Highlights
-
•
The nuclear pore complex is well conserved, with some regions of divergence.
-
•
The nuclear lamina appears quite variable between major supergroups.
-
•
Centrosomes are ancient structures, but with complex evolutionary history.
-
•
There is evidence for prokaryotic ancestors of some nuclear components.
-
•
Analysis of divergent organisms is essential to fully understand nuclear biology and its origins.
Abstract
The nucleus represents a major evolutionary transition. As a consequence of separating translation from transcription many new functions arose, which likely contributed to the remarkable success of eukaryotic cells. Here we will consider what has recently emerged on the evolutionary histories of several key aspects of nuclear biology; the nuclear pore complex, the lamina, centrosomes and evidence for prokaryotic origins of relevant players.
Current Opinion in Cell Biology 2014, 28:8–15
This review comes from a themed issue on Cell nucleus
Edited by Michael P Rout and Gary H Karpen
For a complete overview see the Issue and the Editorial
Available online 6th February 2014
0955-0674/$ – see front matter, © 2014 The Authors. Published by Elsevier Ltd.
Introduction
The nucleus is enclosed by the nuclear envelope (NE) to form a container for most eukaryotic cellular DNA. Contiguous with the endoplasmic reticulum, the NE separates gene expression (transcription, mRNA maturation) from protein synthesis (translation, folding, assembly), but necessitates a channel for bidirectional trafficking (the nuclear pore complex (NPC)), a mechanism of mechanical support (lamins) and of chromosomal positioning and segregation. The NE and NPC also participate in chromosomal positioning, mitosis and transcriptional control. NE origins are linked to the ER and coated vesicles (CV) [1], probably via a proto-NE, that was possibly freely permeable with a sealed state arising subsequently ([2–4], discussed in [5]). Many models have been offered for nuclear origins and the events that underly the acquisition of an endomembrane system [4,6,7,8••] (Figure 1). Here we consider several nuclear-associated systems to provide insights into how the nucleus has evolved, together with evidence for some of the relevant prokaryotic precursors.
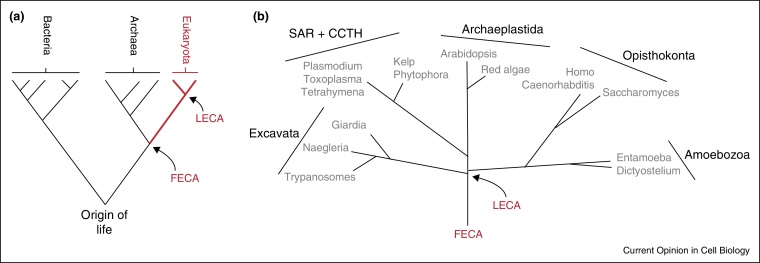
Figure 1.
Phylogenetic tree of the current view of the topology of life and eukaryota. (a) Relationship between prokaryotes and eukaryotes, assuming the three-domain model, whereby the Eukaryota emerged from the Archaea. An alternate two domain model, proposes that the Eukaryotes arose as a lineage within the Archaea, but this remains unresolved [85,86]. LECA/FECA; Last/first eukaryotic common ancestor. (b) Eukaryotic phylogeny, based on discussions provided in [87]. Some relationships, for example within the SAR + CCTH and Excavata clades remain to be fully resolved. Examples of commonly studied and/or organisms familiar to most experimental cell biologists are provided to anchor the reader, and supergroups are indicated by bars. There is a clear emphasis within many clades in the study of pathogenic species, for obvious and fully justified reasons. SAR + CCTH; Stramenopile, Alveolata, Rhizaria + Cryptophyta, Centrohelida, Telonemia and Haptophyta.
The nuclear pore complex: translocator, organiser, regulator
Nucleocytoplasmic transport maintains a distinct composition between the cytoplasm and nucleus to facilitate functional differentiation [9,10] (Figure 2). NPCs with apparently similar morphologies are observed in the NE of many lineages, suggesting that evolutionary changes to the NPC are likely minor in terms of overall composition or architecture, and conservation of the basic mechanisms of transport across eukaryotes is clear. The NPC proteomes for yeast, mammals, trypanosomes, plants and Tetrahymena [11,12••,13–15] provide insights into NPC evolution. The NPC proteins, nucleoporins (Nups), demonstrate greatly divergent amino acid sequences but with retention of secondary structural architectures. However, in silico identification of Nups remains challenging and our understanding of the evolutionary histories of many individual Nups remains unclear [16,17].
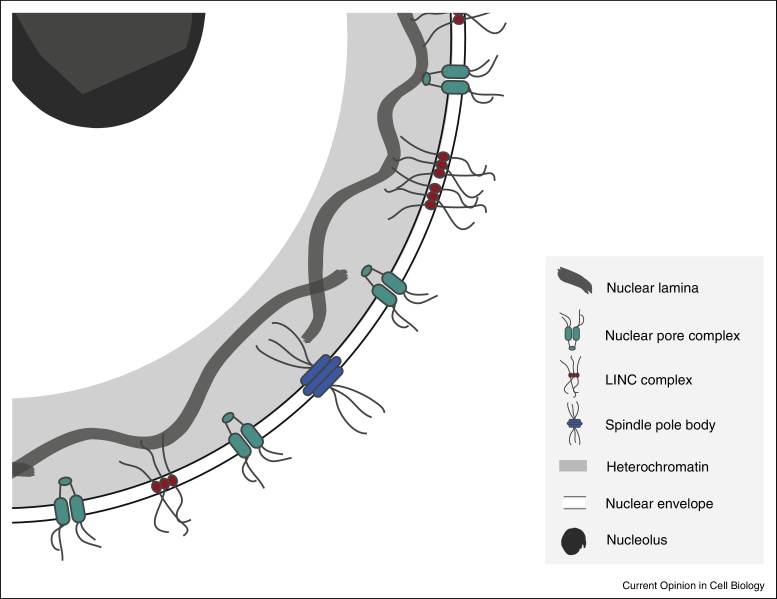
Figure 2.
