Abstract
Background:
Cytochrome P450 2D6 (CYP2D6) metabolizes around 25% of the drugs used in therapeutics and different polymorphisms have been identified in various populations. This study aimed at finding the prevalence of CYP2D6 polymorphisms using dextromethorphan as a probe drug.
Materials and Methods:
Healthy participants were administered 60 mg dextromethorphan after an overnight fast and 5 ml of blood was collected 3 h postdose. A validated laboratory method was used to measure both dextromethorphan and its active metabolite, dextrorphan from plasma. Metabolic ratio (MR) of dextromethorphan to dextrorphan was calculated for each of the participants. Probit analysis was done and antimode was defined. Individuals with log MR equal to or higher than the antimode were classified as poor metabolizers (PMs) and those with values less than antimode were categorized as extensive metabolizers (EMs).
Results:
Data from a total of 149 participants were evaluated and the median (range) of MR was 0.25 (0.03-3.01). The polynomial equation obtained in probit analysis gave an antimode for MR of 1.39. Five (3.36%) participants were PMs and 144 (96.64%) were found to be EMs. One participant had reported mild drowsiness 2 h postdose that subsided spontaneously without any intervention.
Conclusion:
The prevalence of CYP2D6 polymorphism in Western Indian population is low (3.36%) and is similar to other populations.
KEY WORDS: Cytochrome P450 2D6, dextromethorphan, phenotype, probe drug, Western Indians
Introduction
Cytochrome P450 (CYP2D6) group of enzymes is the most widely studied since they metabolize majority of drugs used in clinical practice. CYP2D62D6 metabolizes approximately 25% of drugs[1,2] such as selective serotonin reuptake inhibitors, tricyclic antidepressants, antipsychotics, beta-blockers, and histamine receptor blockers.[3] Genotyping studies of CYP2D6 performed worldwide have shown a wide array of mutant alleles (from *1 to *41) with prevalence ranging from 0% to 8.4%, respectively.[3,4] Of these alleles, *2 is considered to manifest as ultra-rapid metabolizers and *3, *4, *6, *9, *10, *14, *17, *29, and *41 as poor metabolizing types. The remaining types have normal activity.[5]
The function of such polymorphic alleles can be established from phenotype studies, mainly by using a probe drug.[6] Probe drugs are those that are primarily metabolized by one specific enzyme. Hence, by noting the extent of their metabolic conversion (the measurement of their metabolites), the functional/phenotypic activity of that particular enzyme can be delineated. The United States Food and Drug Administration (US FDA) approved probe drug for CYP2D6 is dextromethorphan.[7]
Phenotype studies have shown incidence of around 0-18.8% for poor metabolizer (PM) status of CYP2D6 worldwide[3,8,9] including North and South India.[10,11,12,13,14] Since, there is a lacuna of information from Western Indian population, the present study was envisaged in healthy adults to evaluate the polymorphism of CYP2D6 using dextromethorphan as a probe drug.
Materials and Methods
The study was conducted over a period of 7 months (August 2010 to February 2011) at Department of Clinical Pharmacology, Seth GS Medical College and KEM Hospital after obtaining Institutional Ethics Committee approval (EC/Govt-11/2007). This study was registered with the Clinical Trial Registry of India (CTRI/2010/091/000030). All the participants were recruited by word of mouth. No advertisements were made.
Study Participants
Participants (of either gender between 18 and 45 years) assessed as being healthy based on history and physical examination and who had at least two previous generations permanently residing in Western India (Maharashtra, Gujarat, and Rajasthan) were screened for this study. Those with history of any chronic disease or having consumed any drug or nicotine in the last 7 days, and any history of or suffering from gastrointestinal, renal, liver, cardiovascular, respiratory, central nervous system, and endocrine diseases were excluded from the study. Only those participants who gave written informed consent were recruited for this study.
