SUMMARY
The creation of induced pluripotent stem cells (iPSCs) from somatic cells by ectopic expression of transcription factors has galvanized the fields of regenerative medicine and developmental biology. Here, we report a kinome-wide RNAi-based analysis to identify kinases that regulate somatic cell reprogramming to iPSCs. We prepared 3,686 shRNA lentiviruses targeting 734 kinase genes covering the entire mouse kinome and individually examined their effects on iPSC generation. We identified 59 kinases as barriers to iPSC generation and characterized seven of them further. We found that shRNA-mediated knockdown of the serine/threonine kinases TESK1 or LIMK2 promoted mesenchymal-to-epithelial transition, decreased COFILIN phosphorylation, and disrupted Actin filament structures during reprogramming of mouse embryonic fibroblasts. Similarly, knockdown of TESK1 in human fibroblasts also promoted reprogramming to iPSCs. Our study reveals the breadth of kinase networks regulating pluripotency and identifies a role for cytoskeletal remodeling in modulating the somatic cell reprogramming process.
INTRODUCTION
Generation of induced pluripotent stem cells (iPSCs) by ectopic expression of four transcription factors, Oct4, Sox2, Klf4, and c-Myc (OSKM, referred to here as 4F) (Takahashi and Yamanaka, 2006) has created enthusiasm in regenerative medicine and developmental biology. iPSCs, which exhibit properties similar to embryonic stem cells (ESCs), can be generated from human and mouse cells not only with OSKM (Lowry et al., 2008; Park et al., 2008; Takahashi et al., 2007; Wernig et al., 2007) but also with an alternative set of factors, namely, Oct4, Sox2, Nanog, and Lin28 (Yu et al., 2007). Numerous cell types from different tissues have been successfully reprogrammed, but in each case, heterogeneity, retroviral integration, and low reprogramming efficiency have been the major roadblocks to iPSC derivation and therapeutic use. Recent efforts have focused on screening for small molecules that enhance reprogramming efficiency and/or on developing new methods for iPSC derivation (Ichida et al., 2009; Lyssiotis et al., 2009; Maherali and Hochedlinger, 2009; Shi et al., 2008; Yang et al., 2011). Intriguingly, a recent study reported that a cocktail of seven compounds could generate iPS cells from mouse somatic cells, up to 0.2% efficiency, without any exogenous transcription factors (Hou et al., 2013).
Recent years have seen rapid progress in the development of patient-specific iPSCs, which have created new opportunities not only to understand disease pathophysiology but also to develop therapeutics. Although recent technological advances have increased our understanding of the genomic and proteomic networks involved in reprogramming, relatively little is known about the signaling networks that regulate ESC fate and iPSC generation. Protein kinases regulate signal transduction in all eukaryotic cells and play essential roles in many processes, including cell proliferation, cell cycle progression, metabolic homeostasis, transcriptional activation, neurotransmission, differentiation and development, and aging (Lu and Hunter, 2009). Thus, we hypothesized that kinases would likely play pivotal roles in inducing pluripotency and determining cell fates during differentiation. A recent kinase inhibitors screen identifying small molecules that enhance, or present a barrier to, reprogramming further supports this hypothesis (Li and Rana, 2012). For example, inhibitors of p38, inositol trisphosphate 3-kinase, and Aurora kinase A potently enhanced reprogramming efficiencies and iPSCs achieved a fully reprogrammed state (Li and Rana, 2012). In addition, short hairpin RNA screen targeting 104 ESC-associated phopsphoregulators identified Aurora kinase A as an essential kinase in ESC because depletion of this kinase severely affected self-renewal and differentiation (Lee et al., 2012).
Here, we report a kinome-wide RNAi screen to identify kinases that regulate somatic cell reprogramming to iPSCs. In particular, we uncovered a critical role for cytoskeletal remodeling in iPSC generation and identified two key serine/threonine kinases, TESK1 (testicular protein kinase 1) and LIMK2 (LIM kinase 2), which specifically phosphorylate the actin-binding protein COFILIN (COF) and modulate reorganization of the actin cytoskeleton during reprogramming. Over the past several years, a number of kinases and transcription factors have been discovered to have important functions in reprogramming, but the role of cytoskeletal remodeling in pluripotency and cell fate decisions has not been explored. Our results show that knockdown of TESK1 or LIMK2 in mouse embryonic fibroblasts (MEFs) promotes mesenchymal-to-epithelial (MET) transition, decreases COF phosphorylation, and disrupts the actin cytoskeleton during reprogramming.
RESULTS
A kinome-wide functional analysis identifies kinases regulating reprogramming
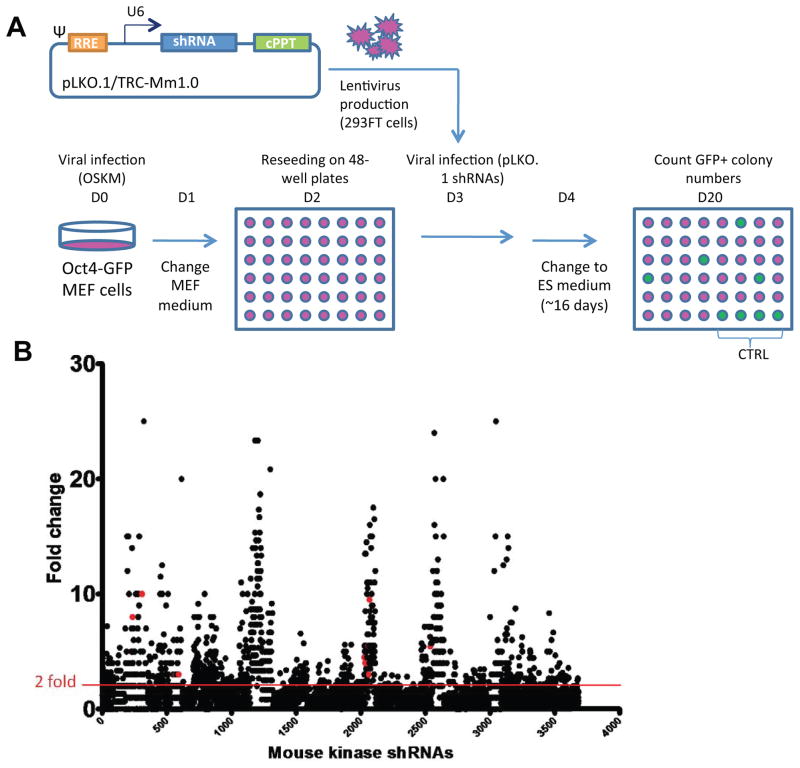
To identify and determine the function of kinases that regulate somatic cell reprogramming to iPSCs, we carried out a whole-kinome RNAi screen (Figure 1A). MEFs harboring a stably integrated Oct4/Pou5f1-driven GFP construct served as a reporter cell line allowing us to monitor iPSC generation quantitatively. Oct4-GFP MEFs were retrovirally transduced with OSKM (4F) and GFP+ colonies, indicative of fully reprogrammed iPSCs. These cells were used to screen a 3,686 lentiviral shRNA library targeting 734 kinase genes covering the entire mouse kinome. Oct4-GFP MEFs were transduced with the shRNA lentiviruses 3 days after 4F infection, and examined in the iPSC generation assay ~13 days later (Figure 1A; Experimental Procedures). A dot-plot representation of the results of the primary screen shows the fold change in GFP+ colony counts after normalization to GFP+ colonies in control pLKO lentivirus-infected cells (Figure 1B).
