Abstract
The majority of non–small cell lung cancer (NSCLC) patients harbor EGFR-activating mutations that can be therapeutically targeted by EGFR tyrosine kinase inhibitors (EGFR-TKI), such as erlotinib and gefitinib. Unfortunately, a subset of patients with EGFR mutations are refractory to EGFR-TKIs. Resistance to EGFR inhibitors reportedly involves SRC activation and induction of epithelial-to-mesenchymal transition (EMT). Here, we have demonstrated that overexpression of CRIPTO1, an EGF-CFC protein family member, renders EGFR-TKI–sensitive and EGFR-mutated NSCLC cells resistant to erlotinib in culture and in murine xenograft models. Furthermore, tumors from NSCLC patients with EGFR-activating mutations that were intrinsically resistant to EGFR-TKIs expressed higher levels of CRIPTO1 compared with tumors from patients that were sensitive to EGFR-TKIs. Primary NSCLC cells derived from a patient with EGFR-mutated NSCLC that was intrinsically erlotinib resistant were CRIPTO1 positive, but gained erlotinib sensitivity upon loss of CRIPTO1 expression during culture. CRIPTO1 activated SRC and ZEB1 to promote EMT via microRNA-205 (miR-205) downregulation. While miR-205 depletion induced erlotinib resistance, miR-205 overexpression inhibited CRIPTO1-dependent ZEB1 and SRC activation, restoring erlotinib sensitivity. CRIPTO1-induced erlotinib resistance was directly mediated through SRC but not ZEB1; therefore, cotargeting EGFR and SRC synergistically attenuated growth of erlotinib-resistant, CRIPTO1-positive, EGFR-mutated NSCLC cells in vitro and in vivo, suggesting that this combination may overcome intrinsic EGFR-inhibitor resistance in patients with CRIPTO1-positive, EGFR-mutated NSCLC.
Introduction
Lung cancer is a major cause of cancer-related mortality worldwide. Non–small cell lung cancer (NSCLC) accounts for about 80% of all lung cancers. In 2004, somatic mutations in the tyrosine kinase domain of EGFR were described in NSCLC; most of those mutations confer sensitivity to the EGFR tyrosine kinase inhibitors (EGFR-TKI) erlotinib (1) and gefitinib (2, 3). EGFR-sensitizing mutations, such as in-frame deletions in exon 19 and L858R missense mutation account for about 90% of EGFR mutations of lung adenocarcinomas (1, 4, 5), and patients with these mutations are highly sensitive to EGFR-TKI treatment (5–7). EGFR-sensitizing mutations have been used for selection of patients with advanced NSCLC for EGFR-TKI treatment. Despite impressive clinical response to EGFR-TKIs, approximately 10% of NSCLC patients harboring EGFR-sensitizing mutations exhibit intrinsic resistance (disease progression) (8) and up to 40% do not attain a major response to treatment. Furthermore, all responding patients invariably acquire resistance following initial response within 10–16 months of therapy (9). Several acquired resistance mechanisms have been uncovered, including secondary EGFR gatekeeper mutation (T790M) (10–12), MET amplification, ERBB3 activation (13), PIK3CA mutation (14), or small cell lung cancer (SCLC) transformation (15). However, the acquired resistance mechanisms remain unknown in about 40% of cases. More recent studies have revealed mechanisms of EGFR-TKI acquired resistance in individuals with EGFR-sensitizing mutations, such as activation of AXL receptor tyrosine kinase (16) and amplification of CRKL oncogene (17). Many of these acquired resistance mechanisms can occur together and may potentially be active in different subclones of the tumor at the same time.
The mechanisms of intrinsic resistance to EGFR-TKIs in the presence of sensitizing mutations, on the other hand, are relatively unknown. The presence of K-Ras mutations confers intrinsic resistance to EGFR-TKIs in NSCLC, but K-RAS and EGFR mutations are usually mutually exclusive (4, 18). The presence of T790M-resistant mutations or other rare exon 20 mutations has been described in only a very small percentage of patients before exposure to EGFR-TKI treatment (19). Several studies showed that many EGFR-mutated NSCLC patients carry a common germline polymorphism of the proapoptotic gene BIM that results in deletion of the death-inducing BH3 domain of BIM and intrinsic resistance to EGFR-TKI therapy (20, 21), although the finding could not be confirmed in another study (22). Moreover, BIM expression is a good marker in predicting TKI resistance (23, 24). A better understanding of intrinsic resistance mechanisms in EGFR-mutated NSCLCs is critical to improving patient stratification and devising new therapeutic strategies.
Human CRIPTO1, also known as teratocarcinoma-derived growth factor 1 (TDGF1), is a glycosylphosphatidyl inositol–linked cell membrane–anchored protein that belongs to the EGF-CFC family (25, 26). CRIPTO1 was originally isolated from human undifferentiated NTERA-2 embryonic carcinoma cells and is not expressed in most adult tissues (27, 28). High levels of CRIPTO1 expression have been reported in a variety of human carcinomas (29) and associated with poor prognosis in gastric (30), colorectal (31), and breast cancer (32) patients. In vivo studies showed that ectopic CRIPTO1 expression induced epithelial-to-mesenchymal transition (EMT), and MMTV-CRIPTO1 transgenic mice developed hyperplasias and tumors in the mammary gland (33). Upon binding to the TGF-β subfamily of proteins NODAL, GDF1 and GDF3, CRIPTO1 functions as a coreceptor of ALK4/7 to activate SMAD2/3/4 and promotes cell proliferation, migration, invasion, and EMT. The latter 3 biological responses to CRIPTO1 probably occur through a GLYPICAN-1/SRC pathway that activates MAPK and PI3K/Akt signaling (34–36). Although CRIPTO1 has not been directly implicated in the resistance to cancer target–specific drugs, EMT and SRC activation are known to associate with EGFR inhibitor resistance of various cancers (37–40). Moreover, it has been reported that inhibition of CRIPTO1 by anti-CRIPTO1 antibodies sensitizes colon cancer and doxorubicin-resistant leukemia cells to cytotoxic drugs (41, 42).
MicroRNAs are involved in a variety of biologic and pathologic processes (43). Notably, the microRNA-200 (miR-200) family and miR-205 are downregulated in TGF-β–induced EMT cells, and ectopic expression of the miR-200 family and miR-205 inhibit TGF-β–induced EMT (44). Known miR-205 targets include ZEB1/ZEB2 (44) and SRC (45), both of which have been implicated in EMT regulation and drug resistance.
In this study, we demonstrate that CRIPTO1 activates both ZEB1 to promote EMT and SRC to stimulate AKT and MEK in the EGFR-mutant lung cancer cells that are resistant to EGFR-TKIs through downregulation of miR-205. The resistance mechanism is mediated through the SRC but not the ZEB1 axis. Higher CRIPTO1 expression correlates with intrinsic EGFR-TKI resistance in lung adenocarcinoma patients harboring EGFR-sensitizing mutations. Targeting EGFR and SRC in combination significantly inhibited the growth of CRIPTO1-positive, erlotinib-resistant NSCLC cells in vitro and in mouse xenograft models.
Results
CRIPTO1 is expressed in NSCLC tumors but not in cell lines.
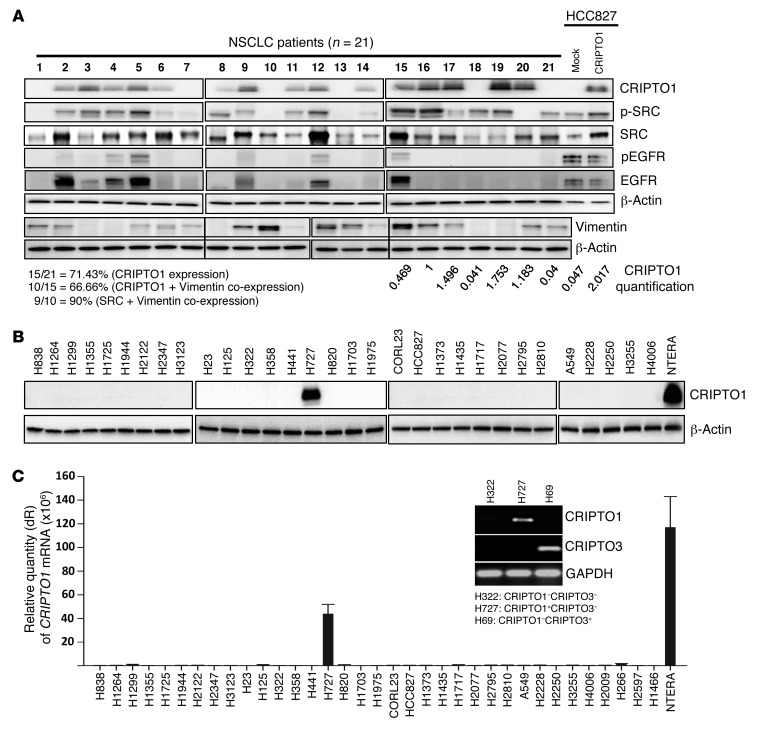
Previous reports established that CRIPTO1 is expressed in various tumors, but not in normal tissues (29). We assessed CRIPTO1 expression in 21 resected snap-frozen NSCLC tumor samples by Western blot. A significant portion (71.4%, 15 out of 21) of NSCLC samples expressed CRIPTO1 protein at various levels (Figure 1A). In contrast to primary NSCLC tumors, only 1 (H727) out of 31 tested NSCLC cell lines expressed CRIPTO1 protein and mRNA by Western blot and RT-PCR analyses, respectively (Figure 1, B and C). The specificity of CRIPTO1 primers for CRIPTO1 mRNA expression analysis is shown in Figure 1C, demonstrating that CRIPTO1-specific primers could only amplify CRIPTO1 in CRIPTO1-positive H727 cells, but not in H69 cells positive for CRIPTO3, a pseudogene that shares significant sequence homology with CRIPTO1 (46). Interestingly, CRIPTO1 expression was clearly detectable in early passages of primary cells derived from NSCLC patients but disappeared in later passaged cells (Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI73048DS1), suggesting that loss of CRIPTO1 expression might be a result of in vitro culture.