Study Procedure
The enrolled participants were asked to refrain from consuming either caffeine or alcohol 48 h prior to the study. On the day of the procedure, they were administered 60 mg (10 ml suspension) of dextromethorphan hydrobromide (Lastuss LA Syrup, FDC Ltd., Pharmaceuticals, batch number LTW0061) orally with 50 ml of water after an overnight fast. At 3 h after, the dosing, 5 ml of blood was collected in a heparinized tube for the estimation of dextromethorphan and its metabolite (dextrorphan). The participants were observed for 8 h postdose for assessing the safety of the drug and then discharged.
Laboratory Methodology
A sensitive method using reverse phase high performance liquid chromatography with fluorescence detector was developed for the simultaneous estimation of dextromethorphan and dextrorphan over the concentration range of 3.21-725.44 ng/ml and 12.40-980.00 ng/ml, respectively.[15] Dextromethorphan, dextrorphan and benzyl benzimidazole (internal standard) were extracted from plasma using simple protein precipitation technique using zinc sulfate (35%). Separation of components were done on μBondapak C18 (10 μm, 3.9 × 300 mm, Waters) using gradient mobile phase A, composed of potassium phosphate buffer 0.01M, pH-3.0:methanol: tetrahydrofuran (68.5:31:0.5 v/v/v) and mobile phase B, containing methanol: tetrahydrofuran (93.25:6.75 v/v). Detection of components was done on excitation wavelength of 230 nm and an emission wavelength of 306 nm. The methodology was standardized as per the US FDA requirements.[16]
Statistical Analysis
The metabolic ratio (MR) of dextromethorphan to dextrorphan[17] for each participant was calculated. Descriptive statistics was used for demographic data and the normality of MR data was assessed by Kolmogorov-Smirnov test. Logarithmic values were obtained for each MR and a frequency histogram plotted. Probit plot was prepared with log MR on X-axis and probit values on Y-axis.[18] Trendlines were added to the plot to get the best linear fit. Based on the selected trendline, a polynomial equation of regression was obtained. Intercept at the X-axis was considered as the antimode. Individuals with log MR equal to or higher than the antimode were classified as PMs and those with values less than antimode were categorized as extensive metabolizers (EMs). These are expressed in proportions with 95% confidence intervals ([]).
Results
Demographics
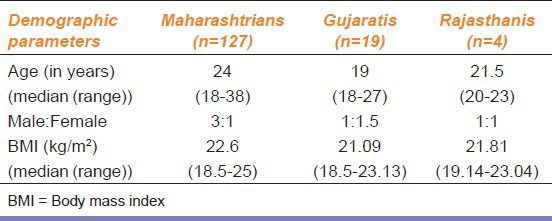
A total of 150 participants (107 males; 43 females) were recruited. The median age was 23 years (range 18-35 years) and body mass index (in kg/m2) was 22.41 (range 18.5-25). A summary of the demographic details has been mentioned in Table 1. A sample from one participant could not be analyzed as there was interference during the laboratory analysis.
Table 1.
Demographic details of the study participants (n=150)
Phenotyping Data
The MR was the ratio of plasma concentration of dextromethorphan to dextrorphan, showing the extent of metabolism by CYP2D6. The median (range) of MR was 0.25 (0.03-3.01). The frequency histogram showed a bimodal distribution of the MR [Figure 1]. The polynomial equation obtained in probit analysis gave an antimode for MR of 1.39 [Figure 1] based on which individuals with an MR of or more than 1.39 were classified as PMs, whereas those with MR <1.39 were classified as EMs.
Figure 1.
