Abstract
Variation in cyanogenesis (hydrogen cyanide release following tissue damage) was first noted in populations of white clover more than a century ago, and subsequent decades of research have established this system as a classic example of an adaptive chemical defence polymorphism. Here, we document polymorphisms for cyanogenic components in several relatives of white clover, and we determine the molecular basis of this trans-specific adaptive variation. One hundred and thirty-nine plants, representing 13 of the 14 species within Trifolium section Trifoliastrum, plus additional species across the genus, were assayed for cyanogenic components (cyanogenic glucosides and their hydrolysing enzyme, linamarase) and for the presence of underlying cyanogenesis genes (CYP79D15 and Li, respectively). One or both cyanogenic components were detected in seven species, all within section Trifoliastrum; polymorphisms for the presence/absence (PA) of components were detected in six species. In a pattern that parallels our previous findings for white clover, all observed biochemical polymorphisms correspond to gene PA polymorphisms at CYP79D15 and Li. Relationships of DNA sequence haplotypes at the cyanogenesis loci and flanking genomic regions suggest independent evolution of gene deletions within species. This study thus provides evidence for the parallel evolution of adaptive biochemical polymorphisms through recurrent gene deletions in multiple species.
Keywords: balanced polymorphism, chemical defence, clover (Trifolium L.), cyanogenic glucosides, copy number variation, molecular adaptation
1. Introduction
Since the Modern Synthesis, a major goal of evolutionary biology has been to understand the connection between genes and their adaptive roles in nature. With recent advances in genomic techniques, it is becoming increasingly possible to study the molecular basis and genomic architecture of phenotypes in wild species [1–3]. New insights are now being gained into such fundamental questions as the roles of cis-regulatory and protein-coding mutations in adaptation and the predictability of parallel adaptive change at the interspecific level [4,5]. However, while genomic data can now be amassed at a rapid rate, the study of adaptation still requires in-depth understanding of a species’ ecology and the relationship between phenotypes and fitness in nature. Thus, species where the adaptive significance of phenotypic variation has been extensively documented can be particularly attractive study systems for understanding the genetic basis of adaptation.
We have focused on one such system in studying the molecular evolution of an adaptive chemical defence polymorphism in white clover (Trifolium repens L., Fabaceae). White clover is polymorphic for cyanogenesis (hydrogen cyanide release following tissue damage), with both cyanogenic and acyanogenic plants occurring in natural populations. This polymorphism was first documented more than a century ago (reviewed in [6,7]), and the selective factors that maintain it have been examined in dozens of studies over the past seven decades (reviewed in [8–10]). In this study, we extend the examination of this adaptive variation to white clover's relatives within the genus Trifolium, with the goal of understanding the importance of conserved versus novel genetic mechanisms in parallel evolution at the interspecific level. Specifically, we assess the occurrence of cyanogenesis polymorphisms in related clover species and the molecular evolutionary forces that have shaped this trans-specific adaptive variation.
(a). The white clover cyanogenesis polymorphism
Trifolium repens is a native species of Eurasia and has become widely naturalized in temperate regions worldwide as a component of lawns, pastures and roadsides. Cyanogenic white clover plants are differentially protected from small, generalist herbivores, including gastropods, voles and insects [11–14]. At the same time, populations show climate-associated clinal variation in cyanogenesis, with acyanogenic plants predominating at higher latitudes and elevations [15,16]. The apparent selective advantage of the acyanogenic form in cooler climates may reflect fitness trade-offs between energetic investment in cyanogenesis versus reproductive output in regions of high and low herbivore pressure [17–19]. Cyanogenesis frequencies also appear to be influenced by aridity, with cyanogenic morphs differentially represented in drier regions [20,21]. Whether primarily shaped by biotic or abiotic factors, the fact that climate-associated cyanogenesis clines have evolved repeatedly in this species—both in native populations [15,16,22–25] and in the introduced species range [9,21,26,27]—suggests that the selective forces maintaining this adaptive polymorphism are strong and geographically pervasive.
The cyanogenic phenotype in clover requires the production of two biochemical components that are separated in intact tissue and brought into contact with cell rupture: cyanogenic glucosides (lotaustralin and linamarin), which are stored in the vacuoles of photosynthetic tissue; and their hydrolysing enzyme, linamarase, which is stored in the cell wall (reviewed in [8]). Acyanogenic clover plants may lack cyanogenic glucosides, linamarase or both components. Inheritance of the two cyanogenic components is controlled by two independently segregating Mendelian genes [28–30]. The gene Ac controls the presence/absence (PA) of cyanogenic glucosides, and Li controls the PA of linamarase; for both genes, the dominant (functional) allele confers the presence of the component. Thus, plants that possess at least one dominant allele at both genes (Ac_, Li_) are cyanogenic, while homozygous recessive genotypes at either or both genes (acac, lili) lack one or more of the required components. The presence or the absence of each component can be determined for individual plants with leaf tissue assays using colorimetric HCN test paper [31] and exogenously added cyanogenic components (method described in [32]). In addition to these discrete cyanogenesis polymorphisms, there is also wide quantitative variation in production of the two cyanogenic components among plants that produce the compounds. This variation is attributable to a combination of factors, including phenotypic plasticity, allelic variation at Ac and Li, and unlinked modifier genes [33,34] (K. Olsen 2014, unpublished observations).
In the past studies, we have documented the molecular basis of the Ac/ac and Li/li biochemical polymorphisms in white clover. The ac and li non-functional alleles correspond, respectively, to gene deletions at two unlinked loci: CYP79D15, which encodes the cytochrome P450 protein catalysing the first dedicated step in cyanogenic glucoside biosynthesis [35]; and Li, which encodes the linamarase protein [32]. Thus, the Ac/ac and Li/li biochemical polymorphisms in white clover arise through two independently segregating gene PA polymorphisms. A recent molecular evolutionary analysis of the genomic sequences flanking these PA polymorphisms indicates that, for both loci, the gene-absence alleles have evolved repeatedly in white clover through recurrent gene deletion events [10].
(b). Cyanogenesis in other clover species
While most studies of clover cyanogenesis have focused on T. repens, the trait has also been examined to a limited extent in related clover species. The legume genus Trifolium includes approximately 255 species found in temperate and subtropical regions worldwide [36], and T. repens falls within section Trifoliastrum, a clade comprising approximately 14 closely related species with a circum-Mediterranean distribution [37]. The presence of one or both cyanogenic components has been previously reported in five other Trifolium species, all within Trifoliastrum: T. isthmocarpum (reportedly monomorphic for AcAc LiLi) [38]; T. nigrescens (primarily AcAc LiLi, with rare occurrence of ac and li alleles reported in T. nigrescens ssp. nigrescens) [38,39]; T. montanum and T. ambiguum (both species lili while polymorphic for Ac/ac) [38] and T. occidentale (primarily AcAc lili, with rare occurrence of ac alleles) [38,40]. Phylogenetic relationships within Trifoliastrum generally lack resolution; the positions of two species, T. montanum and T. ambiguum, are best resolved, with these taxa forming a species-pair that is phylogenetically distinct from other members of the clade [37] (electronic supplementary material, figure S1).
The occurrence of polymorphisms for cyanogenic components in at least four Trifolium species besides white clover raises intriguing questions on the origin and long-term evolution of this adaptive variation. The fact that gene PA polymorphisms underlie both the Ac/ac and Li/li polymorphisms in white clover might suggest that this same molecular basis would occur in related species. On the other hand, null alleles at cyanogenesis genes can easily arise through simple loss-of-function mutations that do not require genomic deletion events [41], which suggests that other mutational mechanisms (e.g. frameshifts, permature stop codons) might also be responsible.