Figure 1. CRIPTO1 expression in NSCLC tumors and cell lines.
(A) Western blot analysis of CRIPTO1, total SRC, phosphorylated SRC, pEGFR, EGFR, and Vimentin in 21 NSCLC patient samples. Relative expression of endogenous CRIPTO1 in tumor cells and exogenous CRIPTO1 in HCC827 cells were shown after normalization against β-actin (lanes 15–21, HCC827 mock, and HCC827/CRIPTO1). Note that lanes 1–14 were not used for comparison, as they were derived from different blots, and that the lanes on 2 sides of the thin black lines were run on the same gel but were noncontiguous. (B) CRIPTO1 expression in 31 NSCLC cell lines by Western blot. (C) CRIPTO1 mRNA expression in 35 NSCLC cell lines by RT-PCR. CRIPTO1 primers could only amplify CRIPTO1 in CRIPTO1-positive/CRIPTO3-negative (H727) cells, but not CRIPTO3 in CRIPTO3-positive/CRIPTO1-negative (H69) cells (inset).
Ectopic CRIPTO1 expression renders EGFR-mutated NSCLC cells resistant to erlotinib and dacomitinib.
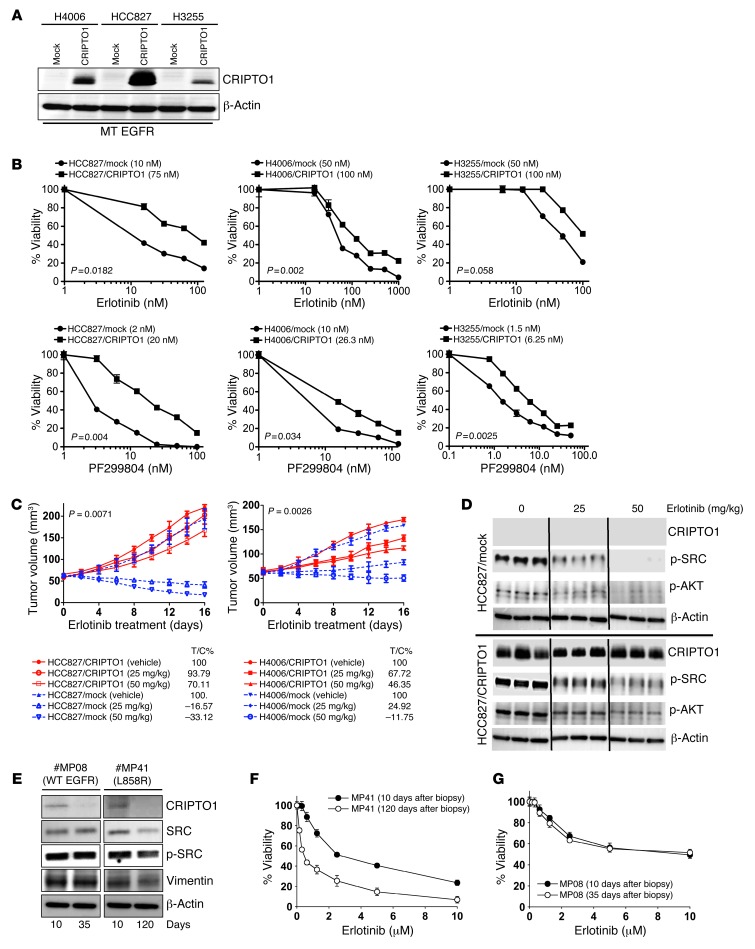
In our NSCLC panel, we noticed a strong correlation between CRIPTO1 expression and SRC phosphorylation in which SRC was phosphorylated in most (93%, 14 out of 15; P = 0.0113) of the CRIPTO1-positive NSCLC samples (Figure 1A). CRIPTO1 can trigger EMT and SRC activation in several in vitro and in vivo models (33, 36). As both SRC and EMT have been implicated in resistance to EGFR-TKIs in several cancer types (40, 47–50), we hypothesized that CRIPTO1 upregulation may render EGFR-mutated NSCLC cells resistant to EGFR-TKIs through activation of EMT and/or SRC signaling. In order to test this hypothesis, we first ectopically expressed CRIPTO1 by stable transfection in 4 CRIPTO1-negative NSCLC cell lines that included 3 EGFR-mutated lines (HCC827 [exon 19 del], H4006 [exon 19 del], and H3255 [L858R], Figure 2A) and one EGFR WT line (H322, Supplemental Figure 2A). Note that the ectopic CRIPTO1 expression level was only marginally higher than the endogenous CRIPTO1 expression of NSCLC tumors (Figure 1A), confirming that the ectopic CRIPTO1 expression level is close to its endogenous counterpart in tumor cells. We also used siRNA to knock down CRIPTO1 in the one EGFR WT cell line H727 that expresses CRIPTO1 (Supplemental Figure 2A). These cells were then exposed to the EGFR inhibitor erlotinib or to the irreversible pan-HER inhibitor dacomitinib/PF299804. Remarkably, CRIPTO1-transfected EGFR-mutated cell lines exhibited higher IC50 than their mock-transfected counterparts in response to erlotinib or dacomitinib treatment, respectively (Figure 2B and Supplemental Table 1A). Caspase 3/7 and cell-cycle assays revealed that the CRIPTO1 attenuated EGFR inhibitor–induced apoptosis (Supplemental Figure 3A) and, to some extent, G1 arrest too (Supplemental Figure 3B), which may account for the CRIPTO1-induced drug resistance. On the other hand, ectopic expression in the EGFR WT cell line H322 or CRIPTO1 knockdown in the EGFR WT cell line H727 had no effect on their sensitivities to erlotinib or dacomitinib (Supplemental Figure 2B and Supplemental Table 1B). To investigate whether CRIPTO1-mediated resistance is specific to EGFR-TKIs (erlotinib and dacomitinib), we treated the CRIPTO1-transfected cells with cisplatin (alkylating agent) and paclitaxel (taxol, microtubule stabilizing drug), both of which are commonly used chemotherapeutics in lung cancer patients (51, 52). Unlike in the erlotinib and dacomitinib experiments, IC50s of cisplatin and paclitaxel were similar between CRIPTO1- and mock-transfected EGFR-mutated HCC827 and H4006 cells (Supplemental Figure 4, A and B). To further verify these findings, we tested the effect of CRIPTO1 in EGFR-mutated NSCLC cells in a mouse xenograft model. As expected, erlotinib significantly attenuated the growth of mock-transfected EGFR-mutated HCC827 and H4006 (HCC827/mock and H4006/mock) xenograft tumors. In contrast, erlotinib had no inhibitory effect on the growth of CRIPTO1-transfected HCC827 and H4006 (HCC827/CRIPTO1 and H4006/CRIPTO1) xenograft tumors (Figure 2C). Taken together, these results suggest that CRIPTO1 renders EGFR-mutated NSCLC cells resistant to EGFR-TKI in vitro and in vivo and that CRIPTO1-mediated drug resistance is EGFR-TKI specific.
Figure 2. CRIPTO1 renders EGFR-mutated NSCLC cells resistant to EGFR-TKIs in vitro, in vivo, and in primary cultures.
(A) Western blot of CRIPTO1 in CRIPTO1-transfected NSCLC cells. MT, mutant. Exogenous CRIPTO1 expression in HCC827/CRIPTO1 cells is the highest among the 3 transfected cell lines and comparable to that in NSCLC samples (Figure 1A), indicating that exogenous CRIPTO1 expression in all the 3 CRIPTO1-transfected cells should be similar to that in the NSCLC samples. (B) Effect of CRIPTO1 on erlotinib and dacomitinib sensitivity of EGFR-mutant NSCLC cells. MTS assays were performed 72 hours after drug treatment. Data represent mean ± SD of triplicate measurements relative to untreated cells. P values were calculated using paired 2-tailed t test. IC50s are reported in parenthesis in graphs. (C) Effect of CRIPTO1 expression on erlotinib sensitivity of HCC827 and H4006 cells in vivo. See Methods for details. P values were significant between vehicle and all the treatment groups (ANOVA). Bars indicate SEM. (D) Western blot of CRIPTO1, pSRC, and pAKT in xenograft tumors. Tumor cells were harvested at day 16 after erlotinib treatment. Lanes were run on the same gel but were noncontiguous. (E) Progressive loss of CRIPTO1 expression during in vitro culture of human primary NSCLC cells. Western blot of CRIPTO1 of primary cells derived from an intrinsic erlotinib-resistant NSCLC patient carrying L858R EGFR (MP41) and an NSCLC patient carrying WT EGFR (MP08) at the indicated days of primary culture. (F and G) Correlation between progressive loss of CRIPTO1 expression and erlotinib sensitivity in MP08 and MP41 cells.