Figure 1. Kinome-wide RNAi screen for regulators of somatic cell reprogramming.
(A) Experimental design. Oct4-GFP MEFs were transduced on day 0 (D0) with retroviruses encoding the four pluripotency factors, Oct4, Sox2, Klf4, and c-Myc (OSKM; 4F), to induce reprogramming. A total of 3,686 lentiviruses carrying shRNAs targeting the entire kinome were produced in 293FT cells. On day 3 after 4F transduction, MEFs were infected with individual lentiviruses in separate wells. ES medium was changed on day 4 and every other day thereafter until GFP+ colonies were quantified on day 20. The pLKO.1/TRC-Mm1.0 vector expresses shRNAs from a U6 promoter and includes downstream central polypurine tracts (cPPT). (B) Identification of barrier kinases from the primary screen. Dot-plot shows the result of the primary screen assessing the effects of 3,686 shRNAs targeting 734 kinase genes. Results are expressed as the fold change in GFP+ colony counts after normalization to the control pLKO lentiviral-infected cells. The red line indicates the two-fold threshold used to select barrier kinases as hits. Validation of 153 genes from the primary screen was performed in duplicate in a 12-well format in the secondary screen. Subsequently, 59 genes were further validated in a tertiary screen in a 12-well format in duplicate and repeated five times. Red dots indicate shRNAs targeting the six kinases that were selected for further investigation.
See also Figure S1 and Tables S1–S3.
In the primary screen, a kinase was classified as a positive hit if: (a) 2–5 shRNA constructs targeting a single kinase caused ≥2-fold increase in the number of GFP+ cells (indicated by a red line in Figure 1B) or (b) a single shRNA targeting a single kinase caused a ≥6-fold increase in GFP+ cells (Figure 1B). The primary screen revealed 153 hits, which underwent a secondary validation screen in a 12-well format (Figure S1A), yielding 59 hits (Figure S1B, Tables S1 and S2). Finally, the 59 hits were further validated in a tertiary screen performed in the same format and repeated five times. Because cell cycle control is one of the key pathways that regulate the reprogramming process (Ruiz et al., 2011), we selected a panel of 15 candidate kinases from the 59 genes for examination of their effects on the cell cycle (Table S3). siRNA-mediated knockdown of most of the 15 genes decreased the frequency of MEFs in the G1 phase of the cell cycle and increased the percentage in G2, similar to the effects of p53 knockdown (Figures S1C–S1F).
Identification of seven kinases that promotes MET in the initial reprogramming step
We used IPA software (Ingenuity System version 8.7) to analyze the molecular functions and canonical pathways that were significantly overrepresented among the 59 identified kinases (right-tailed Fisher’s exact test, P < 0.05), and then analyzed the biological functional networks using the Ingenuity Pathways Knowledge Base (IPKB) system. From this, we identified four networks in which the 59 kinases showed highly interconnected molecular functions: (I) amino acid metabolism, post-translational modification, small molecule biochemistry; (II) gene expression and cellular development; (III) cell cycle, cell signaling, cell death; (IV) cellular growth and proliferation, cancer (Figure S2A). Of the 59 kinases identified as barriers in iPSC generation, five function in the integrin-linked kinase (ILK) signaling network.
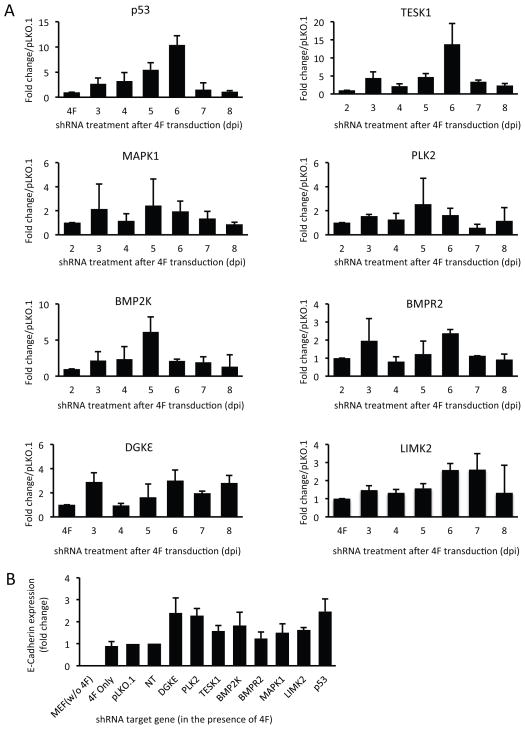
For further study, we selected seven kinases that (i) when knocked down, resulted in a consistent >2-fold increase in GFP+ colony formation in multiple experiments; (ii) were more highly expressed in MEFs than in mESCs; and (iii) were involved in a variety of functions that might relate to development and iPSC biology (Tables S2 and S4). These kinases were DGKε, PLK2, TESK1, LIMK2, BMP2K, BMPR2, and MAPK1. The function of MAPK1 as a barrier to iPSC generation has previously been reported (Nichols et al., 2009). To determine the time frame during which these kinases function in reprogramming, MEFs were transfected with specific shRNAs at various times between 3 and 10 dpi with 4F. As shown in Figure 2, the effect of shRNA-mediated knockdown (KD) of p53 (a positive control) or TESK1 on reprogramming was maximal when shRNA was transduced on 6 dpi. A similar trend was observed when BMP2K expression was reduced, whereas maximal enhancement of reprogramming efficiency by the remaining kinases was obtained when they were knocked down on 3 dpi, and did not change thereafter. The efficiency of shRNA-mediated KD of kinases was confirmed by measuring mRNA on different days (Figure S2B).
Figure 2. Lentivirus-based knockdown of seven kinases at different times during reprogramming reveals the existence of time windows critical for enhancing reprogramming and their role in MET.
(A) The effects of silencing p53 and seven selected kinases on reprogramming efficiency were evaluated at different times after 4F transduction. Oct4-GFP MEFs were transduced with 4F and passaged at 2 days post-infection (dpi). Lentivirus-based shRNA knockdown was performed on the indicated days by adding fresh virus-containing supernatants. GFP+ colonies were counted on 18 dpi. Results show the fold increase in GFP+ colonies relative to numbers obtained with empty pLKO.1. Results are mean ± SD of two independent experiments performed in triplicate. (B) Knockdown of p53 and the seven kinases enhances MET in 4F-transduced MEFs. E-cadherin expression served as a marker for induction of MET during the initial stage of reprogramming. MEFs were transduced with shRNAs targeting p53 or the seven kinases in the presence of 4F. Empty vector or a non-targeting shRNA served as controls. Total RNA was harvested on day 3 after shRNA lentiviral infection and E-cadherin expression was measured by RT-qPCR. See also Figure S2 and Table S4.
Based on recent reports that MET is required at the initial stages of reprogramming (Li et al., 2010; Samavarchi-Tehrani et al., 2010), we examined E-cadherin expression, which is upregulated during MET, to determine whether shRNAs targeting the seven kinases facilitated this step of iPSC generation. Efficient knockdown of all target kinases (Figure S2C) resulted in a 1.5- to 2-fold increase in E-cadherin expression compared to control MEFs infected with non-targeting shRNA vector (Figure 2B), which was similar to that induced by knockdown of p53 (~2.5-fold; Figure 2B). Collectively, these data indicate that knockdown of the selected seven kinases promotes MET at the initial reprogramming step.