A related question concerns the evolutionary persistence of Ac/ac and Li/li alleles. Evolutionary studies of other adaptive PA polymorphisms, particularly those involving plant pathogen resistance genes (R-genes), have revealed signatures of long-term balancing selection, which are consistent with the selective maintenance of ancient gene-presence and -absence alleles [42–44]. For Trifolium, the close phylogenetic relationships within Trifoliastrum suggest that selectively maintained alleles might pre-date the diversification of the clade and be shared across species boundaries. On the other hand, the fact that ac and li gene-deletion alleles have evolved repeatedly within white clover [10] might instead suggest a high enough gene deletion rate that all Ac/ac and Li/li allelic variation would be species-specific. For adaptive PA polymorphisms, genealogical relationships between gene-presence and -absence alleles can be assessed by examining sequences adjacent to the PA locus, as these sequences are present in all plants but are linked to the PA variation [10,42–44].
In this study, we address three specific questions on the origin and persistence of cyanogenesis polymorphisms in Trifolium: (i) What is the distribution of cyanogenic components and polymorphisms for these components among white clover's closest relatives in Trifolium section Trifoliastrum, and more broadly across the genus? (ii) What is the molecular basis of any observed cyanogenesis polymorphisms in these species? Specifically, are these PA polymorphisms, as in white clover, or are other mutational mechanisms involved? and (iii) Does the evolution of these polymorphisms pre-date species diversification, or has there been independent evolution of the polymorphisms within species? Our results suggest that cyanogenesis polymorphisms occur in multiple Trifoliastrum species, that they have evolved independently in the different species in which they occur, and that this parallel evolution has occurred through a conserved mutational mechanism involving gene deletion events.
2. Material and methods
(a). Sampling
Seeds of 139 Trifolium accessions were obtained either through the USDA National Plant Germplasm System or from other sources (see the electronic supplementary material, table S1) and grown in the Washington University greenhouse. Samples included 119 accessions representing 13 of the 14 species within Trifolium sect. Trifoliastrum; remaining accessions included five species in sect. Involucrarium (sister clade to sect. Trifoliastrum), three species in sect. Vesicastrum (sister clade to the clade comprising sects. Involucrarium and Trifoliastrum) and individual species representing more distantly related Trifolium lineages (sect. Trifolium, sect. Trichocephalum and subgenus Chronosemium) (electronic supplementary material, table S1).
Species within sect. Trifoliastrum are native to the Mediterranean and Eurasia and occur across a diverse range of habitats in temperate and subtropical regions [36,37]. Three species in the clade are known to be polyploid (T. ambiguum, T. repens and T. uniflorum). In the case of white clover, which is allotetraploid, the Ac and Li genes occur within only one of its two parental genomes, suggesting that this species may have originated through the hybridization of a cyanogenic and an acyanogenic diploid progenitor [39,45]. Proposed progenitors within Trifoliastrum have included T. occidentale, T. nigrescens ssp. petrisavii, T. pallescens and an unknown lineage [37,39,45,46].
One plant per named accession was used in genetic characterizations unless cyanogenesis assays (described later) revealed intra-accession variation in cyanogenesis phenotype; in those rare cases, representative plants of each cyanogenesis phenotype were included (see the electronic supplementary material, table S1). Because of morphological ambiguities among Trifolium species, the species identity for each individual plant was determined by PCR-amplifying and sequencing the nuclear ITS rDNA region and the cpDNA trnL intron and performing BLAST searches against published data [37]. ITS and trnL sequences are individually diagnostic for most Trifolium species, and the two-locus combination was diagnostic for all species examined in this study. Primers and PCR conditions for ITS and trnL sequencing are described by Ellison et al. [37]. Inferred species identities for all accessions are indicated in the electronic supplementary material, table S1.
(b). Phenotypic and genetic analyses
Cyanogenesis assays to determine the presence or the absence of cyanogenic components (cyanogenic glucosides, linamarase) in each plant were performed using leaf tissue in a modified Feigl-Anger HCN assay, as described previously for white clover [32,35]. For genetic analyses, genomic DNA was extracted from fresh leaf tissue using either Nucleon Phytopure extraction kits (Tepnel Life Sciences, Stamford, CT) or the protocol of Porebski et al. [47]. Two methods were used to screen plants for the presence or the absence of the Li and CYP79D15 loci. First, PCR was performed using primers specific for each gene. For Li, most amplifications used the following primer pair: Lin_01aF: ACATGCTTTTAAACCTCTTCC, Lin_05dR: TGGGCTGGTCCATTTGATTTAAC; an alternative forward primer was used in some reactions: Lin_01eF CCATCACTACTACTCATATCCATGCT. Both primer combinations amplify nearly the entire 3.9 kb Li gene. For CYP79D15, PCR was performed with the following primer pair, which amplifies nearly the entire 1.7 kb gene: CYP_Fb: TGGACTTTTTTGCTTGTTGTGATATT, CYP_Rb: GCAGCCAATCTTGGTTTTGC. PCR conditions are as described previously for Li [32] and CYP79D15 [35]. The absence of a PCR product after three or more attempts provided preliminary evidence suggesting the absence of the corresponding cyanogenesis gene.
As a second method to screen for the presence or the absence of the cyanogenesis genes, Southern hybridizations were performed using probes specific to CYP79D15, Li, or to a gene closely related to Li that encodes a non-cyanogenic glucosidase (‘Li-paralogue’ [32]). The CYP79D15 probe (CYP1) spans approximately half of the gene, including the single intron; the Li probe (L1) corresponds to a 0.9 kb portion of Li intron 2; and the Li-paralogue probe (P1) corresponds to the equivalent intron of the non-cyanogenic glucosidase gene [32,35]. While not involved in cyanogenesis, the Li-paralogue is genetically very similar to Li (94% nucleotide sequence identity), so that there is some cross-hybridization between genes in Southern hybridizations; probing specifically for the Li-paralogue is therefore useful in confirming that weak bands detected in hybridizations of lili plants do not correspond to the Li gene [32]. Protocols for Li and CYP79D15 Southern hybridizations followed those used previously for white clover [32,35]. Genomic DNA digests for Southerns were performed primarily using the restriction enzyme AseI, which is predicted to cut once within the L1 probe and to be a non-cutter within the CYP1 probe; thus, hybridizations would be expected to reveal two bands for Li if it is present as a single-copy gene and one band for CYP79D15 if it is present as a single-copy gene.
For plants where PCR screening and Southern hybridizations indicated the presence of a given cyanogenesis gene, PCR products were cloned into pGEM-T Easy vectors (Promega) and sequenced using reaction conditions and internal primers as described previously [32,35]. A minimum of three clones per PCR product were sequenced (with four or more clones sequenced for many accessions). DNA sequencing was performed using an ABI 3130 capillary sequencer in the Biology Department of Washington University. Creation of contigs and DNA sequence aligning and editing were performed using BioLign v. 4.0.6 [48]. Singletons observed in individual clones were treated as artefacts of polymerase error and removed, yielding one definitive haplotype sequence per accession. DNA sequences are available on GenBank (accession nos. KJ467253–KJ467351).
DNA sequences adjacent to an adaptive PA polymorphism can provide information on the evolution of the linked gene-presence and -absence alleles [10,42–44]. In a previous study of white clover, we used genome-walking to identify sequences immediately flanking CYP79D15 and Li to characterize the boundaries of gene-deletion alleles and to test for molecular signatures of balancing selection [10]. Two downstream regions were identified as occurring within 325 bp of the boundaries of most ac and li gene-deletion alleles: 3CYP-2.34, a 1.14-kb region starting 2.34 kb downstream of the CYP79D15 stop codon; and 3Li-6.65, 0.9-kb region located 6.65 kb downstream of the Li stop codon. For this study, orthologues of these T. repens sequences were targeted in the other Trifoliastrum species to assess phylogenetic relationships of gene-presence and -absence haplotypes within and among species. Amplicons were cloned and sequenced as described above for the cyanogenesis genes. If gene-presence and -absence haplotypes for a given species are more closely related to each other than to haplotypes in other species, this would suggest independent evolution of PA polymorphisms within species.
