Summary
Arabidopsis cell suspension cultures are used as a biological tool to better understand the key role of functional chloroplasts in singlet oxygen-mediated programmed cell death in plants.
Key words: Arabidopsis cell cultures, photosensitizers, programmed cell death, reactive oxygen species, Rose Bengal, singlet oxygen, transcriptional defence responses.
Abstract
Light-grown Arabidopsis thaliana cell suspension culture (ACSC) were subjected to mild photooxidative damage with Rose Bengal (RB) with the aim of gaining a better understanding of singlet oxygen-mediated defence responses in plants. Additionally, ACSC were treated with H2O2 at concentrations that induced comparable levels of protein oxidation damage. Under low to medium light conditions, both RB and H2O2 treatments activated transcriptional defence responses and inhibited photosynthetic activity, but they differed in that programmed cell death (PCD) was only observed in cells treated with RB. When dark-grown ACSC were subjected to RB in the light, PCD was suppressed, indicating that the singlet oxygen-mediated signalling pathway in ACSC requires functional chloroplasts. Analysis of up-regulated transcripts in light-grown ACSC, treated with RB in the light, showed that both singlet oxygen-responsive transcripts and transcripts with a key role in hormone-activated PCD (i.e. ethylene and jasmonic acid) were present. A co-regulation analysis proved that ACSC treated with RB exhibited higher correlation with the conditional fluorescence (flu) mutant than with other singlet oxygen-producing mutants or wild-type plants subjected to high light. However, there was no evidence for the up-regulation of EDS1, suggesting that activation of PCD was not associated with the EXECUTER- and EDS1-dependent signalling pathway described in the flu mutant. Indigo Carmine and Methylene Violet, two photosensitizers unable to enter chloroplasts, did not activate transcriptional defence responses in ACSC; however, whether this was due to their location or to their inherently low singlet oxygen quantum efficiencies was not determined.
Introduction
Singlet oxygen (1O2) is a reactive oxygen species (ROS) that is formed constitutively in photosystem II (PSII) of plant chloroplasts. Plants can cope with the basal production of 1O2 under normal environmental conditions, but high levels of 1O2 are produced in response to excess excitation energy in PSII when photosynthetic activity is inhibited by stress or inhibitors (Mullineaux and Baker, 2010). Overproduction of 1O2 causes damage to lipids and proteins in the neighbourhood of PSII, leading to a decrease in photosynthetic efficiency and an inhibition of plant growth. Damage to the β-carotene molecules of the PSII reaction centre (RC) by 1O2 has also been reported and, interestingly, some of the β-carotene oxidation products have been proposed to be stress signals that mediate gene responses to 1O2 (Ramel et al., 2012a , b ). The dual role of 1O2 as a cytotoxic molecule and a signal molecule is now well established (Kim et al., 2008; Triantaphylidès and Havaux, 2009; Fischer et al., 2013) and much of what is known about the role of 1O2 as a signalling molecule comes from the conditional fluorescence (flu) mutant of Arabidopsis (op den Camp et al., 2003; Danon et al., 2005; Laloi et al., 2007; Lee et al., 2007). The flu mutant contains a mutation in a negative regulator of chlorophyll (Chl) biosynthesis that results in enhanced production of protochlorophyllide (Pchlide)—a potent, natural 1O2 photosensitizer—(Meskauskiene et al., 2001; Kauss et al., 2012). When seedlings of the flu mutant are dark adapted, Pchlide accumulates in thylakoids. After a dark to light shift, a surge in 1O2 production occurs in chloroplasts, eventually leading to cell death. Direct photodamage by 1O2 takes place in chloroplasts of the flu mutant, but the ongoing events responsible for the cell death are genetically mediated by two plastid proteins EXECUTER1 (EX1) and EX2 (Lee et al., 2007; Kim et al., 2008; Kim and Apel, 2013). In other words, programmed cell death (PCD) is the default defence response that becomes active in the flu mutant. The role of the EX proteins has also been extended to wild-type plants, but the defence responses triggered seem different. Kim and co-workers (2012) proposed that EX-dependent signalling induces the formation of microlesions, but not the disintegration of chloroplasts, a finding that was interpreted as an acclimation response that enhances stress resistance in wild-type plants. Another Arabidopsis mutant