Important structures associated with the nuclear envelope. A sector of a generalised nucleus is shown, with various structures drawn as cartoons either embedded within the nuclear envelope or associated with it. Note that the structures are not drawn to an accurate scale.
The NPC has eight spokes surrounding a central channel, and connected by the inner ring facing the channel (Nup170/Nup155 complex in yeast/metazoa), outer rings (Nup84/Nup107-160 complex in yeast/metazoa) and membrane rings (Pom152 in yeast, gp210 in metazoa) [18]. The inner and outer rings represent the structural scaffold, and most of their Nups conform to the protocoatomer architecture, that is possess β-propeller and/or α-solenoid domains, and are well conserved and structurally related to vesicle coats [1,7,19]. Further, structural similarity between some Nups and karyopherins suggest a common origin; Nup188 and Nic96 bind FG-repeats and translocate through NPCs, providing experimental evidence in support of the proposed common origin between the NPC and the soluble nuclear transport machinery [20••,21••]. This may indicate that the Kaps arose as a soluble Nup variant, or potentially vice versa. Some Nups, for example Seh1 and Elys are non-universal while the trypanosome Nup84 complex equivalent may possess additional subunits (S. Obado, MCF, M.P. Rout and B.T. Chait, in preparation). Most remaining Nups are conserved; Aladin for example is widely retained, but lost specifically from yeasts [5]. Clearly the major membrane-deforming/stabilising functionality of the NPC is evolutionary stable, and hence likely mechanistically similar, across the eukaryotes, consistent with a comparatively invariable morphology (Figure 3).
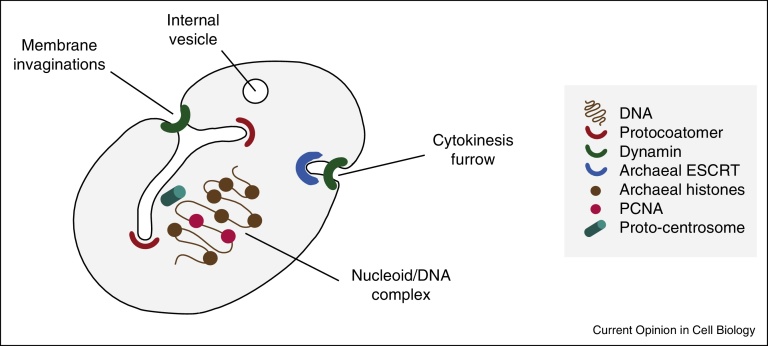
Figure 3.
A number of features associated with prokaryotic cells that are shared with eukaryotes. Note that not all of these features are present in any one lineage. Highlighted endomembrane complexes are putative protocoatomer-like proteins that may associate with membrane in the planctomycetes, bacterial dynamin that is associated with cytokinesis and possibly other membranous structures, the partial ESCRT system found in Archaea and which plays a conserved role in cytokinesis with eukaryotes. Archaea also possess histone-like proteins and a PCNA ortholog, while it is likely that the centrosome was associated with an early membranous structure that gave rise to the nuclear envelope.
The membrane ring displays considerable flexibility, and the sequence divergence between yeast and animal Pom152 and gp210 is well known. Possible orthologs of gp210 and NDC1, but not Pom121, are present in Arabidopsis [13], but in T. brucei no membrane Nups have been identified to date [14]. Therefore the interface between the scaffold and NE may vary between taxa, albeit with unclear consequences, but may also have an association with NPC assembly [22]. FG-repeat Nups serve to provide gating functionality, and the FG/FXFG repeat, if not the precise arrangement within the Nup protein bearing the repeats, appears very widely conserved across eukaryotes. An interesting example of an exception to FG or FXFG repeat architecture comes from Tetrahymena, where the transcriptionally inactive micronucleus possesses Nup98 paralogs with poly-N/NIFN repeats and the transcriptionally active macronucleus, more conventional FG-repeat Nup98 [12••,23]. Significantly, despite variation in sequence and locations of the FG repeats in Nups, the number of FGs and sequence environment within which the FGs are embedded appear to be better conserved, implying conservation of the gating mechanism, although the precise mechanisms by which this operates remain controversial [14,16].
Both cytoplasmic fibrils and the nuclear basket exhibit complex evolutionary patterns [5], likely impacting their interactions with other cellular systems. For example, Nup358 anchors RanGAP at the cytoplasmic fibrils in metazoa but trypanosomes lack Nup358 and an alternate anchor for RanGAP is present in plants, while yeast RanGAP is solely cytoplasmic [13,16,24••]. Amongst the many nuclear basket connections are the transcriptional apparatus, the lamina and protein/RNA transport systems. There is evidence for conserved interactions between the NPC and TREX-2 and SAGA, important in mRNA export and transcription respectively [25] and also Nup-interactions with the spindle and checkpoint proteins [26,27••]. Given that TREX-2 and SAGA subunits are present in many lineages, it is again likely that this is ancient and central to NPC function. Interesting, the inner nuclear NPC components, Tpr in vertebrates and Mlps in yeasts, are orthologs and widely represented across the eukaryotes, whereas the discicristata (Euglenozoa plus Percolozoa) have no detectable Tpr/Mlp homologue, but have two nucleoporins with similar architectures and functions [5,26] (Holden et al., submitted for publication). Significantly, those data suggest that LECA possessed Tpr/Mlp; the presence of analogues in early diverging trypanosomes suggests that a second mechanism was present in LECA or that this replaced an original Tpr/Mlp-based system for interacting with the nuclear interior in the discicristata, possibly as a response to changes in transcriptional mechanisms.