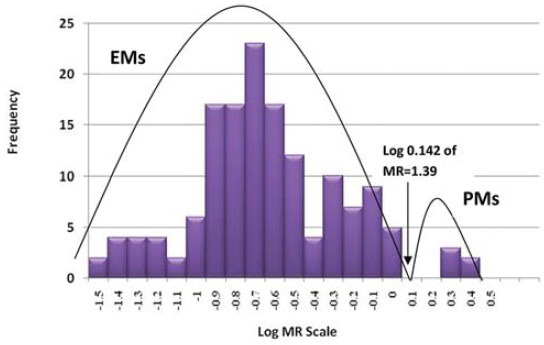
Frequency histogram showing bimodal distribution of log metabolic ratio scale and their frequency (n = 149)
Of the 149, 5 (3.36% [1.4, 7.6]) participants were PMs with median (range) MR of 2.80 (2.32-3.01) and the remaining (96.64% [92.4, 98.6]) were EMs with median (range) MR of 0.25 (0.03-1.29). Gender did not influence the metabolizer status.
Safety
One participant reported mild drowsiness 2 h postdose that subsided spontaneously without any intervention. There were no serious adverse events.
Discussion
This study evaluated the functional activity of CYP2D6 enzyme using MR of dextromethorphan to its metabolite, dextrorphan in adult healthy Western Indian participants. We found that among 149 participants, only 5 (3.36%) were PMs and the drug was tolerated well.
CYP2D6 is the most extensively studied drug metabolizing enzyme that exists in different polymorphisms and the first enzyme that was completely characterized at the molecular level.[19] Three distinct phenotypes have been reported associated with CYP2D6 polymorphisms, namely PMs, extensive and ultra-rapid metabolizers.[20] The prevalence of PMs in Western Indian population was found to be similar to that of other populations including North and South Indians [Figure 2].[3,10,11,12,13,16] In contrast to our study, which shows a low percentage of Western Indians being PMs, studies done in South Africa, Estonia and Poland show a higher proportion of PMs (9.4-18%)[21,22] and South East Asian studies done in China and Korea show an extremely low percentage of PMs (0.6-0.7%).[23,24] In this study, we did not find any ultra-rapid metabolizing phenotype.
Figure 2.
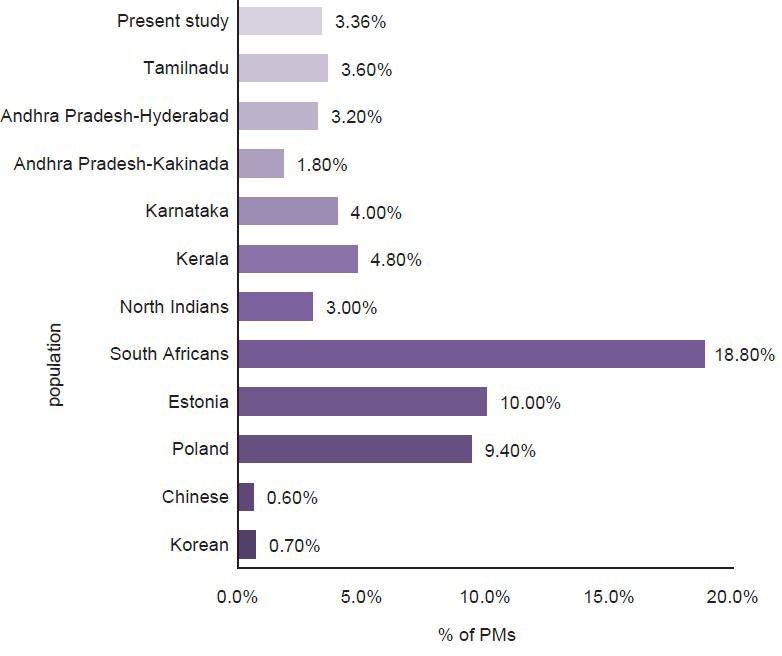
Poor metabolizers in different ethnic groups by phenotyping [3,10,11,12,13,16,21,22,23,24]
Many drug classes such as selective serotonin reuptake inhibitors, tricyclic antidepressants, antipsychotics, beta-blockers, and histamine receptor blockers are metabolized by CYP2D6.[3] Considering the fact that only around 3% of our population has a PM status, it might not be considered cost-effective to evaluate the CYP2D6 status in all the patients before prescribing any of these drugs. In addition, this study was done in healthy participants and the application of this phenotype test in the general population is limited because of the concomitant drugs and disease states that may contraindicate/have interaction with dextromethorphan. Although genotyping of CYP2D6 may have a role in such patients, it is unclear whether they are cost-effective in the current situation. Further studies with a larger sample population are needed in this area to evaluate the cost-effectiveness of evaluating CYP2D6 activity.