Phylogenetic relationships among haplotypes were assessed using maximum-likelihood (ML) analyses for each sequenced locus, with the best-fit model of nucleotide substitution selected in jModelTest v. 2.1.4 [49,50] based on likelihood scores for 88 different models. The GTR model of molecular evolution was employed for all sequence datasets based on jModelTest results. ML trees were generated in PhyML v. 3.0 [51] via the ATGC web platform (http://atgc.lirmm.fr/phyml/), with default settings for tree searching and bootstrap analysis. For T. repens, three representative haplotypes were included from previous analyses for the CYP79D15 and Li datasets [32,35]; for loci flanking the cyanogenesis loci, three haplotypes apiece for gene-presence and -absence alleles were used [10]. Outside of Trifolium, no clear orthologues of the Li gene are known, and the closest putative orthologue of CYP79D15 occurs in Lotus japonicus, where the corresponding gene is unalignable in non-coding regions; therefore, midpoint rooting was used for the Trifolium haplotype trees.
3. Results
(a). Cyanogenesis polymorphisms occur in multiple Trifolium species
Biochemical assays for the presence or absence of cyanogenic glucosides and linamarase revealed a diversity of patterns for the production of cyanogenic components among Trifolium species in section Trifoliastrum. Results of cyanogenesis assays are summarized in table 1. Within this clade, which also includes white clover, one or both cyanogenic components were detected in seven of the 12 species tested. This set of seven species includes all four species where cyanogenesis polymorphisms have been reported previously (T. ambiguum, T. montanum, T. nigrescens and T. occidentale) [38–40], as well as T. isthmocarpum, which was previously reported to be monomorphic for the production of both components [38]. Five of the seven species were found to be polymorphic for the PA of cyanogenic glucosides, and two were polymorphic for linamarase production. Only one species, T. isthmocarpum, was polymorphic at both Ac/ac and Li/li, as is found in white clover. Given the small sample sizes for some of the tested species, it is quite possible that expanded sampling could reveal additional cyanogenesis polymorphisms among species of this clade. In contrast to members of Trifoliastrum, no cyanogenic components were detected in any Trifolium species outside of this clade (electronic supplementary material, table S1).
Table 1.
Cyanogenesis polymorphisms in sampled species of Trifolium sect. Trifoliastrum. Numbers in parentheses indicate sample sizes. Subspecies designations follow the taxonomy of Ellison et al. [37].
| species | Ac_ Li_ | Ac_ lili | acac Li_ | acac lili |
|---|---|---|---|---|
| T. ambiguum (13) | — | 7 | — | 6 |
| T. cernuum (5) | — | — | — | 5 |
| T. glomeratum (5) | — | — | — | 5 |
| T. isthmocarpum (16) | 8 | 7 | — | 1 |
| T. montanum | ||||
| subsp. montanum (17) | — | 9 | — | 8 |
| subsp. humboldtianum (2) | 1 | 1 | ||
| T. nigrescens | ||||
| subsp. nigrescens (7) | 6 | 1 | — | — |
| subsp. petrisavii (7) | 7 | — | — | — |
| subsp. meneghinianum (6) | 6 | — | — | — |
| T. occidentale (9) | — | 9 | — | — |
| T. pallescens (3) | — | — | — | 3 |
| T. retusum (8) | — | — | — | 8 |
| T. suffocatum (5) | — | 1 | — | 4 |
| T. thallii (1) | — | — | — | 1 |
| T. uniflorum (4) | — | 2 | — | 2 |
(b). All cyanogenesis polymorphisms are presence/absence polymorphisms
We successfully PCR-amplified the 1.7-kb CYP79D15 gene in all plants that produce cyanogenic glucosides, and the gene was never amplified in plants lacking these compounds. Similarly, we were able to amplify the entire 3.9 kb Li gene in nearly all plants producing linamarase (only partial gene amplification was successful for four accessions; electronic supplementary material, table S1), while it was never amplified in plants lacking the enzyme.
As the Ac/ac and Li/li biochemical polymorphisms in white clover correspond to gene PA polymorphisms at CYP79D15 and Li, respectively [32,35], these PCR results suggested that PA polymorphisms might also account for the biochemical polymorphisms in other Trifolium species. We therefore used Southern hybridizations to test for a relationship between the production of cyanogenic components and gene PA across Trifoliastrum. In all cases, the presence or the absence of cyanogenic components (table 1) matches the presence or the absence of detectable bands in Southern hybridizations.
Representative results of CYP79D15 Southern hybridizations for AseI genomic DNA digests are shown in figure 1 (see also the electronic supplementary material, figure S2). For every species where the Ac/ac biochemical polymorphism was detected, plants that lack cyanogenic glucosides (acac genotypes) also lack bands corresponding to the 0.9 kb CYP probe. Interestingly, for plants that possess the gene, there appears to be variation in gene copy number, with one to two bands present among individuals of three species (T. montanum, T. ambiguum, T. isthmocarpum; figure 1a,b,d), and up to three clear bands present in T. nigrescens ssp. meneghinianum (electronic supplementary material, figure S2). This band variation is not correlated with ploidy, as only one of these species is a known polyploid (T. ambiguum) [37]. Nor is it attributable to AseI restriction site variation within the probed gene region, as no such nucleotide variation was observed in any CYP79D15 DNA sequences (described later). Within T. nigrescens, the banding pattern for subspecies nigrescens is recognizably distinct from that of the other two subspecies (petrisavii and meneghinianum) (electronic supplementary material, figure S2), consistent with the genomic and morphological divergence of this subspecies from the other two [52]
Figure 1.
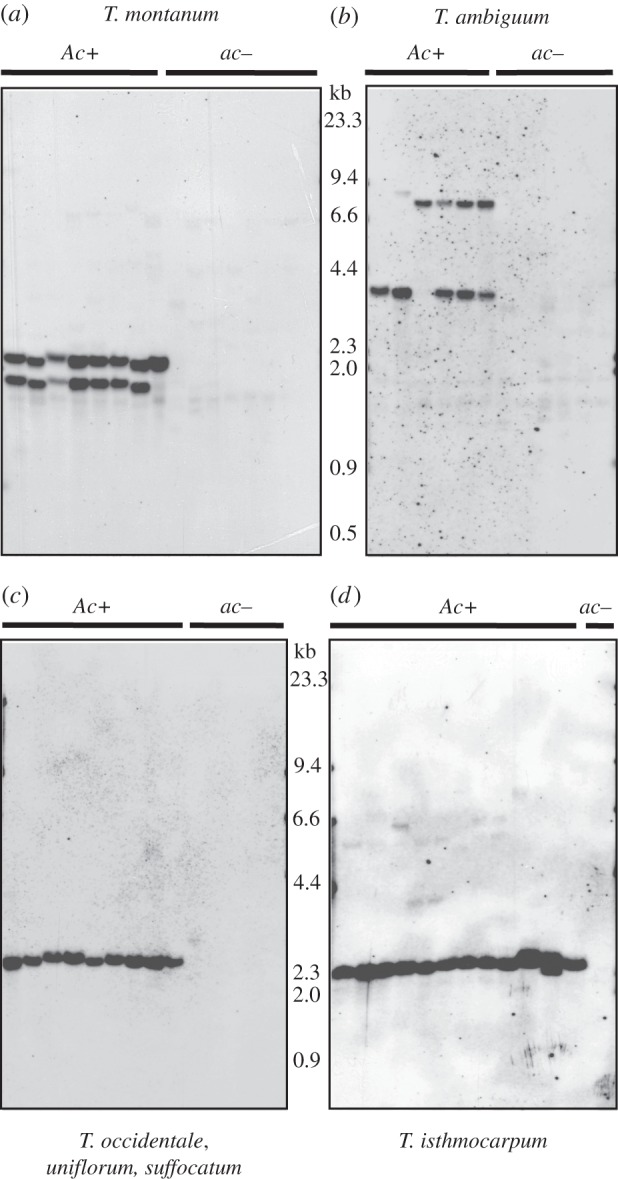
Southern hybridizations for CYP79D15 in Trifolium species with the Ac/ac polymorphism. Ac+ and ac– correspond, respectively, to accessions with and without detectable levels of cyanogenic glucosides in HCN assays. The presence of a single band is consistent with the occurrence of CYP79D15 as a single-copy gene. (a) Trifolium montanum accessions, left to right: PI 234940, PI 542854b, PI 542861, PI 611617, MU-349b, PI 542811, KEW 0010849b, PI 611633, PI 205314, PI 418851, PI 440734, PI 542846, PI 542850, PI 542854a, PI 611651, MU-349a; (b) T. ambiguum accessions, left to right: PI 604743, PI 604720, PI 440712, PI 502614, PI 405124, PI 604749, PI 598987, PI 604752, MU-347, PI 440693, PI 641674, PI 604703; (c) T. occidentale, T. uniflorum and T. suffocatum accessions, left to right: PI 214207 (T. repens positive control), Leca de Palma, exAZ 4720, MU-353, KEW 0055322, PI 641364, PI 641363, PI 351076, PI 369138b, PI 516460, PI 369138c, PI 369138a, PI 369135, MU-084; (d) T. isthmocarpum accessions, left to right: MU-209a, PI 517109, PI 517110, PI 517111, PI 517112, PI 203664, MU-116, MU-209b, PI 338675, PI 535692, PI 535571, PI 202517, PI 653511.