Centrosomes, centrins and spindle poles
Centrosomes serve as the main microtubule-organising centres (MTOCs), and are essential for cell architecture in all organisms using microtubules for organelle positioning. Nuclear-associated bodies (NABs) or spindle pole bodies (SPBs) are centrosomal structures in association with the nucleus, and are best characterised in yeasts and Dictyostelium amoebae. In budding yeast the SPB consists of a stack of three plaques and is permanently inserted into the NE (Figure 2). In Dictyostelium, the NAB also contains a tripartite core; although attached to the NE, the Dictyostelium NAB is cytosolic during interphase, only entering the NE upon centrosome duplication at mitosis, similar to fission yeast [28,29]. The Dictyostelium NAB organises a radial microtubule cytoskeleton very similar to metazoan cells. Centrosomes of animals, yeasts and amoebozoa share a surprisingly small cohort of components: the γ-tubulin small complex (γ-TuSC; γ-tubulin, GCP2, GCP3) required for microtubule nucleation; EB1, TACC and XMAP215 for microtubule dynamics and stabilisation; centrin, Cep192/SPD2, and centrosomin (Cnn) as scaffolding proteins, kinases from the polo, aurora, NIMA and Cdk family regulating duplication and spindle organisation and the dynein motor protein [30–33]. Hence much of the diversity of centrosomal functions is likely a direct result of divergent composition in modern lineages.
The amoeboid cell state has been regarded as ancestral, and acentriolar MTOCs in fungi and amoebozoans were therefore considered to represent the primitive centrosomal form. However, comparative genomics indicates that LECA likely possessed one or two centrioles associated with a cilium, since centrioles are found in all major eukaryotic subgroups [30,34,35] and the LECA was almost definitely flagellated [8••]. The absence of centrioles in higher plants, fungi and most amoebozoans is therefore a secondary loss, and implies that centrosomes likely had original roles in initiating cilium formation while the centriole served as a basal body for microtubule nucleation. Indeed, ciliate centrioles act exclusively as basal bodies and their mitotic spindle poles are devoid of centrioles [36]. Centrioles may have originally exploited spindle association to ensure an equal distribution into daughter cells, rather than having an active role at the spindle [37–39], and this possibility is supported by evidence that centrioles are dispensable for spindle formation [40–42]. Despite this, these same studies found that centrioles are essential for formation of astral microtubules and cilia. The ancestral centrosome may thus have been a membrane/chromatin-associated microtubule nucleation centre with dual centromere/centrosome functions. Subsequently duplicated during eukaryotic evolution, a centrosome remained attached to the plasma membrane while a microtubule nucleation centre attached to proto-endomembranes that later differentiated into the NE [35]. This process could have generated an intranuclear microtubule nucleation centre that organised the spindle and an extra-nuclear centrosome responsible for organising pellicular and flagellum microtubules and significantly this configuration is present in the discicristata, such as Euglena and trypanosomes [43]. These scenarios suggest that the tight association of a nucleus-associated centrosome with clustered centromeres during the entire cell cycle, as in fission yeast and Dictyostelium, is primitive. The hypothesis that nuclear centromeres originally had dual centromere/centrosome functions is supported by observations that both structures remain closely associated with each other during the entire cell cycle, as in fission yeast or Dictyostelium where centromeres cluster close to the inner nuclear membrane and permanently associate with the SPB/centrosome at the cytosolic nuclear face [44–46].
Besides tubulins, centrins (of the calmodulin family of calcium-binding proteins) may be the most ancient centrosomal proteins [47], with general functions in connecting microtubular and membrane-bound structures. Centrins may have been critical to assembly of the primitive centromeric microtubule nucleation complex [35]. In S. cerevisiae Cdc31p (yeast centrin) is a major constituent of the assembly platform for the new SPB upon SPB duplication at the NE. There are several centrin isoforms, which in most species can be grouped into two subfamilies: human centrin-2-like and yeast Cdc31p/centrin-3-like proteins. Since members are present in both unikonts and bikonts, these subfamilies arose early [48], and losses are likely secondary events. By this model, yeasts retained only centrin-3 with its ancient, nuclear functions after loss of cilia for locomotion [35]. However, this is likely too simplistic as flies and nematodes lack centrin-3 and centrin-2 assumes the nuclear role [48]. Further, Dictyostelium CenA and CenB belong to neither subfamily, but both predominantly associate with the nucleus, with CenA concentrated at centromeres and CenB at nuclear internal [49,50]. While an exact function of CenA is unknown, CenB is important for nuclear architecture and centrosome nuclear attachment, the latter function being conserved with S. cerevisiae Cdc31p [51].
Lamins, laminas and LINCs
The NE is subtended in most cells by a morphologically recognisable lamina, first described in amoebozoa [52–54]. The lamina in metazoan cells is comprised of lamins, a family of repetitive coiled coil ∼60–80 kDa proteins [55]. Lamins serve as organisers of heterochromatin, NPCs and multiple additional nuclear structures, reflected in the importance of laminopathies to human disease ([56], compiled in [55,57]). Lamins are targeted to the NE by C-terminal prenylation, and in mammalian cells the distinct isoforms have somewhat differing locations [58,59]. Lamins were assumed to be metazoan specific, suggesting a recent origin. It is clear this is incorrect as lamin orthologs are present in several amoebozoan species, with the best characterised being Dictyostelium NE81, with functions fully compatible with a bona fide lamin [60••,61], pushing the lamin origin to the origin of unikonts and perhaps even earlier. Furthermore, while there are no documented lamins within bikonts, the discicristate NUP-1 protein, and higher plant NMCP proteins assume similar locations and functions, as well as retain a predicted coiled coil architecture [55,62••,63••]. It is unknown if the LECA had a lamina of NUP-1-like, NMCP-like or lamin-like proteins, or if NUP-1/NMCP and lamins are in some manner evolutionarily related. It is formally possible that the LECA had a more complex lamina and that all but one system was subsequently lost, or that the discicristata and plants replaced a lamin-based lamina with NUP-1 or NMCP respectively.