Acknowledgment
We thank Dr. Sanjay Oak, Director (ME and MHs) who gave us permission to conduct the study.
Footnotes
Source of Support: Nil
Conflict Interest: No
References
- 1.Hasler JA, Estabrook R, Murray M. Human cytochrome P450. Mol Aspects Med. 1999;20:1–137. [Google Scholar]
- 2.Gonzalez FJ, Coughtrie M, Tukey RH. Drug metabolism. In: Brunton L, Chabner B, Knollman B, editors. Goodman and Gilman's the Pharmacological Basis of Therapeutics. 12th ed. New York: McGraw Hill; 2011. pp. 123–43. [Google Scholar]
- 3.Abraham BK, Adithan C. Genetic polymorphism of CYP2D6. Indian J Pharmacol. 2011;33:147–69. [Google Scholar]
- 4.Aynacioglu AS, Sachse C, Bozkurt A, Kortunay S, Nacak M, Schröder T, et al. Low frequency of defective alleles of cytochrome P450 enzymes 2C19 and 2D6 in the Turkish population. Clin Pharmacol Ther. 1999;66:185–92. doi: 10.1053/cp.1999.v66.100072001. [DOI] [PubMed] [Google Scholar]
- 5.What are the common genetic polymorphisms to cytochrome P450 2D6 that could impact drug metabolism? [Last accessed on 2014 Feb 24]. Available from: http://www.pharmacologyweekly.com/articles/genetic-polymorphism-CYP-P450-2D6-drug-metabolism .
- 6.Linder MW, Prough RA, Valdes R., Jr Pharmacogenetics: A laboratory tool for optimizing therapeutic efficiency. Clin Chem. 1997;43:254–66. [PubMed] [Google Scholar]
- 7.Frank D, Jaehde U, Fuhr U. Evaluation of probe drugs and pharmacokinetic metrics for CYP2D6 phenotyping. Eur J Clin Pharmacol. 2007;63:321–33. doi: 10.1007/s00228-006-0250-8. [DOI] [PubMed] [Google Scholar]
- 8.Islam SI, Idle JR, Smith RL. The polymorphic 4-hydroxylation of debrisoquine in a Saudi Arab population. Xenobiotica. 1980;10:819–25. doi: 10.3109/00498258009033812. [DOI] [PubMed] [Google Scholar]
- 9.Wolf CR, Smith G. Cytochrome P450 CYP2D6. IARC Sci Publ. 1999;148:209–29. [PubMed] [Google Scholar]
- 10.Lamba V, Lamba JK, Dilawari JB, Kohli KK. Genetic polymorphism of CYP2D6 in North Indian subjects. Eur J Clin Pharmacol. 1998;54:787–91. doi: 10.1007/s002280050552. [DOI] [PubMed] [Google Scholar]
- 11.Abraham BK, Adithan C, Shashindran CH, Vasu S, Alekutty NA. Genetic polymorphism of CYP2D6 in a Keralite (South India) population. Br J Clin Pharmacol. 2000;49:285–6. doi: 10.1046/j.1365-2125.2000.00142b.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abraham BK, Adithan C, Kiran PU, Asad M, Koumaravelou K. Genetic polymorphism of CYP2D6 in Karnataka and Andhra Pradesh population in India. Acta Pharmacol Sin. 2000;21:494–8. [PubMed] [Google Scholar]
- 13.Mamidi RN, Satyavageeswaran S, Vakkalanka SV, Chaluvadi MR, Katneni K, Brahmadevara N, et al. Polymorphism of dextromethorphan oxidation in South Indian subjects. Clin Pharmacol Ther. 1999;66:193–200. doi: 10.1053/cp.1999.v66.99989. [DOI] [PubMed] [Google Scholar]
- 14.Afshar M, Rouini MR, Amini M. Simple chromatography method for simultaneous determination of dextromethorphan and its main metabolites in human plasma with fluorimetric detection. J Chromatogr B Analyt Technol Biomed Life Sci. 2004;802:317–22. doi: 10.1016/j.jchromb.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 15.Guidance for industry. Bioanalytical method validation. [Last accessed on 2010 Aug 27]. Available from: http://www.fda.gov/downloads/Drugs/Guidances/ucm070107 .