To determine whether CRIPTO1-mediated EGFR-TKI resistance has any clinical implications, we examined erlotinib sensitivity of primary tumor cell cultures derived from either NSCLC patients carrying WT (MP08) or mutant EGFR (MP41). As illustrated in Figure 2E and Supplemental Figure 1A, CRIPTO1 expression was prominent in the primary cells at early passages but diminished in all cells at later passages. MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphen yl)-2-(4-sulfophenyl)-2H-tetrazolium) analysis showed that loss of CRIPTO1 expression in the EGFR-mutated MP41 cells correlated with increasing sensitivity to erlotinib (Figure 2F, IC50 changed from 2.5 μM to 300 nM), whereas such correlation was not observed in the EGFR WT MP08 cells (Figure 2G, IC50 is 10 μM). In agreement with this finding, the NSCLC patient from whom the EGFR-mutated MP41 cells were derived showed progressive disease (PD) (increase in lung disease and pleural effusion) after 6 weeks of erlotinib treatment (ClinicalTrials.gov identifier: NCT01306045). This finding suggests that CRIPTO1 expression contributes to intrinsic resistance to erlotinib in NSCLC patients carrying EGFR-sensitizing mutations.
CRIPTO1 activates SRC and induces EMT in EGFR-mutated NSCLCs.
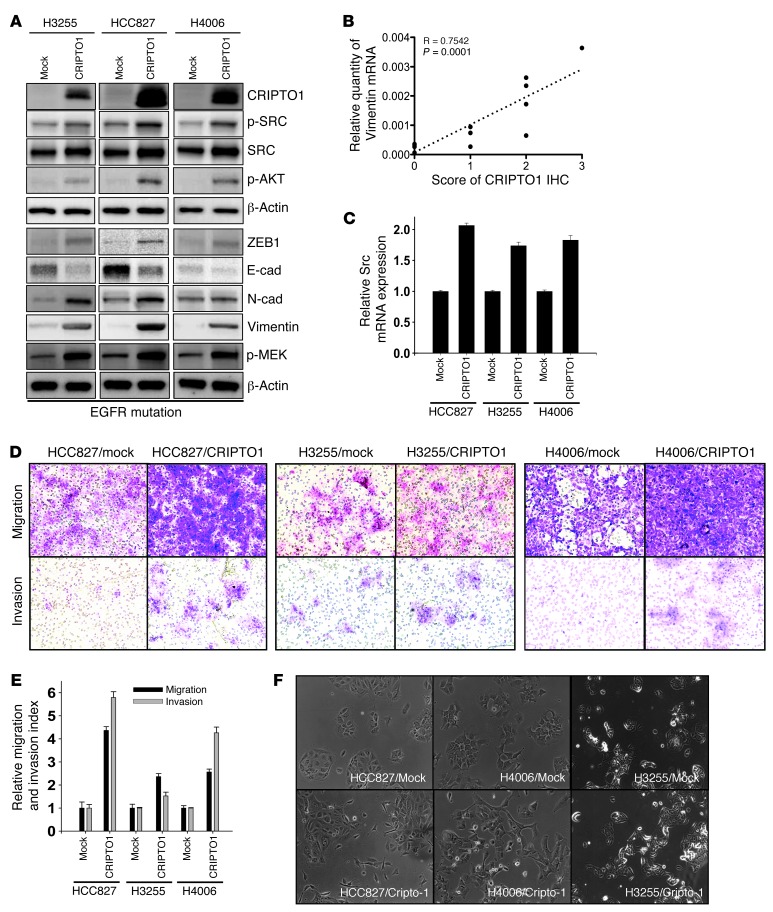
Although CRIPTO1 has been implicated in EMT and SRC activation in gastric cancer, breast cancer, and melanoma (30, 53, 54), the involvement of EMT and SRC activated by CRIPTO1 in EGFR-TKI resistance in EGFR-mutated NSCLC has not been explored. We thus examined EMT markers (ZEB1, E-cadherin, N-cadherin, and Vimentin) and SRC-signaling proteins (phospho-SRC and phospho-AKT) by Western blot in CRIPTO1-transfected HCC827, H4006, H3255, and H322 cells (Figure 3A and Supplemental Figure 5A). We observed that ZEB1 was upregulated in all CRIPTO1-transfected cells compared with their mock-transfected counterparts (Figure 3A), whereas ZEB1 was downregulated in H727 cells with CRIPTO1 siRNA knockdown (Supplemental Figure 5A). Concomitant N-cadherin upregulation and E-cadherin downregulation were also evident in CRIPTO1-transfected H3255 and HCC827 cells (Figure 3A). Consistent with these findings, Vimentin was found to be expressed in the majority of CRIPTO1-positive NSCLC samples (Figure 1A) and a positive correlation was found between the 2 markers (Figure 3B); Vimentin was also expressed in MP41 cells at early passages (Figure 2E), though no change in EMT-like morphology was detected between early and later passages of MP41 cells (Supplemental Figure 1B). Interestingly, CRIPTO1 induced Vimentin expression in EGFR-mutated H3255, HCC827, and H4006 cells (Figure 3A), but no change in Vimentin expression was found in EGFR WT H322/CRIPTO1 and H727/CRIPTO1 siRNA cells (Supplemental Figure 5A). These results suggest that activation of CRIPTO1 triggers EMT signaling in EGFR-mutated NSCLC cells. To further validate this finding, we examined the invasion and migration capacity, the archetypal properties of EMT (55), in CRIPTO1-transfected cells. Boyden chamber migration and invasion assays showed that CRIPTO1 augmented migration and invasion in EGFR-mutated NSCLC cell lines (HCC827, H3255, and H4006), but not in EGFR WT cell line H322 (Figure 3, D and E, and Supplemental Figure 5, C and D). In addition, HCC827/CRIPTO1 and H4006/CRIPTO1 cells exhibited more prominent mesenchymal-like morphology than their mock-transfected counterparts (Figure 3F). No obvious mesenchymal morphology was observed in EGFR WT H322/CRIPTO1 and mock-transfected cells (Supplemental Figure 5E). Similarly, CRIPTO1 knockdown did not significantly alter the migration, invasion, or EMT morphology of EGFR WT H727 cells (Supplemental Figure 5, C–E). Though the mechanism or mechanisms underlying the different EMT response between EGFR WT and mutant cells are currently unclear, the differences correlated with the differential Vimentin expressions induced by CRIPTO1 between EGFR mutant and WT and cell lines (Figure 3A and Supplemental Figure 5A). Our data suggest that CRIPTO1 induces EMT in NSCLC cells, particularly in EGFR-mutated NSCLC cells.
Figure 3. CRIPTO1 activates the SRC pathway and induces EMT in EGFR-mutated NSCLC cells.
(A) Western blot analysis of both SRC and EMT signaling proteins in CRIPTO1 stably transfected NSCLC cell lines. The CRIPTO1 blot is the same blot shown in Figure 2A. (B) Correlation between IHC scores of CRIPTO1 and Vimentin expression in 13 NSCLC patients’ samples. (C) CRIPTO1 increases SRC mRNA expression in EGFR-mutated NSCLC cells. (D and E) CRIPTO1 increases migration and invasion of EGFR-mutated NSCLC cells. Mock and CRIPTO1-transfected NSCLC cell lines were serum starved for 24 hours and then analyzed for cell migration and invasion at 24 and 48 hours, respectively. Cell migration and invasion indexes were measured as described in Methods. Data are representative of 3 independent experiments. (F) Phase-contrast images of CRIPTO1-transfected NSCLC cell lines. Note morphology changes in HCC827/CRIPTO1 and H4006/CRIPTO1 cells. Original magnification, ×200.
To determine whether CRIPTO1 may activate the SRC-signaling pathway in NSCLC cells, we examined SRC, AKT, and MEK phosphorylation in CRIPTO1-overexpressing and -knockdown cells. Similar to the above-mentioned EMT induction results, we found that CRIPTO1-transfected EGFR-mutated NSCLC cells (H3255, HCC827, and H4006) showed increased SRC at both protein and mRNA levels and phosphorylated SRC, AKT, and MEK compared with their mock-transfected counterparts, with the only exception being pMEK, which did not change significantly in H3255 cells (Figure 3, A and C). In contrast, neither CRIPTO1 overexpression nor knockdown significantly altered SRC, AKT, and MEK phosphorylation in EGFR WT NSCLC cells (H322 and H727, Supplemental Figure 5, A and B). Taken together, our results suggest that CRIPTO1 activates both EMT and SRC pathways in EGFR-mutated NSCLCs in vitro and in vivo.