A further group of proteins associated with the NE is SUN and KASH domain proteins [64••,65]. SUN proteins are present in all major eukaryotic groups, except for the discicristata [66•]. SUN proteins are concentrated at the inner NE and interact with KASH-family proteins at the outer NE, forming the LINC complex [64••,65,67]. Different KASH-family proteins manage direct or indirect connections to cytoplasmic microtubules, actin filaments, intermediate filaments and dynein, which in turn maintains the centrosome close to the nucleus through its microtubule minus end-directed motor activity. SUN proteins are linked to lamins [68], required for proper centrosome/nucleus attachment [69]. Although this linkage has been proven only for metazoa, since Dictyostelium NE81 is required for centrosome/nucleus attachment and interference with NE81 causes phenotypes similar to SUN1 disruptions, this likely extends to Amoebozoa [46,60••,70]. Thus, lamins may have co-evolved with SUN-proteins, suggesting the widespread presence of lamins, while the absence of SUN and lamins from the discicristata is compatible with the absence of lamins and substitution by NUP-1. However, as plants also have SUN/KASH and NMCP proteins but not lamins, coevolution is therefore not strictly necessary [71]. Further, a functional connection between lamins and open mitosis also can be discounted [35]. Dictyostelium has a partial closed mitosis, comparable to Aspergillus [72]; as the former has a lamin and the latter apparently does not, these features are not linked. Dictyostelium may have solved the problem of making the NE sufficiently flexible for karyokinesis by partial disassembly of NE81 networks, as NE81 remains associated with the NE throughout the cell cycle [60••]. Hence at present there remains no obvious rationale to underpin the use of a particular set of proteins to build a lamina or the functional implications of these potentially distinct systems.
Prokaryotic origins
Given the clear conservation of many nuclear functions and structures, it is perhaps no surprise that there is growing evidence for origins of several systems and components pre-LECA, and even reaching back to prokaryotes (Table 1). Surprisingly, prokaryotic homologues of proteins with the protocoatomer architecture, that is, related to the NPC scaffold, have been detected only in bacteria belonging to the PVC superphylum [73]. PVC bacteria have a unique endomembrane system that is complex and dynamic [73–75], and it is unclear if this represents an example of convergence, lateral gene transfer or deep evolutionary relationships. Importantly, this may indicate that there is a fundamental aspect to the protocoatomer architecture that is of extreme value to membrane modeling, and further highlights that internal membranes of considerable complexity exist outside eukaryotes. Orthologs of many nuclear proteins and RNAs are present in Archaea, including PCNA, Sm-like, MCM and GINS, encompassing functions from transcription, DNA replication, mRNA processing and telomere construction [76–78]. Similarly, most archaea encode histone variants [79,80] and snoRNA genes [81], all indicating a shared cohort of nuclear genes/RNAs between Archaea and eukaryotes. With improved detection methods eukaryotic features are increasingly being identified in prokaryotes and it is becoming clear that the transition between these two major cellular forms may have been more gradual that previously suspected (Table 1).
Table 1.
Selection of genes that are represented both in prokaryotic and eukaryotic genomes. A small selection of examples is given, to illustrate that both bacteria and Archaea may share genes with eukaryotes which have important roles in the nucleus.
| Protein complex | Functions in | Present in | Reference |
|---|---|---|---|
| MC proteins | Endomembrane system | Bacteria | [73] |
| PCNA | DNA metabolism | Archaea | [76] |
| Sm-like | Small nuclear ribonucleoproteins | Archaea | [77] |
| CMG complex | DNA replication | Archaea | [78] |
| snoRNA | Post-trancriptional modifications | Archaea | [81] |
| Dynamin | Membrane manipulation | Bacteria | [82] |
| ESCRT | Membrane/cell division | Archaea | [83,84] |
Summary
Many nuclear functions, including complex interactions and dynamics, are conserved across eukaryotes, and which engage massive assemblies of proteins with ancient origins. A number of notable, lineage-specific features have been described, most prominently the lamina, and the implications remain to be fully established. Furthermore, centrosomes have complex nuclear evolutionary relationships and even the strict view of an endomembrane system as a eukaryotic feature is challenged by the presence of membranous systems in prokaryotes. The emergence of additional model systems beyond the classical yeasts and animals will continue to contribute to understanding the evolution of nuclear functions and the origins of the nucleus itself.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
Work in our laboratories was supported by the following agencies, and which is gratefully acknowledged; MRC and Wellcome Trust (MR/K008749/1 and 090007/Z/09/Z respectively, to MCF), C2A Junta de Andalucia to DPD and DFG GR1642/4-1 to RG. The authors have no conflict of interest to declare.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.