Kinase-depleted MEFs reprogram to the fully pluripotent state
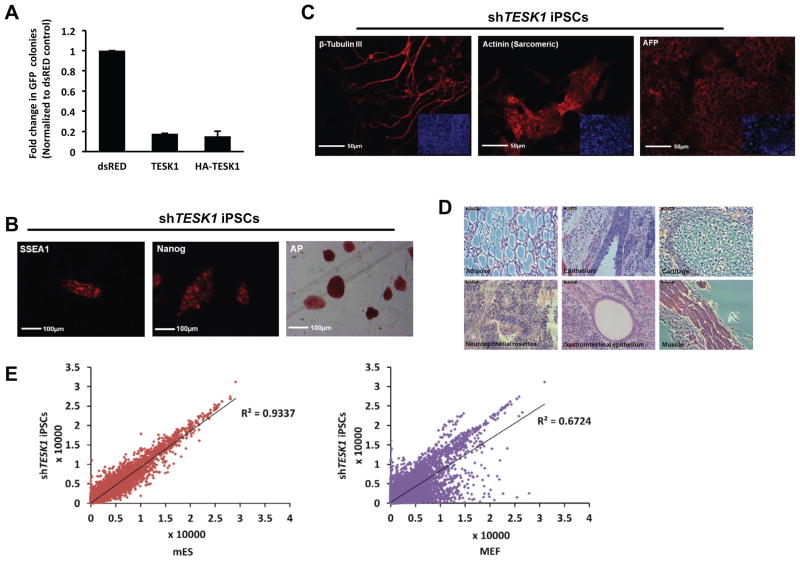
To address if the overexpression of a barrier kinase would compromise reprogramming, we cloned and expressed TESK1 or HA-TESK1 during 4F transduction. Immunoblotting confirmed the protein expression and function by enhancing COF phosphorylation (Figure S4B). We observed that overexpression of TESK1 decreased reprogramming efficiency of MEFs by more than 80% (Figure 3A).
Figure 3. shTESK1-iPSCs exhibit a fully pluripotent state.
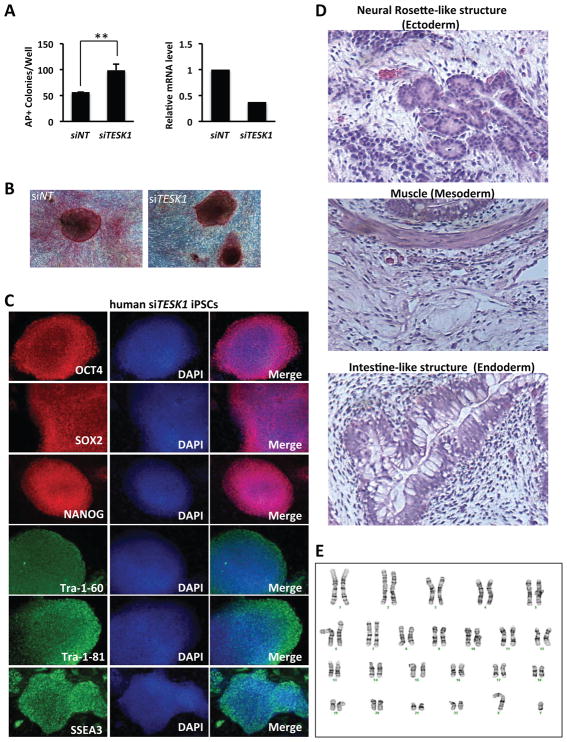
(A) TESK1 overexpression compromises reprogramming efficiency. TESK1 or HA-TESK1 were cloned and expressed during 4F transduction. Immunoblotting confirmed the protein expression and function by enhancing cofilin phosphorylation (Figure S4B). Reprogramming efficiency was decreased more than 80% in TESK1-overexpressing cells compared with pMX-dsRed control-transduced cells. (B) shTESK1-iPSCs switch on endogenous mESC markers. GFP+ shTESK1-iPSCs cultured on feeder layers and collected on 16 dpi show positive staining for SSEA-1, Nanog, and alkaline phosphatase (AP), all indicators of pluripotency. (C) shTESK1-iPSCs can differentiate into three germ layers in vitro. Embryoid bodies formed from shTESK1-iPSCs were collected on day 14, fixed with 4% PFA and immunostained for β-tubulin III (ectoderm), sarcomeric actinin (mesoderm), or α-fetoprotein (AFP; endoderm). Insets show DAPI staining of nuclei (blue) in a wider field of view. (D) shTESK1-iPSCs can differentiate into many lineages in vivo. shTESK1-iPSCs were injected subcutaneously into the backs of Nude mice. Teratomas were removed at 3–4 weeks and stained with H&E. (E) shTESK1-iPSCs show gene expression profiles similar to mESCs. Genome-wide mRNA expression of shTESK1-iPSCs was compared with mESCs and MEF controls. See also Figure S3.
Next, to determine if MEFs could be fully reprogrammed following knockdown of the barrier kinases, we characterized iPSC clones obtained following silencing of each kinase. After confirming that the shRNAs were integrated into the iPSC genomes (Figure S3A), we derived several iPSC clones for each kinase shRNA and examined them for expression of pluripotency markers. All clones were GFP+, indicative of reactivated Oct4 expression, and expressed alkaline phosphatase (AP), Nanog, and SSEA1 (Figures 3B and S3B). To investigate whether shRNA-iPSCs exhibited the full differentiation capacity of mESCs, we evaluated embryoid body (EB) formation. All derived clones showed efficient EB formation, and EBs showed positive staining for the lineage markers β-tubulin III (ectoderm), α-fetoprotein (AFP; endoderm), and α-actinin (mesoderm) (Figures 3C and S3C). We also examined the pluripotency of shTESK1-iPSCs in vivo by examining teratoma formation in Nude mice; indeed, we found that teratomas containing all germ layers were readily formed within 3–4 weeks of cell injection (Figure 3D). Whole-genome mRNA expression profiling also indicated that shTESK1-derived iPSC clones exhibited a gene expression pattern more similar to mESCs than to MEFs (Figure 3E). shTESK1-iPSCs were also able to generate chimeric mice when injected into recipient blastocysts (Figures S3D and S3E). Additional siTESK1-iPSCs were derived and their differentiation capacity was confirmed by formation of teratomas (Figure S3F) and generation of live chimeric mice (Figure S3G). Taken together, these data suggest that the kinase RNAi-derived iPSCs had reached a fully reprogrammed state.