CRIPTO1 activates EMT and SRC pathways through downregulation of miR-205 expression.
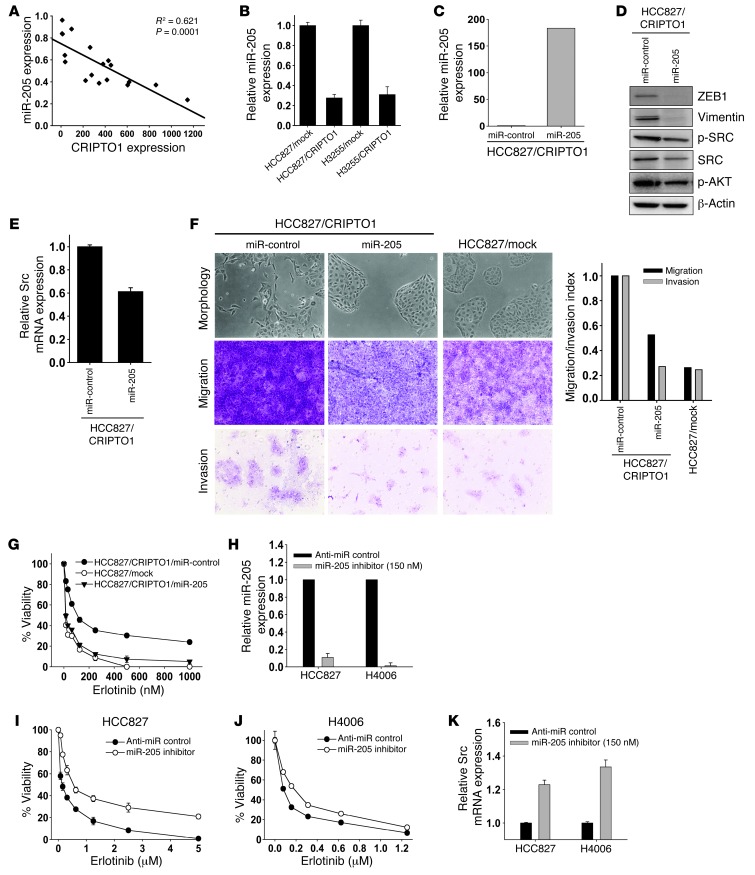
Since both ZEB1 and SRC are direct targets of miR-205 (45, 56–58), we examined whether CRIPTO1 may control EMT and SRC activity through miR-205. Since CRIPTO1 function has not been linked to miR-205 previously, we first studied CRIPTO1 and miR-205 expression in lung adenocarcinoma patient samples (n = 17) by real-time PCR. A significant inverse correlation between the expression of miR-205 and CRIPTO1 was identified (R2 = 0.621, P = 0.001), in which miR-205 was significantly downregulated in samples with high CRIPTO1 expression and upregulated in samples with low CRIPTO1 expression (Figure 4A). Moreover, miR-205 expression in CRIPTO1-transfected HCC827 and H3255 cells was 4-fold lower than in their mock-transfected HCC827 counterparts (Figure 4B). Based on these results, we reasoned that CRIPTO1 might regulate SRC and ZEB1 through downregulation of miR-205. To test this hypothesis, we investigated whether reconstitution of miR-205 expression in CRIPTO1-transfected cells may revert EMT and SRC signaling. HCC827/CRIPTO1 cells were transfected with miR-205, and the level of ectopic miR-205 expression was confirmed by miR-205 quantitative RT-PCR (qRT-PCR) analysis (Figure 4C). As shown in Figure 4, D and E, ectopic miR-205 expression in HCC827/CRIPTO1 cells led to significant downregulation of ZEB1, Vimentin, phosphorylated SRC, total SRC, and SRC mRNA; this was accompanied by a reversion of EMT morphology, and a reduction of cell migration and invasion (Figure 4F). We next tested whether miR-205 could restore erlotinib sensitivity by reverting EMT and SRC signaling in HCC827/CRIPTO1 cells. Indeed, ectopic miR-205 expression reversed erlotinib resistance in HCC827/CRIPTO1 cells (Figure 4G). Conversely, knockdown of miR-205 by miR-205 inhibitor (Figure 4H) rendered HCC827 and H4006 cells resistant to erlotinib (Figure 4, I and J), accompanied by upregulation of SRC (Figure 4K). These results suggest that CRIPTO1 activates ZEB1 and SRC through downregulation of miR-205 and that CRIPTO1-induced erlotinib resistance may be attributed to ZEB1 and SRC activation.
Figure 4. CRIPTO1 activates SRC and EMT pathways through downregulation of miR-205.
(A) Inverse correlation between CRIPTO1 and miR-205 expression in patient NSCLC samples. The graph illustrates a nonlinear regression analysis of CRIPTO1 protein and miR-205 expression in 17 lung adenocarcinoma samples. (B) Real-time PCR analysis of miR-205 expression. miR-205 expression is downregulated by CRIPTO1 in HCC827 and H3255 cells. (C) Real-time PCR analysis of ectopic miR-205 expression in HCC827/CRIPTO1/miR-205 and HCC827/CRIPTO1/mock stable cells. (D) Effect of ectopic miR-205 expression on ZEB1, Vimentin, pSRC, total SRC, and pAKT in HCC827/CRIPTO1 revealed by Western blot analysis. (E) Downregulation of total Src mRNA in miR-205–transfected HCC827/CRIPTO1 cells by real-time PCR analysis. (F) Effect of ectopic miR-205 expression on EMT morphology, migration, and invasion of HCC827/CRIPTO1 cells. Quantitation of migration and invasion is shown at the bottom. Original magnification, ×200. (G) Ectopic miR-205 expression renders HCC827/CRIPTO1 cells sensitive to erlotinib. MTS assays were performed 72 hours after erlotinib treatment of the indicated cells at the different concentrations. Data represent mean ± SD of triplicate experiments relative to untreated cells. (H) Real-time PCR analysis of miR-205 expression in HCC827 and H4006 cells treated with miR-205 inhibitor (AM11015; Ambion). Note that miR-205 knockdown efficiency by miR-205 inhibitor was comparable to CRIPTO1-induced miR-205 downregulation (see Figure 4B). (I and J) miR-205 inhibition renders HCC827 cells resistant to erlotinib. (K) Real-time PCR analysis of SRC expression in miR-205 inhibitor–treated HCC827 and H4006 cells.
CRIPTO1-induced erlotinib resistance is SRC dependent and ZEB1 independent.
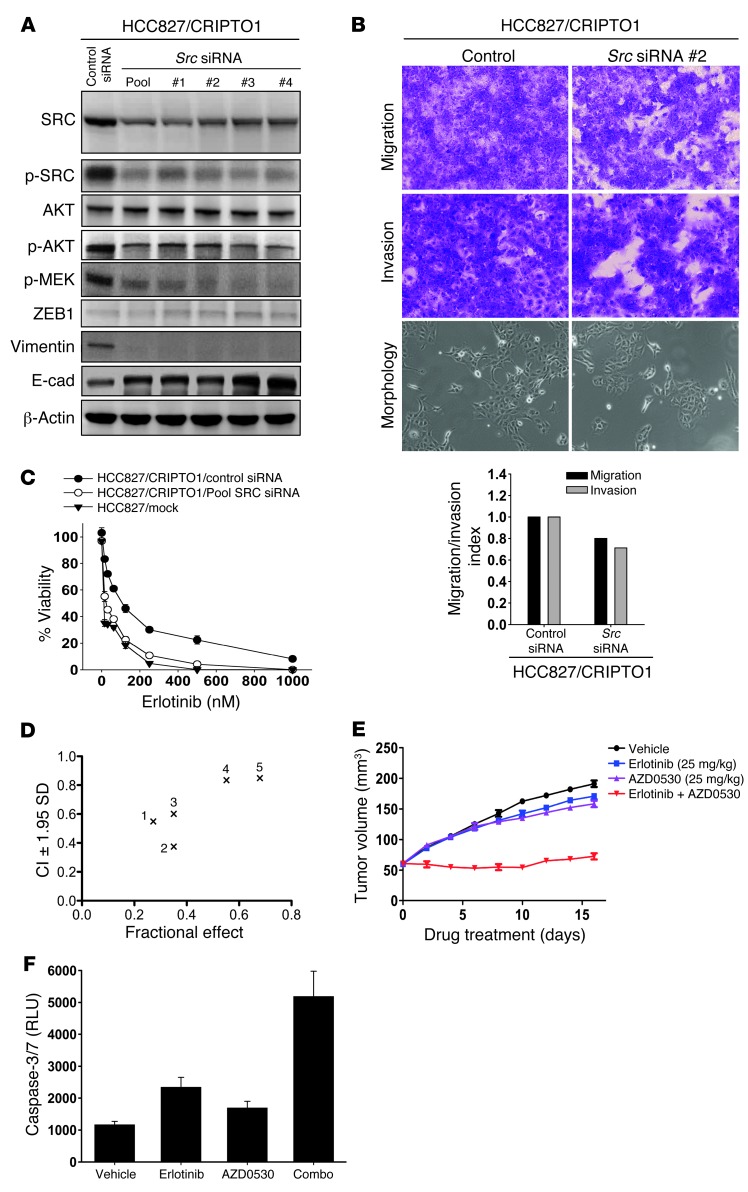
To further dissect the significance of EMT and SRC in CRIPTO1-induced erlotinib resistance, we depleted ZEB1 and SRC in HCC827/CRIPTO1 cells by ZEB1 shRNA and SRC siRNA, respectively. Phosphorylation of SRC, AKT, and MEK was not altered in ZEB1-depleted cells (Supplemental Figure 6A). ZEB1 depletion led to significant reduction of Vimentin and N-cadherin expression accompanied by the loss of EMT morphology and the attenuation of migration and invasion of HCC827/CRIPTO1 cells (Supplemental Figure 6B). Intriguingly, MTS assay showed that the ZEB1-depleted HCC827/CRIPTO1 cells were as resistant to erlotinib as HCC827/CRIPTO1 cells treated with control shRNA (Supplemental Figure 6C), suggesting that CRIPTO1-induced erlotinib resistance is ZEB1-independent.