References
- 1.Devos D., Dokudovskaya S., Alber F., Williams R., Chait B.T., Sali A., Rout M.P. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2004;2:e380. doi: 10.1371/journal.pbio.0020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alberts B., Johnson A., Lewis J., Raff M., Roberts K., Walter P. edn 5. Garland Science; Abingdon, Oxford, UK: 2007. Molecular Biology of the Cell. ISBN-10 0815341059. [Google Scholar]
- 3.Hoelz A., Debler E.W., Blobel G. The structure of the nuclear pore complex. Annu Rev Biochem. 2011;80:613–643. doi: 10.1146/annurev-biochem-060109-151030. [DOI] [PubMed] [Google Scholar]
- 4.Wilson K.L., Dawson S.C. Evolution: functional evolution of nuclear structure. J Cell Biol. 2011;195:171–181. doi: 10.1083/jcb.201103171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Field M., Koreny L., Rout M. Enriching the pore: splendid complexity from humble origins. Traffic. 2014;15((2) Feb):141–156. doi: 10.1111/tra.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Embley T.M., Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- 7.Field M.C., Sali A., Rout M.P. Evolution: on a bender—BARs, ESCRTs, COPs, and finally getting your coat. J Cell Biol. 2011;193:963–972. doi: 10.1083/jcb.201102042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8••.Koumandou V.L., Wickstead B., Ginger M.L., van der Giezen M., Dacks J.B., Field M.C. Molecular paleontology and complexity in the last eukaryotic common ancestor. Crit Rev Biochem Mol Biol. 2013;48:373–396. doi: 10.3109/10409238.2013.821444. [DOI] [PMC free article] [PubMed] [Google Scholar]; An attempted grand synthesis of the structure of the LECA from the viewpoint of multiple cellular systems, including trafficking, nuclear architecture, metabolism, cytoskeletal systems and organellar evolution.
- 9.Tetenbaum-Novatt J., Rout M.P. The mechanism of nucleocytoplasmic transport through the nuclear pore complex. Cold Spring Harb Symp Quant Biol. 2010;75:567–584. doi: 10.1101/sqb.2010.75.033. [DOI] [PubMed] [Google Scholar]
- 10.Cook A., Bono F., Jinek M., Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- 11.Rout M.P., Aitchison J.D., Suprapto A., Hjertaas K., Zhao Y., Chait B.T. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12••.Iwamoto M., Mori C., Kojidani T., Bunai F., Hori T., Fukagawa T., Hiraoka Y., Haraguchi T. Two distinct repeat sequences of Nup98 nucleoporins characterize dual nuclei in the binucleated ciliate Tetrahymena. Curr Biol. 2009;19:843–847. doi: 10.1016/j.cub.2009.03.055. [DOI] [PubMed] [Google Scholar]; An example of nuclear differentiation based on the differential targeting of Nup paralogs. In this case Nup98 exists as four paralogs in Tetrahymena, two targeted to each nucleus. The transcriptionally silent nucleus has Nup98 paralogs with unusual non-FG repeats and which correlates with differences in nucleocytoplasmic trafficking potential, and which may safeguard the micronucleus.
- 13.Tamura K., Fukao Y., Iwamoto M., Haraguchi T., Hara-Nishimura I. Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant Cell. 2010;22:4084–4097. doi: 10.1105/tpc.110.079947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Degrasse J.A., Dubois K.N., Devos D., Siegel T.N., Sali A., Field M.C., Rout M.P., Chait B.T. Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol Cell Proteomics. 2009;8:2119–2130. doi: 10.1074/mcp.M900038-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cronshaw J.M., Krutchinsky A.N., Zhang W., Chait B.T., Matunis M.J. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neumann N., Lundin D., Poole A.M. Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor. PLoS ONE. 2010;5:e13241. doi: 10.1371/journal.pone.0013241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mans B.J., Anantharaman V., Aravind L., Koonin E.V. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle. 2004;3((12) Dec):1612–1637. doi: 10.4161/cc.3.12.1316. [DOI] [PubMed] [Google Scholar]
- 18.Alber F., Dokudovskaya S., Veenhoff L.M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B.T. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 19.Devos D., Dokudovskaya S., Williams R., Alber F., Eswar N., Chait B.T., Rout M.P., Sali A. Simple fold composition and modular architecture of the nuclear pore complex. Proc Natl Acad Sci U S A. 2006;103:2172–2177. doi: 10.1073/pnas.0506345103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20••.Flemming D., Devos D.P., Schwarz J., Amlacher S., Lutzmann M., Hurt E. Analysis of the yeast nucleoporin Nup188 reveals a conserved S-like structure with similarity to karyopherins. J Struct Biol. 2011 doi: 10.1016/j.jsb.2011.11.008. [DOI] [PubMed] [Google Scholar]; First evidence that the solenoid region of a nucleoporin may adopt a more complex tertiary structure, and which then goes on to note a potential similarity between that conformation and the known structures of nuclear transport receptors.
- 21••.Andersen K.R., Onischenko E., Tang J.H., Kumar P., Chen J.Z., Ulrich A., Liphardt J.T., Weis K., Schwartz T.U. Scaffold nucleoporins Nup188 and Nup192 share structural and functional properties with nuclear transport receptors. eLife. 2013;2:e00745. doi: 10.7554/eLife.00745. [DOI] [PMC free article] [PubMed] [Google Scholar]; Provides experimental evidence that the solenoid fold of nucleoporins adpits an S-shaped superstructure that is highly similar to that for the nuclear transport receptors of the karyopherin family. May provide a link between these two systems.
- 22.Doucet C.M., Talamas J.A., Hetzer M.W. Cell cycle-dependent differences in nuclear pore complex assembly in metazoa. Cell. 2010;141:1030–1041. doi: 10.1016/j.cell.2010.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malone C.D., Falkowska K.A., Li A.Y., Galanti S.E., Kanuru R.C., LaMont E.G., Mazzarella K.C., Micev A.J., Osman M.M., Piotrowski N.K. Nucleus-specific importin alpha proteins and nucleoporins regulate protein import and nuclear division in the binucleate Tetrahymena thermophila. Eukaryot Cell. 2008;7:1487–1499. doi: 10.1128/EC.00193-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24••.Meier I., Zhou X., Brkljacić J., Rose A., Zhao Q., Xu X.M. Targeting proteins to the plant nuclear envelope. Biochem Soc Trans. 2010;38:733–740. doi: 10.1042/BST0380733. [DOI] [PubMed] [Google Scholar]; Identification of LINC protein orthologs in plants, and providing evidence that the entire complex may well be conserved in this lineage.