TESK1 regulates reprogramming through effects on the cytoskeleton
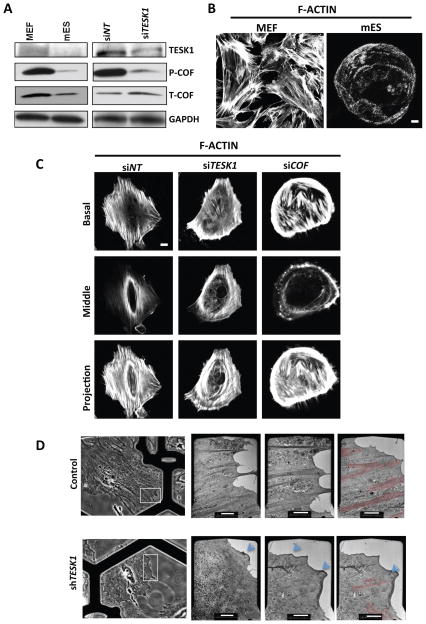
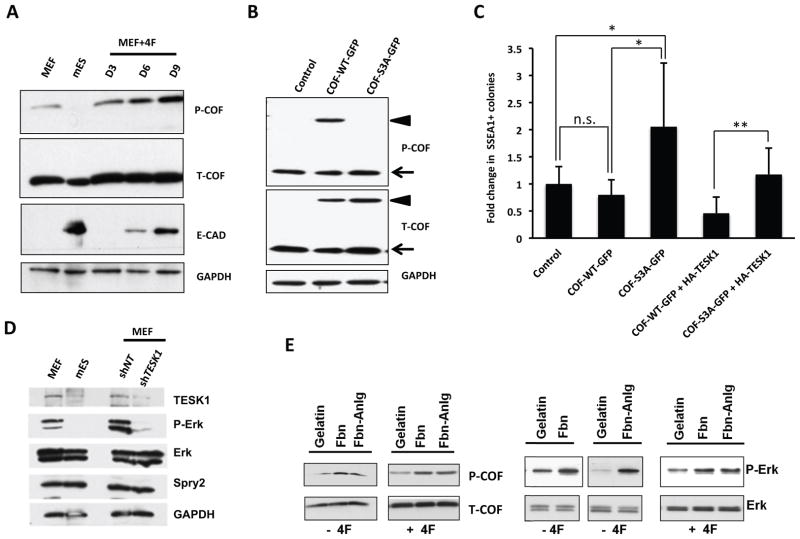
The functional activity and mechanism of two kinases was examined in further detail. TESK1 and LIMKs specifically phosphorylate the Actin-binding protein COF at serine 3 (Ser-3) in vitro and in vivo, and TESK1 stimulates formation of both actin stress fibers and focal adhesions (Toshima et al., 2001). Cofilin binding depolymerizes Actin, an activity inhibited by Ser-3 phosphorylation (Agnew et al., 1995). LIMK1 phosphorylation of COF has been suggested to play a role in Rac-mediated reorganization of the actin cytoskeleton (Yang et al., 1998). Because our analysis of the ILK signaling network by the IPKB system (Figure S5A) suggested that two of our identified kinases, TESK1 and LIMK2, phosphorylate COF, we hypothesized that knockdown of TESK1 and LIMK2 might enhance iPSC generation through effects on the actin cytoskeleton. Consistent with this, MEFs expressed higher levels of TESK1and phosphorylated cofilin (P-COF) than mESCs (Figure 4A), and exhibited a more highly organized actin cytoskeleton under confocal microscopy (Figure 4B). Quantitative analysis of band intensities revealed that mES cells had ~ 70% lower P-COF/T-COF levels compared to MEFs (Figure 4A).
Figure 4. TESK1 regulates cytoskeletal remodeling during reprogramming.
(A) TESK1 is highly expressed in MEFs and regulates cofilin phosphorylation. TESK1, phosphorylated (P-) cofilin, and total (T-) cofilin expression levels were detected by immunoblotting of extracts from Oct4-GFP MEFs, mESCs, or MEFs transfected with TESK1-targeting siRNA (siTESK1) or control siRNA (siNT). GAPDH served as an internal control. (B) Visualization of polynucleation of actin filaments. Actin filaments were detected by staining MEFs and mESCs with rhodamine-labeled phalloidin. (C) TESK1 knockdown disrupts actin filamentous structure. Actin cytoskeleton organization was visualized by confocal microscopy of MEFs transfected with control siRNA or sRNA targeting TESK1 or COF. The top, middle, and bottom panels show the basal section, middle section, and Z-projection, respectively. (D) TESK1 regulates filamentous actin network organization. MEFs expressing non-targeting shRNA (top row) or shTESK1 (bottom row) were imaged by light microscopy (column 1) or TEM (columns 2–4). The white boxes in the left panel mark the areas shown enlarged in the right panels. On top the view is rotated by ~45 degrees anticlockwise from the left panel to the right panels, on the bottom the view is rotated by ~20 degrees anticlockwise. Blue arrows indicate an isotropic network and small vertical ruffles, red highlighting indicates actin bundles. The experiment was repeated at least three times for each condition, with similar results. Scale bars = 20 μm in B and D and 5 μm in C. See also Figure S4.
To determine the role of TESK1 in stabilization of the cytoskeleton, we evaluated actin organization in MEFs treated with control siRNA or siTESK1. TESK1-KD MEFs showed decreased levels of P-COF, but not total cofilin (T-COF) (Figure 4A, right), and clear perturbation of the actin cytoskeleton, visualized by confocal microscopy (Figure 4C). Whereas MEFs expressing control siRNA exhibited polynucleation of actin filaments, this filamentous structure was disrupted in TESK1-KD and COF-KD MEFs (Figure 4C). These findings were supported by electron microscopic (EM) analysis of whole-mount MEFs cells expressing control or TESK1 shRNA (Figure 4D). This analysis allowed us to directly image the distinct morphologies of the leading edges of the cells and their associated transverse Actin bundles (Figure 4D dark grey or highlighted in red). In control cells, dense, directional stress fibers were found at the cell edge interspersed with isotropic actin networks. In contrast, the morphology of TESK1-KD MEFs revealed a loss of filaments in the ordered array of bundles, but the homogenous, isotropic filament networks were maintained. A few ruffles were also visible at the leading edge area (Figure 4D, arrows), and the few bundles present in the TESK1-KD cells were superimposed by a dense, homogeneous filament array characteristic of a lamella network. These structurally distinct networks are also defined by their molecular, kinetic, and kinematic signatures.
We next examined cofilin phosphorylation and structural changes in the actin cytoskeleton structures during reprogramming. For this, MEFs were transduced with OSKM followed by shTESK1, shLIMK2, or control shRNA on various days (D2, D4, and D6). TESK1 and LIMK2 knockdown MEFs showed decreased levels of P-COF compared with control MEFs, and these changes correlated well with the enhanced remodeling of the actin cytoskeleton in TESK1- and LIMK2-KD cells (Figure S4A). Collectively, these findings suggest that TESK1 and LIMK2 play important roles in remodeling of the actin cytoskeleton during iPSC generation.
To probe this further, we examined the effects of retroviral-mediated overexpression of unmodified or HA-tagged TESK1 protein in MEFs (confirmed by immunoblotting; Figure S4B). Notably, overexpression of TESK1 resulted in a corresponding increase in phosphorylated, but not total, COF (Figure S4B). Moreover, overexpression of TESK1 decreased reprogramming efficiency of MEFs by more than 80% (Figure 3A) and decreased pluripotent marker expression in CCE mESCs (Figure S5E). These data confirm that TESK1 acts as a barrier kinase for iPSC generation and further suggest that TESK1 and LIMK2 modulate reprogramming through effects on COF phosphorylation and cytoskeletal remodeling.