SRC phosphorylation was less affected by erlotinib treatment in erlotinib-resistant HCC827/CRIPTO1 xenografts than in erlotinib-sensitive HCC827/mock xenografts (Figure 2D), implying that activation of SRC signaling may play a role in CRIPTO1-induced erlotinib resistance. To test this hypothesis, we performed SRC siRNA knockdown experiments in both HCC827/CRIPTO1 (Figure 5) and HCC4006/CRIPTO1 cells (Supplemental Figure 7). SRC depletion in HCC827/CRIPTO1 cells led to downregulation of pAKT and pMEK, reduced Vimentin (Figure 5A and Supplemental Figure 7A), and increased E-cadherin expression without apparent alteration of ZEB1 expression (Figure 5A); this was accompanied by a moderate attenuation of migration, invasion, and EMT morphology (Figure 5B). This is in agreement with previous reports in which SRC was found to modulate EMT through a ZEB1-independent mechanism (59, 60). MTS assay showed that siRNA knockdown of SRC restored erlotinib sensitivity in HCC827/CRIPTO1 and H4006/CRIPTO1 cells, suggesting that CRIPTO1-induced erlotinib resistance is SRC dependent (Figure 5C and Supplemental Figure 7B). These results prompted us to assess whether combined inhibition of SRC and EGFR may be a valuable strategy to overcome CRIPTO1-mediated erlotinib resistance. The combination activity was assessed by the Chou-Talalay method (61) using different concentrations of the SRC inhibitor AZD0530 with erlotinib in HCC827/CRIPTO1 and H4006/CRIPTO1 cells. In line with the SRC siRNA experiments, AZD0530 rendered HCC827/CRIPTO1 and H4006/CRIPTO1 cells sensitive to erlotinib, with significant synergy (CI < 1) observed at all concentrations tested (Figure 5D, Supplemental Figure 7C, and Supplemental Table 2, A and B). In addition, xenograft experiments with HCC827/CRIPTO1 cells showed that AZD0530 and erlotinib combination reverted CRIPTO1-induced erlotinib resistance and significantly attenuated tumor growth in comparison with either drug alone (Figure 5E). This observed synergy was partly attributed to the increased apoptotic cells, as measured by the caspase 3/7 activity assay (Figure 5F). In summary, our studies indicate that CRIPTO1-induced erlotinib resistance may be mediated through SRC but not ZEB1 signaling and that an EGFR/SRC inhibitor combination may represent a new therapeutic strategy to overcome erlotinib resistance in CRIPTO1-positive, EGFR-mutated NSCLC patients.
Figure 5. CRIPTO1-induced EGFR-TKI resistance is SRC dependent.
(A) Western blot analysis of HCC827/CRIPTO1 cells 72 hours after SRC siRNA transfection. β-actin was used as loading control. (B) SRC knockdown decreases migration and invasion of HCC827/CRIPTO1 stable cells. Control and SRC siRNA–transfected NSCLC cell lines were serum starved for 24 hours and then analyzed for cell migration and invasion at 24 and 48 hours, respectively. Cell migration and invasion indexes are shown at the bottom. Original magnification, ×200. (C) SRC siRNA reinstated erlotinib sensitivity of HCC827/CRIPTO1 cells. Two days after transfection, the indicated cell lines were treated with erlotinib for 3 days, followed by MTS assay. Data represent triplicate experiments. (D) Synergistic effect of AZD0530 and erlotinib combination. MTS assays were performed in HCC827/CRIPTO1 cells treated with 100 nM of erlotinib plus increasing concentrations of AZD0530 (10, 50, 100, 500, and 1000 nM) for 3 days (see Supplemental Table 2A for detail). (E) AZD0530 sensitized HCC827/CRIPTO1 cells to erlotinib in vivo. Mice were implanted subcutaneously with HCC827/CRIPTO1 cell lines. Each point represents the mean ± SEM of tumor volumes of 5 mice in each group. Mice were treated with vehicle (saline), AZD0530 at 25 mg/kg every day for 16 days, erlotinib at 25 mg/kg every 2 days for 16 days, or the combination of the 2 drugs with AZD0530 treatment for 16 days. (F) Caspase 3/7 activity of HCC827/CRIPTO1 xenograft tumors derived from single (erlotinib or AZD0530) or combination (combo) treatment groups.
High CRIPTO1 expression correlates with intrinsic resistance to EGFR-TKI in NSCLC carrying EGFR-sensitizing mutations.
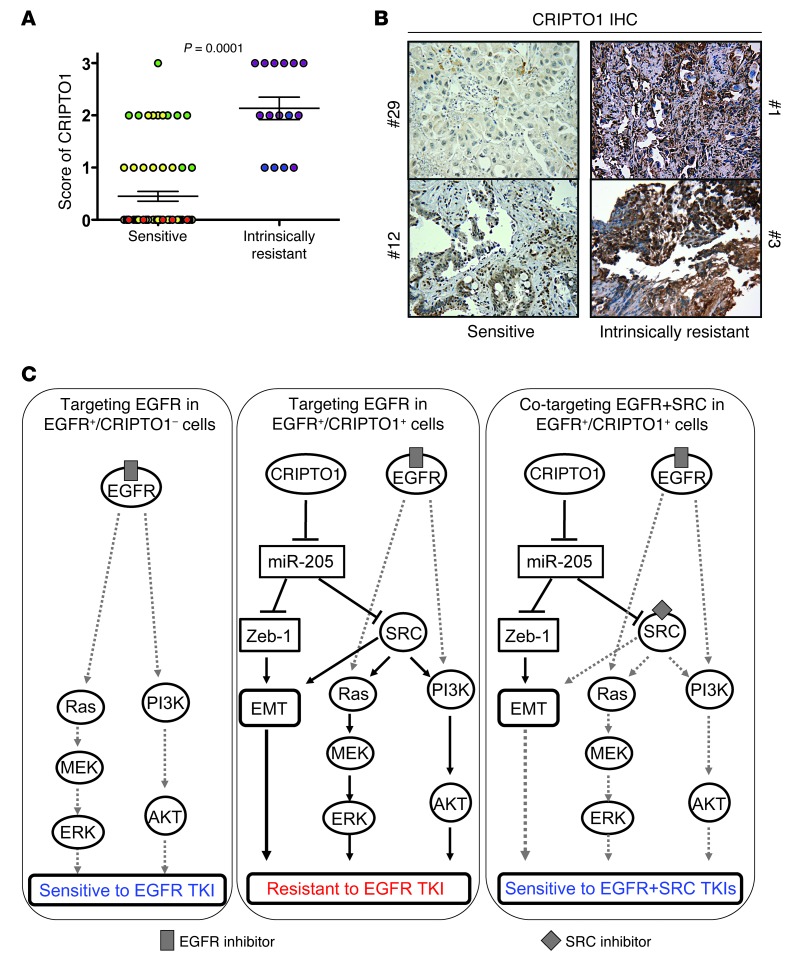
In order to test the clinical significance of our finding, we analyzed CRIPTO1 expression by immunohistochemistry (IHC) in 85 FFPE EGFR-mutated NSCLC specimens (Supplemental Table 3) using the validated CRIPTO1-specific antibody (Supplemental Figure 8, A–E), which was confirmed by RT-PCR with CRIPTO1-specific primers (Supplemental Figure 8, F and G). These are patients who were either sensitive (those with partial response [PR], complete response [CR], or stable disease [SD] of more than 4-month duration, n = 70) or intrinsically resistant (those with PD or SD of 4-month duration or less, n = 15) to erlotinib or gefitinib treatment (Supplemental Table 3). CRIPTO1 expression was significantly higher in intrinsically resistant patients than in sensitive patients (P = 0.0001, Figure 6, A and B). Nearly 70% (49/70) of the sensitive tumors showed no CRIPTO1 expression, whereas all (15/15) intrinsically resistant tumors exhibited some degree of CRIPTO1 expression. These data suggest that CRIPTO1 overexpression is associated with intrinsic erlotinib or gefitinib resistance in EGFR-mutated NSCLC patients.