- 25.Lu Q., Tang X., Tian G., Wang F., Liu K., Nguyen V., Kohalmi S.E., Keller W.A., Tsang E.W.T., Harada J.J. Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J Cell Mol Biol. 2010;61:259–270. doi: 10.1111/j.1365-313X.2009.04048.x. [DOI] [PubMed] [Google Scholar]
- 26.Xu X.M., Rose A., Muthuswamy S., Jeong S.Y., Venkatakrishnan S., Zhao Q., Meier I. Nuclear pore anchor, the Arabidopsis homolog of Tpr/Mlp1/Mlp2/megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development. Plant Cell. 2007;19:1537–1548. doi: 10.1105/tpc.106.049239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27••.Ding D., Muthuswamy S., Meier I. Functional interaction between the Arabidopsis orthologs of spindle assembly checkpoint proteins MAD1 and MAD2 and the nucleoporin NUA. Plant Mol Biol. 2012;79:203–216. doi: 10.1007/s11103-012-9903-4. [DOI] [PubMed] [Google Scholar]; The paper indicates that the control of spindle assembly in plants requires interactions between the NPC and the MAD proteins. This is significant as this indicates that the same mechanism is being used here as in animals and fungi, and implies that the mechanism is therefore very old.
- 28.Ding R., West R.R., Morphew D.M., Oakley B.R., McIntosh J.R. The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol Biol Cell. 1997;8:1461–1479. doi: 10.1091/mbc.8.8.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ueda M., Schliwa M., Euteneuer U. Unusual centrosome cycle in Dictyostelium: correlation of dynamic behavior and structural changes. Mol Biol Cell. 1999;10:151–160. doi: 10.1091/mbc.10.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carvalho-Santos Z., Machado P., Branco P., Tavares-Cadete F., Rodrigues-Martins A., Pereira-Leal J.B., Bettencourt-Dias M. Stepwise evolution of the centriole-assembly pathway. J Cell Sci. 2010;123:1414–1426. doi: 10.1242/jcs.064931. [DOI] [PubMed] [Google Scholar]
- 31.Carvalho-Santos Z., Azimzadeh J., Pereira-Leal J.B., Bettencourt-Dias M. Evolution: tracing the origins of centrioles, cilia, and flagella. J Cell Biol. 2011;194:165–175. doi: 10.1083/jcb.201011152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Still I.H., Vettaikkorumakankauv A.K., DiMatteo A., Liang P. Structure–function evolution of the transforming acidic coiled coil genes revealed by analysis of phylogenetically diverse organisms. BMC Evol Biol. 2004;4:16. doi: 10.1186/1471-2148-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuhnert O., Baumann O., Meyer I., Gräf R. Functional characterization of CP148, a novel key component for centrosome integrity in Dictyostelium. Cell Mol Life Sci. 2012;69:1875–1888. doi: 10.1007/s00018-011-0904-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hodges M.E., Scheumann N., Wickstead B., Langdale J.A., Gull K. Reconstructing the evolutionary history of the centriole from protein components. J Cell Sci. 2010;123:1407–1413. doi: 10.1242/jcs.064873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cavalier-Smith T. Origin of the cell nucleus, mitosis and sex: roles of intracellular coevolution. Biol Direct. 2010;5:7. doi: 10.1186/1745-6150-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pearson C.G., Winey M. Basal body assembly in ciliates: the power of numbers. Traffic. 2009;10:461–471. doi: 10.1111/j.1600-0854.2009.00885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Friedländer M., Wahrman J. The spindle as a basal body distributor. A study in the meiosis of the male silkworm moth, Bombyx mori. J Cell Sci. 1970;7:65–89. doi: 10.1242/jcs.7.1.65. [DOI] [PubMed] [Google Scholar]
- 38.Pickett-Heaps J. The autonomy of the centriole: fact or fallacy? Cytobios. 1971;3:205–214. [Google Scholar]
- 39.Debec A., Sullivan W., Bettencourt-Dias M. Centrioles: active players or passengers during mitosis? Cell Mol Life Sci. 2010;67:2173–2194. doi: 10.1007/s00018-010-0323-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khodjakov A., Cole R.W., Oakley B.R., Rieder C.L. Centrosome-independent mitotic spindle formation in vertebrates. Curr Biol. 2000;10:59–67. doi: 10.1016/s0960-9822(99)00276-6. [DOI] [PubMed] [Google Scholar]
- 41.Khodjakov A., Rieder C.L. Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. J Cell Biol. 2001;153:237–242. doi: 10.1083/jcb.153.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Basto R., Lau J., Vinogradova T., Gardiol A., Woods C.G., Khodjakov A., Raff J.W. Flies without centrioles. Cell. 2006;125:1375–1386. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 43.Ratcliffe H. Mitosis and cell division in Euglena spirogyra Ehrenberg. Biol Bull. 1927;53:109–122. [Google Scholar]
- 44.Kaller M., Euteneuer U., Nellen W. Differential effects of heterochromatin protein 1 isoforms on mitotic chromosome distribution and growth in Dictyostelium discoideum. Eukaryot Cell. 2006;5:530–543. doi: 10.1128/EC.5.3.530-543.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.King M.C., Drivas T.G., Blobel G. A network of nuclear envelope membrane proteins linking centromeres to microtubules. Cell. 2008;134:427–438. doi: 10.1016/j.cell.2008.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schulz I., Baumann O., Samereier M., Zoglmeier C., Gräf R. Dictyostelium Sun1 is a dynamic membrane protein of both nuclear membranes and required for centrosomal association with clustered centromeres. Eur J Cell Biol. 2009;88:621–638. doi: 10.1016/j.ejcb.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 47.Hartman H., Fedorov A. The origin of the eukaryotic cell: a genomic investigation. Proc Natl Acad Sci U S A. 2002;99:1420–1425. doi: 10.1073/pnas.032658599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bornens M., Azimzadeh J. Origin and evolution of the centrosome. Adv Exp Med Biol. 2007;607:119–129. doi: 10.1007/978-0-387-74021-8_10. [DOI] [PubMed] [Google Scholar]