For the two species where linamarase production was detected, results of Southern hybridizations for the Li gene are shown in figure 2 (see also the electronic supplementary material, figure S3). The L1 probe, designed to be specific to the Li gene, contains one AseI restriction site, so that the occurrence of two bands on a Southern blot is consistent with the occurrence of a single Li gene copy. Because of high nucleotide similarity between Li and the non-cyanogenic Li-paralogue, hybridizations using the L1 probe are not entirely specific to the Li gene. Therefore, we also performed hybridizations using the P1 probe, designed to be specific to the Li-paralogue, as a way of identifying bands that do not correspond to Li (see Material and methods). For T. nigrescens, where a single lili individual was observed, the two bands that are evident in all Li_ individuals (at approx. 0.9 and 1.4 kb) are absent in this individual (figure 2a), and the one band that is present (at approx. 2 kb) shows strong hybridization to the P1 probe (figure 2b). Thus, the bands corresponding to the Li gene are not present in the lili accession. Similarly, for T. isthmocarpum, all individuals with detectable linamarase production show two bands at approximately 0.5 kb and approximately 0.8 kb that are absent in lili plants; weaker bands are also present in Li_ plants at approximately 2 kb as well as at various sizes in lili plants (figure 2c). Hybridization with the P1 probe indicates that these weaker bands are more similar to the Li-paralogue sequence (figure 2d). These patterns confirm that, as with T. nigrescens, there are no bands present in lili accessions that correspond to the Li gene.
Figure 2.
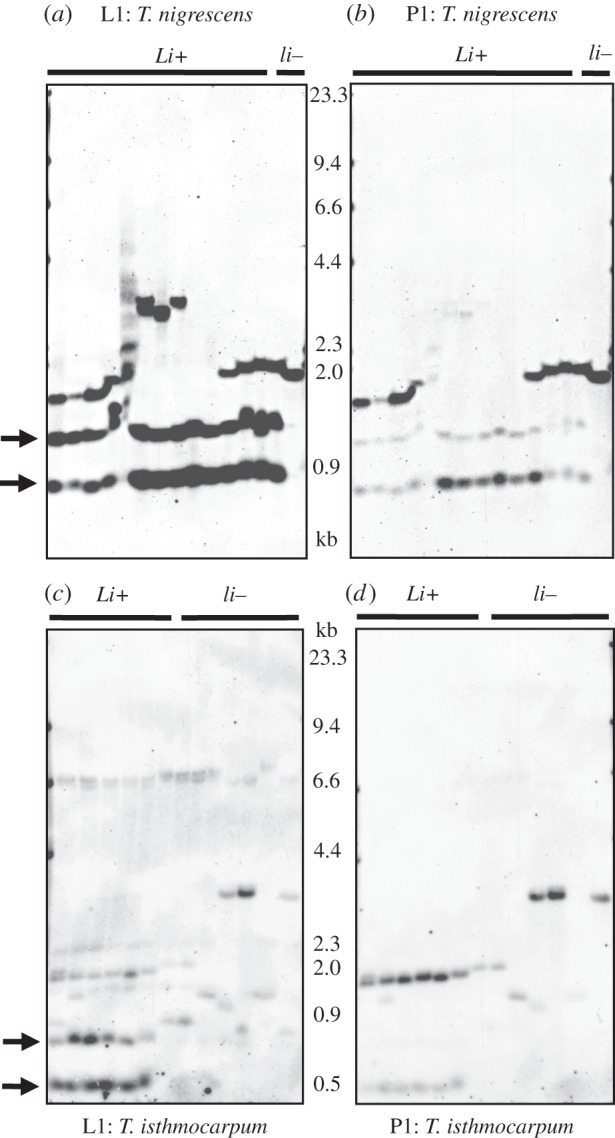
Southern hybridizations for the Li gene and Li-paralogue in Trifolium species with the Li/li polymorphism. Li+ and li– correspond, respectively, to accessions with and without detectable levels of linamarase in HCN assays. The L1 and P1 probes correspond, respectively, to Li and to the non-cyanogenic Li-paralogue. The presence of two bands with the L1 probe is consistent with the occurrence of Li as a single-copy gene (see Material and methods). Arrows indicate expected locations of bands corresponding to the Li gene. For (a) (L1 probe) and (b) (P1 probe), T. nigrescens accessions include five plants per subspecies, with subspecies petrisavii, meneghinianum and nigrescens from left to right as follows: PI 120132, PI 249855, PI 298478, PI 583447, PI 583448, PI 120120, PI 120139, PI 238156, PI 287173, PI 304380, PI 210354, PI 233723, PI 419310, PI 419408, PI 591672. For (c) (L1 probe) and (d) P1 probe, T. isthmocarpum accessions are as follows, from left to right: MU-209a, PI 517109, PI 517110, PI 517111, PI 517112, PI 203664, MU-116, MU-209b, PI 338675, PI 535692, PI 535571, PI 202517, PI 653511.
In a pattern similar to CYP79D15, there appears to be some Li gene copy number variation (CNV) among plants that carry the gene. Most notably, three individuals of T. nigrescens ssp. meneghinianum show extra bands at approximately 4.0 kb that hybridize strongly to the L1 probe with negligible cross-hybridization to P1 (figure 2a,b), consistent with the presence of additional Li gene copies. As with CYP79D15, this banding variation is not obviously attributable to AseI restriction site variation or polyploidy.