Figure 6. High CRIPTO1 expression correlates with intrinsic EGFR-TKI resistance in EGFR-mutated NSCLC patients.
(A) High CRIPTO1 expression correlated with intrinsic gefitinib or erlotinib resistance (Student’s t test, P = 0.0001) in EGFR-mutated NSCLC patients (n = 85, see Supplemental Table 3 for reference). Levels of expression (scores 0–3) of CRIPTO1 IHC by a pathologist (M. Raffeld) are shown. Sensitive patients (those with PRs [yellow], CRs [red], or SD [green] of more than 4-month duration, n = 70) group and patients intrinsically resistant (those with PD [purple] or SD [blue] of 4-month duration or less, n = 15) to erlotinib or gefitinib treatment. (B) Representative CRIPTO1 IHC images in sensitive (tumor nos. 29 and 12) and intrinsically resistant (tumor nos. 1 and 3) EGFR-mutated NSCLC patients who received gefitinib and erlotinib treatment (See Supplemental Table 2). Original magnification, ×400. (C) Schemas illustrating the identified mechanisms of CRIPTO1-mediated resistance to EGFR inhibitions. Left: Targeting EGFR in EGFR-mutated (EGFR+)/CRIPTO1-negative cells. Middle: Targeting EGFR in EGFR-mutated/CRIPTO1-positive cells. Right: Cotargeting EGFR and SRC in EGFR-mutated/CRIPTO1-positive cells. Solid lines indicate the effects of CRIPTO1 on its downstream effectors. Gray lines depict to what extent the signaling pathways are blocked by target specific inhibitors.
Discussion
Intrinsic resistance to EGFR-TKIs occurs in approximately 10% of EGFR-mutated NSCLC patients, and up to 40% do not achieve a major response to erlotinib or gefitinib. In this study, we found that CRIPTO1 expression in EGFR-mutated NSCLC is likely to be one of the major mechanisms that confer intrinsic resistance to EGFR-TKIs. Our data suggest the following model: in CRIPTO1-negative, EGFR-mutated NSCLC cells, EGFR-TKI is sufficient to inhibit AKT and MEK activation and cell proliferation (Figure 6C); expression of CRIPTO1 downregulates miR-205, which leads to activation of SRC signaling, thereby offsetting the impact of EGFR-TKI on cell viability (Figure 6C). In this latter case, the efficacy of EGFR inhibition can be improved dramatically by coinhibition of SRC, which reduces cell viability by blocking both the SRC-mediated EMT and survival signaling (Figure 6C). Our model suggests that cotargeting EGFR and SRC may be an effective strategy for overcoming CRIPTO1-induced intrinsic resistance to EGFR-TKIs in CRIPTO1-positive, EGFR-mutated NSCLC patients.
CRIPTO1 expression was significantly higher in EGFR-mutated tumors of NSCLC patients who did not respond (SD of short duration and PD) to EGFR-TKI treatment compared with those patients who responded (PR, CR, and SD of long duration), supporting the notion that high CRIPTO1 expression is causal to intrinsic EGFR-TKI resistance. Importantly, whereas CRIPTO1 is expressed in many NSCLC tumor specimens examined here, it is absent in most of the established NSCLC cell lines. In agreement with this, we found that a primary tumor cell culture (MP41) from an intrinsic erlotinib-resistant EGFR-mutated NSCLC patient progressively lost CRIPTO1 expression during in vitro culture and concomitantly gained sensitivity to erlotinib. This finding suggests that CRIPTO1 expression contributes to intrinsic erlotinib resistance in NSCLC patients carrying sensitizing EGFR mutations.
In this study, we also assessed whether CRIPTO1 could contribute to acquired EGFR-TKI resistance. In acquired erlotinib-resistant clones (HCC827ER and H4006ER) generated by chronic, repeated exposure to erlotinib in vitro, induction of CRIPTO1 expression was not observed (Supplemental Figure 9A). In addition, unlike in HCC827/CRIPTO1 and H4006/CRIPTO1 cells, coinhibition of EGFR and SRC did not elicit any synergistic effect in HCC827ER and H4006ER cells (Supplemental Figure 9, B and C, Supplemental Table 4, A and B). Thus, under our in vitro selection conditions, CRIPTO1 expression may not contribute to acquired resistance to erlotinib. However, this may not reflect the genuine impact of CRIPTO1 on acquired erlotinib resistance in CRIPTO1-positive/EGFR-mutated NSCLC patients, as HCC827ER and H4006ER cells employed here are CRIPTO1 negative. Thus, in addition to the role of CRIPTO1 in intrinsic erlotinib resistance, further study is necessary to determine whether CRIPTO1 also contributes to acquired resistance to EGFR-TKIs in patients.
miR-205 has been shown to be an upstream regulator of EMT and SRC signaling (44, 45). We showed that ectopic CRIPTO1 expression downregulates miR-205 expression, and CRIPTO1-mediated ZEB1, SRC signaling, and erlotinib resistance is miR-205 dependent. Ectopic miR-205 downregulates SRC and ZEB1 expression, which may be achieved by miR-205–mediated inhibition of SRC and ZEB1, as both ZEB1 and SRC 3′ UTRs contain putative miR-205 target sequences (44, 45). It has been shown that CRIPTO1 activates SRC upon binding to glypican-1 (36). Our study suggests that CRIPTO1 may activate SRC through downregulation of miR-205 as an alternative mechanism. Further study will be required to elucidate the mechanisms by which CRIPTO1 regulates miR-205 expression.
Both SRC and EMT have been implicated in EGFR-TKI resistance in several cancers (37–40, 50). SRC also regulates E-cadherin membrane localization and drives ZEB1-independent EMT through integrin signaling and FAK phosphorylation (59, 60). In line with this finding, Strizzi et al. have shown that in CRIPTO1-driven transgenic mouse mammary tumors, there is a significant increase in both pSRC/pFAK and integrin signaling as compared with normal mammary tissue (33). These pathways may therefore be engaged in CRIPTO1-overexpressing NSCLC cells harboring EGFR mutations and contribute to resistance (62). In our study, we showed that ectopic expression of CRIPTO1 led to both induction of EMT and SRC activation. It is possible that both SRC and ZEB1 are required for CRIPTO1-induced EMT. Intriguingly, blocking SRC but not ZEB1 reversed erlotinib resistance in CRIPTO1-positive, EGFR-mutated NSCLC cells. One plausible explanation is that blocking SRC inhibits both CRIPTO1-mediated AKT and p44/42 MAPK signaling and EMT, both of which are required for the reversal of CRIPTO1-induced erlotinib resistance. In contrast, although blocking ZEB1 compromises EMT, SRC-mediated AKT and p44/42 MAPK signaling may be sufficient to sustain erlotinib resistance in CRIPTO1-positive, EGFR-mutated NSCLC cells. This is consistent with the notion that EGFR-TKI resistance is often associated with AKT and p44/42 MAPK activation through alternative means. Consistent with the in vitro finding that inhibition of SRC by siRNA or AZD0530 sensitizes CRIPTO1-positive, EGFR-mutated NSCLC cells toward erlotinib-induced cytotoxicity, AZD0530 and erlotinib combination significantly reduced the growth of CRIPTO1-positive, EGFR-mutated NSCLC xenograft tumors, establishing the basis for a rational combination therapy to treat this class of NSCLCs. It should also be noted that Nodal has recently been demonstrated to promote invasiveness and metastasis in breast cancer cells through an EMT program that is due to activation of the p44/42 MAPK pathway (63). It has yet to be investigated whether Nodal and/or other GDF ligands are cofunctioning with CRIPTO1 to generate EGFR-TKI resistance in EGFR-mutated NSCLC cells.
In conclusion, we demonstrate that CRIPTO1 overexpression potentially represents a novel mechanism of intrinsic EGFR-TKI resistance in NSCLC-harboring sensitizing EGFR mutations. Our study suggests cotargeting of EGFR and SRC as a potential rational approach to the treatment of erlotinib-resistant, CRIPTO1-positive, EGFR-mutated NSCLC patients.
Methods
Cancer cell lines, drugs, and antibodies.
All NSCLC cell lines were purchased from ATCC and were grown in medium with 10% FBS and 1× antibiotics (Gibco; Invitrogen). MP08 and MP41 cells were cultured in a conditioned medium as described by Liu et al. (64). Erlotinib (EGFR inhibitor) was purchased from LC Laboratories, PF299804 (second generation EGFR-TKI, dacomitinib) was obtained from Pfizer, and AZD0530 (SRC inhibitor) was purchased from Selleck. All antibodies except antibodies to CRIPTO1 (Epitomics and Rockland), ZEB1 (Santa Cruz Biotechnology Inc.), and β-actin (Sigma-Aldrich) were purchased from Cell Signaling for Western blot analysis. For Western blot and IHC, all antibodies were diluted according to the manufacturers’ instructions.
Xenografts.