- 49.Daunderer C., Schliwa M., Gräf R. Dictyostelium centrin-related protein (DdCrp), the most divergent member of the centrin family, possesses only two EF hands and dissociates from the centrosome during mitosis. Eur J Cell Biol. 2001;80:621–630. doi: 10.1078/0171-9335-00198. [DOI] [PubMed] [Google Scholar]
- 50.Mana-Capelli S., Gräf R., Larochelle D.A. Dictyostelium centrin B localization during cell cycle progression. Commun Integr Biol. 2010;3:39–41. doi: 10.4161/cib.3.1.9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li S., Sandercock A.M., Conduit P., Robinson C.V., Williams R.L., Kilmartin J.V. Structural role of Sfi1p-centrin filaments in budding yeast spindle pole body duplication. J Cell Biol. 2006;173:867–877. doi: 10.1083/jcb.200603153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feldherr C.M. Nucleocytoplasmic exchanges during early interphase. J Cell Biol. 1968;39:49–54. doi: 10.1083/jcb.39.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leeson T.S., Bhatnagar R. Amoeba proteus: the nuclear periphery. Cell Differ. 1975;4:79–86. doi: 10.1016/0045-6039(75)90020-2. [DOI] [PubMed] [Google Scholar]
- 54.Schmidt M., Grossmann U., Krohne G. The nuclear membrane-associated honeycomb structure of the unicellular organism Amoeba proteus: on the search for homologies with the nuclear lamina of metazoa. Eur J Cell Biol. 1995;67:199–208. [PubMed] [Google Scholar]
- 55.Dittmer T.A., Misteli T. The lamin protein family. Genome Biol. 2011;12:222. doi: 10.1186/gb-2011-12-5-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dahl K.N., Scaffidi P., Islam M.F., Yodh A.G., Wilson K.L., Misteli T. Distinct structural and mechanical properties of the nuclear lamina in Hutchinson–Gilford progeria syndrome. Proc Natl Acad Sci U S A. 2006;103:10271–10276. doi: 10.1073/pnas.0601058103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Worman H.J. Nuclear lamins and laminopathies. J Pathol. 2012;226:316–325. doi: 10.1002/path.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shimi T., Pfleghaar K., Kojima S., Pack C.-G., Solovei I., Goldman A.E., Adam S.A., Shumaker D.K., Kinjo M., Cremer T. The A- and B-type nuclear lamin networks: microdomains involved in chromatin organization and transcription. Genes Dev. 2008;22:3409–3421. doi: 10.1101/gad.1735208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hozák P., Sasseville A.M., Raymond Y., Cook P.R. Lamin proteins form an internal nucleoskeleton as well as a peripheral lamina in human cells. J Cell Sci. 1995;108(Pt 2):635–644. doi: 10.1242/jcs.108.2.635. [DOI] [PubMed] [Google Scholar]
- 60••.Krüger A., Batsios P., Baumann O., Luckert E., Schwarz H., Stick R., Meyer I., Gräf R. Characterization of NE81, the first lamin-like nucleoskeleton protein in a unicellular organism. Mol Biol Cell. 2012;23:360–370. doi: 10.1091/mbc.E11-07-0595. [DOI] [PMC free article] [PubMed] [Google Scholar]; First identification of a lamin ortholog outside of the metazoa. NE81 bears remarkably conserved sequence features similar to lamins, and demonstrates nuclear envelope association throughout the cell cycle.
- 61.Batsios P., Peter T., Baumann O., Stick R., Meyer I., Gräf R. A lamin in lower eukaryotes? Nucleus. 2012;3:237–243. doi: 10.4161/nucl.20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62••.Ciska M., Masuda K., Moreno Díaz de la Espina S. Lamin-like analogues in plants: the characterization of NMCP1 in Allium cepa. J Exp Bot. 2013;64:1553–1564. doi: 10.1093/jxb/ert020. [DOI] [PMC free article] [PubMed] [Google Scholar]; Identification of NMCP as a possible lamina analogue in higher plants. The experimental data are based on localisation and behaviour of the NMCP protein, the best candidate for such a factor.
- 63••.DuBois K.N., Alsford S., Holden J.M., Buisson J., Swiderski M., Bart J.-M., Ratushny A.V., Wan Y., Bastin P., Barry J.D. NUP-1 is a large coiled-coil nucleoskeletal protein in trypanosomes with lamin-like functions. PLoS Biol. 2012;10:e1001287. doi: 10.1371/journal.pbio.1001287. [DOI] [PMC free article] [PubMed] [Google Scholar]; Identification of a protein that is a probable analogue of lamins. The NUP-1 protein retains roles in telomeric silencing, nuclear structural integrity and NPC positioning, despite the protein being five times larger than mammalian lamins.
- 64••.Rothballer A., Kutay U. The diverse functional LINCs of the nuclear envelope to the cytoskeleton and chromatin. Chromosoma. 2013;122:415–429. doi: 10.1007/s00412-013-0417-x. [DOI] [PMC free article] [PubMed] [Google Scholar]; The LINC protein family is complex, and has many members, significantly the differentiation between many of these isoforms remains unclear.