Recently, Wiggan et al. (Wiggan et al., 2012) reported that from unicellular organisms to humans, ADF/cofilins have a conserved function to inhibit Myosin II-Actin binding. Since phosphorylation of cofilin inactivates its interaction with Actin, we reasoned that TESK1 or cofilin depletion would regulate the physiological and competitive equilibrium between actin-cofilin and myosin-actin interactions, thus providing further insight into the TESK1 role in cytoskeletal reorganization. To address this hypothesis, we treated MEF cells with siRNAs targeting TESK1, COFILIN (COF), or siNT and untreated MEF cells were used as controls. Confocal images of MEF cells immunostained for Myo IIb (green) and F-actin using Rhodamine phalloidin (red) were obtained (Figure S4E). In control cells MyoIIb was found on the Actin cables (marked by broken white line). Similarly, in siCOF treated cells, MyoIIb was also located on the actin cables (marked by broken lines). On the other hand, in siTESK1 treated cells, MyoIIb was found dispersed without forming obvious foci on the Actin cables. In TESK1 knockdown cells, which have less P-COF than control NT cells and hence increased levels of active COF, we found decreased colocalization of Myosin on the Actin cables compared to either control or COF knockdown cells. These findings further support our results presented above suggesting that TESK1 regulates the Actin cytoskeleton through COF phosphorylation and TESK1 indeed plays a role in maintaining the equilibrium between active and phospho-COF levels in MEF cells.
Cofilin phosphorylation modulates reprogramming
To determine the dynamics of cofilin phosphorylation (P-COF) during reprogramming, MEFs were transduced with OSKM, cells were harvested on various days (D3, D6, and D9), and P-COF and total cofilin (T-COF) levels were analyzed by immunoblotting. When compared with mES and MEF cells, P-COF was gradually increased during reprogramming from D3 to D9 while T-COF remained unchanged (Figure 5A). We also analyzed E-CAD expression levels that served as an indicator of MET and ES cell states (Fig 5A).
Figure 5. Cofilin phosphorylation modulates reprogramming.
(A) Cofilin phosphorylation (P-COF) is dynamically regulated during the reprogramming process. 4F transduced MEFs were harvested on different days (D3, D6 and D9) and the cell lysates were used to analyze P-COF and total cofilin (T-COF) levels by immunoblotting. GAPDH served as loading control while E-CAD served as marker for MET and ES cell states. (B) Immunoblot analysis of MEF cells overexpressing GFP-fusion proteins, wild-type cofilin (COF-WT-GFP) or a mutant cofilin (COF-S3A-GFP), in the presence of non-targeting (siNT) or cofilin-targeting (siCOF) siRNA. Empty expression vector was used as a transfection control and GAPDH was used as loading control. Arrowhead indicates exogenously expressed cofilin-GFP and arrow indicates endogenous cofilin. (C) Phosphorylation incompetent cofilin promotes iPSC generation. Quantification of SSEA1+ iPSC colonies obtained from MEFs transduced with OSKM plus empty vector, wild-type cofilin (COF-WT-GFP), mutant cofilin (COF-S3A-GFP), COF-WT-GFP+HA-TESK1 or COF-S3A-GFP+HA-TESK1. Results are the means ± SD of three independent experiments. (D) Knockdown of TESK1 decreases ERK phosphorylation. Immunoblotting of TESK1, ERK1/2, phospho-ERK1/2, and Spry2 in Oct4-GFP MEFs, mESCs, and non-targeting (NT) shRNA- and shTESK1-transduced Oct4-GFP MEFs 4 days post-transduction. GAPDH served as an internal control. (E) Activation of the ILK signaling pathway results in ERK and cofilin phosphorylation. MEFs were grown on plates coated with fibronectin (Fbn) or a fibronectin analogue (Fbn-Anlg) to induce the ILK signaling pathway. MEFs seeded on gelatin-coated plates served as controls. Immunoblot analysis of phospho- and total ERK and cofilin in cell lysates prepared on 4 dpi. See also Figure S5.
To determine whether cofilin phosphorylation is specifically required to induce cytoskeletal remodeling and affect reprogramming, we transduced MEF cells with one of two retrorviral GFP constructs encoding wild-type cofilin (COF-WT-GFP) or a nonphosphorylatable serine 3 point mutant (COF-S3A-GFP). Extracts of these cells were prepared and analyzed by immunoblotting to verify protein expression and confirm that COF-WT-GFP but not COF-S3A-GFP was phosphorylated in MEFs (Figure 5B). Next, we analyzed reprogramming of MEFs expressing COF-WT-GFP or COF-S3A-GFP in the presence and absence of TESK1. Since TESK1 and LIMK2 are not limiting in cells, we reasoned that overexpression of wild type cofilin would lead to increased amount of COF-P, unlike in the situation where overexpression of mutant COF that cannot be phosphorylated. In addition, overexpression of WT-COF may not competitively perturb the balance between the non-phosphorylated and P-COF while mutant COF overexpression would tilt the balance towards the non-phosphorylated-COF. Thus, an increase in iPS generation should be observed when mutant COF was overexpressed and not with COF-WT overexpression. Consistent with our rationale, overexpression of the non-phosphorylatable COF-S3A-GFP mutant protein showed enhanced iPSC production mimicking the TESK1 knockdown effects, while the overexpression of its wild-type counterpart showed insignificant reduction in the iPS colony formation (Figure 5C). Furthermore, overexpression of the HA-TESK1 along with either COF-S3A-GFP or COF-WT-GFP rescues the phenotype shown by overexpression of these constructs in isolation. Taken together, these results establish the specificity of cofilin phosphorylation and its role in iPSC production and further support the notion that TESK1 acts as a barrier to reprogramming by promoting cofilin phosphorylation.
Crosstalk between enzymes of the ILK and TGFβ pathways regulates cytoskeletal remodeling
Cofilin phosphorylation and actin cytoskeleton remodeling are observed following activation of both the ILK pathway enzyme, TESK1 (Toshima et al., 2001) and the TGFβ pathway enzyme, LIMK2 (Vardouli et al., 2005). Therefore, we next asked whether there might be crosstalk between these pathways during reprogramming. The involvement of TGFβ and its downstream effector protein ERK1/2 in reprogramming of somatic cells is well established. Inhibition of the TGFβ pathway activates reprogramming (Ichida et al., 2009; Li et al., 2009; Lyssiotis et al., 2009; Maherali and Hochedlinger, 2009; Shi et al., 2008), and blocking ERK activity can lead to reprogramming of neural stem cells (Silva et al., 2008). Mouse ESC self-renewal is promoted by suppression of the phosphatase SHP-2 and ERK1/2, and conversely, is impaired by ERK activation (Burdon et al., 1999). As reported previously, we observed that ERK phosphorylation is downregulated in mESCs compared with MEFs (Figure 5D), which secures the ground state of ESC self-renewal (Nichols et al., 2009). Interestingly, we found that expression of phosphorylated ERK1/2 was decreased in shTESK1-KD MEFs, whereas total ERK 1/2 and Spry2 levels were unaffected (Figure 5D).