- 16.Abraham BK, Adithan C, Mohanasundaram J, Shashindran CH, Koumaravelou K, Asad M. Genetic polymorphism of CYP2D6 in Tamil population. Eur J Clin Pharmacol. 2001;56:849–50. doi: 10.1007/s002280000231. [DOI] [PubMed] [Google Scholar]
- 17.Jurica J, Bartecek R, Zourkova A, Pindurova E, Sulcova A, Kasparek T, et al. Serum dextromethorphan/dextrorphan metabolic ratio for CYP2D6 phenotyping in clinical practice. J Clin Pharm Ther. 2012;37:486–90. doi: 10.1111/j.1365-2710.2012.01333.x. [DOI] [PubMed] [Google Scholar]
- 18.Varshney E, Saha N, Tandon M, Shrivastava V, Ali S. Prevalence of poor and rapid metabolizers of drugs metabolized by CYP2B6 in North Indian population residing in Indian national capital territory. Springerplus. 2012;1:34. doi: 10.1186/2193-1801-1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ingelman-Sundberg M. Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6): Clinical consequences, evolutionary aspects and functional diversity. Pharmacogenomics J. 2005;5:6–13. doi: 10.1038/sj.tpj.6500285. [DOI] [PubMed] [Google Scholar]
- 20.Wang L, Weinshilbourn RM. Pharmacogenomics. In: Golan DE, Tashjian AH, Armstrong EJ, Armstrong AW, editors. Principles of Pharmacology the Pathophysiologic Basis of Drug Therapy. 3rd ed. Philadelphia: Lippincott Williams and Wilkins; 2012. pp. 72–5. [Google Scholar]
- 21.Wojtczak A, Rychlik-Sych M, Krochmalska-Ulacha E, Skretkowicz J. CYP2D6 phenotyping with dextromethorphan. Pharmacol Rep. 2007;59:734–8. [PubMed] [Google Scholar]
- 22.Marandi T, Dahl ML, Kiivet RA, Rägo L, Sjöqvist F. Debrisoquin and S-mephenytoin hydroxylation phenotypes and CYP2D6 genotypes in an Estonian population. Pharmacol Toxicol. 1996;78:303–7. doi: 10.1111/j.1600-0773.1996.tb01379.x. [DOI] [PubMed] [Google Scholar]
- 23.Sohn DR, Shin SG, Park CW, Kusaka M, Chiba K, Ishizaki T. Metoprolol oxidation polymorphism in a Korean population: Comparison with native Japanese and Chinese populations. Br J Clin Pharmacol. 1991;32:504–7. doi: 10.1111/j.1365-2125.1991.tb03939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lane HY, Deng HC, Huang SM, Hu WH, Chang WH, Hu OY. Low frequency of dextromethorphan O-demethylation deficiency in a Chinese population. Clin Pharmacol Ther. 1996;60:696–8. doi: 10.1016/S0009-9236(96)90219-2. [DOI] [PubMed] [Google Scholar]