(c). Cyanogenesis polymorphisms have evolved independently within species
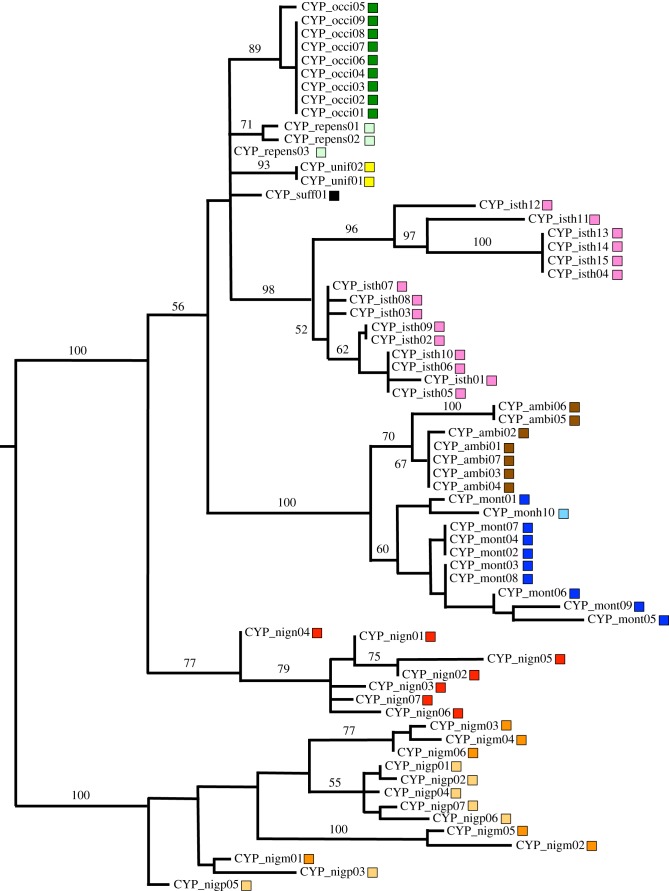
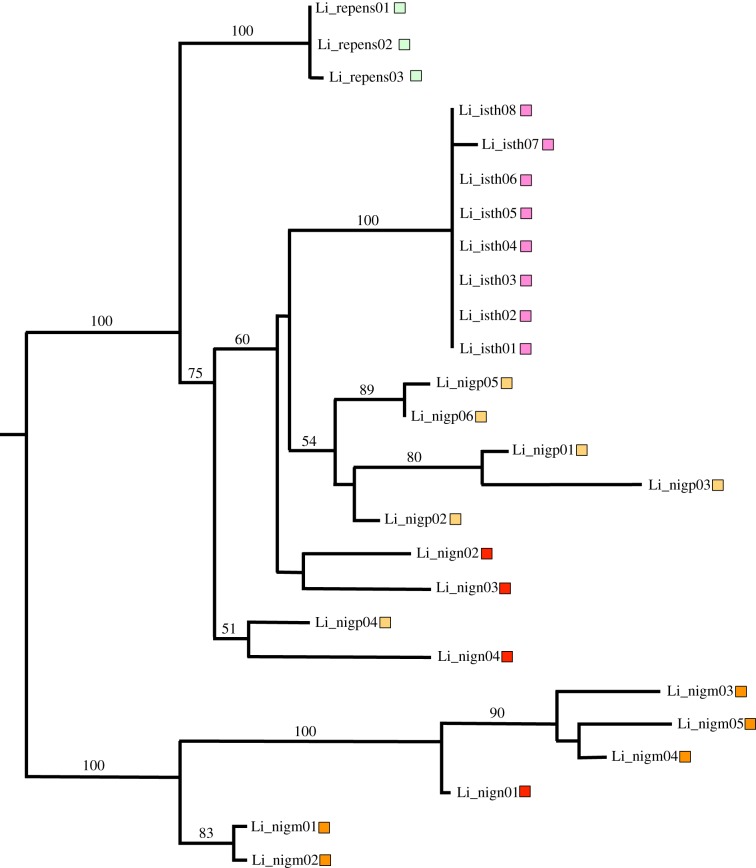
The two cyanogenesis genes were PCR-amplified, cloned and sequenced for all Ac_ plants and all but four Li_ plants (electronic supplementary material, table S1). ML trees for the two genes are shown in figures 3 and 4. No evidence of paralogous gene sequences was observed for either gene; this suggests that the gene CNV detected in Southerns reflects tandem copies that are evolving in concert. For both CYP79D15 and Li, the haplotypes are generally grouped by species with high bootstrap support. This pattern is especially evident for CYP79D15, where eight species are represented in the tree (figure 3). While phylogenetic relationships within Trifoliastrum are incompletely resolved [37], the CYP79D15 tree is also compatible with known species relationships. For example, T. ambiguum and T. montanum are grouped as a species-pair with 100% bootstrap support, consistent with neutral gene phylogenies [37] (electronic supplementary material, figure S1). The CYP79D15 tree also confirms the genetic distinctness of T. nigrescens ssp. nigrescens as observed in Southern hybridizations (see above); haplotypes of this subspecies form a distinct clade with 77% bootstrap support. For Li, where there are only three species with linamarase production, haplotypes for two of the species, T. isthmocarpum and T. repens, form species-specific clades, each with 100% bootstrap support; these clades are nested within T. nigrescens haplotypes on the midpoint-rooted tree (figure 4). For both CYP79D15 and Li sequences, there is no sharing of haplotypes among species, as would be expected if they predated the diversification of the clade.
Figure 3.
CYP79D15 ML haplotype tree. Numbers along branches indicate percentage bootstrap support; only values more than 50% are shown. Haplotype labels correspond to accessions as indicated in the electronic supplementary material, table S1; colours indicate taxa, including subspecies designations (table 1). Taxon abbreviations are as follows: T. ambiguum (ambi; brown), T. isthmocarpum (isth; pink), T. montanum ssp. humboldtianum (monh; light blue), T. montanum ssp. montanum (mont; dark blue), T. nigrescens ssp. meneghinianum (nigm; orange), T. nigrescens ssp. nigrescens (nign; red), T. nigrescens ssp. petrisavii (nigp; beige), T. occidentale (occi; dark green), T. repens (repens; light green); T. suffocatum (suff; black), T. uniflorum (unif; yellow).
Figure 4.
Li ML haplotype tree. Numbers along branches indicate percentage bootstrap support; only values more than 50% are shown. Haplotype labels correspond to accessions as indicated in the electronic supplementary material, table S1. Taxon abbreviations are as follows: T. isthmocarpum (isth; pink), T. nigrescens ssp. meneghinianum (nigm; orange), T. nigrescens ssp. nigrescens (nign; red), T. nigrescens ssp. petrisavii (nigp; beige), T. repens (repens; light green).
By definition, Li and CYP79D15 sequences represent only the gene-presence alleles for each locus. By contrast, sequences flanking a PA polymorphism can be used to directly assess evolutionary relationships between gene-presence and -absence alleles [10,42–44]. If gene-deletion alleles have evolved independently within species, then flanking sequences for gene-presence and -absence alleles would be expected to group by species. To test this hypothesis, we targeted sequences immediately downstream of the Li and CYP79D15 PA polymorphisms for PCR and sequencing. The targeted loci, 3CYP-2.34 (an approx. 1.4 kb region located 2.34 kb downstream of the CYP79D15 stop codon) and 3Li-6.65 (an approx. 0.9 kb region located 6.65 kb downstream of the Li stop codon) are located at the boundaries of the most common cyanogenesis gene-deletion alleles for each gene in white clover [10].
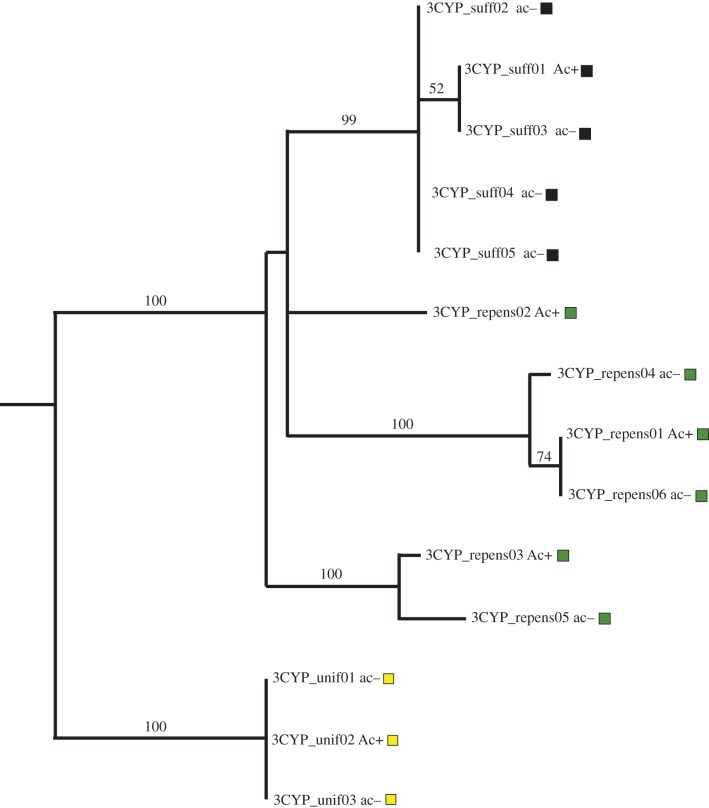
For Li, we were unable to PCR-amplify the targeted flanking sequence in lili accessions of the two species besides white clover that produce linamarase (T. isthmocarpum, T. nigrescens; table 1); therefore, we could not assess genealogical relationships between gene-presence and -absence haplotypes. By contrast, we successfully amplified the targeted CYP79D15-flanking sequence in plants with and without cyanogenic glucoside production in two species besides white clover that are polymorphic at Ac/ac (T. suffocatum, T. uniflorum). For both species, haplotypes are grouped by species with high bootstrap support (figure 5), providing strong evidence that the PA polymorphisms have evolved independently in each species. Taken together with the observations that all observed biochemical polymorphisms correspond to gene PA polymorphisms (figures 1 and 2), and that ac and li alleles have evolved recurrently within white clover [10], this finding suggests that the cyanogenesis polymorphisms in Trifoliastrum have evolved independently in each species where they occur, and that they have done so through the parallel evolution of gene-deletion alleles.