To determine if crosstalk between the ILK and TGFβ pathways might contribute to regulation of the cytoskeletal structure in MEFs, cells were seeded on control plates coated with gelatin, or plates coated with the integrin ligand fibronectin or a fibronectin analogue. Cells were untreated or 4F-transduced and then harvested on different days. Immunoblot analysis of total cell lysates showed that incubation of cells on fibronectin or the analogue induced ERK1/2 and cofilin phosphorylation but did not affect total ERK and cofilin levels (Figure 5E). Actin cytoskeletal remodeling was also observed upon treatment with the ligands. Both fibronectin and its analogue induced an extensive polynucleated mesh-like network of the actin cytoskeleton in the presence and the absence of 4F (Figure S5B). Treatment of cells with the TGFβR1 inhibitor accelerated dissolution of the filamentous structure, and this was evident as early as day 3 (data not shown). These data altogether suggest the existence of crosstalk between the ILK and TGFβ pathways at the level of ERK and cofilin phosphorylation, which is supported by the observations on actin cytoskeleton remodeling.
Expression of ILK pathway genes and TESK1 is altered during reprogramming
To determine how the expression of ILK pathway genes changes during reprogramming, we examined expression in MEFs on different days after 4F transduction and compared the pattern with that in MEFs and mESCs. Virtually all of the ILK pathway genes examined were severely downregulated in CCE mESCs compared with MEFs (Figure S5C). However, the expression patterns of individual genes varied over the course of reprogramming. Some genes were downregulated as early as 3 dpi (Integrin β3, ILK, β-actin), others were up- or downregulated during the reprogramming process and were then extinguished at the fully pluripotent stage (Integrin α6 and β1), and some showed very little change in expression during reprogramming (Rac and LIMK2). Interestingly, TESK1 and LIMK1 were upregulated soon after 4F transduction and were fully downregulated only at the ESC stage (Figure S5C). We also examined which transcription factor(s) play critical roles in regulating expression of individual genes by transducing MEFs with the four factors in all possible combinations. qRT-PCR analysis of cells on 8 dpi identified Klf4 as the key regulator of TESK1 expression, although other factors also contributed (Figure S5D). Furthermore, overexpression of HA-TESK1 in ES cells reduced the levels of pluripotency marker genes (Figure S5E). Klf4 also regulated the expression of LIMK1 and β-actin. For LIMK2, Rac, ILK, and Integrin β1 expression, no single factor appeared to influence their expression preferentially. Integrin α6 and β3 were upregulated by Klf4 but downregulated by other factors (Figure S5F). Taken together, these data indicate that the expression of ILK pathway genes and TESK1 is regulated by all four reprogramming factors, with Klf4 playing the most significant role.
TESK1 knockdown promotes reprogramming of human fibroblasts
To assess whether TESK1 knockdown could promote human iPSC generation, human foreskin fibroblast (BJ) cells were transfected with control or TESK1-targeting siRNA and reprogrammed to iPSCs using episomal DNA vectors. TESK1 siRNA effectively reduced TESK1 mRNA levels and resulted in an increase in AP+ colonies (Figure 6A). iPSCs derived from control and siTESK1 fibroblasts exhibited similar morphology (Figure 6B). Further characterization of human siTESK1-iPSCs by immunohistochemistry showed that these cells expressed human ESC markers, including OCT4, SOX2, NANOG, Tra-160, Tra-1-81, and SSEA3 (Figure 6C), indicating that these cells have achieved an ESC-like pluripotent state. Next, we determined the differentiation capabilities of siTESK1-iPSCs in vitro with EB formation assays and in vivo by teratoma formation in mice. EBs expressed markers of three germ layers, including SOX17, FOXA2, GATA4, NESTIN, and PAX6, on day 14 after inducing differentiation (Figure S6). Similarly, teratomas formed 8 weeks after transplantation of human iPSCs into the dorsal flanks of SCID mice contained tissues from three germ layers, including neural rosette-like structures (ectoderm), striated muscle (mesoderm), and intestine-like structures (endoderm) (Figure 6D). Further, karyotyping of the siTESK1-iPSCs showed normal chromosome structures (Figure 6E). Together, these results demonstrate that TESK1 KD promotes the generation of human iPSCs that are pluripotent and have a normal karyotype.
Figure 6. TESK1 knockdown promotes reprogramming of human fibroblasts.
(A) Left panel, number of AP+ clones derived from human BJ fibroblasts transfected with non-targeting (siNTC) or TESK1-targeting (siTESK1) siRNA. Right panel, RT-qPCR of TESK1 mRNA levels in control and TESK1-KD cells. (B) Representative AP+ clones derived from reprogramming of human BJ cells expressing siNT or siTESK1. (C) Expression of ESC pluripotent markers in iPSCs derived from human BJ cells expressing siTESK1. Staining for SSEA-3, NANOG, SOX2, Tra-1-81, Tra-1-60, and OCT4 was performed. (D) Teratoma formation shows the pluripotency of iPSCs. Cells were injected into SCID mice and tumors were harvested 8 weeks later and stained with H&E. (E) Normal karyotype of a human siTESK1-iPSC clone.
See also Figure S6.
DISCUSSION
Using Oct4-GFP MEFs and standard OSKM reprogramming protocols, we performed a kinome-wide functional genomics screen to identify the role of kinases in iPSC generation. In particular, we were interested in identifying signaling mechanisms or networks that could act as barriers to reprogramming. Induced reprogramming is an unnatural phenomena governed by the addition of transcription factors, RNA, or small molecules where one cell type is changed to another. In our case, we reprogrammed MEFs to iPSCs by using OSKM transcription factors. Therefore, addition of four factors in MEFs would upregulate resistance mechanisms for cell fate changes that we denote as “barrier pathways”. We reasoned that knockdown of kinases that lower these barrier signaling mechanisms would enhance reprogramming.
Our kinome wide RNAi screen identified 59 kinases involved in diverse highly interconnected molecular functions that acted as barriers to iPSC generation. Interestingly, of the 59 kinases identified as barriers in iPSC generation, five function in the integrin-linked kinase (ILK) signaling network. After in-depth analysis of seven of the kinases, we observed that of TESK1 or LIMK2 silencing in MEFs promoted MET, decreased cofilin phosphorylation, and disrupted actin filament structures during reprogramming. The morphology of TESK1-KD MEFs visualized by both fluorescence confocal microscopy and EM showed disruption of actin filaments in the ordered array of bundles. Using integrin ligands and small molecule inhibitors of TGFβR1, we found the existence of a crosstalk between the ILK and TGFβ pathways to regulate cytoskeletal remodeling.
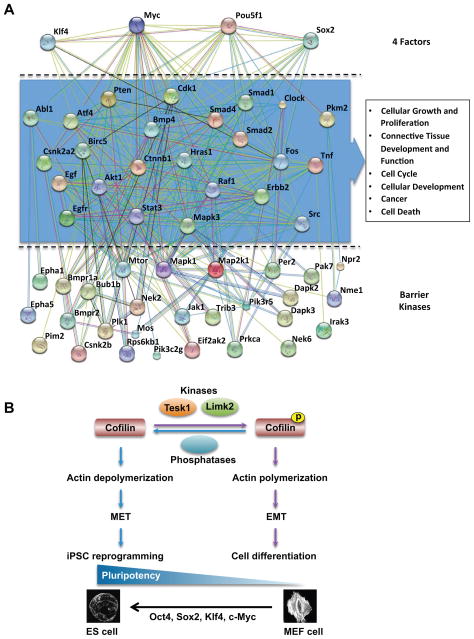
To build a kinome interaction network for iPSC generation and to understand the functions of the 59 barrier kinases and OSKM, the known and predicted protein interactions of barrier kinases and transcription factors were derived from the STRING database with high confidence score. We first analyzed the distribution and number of interacting proteins for each kinase because we reasoned that the proteins functionally interacting with both kinases and transcription factors would be likely to play important roles in iPSC generation. To narrow down the many interactions generated from the prediction and to increase the reliability of the interpretation, our analysis included only those proteins having functional associations with two or more kinases and at least one transcription factor. A total of 24 bridge proteins, listed in Table S4, were found to connect 28 barrier kinases and four transcription factors in iPSC generation. The interactome network showing the bridge proteins clustered in the middle tier is illustrated in Figure 7A. The analysis of canonical pathways indicated that most of the bridge proteins are components of signaling and cell communication pathways that trigger and regulate the EMT (Table S5). The top-ranked functional annotation of the bridge proteins in the enrichment analysis by Ingenuity IPA also suggested that these proteins were involved in the EMT and/or MET processes (Figure 7A).