- 65.Starr D.A., Fridolfsson H.N. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol. 2010;26:421–444. doi: 10.1146/annurev-cellbio-100109-104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66•.Field M.C., Horn D., Alsford S., Koreny L., Rout M.P. Telomeres, tethers and trypanosomes. Nucleus. 2012;3:478–486. doi: 10.4161/nucl.22167. [DOI] [PMC free article] [PubMed] [Google Scholar]; Presents in silico evidence for the absence of SUN-domain proteins from the nuclei of several lineages, most prominently the trypanosomatids.
- 67.Stewart-Hutchinson P.J., Hale C.M., Wirtz D., Hodzic D. Structural requirements for the assembly of LINC complexes and their function in cellular mechanical stiffness. Exp Cell Res. 2008;314:1892–1905. doi: 10.1016/j.yexcr.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Haque F., Lloyd D.J., Smallwood D.T., Dent C.L., Shanahan C.M., Fry A.M., Trembath R.C., Shackleton S. SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol Cell Biol. 2006;26:3738–3751. doi: 10.1128/MCB.26.10.3738-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schneider M., Lu W., Neumann S., Brachner A., Gotzmann J., Noegel A.A., Karakesisoglou I. Molecular mechanisms of centrosome and cytoskeleton anchorage at the nuclear envelope. Cell Mol Life Sci. 2011;68:1593–1610. doi: 10.1007/s00018-010-0535-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xiong H., Rivero F., Euteneuer U., Mondal S., Mana-Capelli S., Larochelle D., Vogel A., Gassen B., Noegel A.A. Dictyostelium Sun-1 connects the centrosome to chromatin and ensures genome stability. Traffic. 2008;9:708–724. doi: 10.1111/j.1600-0854.2008.00721.x. [DOI] [PubMed] [Google Scholar]
- 71.Zhou X., Meier I. How plants LINC the SUN to KASH. Nucleus. 2013;4:206–215. doi: 10.4161/nucl.24088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.De Souza C.P.C., Osmani S.A. Mitosis, not just open or closed. Eukaryot Cell. 2007;6:1521–1527. doi: 10.1128/EC.00178-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Santarella-Mellwig R., Franke J., Jaedicke A., Gorjanacz M., Bauer U., Budd A., Mattaj I.W., Devos D.P. The compartmentalized bacteria of the planctomycetes-verrucomicrobia-chlamydiae superphylum have membrane coat-like proteins. PLoS Biol. 2010;8:e1000281. doi: 10.1371/journal.pbio.1000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Santarella-Mellwig R., Pruggnaller S., Roos N., Mattaj I.W., Devos D.P. Three-dimensional reconstruction of bacteria with a complex endomembrane system. PLoS Biol. 2013;11:e1001565. doi: 10.1371/journal.pbio.1001565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee K.-C., Webb R., Fuerst J. The cell cycle of the planctomycete Gemmata obscuriglobus with respect to cell compartmentalization. BMC Cell Biol. 2009;10:4. doi: 10.1186/1471-2121-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pan M., Kelman L.M., Kelman Z. The archaeal PCNA proteins. Biochem Soc Trans. 2011;39:20–24. doi: 10.1042/BST0390020. [DOI] [PubMed] [Google Scholar]
- 77.Salgado-Garrido J., Bragado-Nilsson E., Kandels-Lewis S., Séraphin B. Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J. 1999;18:3451–3462. doi: 10.1093/emboj/18.12.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Makarova K.S., Koonin E.V., Kelman Z. The CMG (CDC45/RecJ, MCM, GINS) complex is a conserved component of the DNA replication system in all archaea and eukaryotes. Biol Direct. 2012;7:7. doi: 10.1186/1745-6150-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sandman K., Reeve J.N. Archaeal histones and the origin of the histone fold. Curr Opin Microbiol. 2006;9:520–525. doi: 10.1016/j.mib.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 80.Pereira S.L., Grayling R.A., Lurz R., Reeve J.N. Archaeal nucleosomes. Proc Natl Acad Sci U S A. 1997;94:12633–12637. doi: 10.1073/pnas.94.23.12633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Omer A.D., Lowe T.M., Russell A.G., Ebhardt H., Eddy S.R., Dennis P.P. Homologs of small nucleolar RNAs in Archaea. Science. 2000;288:517–522. doi: 10.1126/science.288.5465.517. [DOI] [PubMed] [Google Scholar]
- 82.Low H.H., Löwe J. A bacterial dynamin-like protein. Nature. 2006;444:766–769. doi: 10.1038/nature05312. [DOI] [PubMed] [Google Scholar]
- 83.Samson R.Y., Obita T., Freund S.M., Williams R.L., Bell S.D. A role for the ESCRT system in cell division in archaea. Science. 2008;322:1710–1713. doi: 10.1126/science.1165322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lindås A.C., Karlsson E.A., Lindgren M.T., Ettema T.J., Bernander R. A unique cell division machinery in the Archaea. Proc Natl Acad Sci U S A. 2008;105:18942–18946. doi: 10.1073/pnas.0809467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pace N.R. Time for a change. Nature. 2006;441:289. doi: 10.1038/441289a. [DOI] [PubMed] [Google Scholar]
- 86.Gribaldo S., Poole A.M., Daubin V., Forterre P., Brochier-Armanet C. The origin of eukaryotes and their relationship with the Archaea: are we at a phylogenomic impasse? Nat Rev Microbiol. 2010;8((10) Oct):743–752. doi: 10.1038/nrmicro2426. [DOI] [PubMed] [Google Scholar]
- 87.Adl S.M., Simpson A.G.B., Lane C.E., Lukeš J., Bass D., Bowser S.S., Brown M.W., Burki F., Dunthorn M., Hampl V. The revised classification of eukaryotes. J Eukaryot Microbiol. 2012;59:429–493. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]