Figure 5.
3CYP-2.34 ML haplotype tree. Numbers along branches indicate percentage bootstrap support; only values more than 50% are shown. Haplotype labels correspond to accessions as indicated in the electronic supplementary material, table S1. Taxon abbreviations are as follows: T. repens (repens; dark green); T. suffocatum (suff; black), T. uniflorum (unif; yellow). Ac+ and ac– correspond, respectively, to plants that do or do not possess the CYP79D15 gene.
4. Discussion
Variation for cyanogenesis was first identified in white clover more than a century ago [6,7]. Subsequent decades of ecological and genetic research have established this polymorphism as a textbook example of adaptive variation maintained by opposing selective forces [53,54]. In this study, we have documented that polymorphisms for cyanogenic components also occur in several of white clover's relatives in Trifolium section Trifoliastrum (table 1). Moreover, our data suggest that these polymorphisms have evolved independently in each species, through recurrent gene deletion events giving rise to gene PA polymorphisms (figures 1–5).
(a). Distribution of cyanogenic components among Trifolium species
The observed distribution of cyanogenic components in Trifoliastrum raises intriguing questions about the adaptive function of these compounds outside of T. repens. While cyanogenic glucosides were detected in seven of the 12 species tested, linamarase was only detected in two of these species (table 1). Thus, there are several species where one of the two required components for cyanogenesis is present, but where the other component is either absent or too rare to be detected in our sampling. Kakes & Chardonnens [40] observed a similar pattern in their extensive sampling of more than 750 T. occidentale plants; no plants with linamarase production were detected in this species although more than 75% of samples produced cyanogenic glucosides. These patterns suggest a potential selective advantage for the production of cyanogenic glucosides in the absence of the cyanogenic phenotype, at least under some environmental conditions.
Two potential explanations, which are not mutually exclusive, could most easily account for this asymmetric distribution of cyanogenic components. The first is that the adaptive role of cyanogenic glucosides as a chemical defence may not always require the presence of linamarase. Gastropods, which are major clover herbivores, possess glucosidase enzymes in their guts that are capable of hydrolysing cyanogenic glucosides [11,17]. Thus, if post-ingestion cyanogenesis (rather than cyanogenesis at the initial leaf-tasting stage) is sufficient to deter further herbivore damage, cyanogenic glucosides alone could serve as an effective defence against this class of herbivores. Empirical data are inconclusive regarding this hypothesis. In controlled snail grazing experiments, Kakes [17] observed fivefold higher survivorship for white clover seedlings that produce cyanogenic glucosides relative to those without them, with the presence or the absence of linamarase having no effect on herbivore deterrence. By contrast, Dirzo & Harper [11] found that both cyanogenic glucosides and enzyme are required for differential protection against slug herbivory.
A second explanation for the asymmetric distribution is that cyanogenic glucosides may serve adaptive functions unrelated to herbivore deterrence. Cyanogenic glucosides can be metabolized in plants without the release of hydrogen cyanide, and there is evidence that they can serve as nitrogen storage and transport compounds (reviewed in [55]) and as signalling regulators in stress response [56]. Consistent with this hypothesis, recent data from white clover populations suggest that regional variation in aridity may act as a selective factor in maintaining the Ac/ac polymorphism, independent of the Li/li polymorphism [20,21]. Several Trifoliastrum species span a wide range of habitats, including alpine meadows, temperate forest edges, steppes, pastures and coastal cliffs [36]. Thus, it is plausible that for some species, regional variation in selective pressures that are unrelated to HCN release may play a role in maintaining the Ac/ac polymorphism. Ecological genetic studies of the sort employed in white clover will be useful in testing this hypothesis. Beyond these adaptive explanations, it is also possible that neutral processes, such as mutational biases leading to repeated gene loss, could contribute to the observed cyanogenesis distributions.
(b). Molecular evolution of cyanogenesis polymorphisms
A striking finding from this study is that the cyanogenesis polymorphisms have apparently evolved multiple times in species of Trifoliastrum, and that in all cases they have evolved through gene deletions (figures 1, 2 and 5). To the best of our knowledge, this study represents the first documented case of the recurrent, parallel evolution of putatively adaptive PA polymorphisms across a group of related species. Following the discovery that the unlinked Ac/ac and Li/li polymorphisms in white clover both correspond to gene PA polymorphisms, we had previously proposed that the pattern might be attributable to that species’ allotetraploid origin, as genomic deletions are common following polyploidization events (discussed in [35]). Subsequent analyses in white clover called into question that hypothesis, as molecular signatures in flanking sequences indicate recurrent gene deletions within this species rather than long-term balancing selection [10]. Results of the present study further refute a role for polyploidization in the evolution of these polymorphisms; only two of the six polymorphic species in table 1 are polyploid (T. ambiguum, T. uniflorum) [37]. The present findings instead suggest that there is some underlying lability in the genomic regions containing CYP79D15 and Li across species of Trifoliastrum, which has allowed for the repeated evolution of gene PA variation (e.g. [57]). As has been discussed in the context of T. repens cyanogenesis gene deletions [10], tandemly repeated sequences—potentially including gene CNV at the cyanogenesis genes themselves—may be a critical causal mechanism underlying this process.
(c). Evolutionary origins of Trifolium cyanogenesis
Both cyanogenesis genes show high nucleotide similarity across Trifoliastrum. The two most divergent CYP79D15 haplotypes (between two accessions of T. isthmocarpum and T. nigrescens; figure 3) are approximately 95% identical with all silent variation considered; similarly, the two most divergent Li sequences (also between T. isthmocarpum and T. nigrescens) are approximately 98% identical across all sites (figure 4). This close genetic similarity strongly suggests that the gene sequences are orthologous across the clade, and that the presence of cyanogenesis is therefore ancestral for Trifoliastrum. Interestingly, CYP79D15 also appears to be orthologous to the functionally equivalent gene of a somewhat distantly related cyanogenic legume, Lotus japonicus [58], a species that falls outside the large vicioid legume clade to which Trifolium belongs [37]. Exons of the T. repens CYP79D15 sequence are 93% identical to those of the L. japonicus CYP79D3 gene. This close sequence similarity suggests that cyanogenesis may have existed in the shared common ancestor of these taxa, predating the divergence of the vicioid clade (comprising at least 11 genera [37]) from other legume lineages.
In marked contrast to its possible ancestral state among vicioid legumes, cyanogenesis appears to be absent in most clades within Trifolium, as well as in closely related genera. We detected no cyanogenic component production in Trifolium species outside of Trifoliastrum (electronic supplementary material, table S1; see also [38]), and PCR screening and Southern hybridizations for species outside of this clade have revealed no evidence of the underlying cyanogenesis genes (K. Olsen 2014, unpublished observations). Similarly, reports of cyanogenesis in closely related vicioid legume genera (e.g. Melilotus, Trigonella, Medicago) are sporadic or absent [59], and BLAST analyses of the Trifolium cyanogenesis gene sequences against the reference genome of Medicago truncatula reveal no clear orthologues. Thus, if cyanogenesis is ancestral among vicioid legumes, it has apparently been lost repeatedly on a macroevolutionary time scale, a pattern that echoes our findings for the cyanogenesis genes within Trifoliastrum.
