on the volcano plot (panel B), whereas distinguished overall metabolic profiles are more prominent in the heatmap (panel C).
Supplementary Fig. S4. Quantification of decanoylcarnitine and octanoylcarnitine revealed no statistically significant differences (Kolmogorov-Smirnov test). However, trends of increased decanoylcarnitine excretion and decreased octanoylcarnitine excretion following exposure to ionizing radiation are evident in both males and females. Results on the left hand panels are presented as mean of log (µM of metabolite/µM excreted creatinine) ± SEM and on the right panels lines signify the median.
Supplementary Fig. S5. Quantification of energy metabolism metabolites, acetylcarnitine, decanoylcarnitine, octanoylcarnitine and trimethyl-l-lysine. Male and female have been grouped under each time point. A general trend of decreased excretion is observed after exposure to 1.25 Gy of ionizing radiation, whereas decanoylcarnitine levels are higher in the exposed group. Results are presented as mean of log (µM of metabolite/µM excreted creatinine) ± SEM. TML is the only metabolite exhibiting statistically significant differences with a P value of <0.05 (Kolmogorov-Smirnov test).
Supplementary Fig. S6. Quantification of three purine catabolism metabolites, hypoxanthine, xanthine and uric acid. Males and females have been grouped together under each time point (pre-exposure vs. 6 h post-exposure). Results are presented as mean of log (µM of metabolite/µM excreted creatinine) ± SEM. Increased excretion of these metabolites is observed after exposure to ionizing radiation, with xanthine being statistically significant with a P value of <0.05 (Kolmogorov-Smirnov test).