where the 1O2-mediated transcriptional responses have been investigated in detail is the double mutant npq1lut2 that lacks zeaxanthin and lutein (Alboresi et al., 2011). In npq1lut2, the accumulation of 1O2 is selectively enhanced; however, the transcriptional response does not initiate PCD, but rather the induction of genes whose function is to protect chloroplasts against the damaging effect of ROS. In two other mutants, where 1O2 production is selectively enhanced, vte1 npq1, a double mutant deficient in α-tocopherol and zeaxanthin, and ch1, a mutant deficient in Chl b, high light (HL) irradiance led to a large increase of 1O2 that induced cell death as a consequence of a direct 1O2 cytotoxic effect (Triantaphylidès et al., 2008), although it has recently been recognized that ch1 can acclimate to 1O2 if exposed to mild light stress first (Ramel et al., 2013). 1O2-mediated transcriptional responses at HL irradiance were also studied in wild-type plants and cell suspension cultures of Arabidopsis (González-Pérez et al., 2011; Gutiérrez et al., 2011; Ramel et al., 2012b ). HL stress revealed a set of transcripts that overlapped significantly with those listed in the flu mutant after the dark to light shift; however, in contrast to the flu mutant, an acclimatory response responsible for an increased tolerance against a more severe photooxidative stress was activated instead of PCD. It is worth noting that in wild-type plants 1O2 is produced at the heart of PSII (i.e. the PSII RC) and that the above studies provided evidence for 1O2 production in the PSII RC, for example non-enzymatic oxidation of the β-carotene of the PSII RC (Arellano et al., 2011; Ramel et al., 2012a ) and photodamage to the D1 protein of the PSII RC (González-Pérez et al., 2011). Furthermore, Ramel and co-workers (2012b ) established that the differential transcriptional expression induced by β-carotene oxidation products was not mediated by the EX1 protein, and González-Pérez and co-workers (2011) showed no evidence for the up-regulation of EDS1 encoding the ENHANCED DISEASE SUSCEPTIBILITY PROTEIN 1, also a gene key for understanding the 1O2-mediated cell death response (Ochsenbein et al., 2006). At present, it is not well established whether differences in the type of 1O2-mediated transcriptional defence responses depend on the source of 1O2 within the chloroplasts (i.e. PSII RC, thylakoid-bound Chl precursors, infiltrated artificial photosensitizers, etc.), the intensity of the stress (time, concentration, etc.), or a combination of both, and questions about this issue have been raised in the past few years (Galvez-Valdivieso and Mullineaux, 2010; Alboresi et al., 2011; Gutiérrez et al., 2011; Ramel et al., 2012a; Kim and Apel, 2013).
In an attempt to shed more light on the 1O2-mediated cell death response in plants, Rose Bengal (RB)—a potent, artifical 1O2 photosensitizer—and dark-grown and light-grown Arabidopsis cells, the former containing proplastids, while the latter have functional chloroplasts, were used here. RB is an 1O2 elicitor that accumulates inside chloroplasts and has been used in several studies with the model alga Chlamydomonas reinhardtii to investigate the role of 1O2 in chloroplast to nucleus retrograde signalling and acclimatory responses to 1O2 stress (Leisinger et al., 2001; Fischer et al., 2007; Ledford et al., 2007). In these studies, when cells were exposed to light, RB induced (in a similar fashion to HL stress) the transcription of GLUTATHIONE PEROXIDASE HOMOLOGOUS (GPXH), a gene with a key role in defending against photooxidative stress. In addition, constitutive overexpression of GPXH along with GLUTATHIONE-S-TRANSFERASE 1 (GSTS1) was sufficient to augment 1O2 resistance. As with C. reinhardtii, Arabidopsis cell suspension cultures (ACSC) are an excellent plant model system to investigate 1O2 elicitor-mediated transcriptional defence responses and cell viability (or PCD) following chemical treatments. Quantification of PCD rates in cells in vivo can be rather difficult when infiltration of chemicals is required as the defence responses often occur in a small group of inaccessible cells buried in a bulk of surrounding healthy cells. Consequently, plant cell cultures can offer a more suitable means of investigating PCD due to their accessibility, reduced complexity, and uniformity (McCabe and Leaver, 2000). Additionally, ACSC can also be grown in the dark without difficulty, which in turn represents another advantage when chloroplast-dependent physiological responses are under study (Doyle et al., 2010).