(d). Conclusion
For the clover cyanogenesis system, it remains to be seen whether there are particular structural features of the Trifolium genome that have facilitated the repeated, parallel evolution of gene deletions at the CYP79D15 and Li loci. Similarly, much remains to be learned about the selective factors that maintain the Ac/ac and Li/li polymorphisms among Trifoliastrum species, and the extent to which these factors differ from those shaping the classic white clover adaptive polymorphism. Regardless of the specific mechanisms at play, our observations that the cyanogenesis polymorphisms occur across multiple ecologically and geographically diverse clover species suggest that the forces maintaining this variation are long-standing and present across a wide range of environments.
Supplementary Material
Supplementary Material
Acknowledgements
The authors express sincere thanks to Mike Vincent (Miami University of Ohio) and Nick Ellison (AgResearch New Zealand) for generously providing seed samples; to Mike Dyer and staff of the Washington University greenhouse for their expertise in germinating and maintaining clover samples; and to members of the Olsen laboratory for helpful comments and discussion.
Funding statement
Funding for this project was provided through a National Science Foundation CAREER award to K.M.O. (DEB-0845497).
References
- 1.Orsini L, Andrew R, Eizaguirre C. 2013. Evolutionary ecological genomics. Mol. Ecol. 22, 527–531. ( 10.1111/mec.12177) [DOI] [PubMed] [Google Scholar]
- 2.Narum SR, Buerkle CA, Davey JW, Miller MR, Hohenlohe PA. 2013. Genotyping-by-sequencing in ecological and conservation genomics. Mol. Ecol. 22, 2841–2847. ( 10.1111/mec.12350) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Primmer CR, Papakostas S, Leder EH, Davis MJ, Ragan MA. 2013. Annotated genes and nonannotated genomes: cross-species use of Gene Ontology in ecology and evolution research. Mol. Ecol. 22, 3216–3241. ( 10.1111/mec.12309) [DOI] [PubMed] [Google Scholar]
- 4.Stern DL, Orgogozo V. 2008. The loci of evolution: how predictable is genetic evolution? Evolution 62, 2155–2177. ( 10.1111/j.1558-5646.2008.00450.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stern DL. 2013. The genetic causes of convergent evolution. Nat. Rev. Genet. 14, 751–764. ( 10.1038/nrg3483) [DOI] [PubMed] [Google Scholar]
- 6.Mirande MM. 1912. Sur la présence de l'acide cyanhydrique dans le trèfle rampant (Trifolium repens L.). C. R Acad. Sci. Paris 155, 651–653. [Google Scholar]
- 7.Armstrong H, Armstrong E, Horton E. 1913. Herbage studies. II—Variation in Lotus corniculatus and Trifolium repens: (cyanophoric plants). Proc. R. Soc. Lond. B 86, 262–269. ( 10.1098/rspb.1913.0021) [DOI] [Google Scholar]
- 8.Hughes MA. 1991. The cyanogenic polymorphism in Trifolium repens L. (white clover). Heredity 66, 105–115. ( 10.1038/hdy.1991.13) [DOI] [Google Scholar]
- 9.Kooyers NJ, Olsen KM. 2012. Rapid evolution of an adaptive cyanogenesis cline in introduced North American white clover (Trifolium repens L.). Mol. Ecol. 21, 2455–2468. ( 10.1111/j.1365-294X.2012.05486.x) [DOI] [PubMed] [Google Scholar]
- 10.Olsen KM, Kooyers NJ, Small LL. 2013. Recurrent gene deletions and the evolution of adaptive cyanogenesis polymorphisms in white clover (Trifolium repens L.). Mol. Ecol. 22, 724–738. ( 10.1111/j.1365-294X.2012.05667.x) [DOI] [PubMed] [Google Scholar]
- 11.Dirzo R, Harper J. 1982. Experimental studies on slug–plant interactions: III. Differences in the acceptability of individual plants of Trifolium repens to slugs and snails. J. Ecol. 70, 101–117. ( 10.2307/2259867) [DOI] [Google Scholar]
- 12.Pederson GA, Brink GE. 1998. Cyanogenesis effect on insect damage to seedling white clover in a bermudagrass sod. Agron. J. 90, 208 ( 10.2134/agronj1998.00021962009000020015x) [DOI] [Google Scholar]
- 13.Saucy F, Studer J, Aerni V, Schneiter B. 1999. Preference for acyanogenic white clover (Trifolium repens) in the vole Arvicola terrestris: I. Experiments with two varieties. J. Chem. Ecol. 25, 1441–1454. ( 10.1023/A:1020943313142) [DOI] [Google Scholar]
- 14.Viette M, Tettamanti C, Saucy F. 2000. Preference for acyanogenic white clover (Trifolium repens) in the vole Arvicola Terrestris. II. Generalization and further investigations. J. Chem. Ecol. 26, 101–122. ( 10.1023/A:1005441528235) [DOI] [Google Scholar]
- 15.Daday H. 1954. Gene frequencies in wild populations of Trifolium repens II. Distribution by altitude. Heredity 8, 377–384. ( 10.1038/hdy.1954.40) [DOI] [Google Scholar]
- 16.Daday H. 1954. Gene frequencies in wild populations of Trifolium repens I. Distribution by latitude. Heredity 8, 61–78. ( 10.1038/hdy.1954.5) [DOI] [Google Scholar]
- 17.Kakes P. 1989. An analysis of the costs and benefits of the cyanogenic system in Trifolium repens L. Theoret. Appl. Genet. 77, 111–118. ( 10.1007/BF00292324) [DOI] [PubMed] [Google Scholar]
- 18.Pennings SC, Silliman BR. 2005. Linking biogeography and community ecology: latitudinal variation in plant–herbivore interaction strength. Ecology 86, 2310–2319. ( 10.1890/04-1022) [DOI] [Google Scholar]
- 19.Salazar D, Marquis RJ. 2012. Herbivore pressure increases toward the equator. Proc. Natl Acad. Sci. USA 109, 12 616–12 620. ( 10.1073/pnas.1202907109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kooyers NJ, Gage LR, Al-Lozi A, Olsen KM. 2014. Aridity shapes cyanogenesis cline evolution in white clover (Trifolium repens L.). Mol. Ecol. 23, 1053–1070. ( 10.1111/mec.12666) [DOI] [PubMed] [Google Scholar]
- 21.Kooyers NJ, Olsen KM. 2013. Searching for the bull's eye: agents and targets of selection vary among geographically disparate cyanogenesis clines in white clover (Trifolium repens L.). Heredity 111, 495–504. ( 10.1038/hdy.2013.71) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Till-Bottraud I, Kakes P, Dommée B. 1988. Variable phenotypes and stable distribution of the cyanotypes of Trifolium repens L. in Southern France. Acta Oecol. 9, 393–404. [Google Scholar]
- 23.De Araújo AM. 1976. The relationship between altitude and cyanogenesis in white clover (Trifolium repens L.). Heredity 37, 291–293. ( 10.1038/hdy.1976.89) [DOI] [Google Scholar]
- 24.Pederson GA, Fairbrother TE, Greene SL. 1996. Cynogenesis and climatic relationships in U.S. white clover germplasm collection and core subset. Crop Sci. 36, 427–433. ( 10.2135/cropsci1996.0011183X003600020035x) [DOI] [Google Scholar]
- 25.Majumdar S, De KK, Banerjee S. 2004. Influence of two selective factors on cyanogenesis polymorphism of Trifolium repens L. in Darjeeling Himalaya. J. Plant Biol. 47, 124–128. ( 10.1007/BF03030642) [DOI] [Google Scholar]
- 26.Daday H. 1958. Gene frequencies in wild populations of Trifolium repens L. III. World distribution. Heredity 12, 169–184. ( 10.1038/hdy.1958.22) [DOI] [Google Scholar]