Figure 7. Interactome network of kinases and cytoskeletal remodeling in reprogramming.
(A) Interactome network of kinases and transcription factors in iPSC generation. The interactome network depicts the functional associations of 24 bridge proteins that connect 28 barrier kinases and four transcription factors. The predicted network was generated by STRING (version 9.0) on the basis of protein interactions of high confidence (score >0.7). Each bridge protein is associated with at least two barrier kinases and one transcription factor. Top-ranked functional annotations generated by Ingenuity IPA suggests the bridge proteins are likely to be involved in the EMT and/or MET programs. Nodes and edges represent proteins and predicted functional associations, respectively. The colored lines indicate the seven types of evidence used in predicting the associations: red for fusion, green for neighborhood, blue for co-occurrence, purple for experimental, yellow for text mining, light blue for database, and black for coexpression evidence (see Table S5). (B) Proposed mechanism for TESK1 and LIMK2 function in cytoskeletal remodeling and iPSC generation. TESK1 or LIMK2 phosphorylates cofilin to promote the formation and/or stabilization of the cytoskeletal structures involved in EMT and cellular differentiation. RNAi-mediated silencing of either kinase inhibits cofilin phosphorylation, which in turn disrupts actin polynucleation, promotes the MET transition step of reprogramming in 4F-infected MEFs, and thus enhances iPSC generation.
Our results suggest a model in which TESK1 and LIMK2 regulate cytoskeletal changes during reprogramming through phosphorylation of cofilin and/or stabilization of the actin cytoskeleton. In the cell, phosphorylated and non-phosphorylated forms of cofilin exist in dynamic equilibrium maintained by kinases and phosphatases. Two cofilin-specific phosphatases, slingshot and chronophin, dephosphorylate cofilin and induce actin depolymerization in response to a number of stimuli (Kligys et al., 2007). Our data suggest a sequence of events in which RNAi-mediated silencing of TESK1 or LIMK2 in MEFs inhibits cofilin phosphorylation, disrupts actin polymerization, increases E-cadherin expression, promotes the MET step of reprogramming, and thus enhances iPSC generation (Figure 7B). Indeed, knockdown of the slingshot protein SSH1 in MEFs with OSKM, hyperphosphorylated cofilin and decreased the reprogramming efficiency by 40% (Fig S4C and D), further supporting the role of cofilin dephosphorylation in reprogramming. iPSC generation is not an efficient process and large heterogeneous populations of cells are created at various stages of reprogramming, most of which do not successfully attain the pluripotent state. As such, partially reprogrammed cells, which represent the large majority of cells, exhibit increased TESK1 expression, consistent with it being a barrier to reprogramming. Cells which are successfully reprogrammed, however, must have actin depolymerization to achieve their well established ES-like morphology as shown in Figure 4B. Reorganization of the actin cytoskeleton is critical in many cellular functions, including motility, adhesion, morphogenesis, and cytokinesis. Another intriguing possibility is that reorganization of the actin cytoskeleton following TESK1 or LIMK2 depletion alters the extracellular matrix structural framework to create an ESC niche, thereby facilitating iPSC generation. Future studies should shed light on these mechanisms. Overall, our study describes kinase regulators and networks involved in iPSC creation. These findings should foster the development of new technologies and increase our understanding of the mechanisms underlying somatic cell reprogramming.
EXPERIMENTAL PROCEDURES
MEF reprogramming
MEFs were isolated from E13.5 embryos derived from Oct4-GFP mice (Jackson Lab, #008214) (Lengner et al., 2007) and used (up to passage 4) for induction experiments. Oct4-GFP MEFs were maintained in growth medium (DMEM, 10% FBS, L-glutamine, MEM-NEAA). With slight modification, retrovirus production and induction of iPSCs were performed following the Takahashi et al. protocol (Takahashi et al., 2007). Lentivirus-containing media collected from infected 293FT cell cultures were added to the MEF plates on day 3. On the following day, medium was replaced with fresh MEF medium; from day 5 onward, the medium was replaced with mESC growth medium (DMEM, 15% FBS, LIF, monothioglycerol [MTG], L-glutamine, MEM-NEAA) every other day until GFP+ colony counting and picking (day 14 to 18). GFP+ iPSC colonies were trypsinized, resuspended in mESC medium, and plated on 12-well plates with an irradiated MEF feeder layer. iPSC clones were seeded on 24-well plates with feeder layers for immunostaining. For time course experiments, lentivirus-containing media were prepared fresh for each time point and were added to the MEF plates on indicated dpi. The medium was replaced with mESC growth medium on the following day and then every other day until counting of GFP+ colonies.
Teratoma formation by mouse iPSCs
iPSCs were trypsinized and resuspended in EB medium at 1 × 106 cells/ml. Nude mice were anesthetized with Avertin and 150 μl of iPSCs were injected subcutaneously into the back. Teratomas were visible after 1 week and surgically removed at 3–4 weeks. Tissues were fixed in zinc formalin solution overnight, washed three times with PBS, and stained with H&E. All animal work was approved by the Institutional Review Board and was performed following Institutional Animal Care and Use Committee guidelines.
mRNA microarray analysis
Total RNA was extracted from derived iPSCs using Trizol. mRNA microarray analysis was carried out by the microarray facility at the Sanford-Burnham Medical Research Institute. ArrayExpress accession: E-MTAB-2258. A scatter plot was used to compare the genome-wide mRNA expression profiles of iPSCs, MEFs, and mESCs.
Interaction network analysis
The protein-protein interactions of barrier kinases and four transcription factors, Myc, Klf4, Pou5f1, and Sox2, were collected by searching the STRING database (Szklarczyk et al., 2011), version 9.0, with confidence score >0.7. The enrichment analysis of the bridge proteins between barrier kinases and transcription factors was performed using Ingenuity IPA. The annotation of KEGG canonical pathways was downloaded from the Molecular Signatures Database (MSigDB) (Subramanian et al., 2005), version 3.0.
Confocal microscopy
The cells were treated with siRNAs and seeded onto the micro well dishes, 3 and one days prior to immunostaining, respectively. The cells were washed with PBS and fixed in 4% paraformaldehyde for 20 min at rt followed by a PBS wash and permeabilizing with PBST (PBS with 0.1% tritonX100) for 5 min. Cells were blocked with 5% BSA in PBST for 1 h, incubated with primary antibodies diluted in 2.5% BSA in PBST for 1 h at rt, and washed with PBST. After incubating with secondary antibodies (1:400) for 1 h at rt (diluted in 2.5% BSA in PBST), cells were washed with PBST and rinsed twice with PBS. For F-actin, the cells were stained with Rhodamine Phalloidin at 1:40 dilution in PBST for 20 min at rt. The Myosin antibodies were obtained from Cell signaling (cat # 5144). The images were acquired on Zeiss LSM710 confocal microscope and processed using Image J and Photoshop.