Five million CRIPTO1 stably transfected cells (HCC827/CRIPTO1 and H4006/CRIPTO1) were subcutaneously injected into the flanks of athymic nude mice (National Cancer Institute [NCI] at Frederick Animal Production Program). When tumor volumes reached about 50–70 mm3, 5 mice per group were given daily oral doses of erlotinib at 12.5, 25, and 50 mg/kg every 2 days for 16 days. Both P values and percentage of tumor growth inhibition ratio (T/C) values were calculated at the end of the experiment. Tumor volume (V) was measured 3 times a week using the following formula: V = 1/2 (L × W2), where L equals length, and W equals width. The percentage of T/C value was calculated with the following formula: %T/C = (Δtreated tumor volume/Δcontrol tumor volume) × 100, where Δtreated tumor volume represents the mean tumor volume on the evaluation day minus the mean tumor volume at the start of the experiment. In the case of combination treatment experiments, mice were divided into 4 groups when tumors reached approximately 50 to 70 mm3 and treated with (a) saline (vehicle) daily for 16 days; (b) AZD0530 at 25 mg/kg daily for 16 days; (c) erlotinib at 25 mg/kg every 2 days for 16 days; and (d) AZD0530 at 25 mg/kg daily and erlotinib at 25 mg/kg every 2 days for 16 days. The doses and schedules used for combination studies were the same as for single agents.
Combination index analysis.
Combination effects were evaluated by MTS assay in cells treated with 100 nM erlotinib plus increasing concentrations of AZD0530 (10, 50, 100, 500, 1000 nM). The fraction affected (Fa) and combination indexes (CI) were processed using CalcuSyn software (Biosoft). CI of less than 1.0, 1.0, or more than 1.0 were taken to indicate synergistic, additive, and antagonistic effects, respectively.
IHC in NSCLC clinical specimens.
The AJCC Cancer Staging Manual (6th edition. Revised 2009) was used for tumor, lymph node, metastasis (TNM) classification. Specimens were formalin-fixed, paraffin-embedded (FFPE) tumor tissues that were examined to ensure greater than 75% tumor content by a pathologist (M. Raffeld). IHC was performed using Antigen Retrieval Dako Target Retrieval Solution (pH 6.0) and Histostain-Plus 3rd Gen IHC Detection Kit (Invitrogen) on FFPE slides according to the manufacturer’s protocols. Deparaffinized tissue sections were stained with human rabbit CRIPTO1 antibody (Rockland) at a dilution of 1:2500 for xenograft tumors and human specimens. Samples were scored by a pathologist (M. Raffeld) according to overall intensity of staining using a 0–3 scoring system of tumor cell staining intensity.
Statistics.
Differences were examined using 2-tailed Student’s t test. A P value of less than 0.05 was considered statistically significant.
Study approval.
All specimens of both EGFR-resistant and -sensitive cases were obtained from tissue resource committees at the Center for Cancer Research (CCR)/NCI/NIH, Kinki University, Hospital Germans Trias i Pujol, the Prince of Wales Hospital (Sha Tin, Hong Kong), and Georgetown University from individuals with NSCLC under the auspices of IRB-approved clinical protocols at each hospital; subjects gave informed consent. All animal studies were conducted according to guidelines of the Research Animal Resource Center of the NIH and were reviewed and approved by the NIH.
Cell viability assay, caspase 3/7 assay, cell cycle analysis, Western blot, RT-PCR, transfection method, and migration/invasion assay are described in Supplemental Methods.
Supplementary Material
Acknowledgments
The study was supported by the NCI Intramural Research Program, and the Lombardi Comprehensive Cancer Center, Georgetown University.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article:J Clin Invest. 2014;124(7):3003–3015. doi:10.1172/JCI73048.
References
- 1.Pao W, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101(36):13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 3.Paez JG, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 4.Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res. 2004;64(24):8919–8923. doi: 10.1158/0008-5472.CAN-04-2818. [DOI] [PubMed] [Google Scholar]
- 5.Riely GJ, Politi KA, Miller VA, Pao W. Update on epidermal growth factor receptor mutations in non-small cell lung cancer. Clin Cancer Res. 2006;12(24):7232–7241. doi: 10.1158/1078-0432.CCR-06-0658. [DOI] [PubMed] [Google Scholar]
- 6.Inoue A, et al. Prospective phase II study of gefitinib for chemotherapy-naive patients with advanced non-small-cell lung cancer with epidermal growth factor receptor gene mutations. J Clin Oncol. 2006;24(21):3340–3346. doi: 10.1200/JCO.2005.05.4692. [DOI] [PubMed] [Google Scholar]
- 7.Mitsudomi T, et al. Mutations of the epidermal growth factor receptor gene predict prolonged survival after gefitinib treatment in patients with non-small-cell lung cancer with postoperative recurrence. J Clin Oncol. 2005;23(11):2513–2520. doi: 10.1200/JCO.2005.00.992. [DOI] [PubMed] [Google Scholar]
- 8.Takeda M, et al. De novo resistance to epidermal growth factor receptor-tyrosine kinase inhibitors in EGFR mutation-positive patients with non-small cell lung cancer. J Thorac Oncol. 2010;5(3):399–400. doi: 10.1097/JTO.0b013e3181cee47e. [DOI] [PubMed] [Google Scholar]
- 9.Maemondo M, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362(25):2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 10.Kwak EL, et al. Irreversible inhibitors of the EGF receptor may circumvent acquired resistance to gefitinib. Proc Natl Acad Sci U S A. 2005;102(21):7665–7670. doi: 10.1073/pnas.0502860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi S, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352(8):786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 12.Pao W, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2(3):e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engelman JA, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316(5827):1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 14.Engelman JA, et al. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J Clin Invest. 2006;116(10):2695–2706. doi: 10.1172/JCI28656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sequist LV, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3(75):75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Z, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet. 2012;44(8):852–860. doi: 10.1038/ng.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheung HW, et al. Amplification of CRKL induces transformation and epidermal growth factor receptor inhibitor resistance in human non-small cell lung cancers. Cancer Discov. 2011;1(7):608–625. doi: 10.1158/2159-8290.CD-11-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pao W, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2(1):e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sequist LV, Bell DW, Lynch TJ, Haber DA. Molecular predictors of response to epidermal growth factor receptor antagonists in non-small-cell lung cancer. J Clin Oncol. 2007;25(5):587–595. doi: 10.1200/JCO.2006.07.3585. [DOI] [PubMed] [Google Scholar]
- 20.Ng KP, et al. A common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancer. Nat Med. 2012;18(4):521–528. doi: 10.1038/nm.2713. [DOI] [PubMed] [Google Scholar]
- 21.Nakagawa T, et al. EGFR-TKI resistance due to BIM polymorphism can be circumvented in combination with HDAC inhibition. Cancer Res. 2013;73(8):2428–2434. doi: 10.1158/0008-5472.CAN-12-3479. [DOI] [PubMed] [Google Scholar]
- 22.Lee JK, et al. Primary resistance to epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) in patients with non-small-cell lung cancer harboring TKI-sensitive EGFR mutations: an exploratory study. Ann Oncol. 2013;24(8):2080–2087. doi: 10.1093/annonc/mdt127. [DOI] [PubMed] [Google Scholar]
- 23.Faber AC, et al. BIM expression in treatment-naive cancers predicts responsiveness to kinase inhibitors. Cancer Discov. 2011;1(4):352–365. doi: 10.1158/2159-8290.CD-11-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Costa C, et al. The impact of EGFR T790M mutations and BIM mRNA expression on outcome in patients with EGFR-mutant NSCLC treated with erlotinib or chemotherapy in the randomized phase III EURTAC trial. Clin Cancer Res. 2014;20(7):2001–2010. doi: 10.1158/1078-0432.CCR-13-2233. [DOI] [PubMed] [Google Scholar]
- 25.Bianco C, Strizzi L, Normanno N, Khan N, Salomon DS. Cripto-1: an oncofetal gene with many faces. Curr Top Dev Biol. 2005;67:85–133. doi: 10.1016/S0070-2153(05)67003-2. [DOI] [PubMed] [Google Scholar]
- 26.Strizzi L, Bianco C, Normanno N, Salomon D. Cripto-1: a multifunctional modulator during embryogenesis and oncogenesis. Oncogene. 2005;24(37):5731–5741. doi: 10.1038/sj.onc.1208918. [DOI] [PubMed] [Google Scholar]