- 27.Ganders FR. 1990. Altitudinal clines for cyanogenesis in introduced populations of white clover near Vancouver, Canada. Heredity 64, 387–390. ( 10.1038/hdy.1990.48) [DOI] [Google Scholar]
- 28.Coop IE. 1940. Cyanogenesis in white clover (Trifolium repens L.). III. A study of linamarase, the enzyme which hydrolyses lotaustralin. NZ J. Sci. Technol. A 22, 71B–83B. [Google Scholar]
- 29.Corkill L. 1942. Cyanogenesis in white clover (Trifolium repens L.) V. The inheritance of cyanogenesis. NZ J. Sci. Technol. B 23, 178–193. [Google Scholar]
- 30.Melville J, Doak BW. 1940. Cyanogenesis in white clover (Trifolium repens L.). II. Isolation of the glucosidal constituents. NZ J. Sci. Technol. A 22, 67B–71B. [Google Scholar]
- 31.Feigl F, Anger V. 1966. Replacement of benzidine by copper ethylacetoacetate and tetra base as spot-test reagent for hydrogen cyanide and cyanogen. Analyst 91, 282–284. ( 10.1039/an9669100282) [DOI] [PubMed] [Google Scholar]
- 32.Olsen KM, Sutherland BL, Small LL. 2007. Molecular evolution of the Li/li chemical defence polymorphism in white clover (Trifolium repens L.). Mol. Ecol. 16, 4180–4193. ( 10.1111/j.1365-294X.2007.03506.x) [DOI] [PubMed] [Google Scholar]
- 33.Hughes MA, Stirling JD, Collinge DB. 1984. The inheritance of cyanoglucoside content in Trifolium repens L. Biochem. Genet. 22, 139–151. ( 10.1007/BF00499294) [DOI] [PubMed] [Google Scholar]
- 34.Hayden KJ, Parker IM. 2002. Plasticity in cyanogenesis of Trifolium repens L.: inducibility, fitness costs and variable expression. Evol. Ecol. Res. 4, 155–168. [Google Scholar]
- 35.Olsen KM, Hsu S-C, Small LL. 2008. Evidence on the molecular basis of the Ac/ac adaptive cyanogenesis polymorphism in white clover (Trifolium repens L.). Genetics 179, 517–526. ( 10.1534/genetics.107.080366) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zohary M, Heller D. 1984. The genus Trifolium. Jerusalem: Israel Academy of Sciences and Humanities. [Google Scholar]
- 37.Ellison NW, Liston A, Steiner JJ, Williams WM, Taylor NL. 2006. Molecular phylogenetics of the clover genus (Trifolium—Leguminosae). Mol. Phylogenet. Evol. 39, 688–705. ( 10.1016/j.ympev.2006.01.004) [DOI] [PubMed] [Google Scholar]
- 38.Gibson PB, Barnett OW, Gillingham JT. 1972. Cyanoglucoside and hydrolyzing enzyme in species related to Trifolium repens. Crop Sci. 12, 708–709. ( 10.2135/cropsci1972.0011183X001200050051x) [DOI] [Google Scholar]
- 39.Williams WM, Williamson ML. 2001. Genetic polymorphism for cyanogenesis and linkage at the linamarase locus in Trifolium nigrescens Viv. subsp. nigrescens. Theoret. Appl. Genet. 103, 1211–1215. ( 10.1007/s001220100612) [DOI] [Google Scholar]
- 40.Kakes P, Chardonnens AN. 2000. Cyanotypic frequencies in adjacent and mixed populations of Trifolium occidentale Coombe and Trifolium repens L. are regulated by different mechanisms. Biochem. Syst. Ecol. 28, 633–649. ( 10.1016/S0305-1978(99)00110-6) [DOI] [PubMed] [Google Scholar]
- 41.Takos A, et al. 2010. Genetic screening identifies cyanogenesis-deficient mutants of Lotus japonicus and reveals enzymatic specificity in hydroxynitrile glucoside metabolism. Plant Cell 22, 1605–1619. ( 10.1105/tpc.109.073502) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tian D, Araki H, Stahl E, Bergelson J, Kreitman M. 2002. Signature of balancing selection in Arabidopsis. Proc. Natl Acad. Sci. USA 99, 11 525–11 530. ( 10.1073/pnas.172203599) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stahl EA, Dwyer G, Mauricio R, Kreitman M, Bergelson J. 1999. Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature 400, 667–671. ( 10.1038/23260) [DOI] [PubMed] [Google Scholar]
- 44.Shen J, Araki H, Chen L, Chen J-Q, Tian D. 2006. Unique evolutionary mechanism in R-genes under the presence/absence polymorphism in Arabidopsis thaliana. Genetics 172, 1243–1250. ( 10.1534/genetics.105.047290) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Badr A, Sayed-Ahmed H, El-Shanshouri A, Watson IE. 2002. Ancestors of white clover (Trifolium repens L.), as revealed by isozyme polymorphisms. Theoret. Appl. Genet. 106, 143–148. ( 10.1007/s00122-002-1010-5) [DOI] [PubMed] [Google Scholar]
- 46.Hand ML, et al. 2008. Identification of homologous, homoeologous and paralogous sequence variants in an outbreeding allopolyploid species based on comparison with progenitor taxa. Mol. Genet. Genomics 280, 293–304. ( 10.1007/s00438-008-0365-y) [DOI] [PubMed] [Google Scholar]
- 47.Porebski S, Bailey LG, Baum B. 1997. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 15, 8–15. ( 10.1007/BF02772108) [DOI] [Google Scholar]
- 48.Hall T. 2001. BioLign alignment and multiple contig editor See http://en.bio-soft.net/dna/BioLign.html.
- 49.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704. ( 10.1080/10635150390235520) [DOI] [PubMed] [Google Scholar]
- 50.Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772 ( 10.1038/nmeth.2109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. ( 10.1093/sysbio/syq010) [DOI] [PubMed] [Google Scholar]
- 52.Williams W, Ansari HA, Ellison NW, Hussain SW. 2001. Evidence of three subspecies in Trifolium nigrescens Viv. Ann. Bot. 87, 683–691. ( 10.1006/anbo.2001.1399) [DOI] [Google Scholar]
- 53.Dirzo R, Sarukhan J. 1984. Perspectives on plant population ecology. Sunderland, MA: Sinauer. [Google Scholar]
- 54.Silvertown J, Charlesworth D. 2001. Introduction to plant population biology, 4th edn Oxford, UK: Wiley-Blackwell. [Google Scholar]
- 55.Møller BL. 2010. Functional diversifications of cyanogenic glucosides. Curr. Opin Plant Biol. 13, 338–347. ( 10.1016/j.pbi.2010.01.009) [DOI] [PubMed] [Google Scholar]
- 56.Siegień I, Bogatek R. 2006. Cyanide action in plants—from toxic to regulatory. Acta Physiol. Plantarum 28, 483–497. ( 10.1007/BF02706632) [DOI] [Google Scholar]
- 57.Kern AD, Begun DJ. 2008. Recurrent deletion and gene presence/absence polymorphism: telomere dynamics dominate evolution at the tip of 3L in Drosophila melanogaster and D. simulans. Genetics 179, 1021–1027. ( 10.1534/genetics.107.078345) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Forslund K, Morant M, Jørgensen B, Olsen CE, Asamizu E, Sato S. 2004. Biosynthesis of the nitrile glucosides rhodiocyanoside A and D and the cyanogenic glucosides lotaustralin and linamarin in Lotus japonicus. Plant Physiol. 135, 71–84. ( 10.1104/pp.103.038059) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seigler D, Maslin B, Conn E. 1989. Cyanogenesis in the Leguminosae. In Advances in Legume Biology: Proceedings of the Second International Legume Conference, St. Louis, MO, USA, 23–27 June 1986, held under the auspices of the Missouri Botanical Garden and the Royal Botanic Gardens, Kew (eds C Stirton, J Zarucchi), pp. 645–672. St. Louis, MO. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.