Supplementary Material
Highlights.
A Kinome-wide RNAi screen identifies kinases that regulate reprogramming
Four networks of 59 kinases show highly interconnected molecular functions
TESK1 and LIMK2 modulate actin cytoskeleton through phosphorylation of COFILIN
Crosstalk between ILK and TGFb pathways enzymes regulate cytoskeletal remodeling
Acknowledgments
We are grateful for the following shared resource facilities at the Sanford-Burnham Medical Research Institute: genomics and informatics, and data management cores for help with mRNA array experiments and data analysis; the animal facility for generation of teratomas and chimeric mice; the histology and molecular pathology core for characterization of tissues; and the cell imaging core for confocal microscopy. This work was supported in part by National Institutes of Health (NIH) grants (to TMR and CD). Stem cell and iPS core is supported in part by a grant from California Institute for Regenerative Medicine (CIRM). The correlative light and electron microscopy studies were supported by NIH grant P01-GM098412 (to DH).
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no conflicts of interest.
Supplemental Information includes Extended Experimental Procedures, six figures, and five tables can be found with this article online at http://XX.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agnew BJ, Minamide LS, Bamburg JR. Reactivation of phosphorylated actin depolymerizing factor and identification of the regulatory site. The Journal of biological chemistry. 1995;270:17582–17587. doi: 10.1074/jbc.270.29.17582. [DOI] [PubMed] [Google Scholar]
- Burdon T, Stracey C, Chambers I, Nichols J, Smith A. Suppression of SHP-2 and ERK signalling promotes self-renewal of mouse embryonic stem cells. Developmental biology. 1999;210:30–43. doi: 10.1006/dbio.1999.9265. [DOI] [PubMed] [Google Scholar]
- Hou P, Li Y, Zhang X, Liu C, Guan J, Li H, Zhao T, Ye J, Yang W, Liu K, et al. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013;341:651–654. doi: 10.1126/science.1239278. [DOI] [PubMed] [Google Scholar]
- Ichida JK, Blanchard J, Lam K, Son EY, Chung JE, Egli D, Loh KM, Carter AC, Di Giorgio FP, Koszka K, et al. A small-molecule inhibitor of tgf-Beta signaling replaces sox2 in reprogramming by inducing nanog. Cell stem cell. 2009;5:491–503. doi: 10.1016/j.stem.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kligys K, Claiborne JN, DeBiase PJ, Hopkinson SB, Wu Y, Mizuno K, Jones JC. The slingshot family of phosphatases mediates Rac1 regulation of cofilin phosphorylation, laminin-332 organization, and motility behavior of keratinocytes. The Journal of biological chemistry. 2007;282:32520–32528. doi: 10.1074/jbc.M707041200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DF, Su J, Ang YS, Carvajal-Vergara X, Mulero-Navarro S, Pereira CF, Gingold J, Wang HL, Zhao R, Sevilla A, et al. Regulation of embryonic and induced pluripotency by aurora kinase-p53 signaling. Cell stem cell. 2012;11:179–194. doi: 10.1016/j.stem.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengner CJ, Camargo FD, Hochedlinger K, Welstead GG, Zaidi S, Gokhale S, Scholer HR, Tomilin A, Jaenisch R. Oct4 expression is not required for mouse somatic stem cell self-renewal. Cell stem cell. 2007;1:403–415. doi: 10.1016/j.stem.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell stem cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Li W, Wei W, Zhu S, Zhu J, Shi Y, Lin T, Hao E, Hayek A, Deng H, Ding S. Generation of rat and human induced pluripotent stem cells by combining genetic reprogramming and chemical inhibitors. Cell stem cell. 2009;4:16–19. doi: 10.1016/j.stem.2008.11.014. [DOI] [PubMed] [Google Scholar]
- Li Z, Rana TM. A kinase inhibitor screen identifies small-molecule enhancers of reprogramming and iPS cell generation. Nature communications. 2012;3:1085. doi: 10.1038/ncomms2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry WE, Richter L, Yachechko R, Pyle AD, Tchieu J, Sridharan R, Clark AT, Plath K. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci U S A. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Hunter T. Degradation of activated protein kinases by ubiquitination. Annual review of biochemistry. 2009;78:435–475. doi: 10.1146/annurev.biochem.013008.092711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyssiotis CA, Foreman RK, Staerk J, Garcia M, Mathur D, Markoulaki S, Hanna J, Lairson LL, Charette BD, Bouchez LC, et al. Reprogramming of murine fibroblasts to induced pluripotent stem cells with chemical complementation of Klf4. Proc Natl Acad Sci U S A. 2009;106:8912–8917. doi: 10.1073/pnas.0903860106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N, Hochedlinger K. Tgfbeta signal inhibition cooperates in the induction of iPSCs and replaces Sox2 and cMyc. Current biology: CB. 2009;19:1718–1723. doi: 10.1016/j.cub.2009.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Silva J, Roode M, Smith A. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development. 2009;136:3215–3222. doi: 10.1242/dev.038893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Ruiz S, Panopoulos AD, Herrerias A, Bissig KD, Lutz M, Berggren WT, Verma IM, Izpisua Belmonte JC. A high proliferation rate is required for cell reprogramming and maintenance of human embryonic stem cell identity. Curr Biol. 2011;21:45–52. doi: 10.1016/j.cub.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell stem cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Shi Y, Desponts C, Do JT, Hahm HS, Scholer HR, Ding S. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell stem cell. 2008;3:568–574. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Silva J, Barrandon O, Nichols J, Kawaguchi J, Theunissen TW, Smith A. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008;6:e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Toshima J, Toshima JY, Amano T, Yang N, Narumiya S, Mizuno K. Cofilin phosphorylation by protein kinase testicular protein kinase 1 and its role in integrin-mediated actin reorganization and focal adhesion formation. Mol Biol Cell. 2001;12:1131–1145. doi: 10.1091/mbc.12.4.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardouli L, Moustakas A, Stournaras C. LIM-kinase 2 and cofilin phosphorylation mediate actin cytoskeleton reorganization induced by transforming growth factor-beta. J Biol Chem. 2005;280:11448–11457. doi: 10.1074/jbc.M402651200. [DOI] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Wiggan O, Shaw AE, DeLuca JG, Bamburg JR. ADF/cofilin regulates actomyosin assembly through competitive inhibition of myosin II binding to F-actin. Developmental cell. 2012;22:530–543. doi: 10.1016/j.devcel.2011.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CS, Lopez CG, Rana TM. Discovery of nonsteroidal anti-inflammatory drug and anticancer drug enhancing reprogramming and induced pluripotent stem cell generation. Stem cells. 2011;29:1528–1536. doi: 10.1002/stem.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Higuchi O, Ohashi K, Nagata K, Wada A, Kangawa K, Nishida E, Mizuno K. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature. 1998;393:809–812. doi: 10.1038/31735. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.