- 27.Saeki T, et al. Differential immunohistochemical detection of amphiregulin and cripto in human normal colon and colorectal tumors. Cancer Res. 1992;52(12):3467–3473. [PubMed] [Google Scholar]
- 28.Panico L, et al. Differential immunohistochemical detection of transforming growth factor alpha, amphiregulin and CRIPTO in human normal and malignant breast tissues. Int J Cancer. 1996;65(1):51–56. doi: 10.1002/(SICI)1097-0215(19960103)65:1<51::AID-IJC9>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 29.Bianco C, Salomon DS. Targeting the embryonic gene Cripto-1 in cancer and beyond. Expert Opin Ther Pat. 2010;20(12):1739–1749. doi: 10.1517/13543776.2010.530659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhong XY, et al. Positive association of up-regulated Cripto-1 and down-regulated E-cadherin with tumour progression and poor prognosis in gastric cancer. Histopathology. 2008;52(5):560–568. doi: 10.1111/j.1365-2559.2008.02971.x. [DOI] [PubMed] [Google Scholar]
- 31.Miyoshi N, Ishii H, Mimori K, Sekimoto M, Doki Y, Mori M. TDGF1 is a novel predictive marker for metachronous metastasis of colorectal cancer. Int J Oncol. 2010;36(3):563–568. doi: 10.3892/ijo_00000530. [DOI] [PubMed] [Google Scholar]
- 32.Gong YP, et al. Overexpression of Cripto and its prognostic significance in breast cancer: a study with long-term survival. Eur J Surg Oncol. 2007;33(4):438–443. doi: 10.1016/j.ejso.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 33.Strizzi L, et al. Epithelial mesenchymal transition is a characteristic of hyperplasias and tumors in mammary gland from MMTV-Cripto-1 transgenic mice. J Cell Physiol. 2004;201(2):266–276. doi: 10.1002/jcp.20062. [DOI] [PubMed] [Google Scholar]
- 34.Bianco C, et al. Cripto-1 activates nodal- and ALK4-dependent and -independent signaling pathways in mammary epithelial Cells. Mol Cell Biol. 2002;22(8):2586–2597. doi: 10.1128/MCB.22.8.2586-2597.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeo C, Whitman M. Nodal signals to Smads through Cripto-dependent and Cripto-independent mechanisms. Mol Cell. 2001;7(5):949–957. doi: 10.1016/S1097-2765(01)00249-0. [DOI] [PubMed] [Google Scholar]
- 36.Bianco C, et al. A Nodal- and ALK4-independent signaling pathway activated by Cripto-1 through Glypican-1 and c-Src. Cancer Res. 2003;63(6):1192–1197. [PubMed] [Google Scholar]
- 37.Witta SE, et al. Restoring E-cadherin expression increases sensitivity to epidermal growth factor receptor inhibitors in lung cancer cell lines. Cancer Res. 2006;66(2):944–950. doi: 10.1158/0008-5472.CAN-05-1988. [DOI] [PubMed] [Google Scholar]
- 38.Hiscox S, Morgan L, Green TP, Barrow D, Gee J, Nicholson RI. Elevated Src activity promotes cellular invasion and motility in tamoxifen resistant breast cancer cells. Breast Cancer Res Treat. 2006;97(3):263–274. doi: 10.1007/s10549-005-9120-9. [DOI] [PubMed] [Google Scholar]
- 39.Leung EL, et al. SRC promotes survival and invasion of lung cancers with epidermal growth factor receptor abnormalities and is a potential candidate for molecular-targeted therapy. Mol Cancer Res. 2009;7(6):923–932. doi: 10.1158/1541-7786.MCR-09-0003. [DOI] [PubMed] [Google Scholar]
- 40.Thomson S, et al. Epithelial to mesenchymal transition is a determinant of sensitivity of non-small-cell lung carcinoma cell lines and xenografts to epidermal growth factor receptor inhibition. Cancer Res. 2005;65(20):9455–9462. doi: 10.1158/0008-5472.CAN-05-1058. [DOI] [PubMed] [Google Scholar]
- 41.Xing PX, Hu XF, Pietersz GA, Hosick HL, McKenzie IF. Cripto: a novel target for antibody-based cancer immunotherapy. Cancer Res. 2004;64(11):4018–4023. doi: 10.1158/0008-5472.CAN-03-3888. [DOI] [PubMed] [Google Scholar]
- 42.Hu XF, Li J, Yang E, Vandervalk S, Xing PX. Anti-Cripto Mab inhibit tumour growth and overcome MDR in a human leukaemia MDR cell line by inhibition of Akt and activation of JNK/SAPK and bad death pathways. Br J Cancer. 2007;96(6):918–927. doi: 10.1038/sj.bjc.6603641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soifer HS, Rossi JJ, Saetrom P. MicroRNAs in disease and potential therapeutic applications. Mol Ther. 2007;15(12):2070–2079. doi: 10.1038/sj.mt.6300311. [DOI] [PubMed] [Google Scholar]
- 44.Gregory PA, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 45.Majid S, et al. MicroRNA-205 inhibits Src-mediated oncogenic pathways in renal cancer. Cancer Res. 2011;71(7):2611–2621. doi: 10.1158/0008-5472.CAN-10-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun C, et al. CRIPTO3, a presumed pseudogene, is expressed in cancer. Biochem Biophys Res Commun. 2008;377(1):215–220. doi: 10.1016/j.bbrc.2008.09.113. [DOI] [PubMed] [Google Scholar]
- 47.Stabile LP, et al. c-Src activation mediates erlotinib resistance in head and neck cancer by stimulating c-Met. Clin Cancer Res. 2013;19(2):380–392. doi: 10.1158/1078-0432.CCR-12-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frederick BA, et al. Epithelial to mesenchymal transition predicts gefitinib resistance in cell lines of head and neck squamous cell carcinoma and non-small cell lung carcinoma. Mol Cancer Ther. 2007;6(6):1683–1691. doi: 10.1158/1535-7163.MCT-07-0138. [DOI] [PubMed] [Google Scholar]
- 49.Rho JK, et al. Epithelial to mesenchymal transition derived from repeated exposure to gefitinib determines the sensitivity to EGFR inhibitors in A549, a non-small cell lung cancer cell line. Lung Cancer. 2009;63(2):219–226. doi: 10.1016/j.lungcan.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 50.Boerner JL. Role of Src family kinases in acquired resistance to EGFR therapies in cancer. Cancer Biol Ther. 2009;8(8):704–706. doi: 10.4161/cbt.8.8.8183. [DOI] [PubMed] [Google Scholar]
- 51.Ettinger DS. Taxol in the treatment of lung cancer. J Natl Cancer Inst Monogr. 1993;(15):177–179. [PubMed] [Google Scholar]
- 52.Naito Y, et al. Concurrent chemoradiotherapy with cisplatin and vinorelbine for stage III non-small cell lung cancer. J Thorac Oncol. 2008;3(6):617–622. doi: 10.1097/JTO.0b013e3181753b38. [DOI] [PubMed] [Google Scholar]
- 53.Normanno N, et al. Cripto-1 overexpression leads to enhanced invasiveness and resistance to anoikis in human MCF-7 breast cancer cells. J Cell Physiol. 2004;198(1):31–39. doi: 10.1002/jcp.10375. [DOI] [PubMed] [Google Scholar]
- 54.De Luca A, et al. Expression and functional role of Cripto-1 in cutaneous melanoma. Br J Cancer. 2011;105(7):1030–1038. doi: 10.1038/bjc.2011.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Christiansen JJ, Rajasekaran AK. Reassessing epithelial to mesenchymal transition as a prerequisite for carcinoma invasion and metastasis. Cancer Res. 2006;66(17):8319–8326. doi: 10.1158/0008-5472.CAN-06-0410. [DOI] [PubMed] [Google Scholar]
- 56.Burk U, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9(6):582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tellez CS, et al. EMT and stem cell-like properties associated with miR-205 and miR-200 epigenetic silencing are early manifestations during carcinogen-induced transformation of human lung epithelial cells. Cancer Res. 2011;71(8):3087–3097. doi: 10.1158/0008-5472.CAN-10-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lebanony D, et al. Diagnostic assay based on hsa-miR-205 expression distinguishes squamous from nonsquamous non-small-cell lung carcinoma. J Clin Oncol. 2009;27(12):2030–2037. doi: 10.1200/JCO.2008.19.4134. [DOI] [PubMed] [Google Scholar]
- 59.Avizienyte E, et al. Src-induced de-regulation of E-cadherin in colon cancer cells requires integrin signalling. Nat Cell Biol. 2002;4(8):632–638. doi: 10.1038/ncb829. [DOI] [PubMed] [Google Scholar]
- 60.Serrels A, Canel M, Brunton VG, Frame MC. Src/FAK-mediated regulation of E-cadherin as a mechanism for controlling collective cell movement: insights from in vivo imaging. Cell Adh Migr. 2011;5(4):360–365. doi: 10.4161/cam.5.4.17290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raje N, et al. Combination of the mTOR inhibitor rapamycin and CC-5013 has synergistic activity in multiple myeloma. Blood. 2004;104(13):4188–4193. doi: 10.1182/blood-2004-06-2281. [DOI] [PubMed] [Google Scholar]
- 62.Quail DF, Siegers GM, Jewer M, Postovit LM. Nodal signalling in embryogenesis and tumourigenesis. Int J Biochem Cell Biol. 2013;45(4):885–898. doi: 10.1016/j.biocel.2012.12.021. [DOI] [PubMed] [Google Scholar]
- 63.Quail DF, Zhang G, Findlay SD, Hess DA, Postovit LM. Nodal promotes invasive phenotypes via a mitogen-activated protein kinase-dependent pathway. Oncogene. 2014;33(4):461–473. doi: 10.1038/onc.2012.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu X, et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am J Pathol. 2012;180(2):599–607. doi: 10.1016/j.ajpath.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.