In this study, ACSC were subjected to mild photooxidative conditions by adding RB at submicromolar concentrations to avoid side effects that were not directly related to 1O2 production. After the treatment with RB in the light, 1O2-mediated PCD was only observed in Arabidopsis cells containing functional chloroplasts, and their transcriptional defence responses resembled those induced by 1O2 in the flu mutant. The comparative analysis of the transcriptional profiles of ACSC, treated with RB and H2O2 in the light, provided some clues about transcripts with a key role in PCD. In order to test if cellular location was an important factor in responses induced by 1O2, two other 1O2 elicitors were used in this study, Methylene Violet (MV) and Indigo Carmine (IC), which accumulate differently in plant cells (Hideg et al., 2011). However, neither was able to induce any statistically significant change in transcript expression. The poor 1O2 quantum yield of MV and IC in comparison with that of RB (Kovács et al., 2013) and, consequently, the need for higher concentrations to match equal levels of 1O2 production means that it cannot be established whether lack of change in transcript expression was due to different cellular location or possible limitations in their suitability as photosensitizers in studies where the matter of interest is the effect of 1O2 production by them, but not their own chemical toxicity.
Materials and methods
Growth conditions for Arabidopsis cell suspension cultures
Light-grown Arabidopsis thaliana L. (Columbia ecotype) cell suspension cultures (ACSC) were maintained in 200ml of liquid growth medium (Jouanneau and Péaud-Lenoël, 1967; Axelos et al., 1992) as previously described (González-Pérez et al., 2011). For comparative purposes, Arabidopsis cells were also grown in the dark when cell viability was under investigation (see below).
Cellular location of 1O2 photosensitizers
Before proceeding with the photooxidative treatments, the location of RB, MV, and IC inside the cells of Arabidopsis was investigated by confocal microscopy. The absorption and emission spectral features of these three 1O2 photosensitizers have been previously described (Morrison et al., 1997; Roberts et al., 1998; Chang et al., 2008). A 1ml aliquot of ACSC, previously treated with 1mM of each photosensitizer for 10min in the dark, was centrifuged at 120 g for 3min at 4 °C, washed three times with 2ml of 2.7mM KCl, 147.3mM NaCl, and 10mM sodium phosphate [phosphate-buffered salin (PBS)] pH 7.4, and finally resuspended in 1ml of 10mM PBS pH 7.4. The fluorescence emission was then monitored with a confocal microscope (model DM IRB; Leica Microsystems) following excitation of the three photosensitizers and Chl at 543nm and 633nm with an argon laser and a Triplet Dichroic 488/543/633 excitation beam splitter. The fluorescence emission of the selected photosensitizers and chloroplast Chl-binding proteins was collected at 560–620nm and at 680–700nm, respectively.
Photooxidative treatments
Nine-day-old cultures with a cell density of ~125–150mg ml–1 fresh weight were split into two batches, acclimatized before the chemical treatment for 2h in the dark or continuous illumination at 150 μE m–2 s–1 at 24 ºC with gentle agitation, and then treated with the photosensitizers at 0.5 μM and 5 μM. Cultures were also treated with 0.5mM and 5mM H2O2 for comparative purposes. After 30min of chemical treatment, samples were collected, filtered, frozen in liquid nitrogen, and stored at –80 °C until further analysis. Light and dark treatments were carried out simultaneously, performed in duplicate, and replicated three times. Control cultures were also exposed to the same conditions either at 150 μE m–2 s–1 or in the dark.
Photosynthetic activity
In order to determine the photosynthetic activity, the rate of oxygen production was measured polarographically using a Chlorolab 2 system (Hansatech Instruments, Norfolk, UK) at 20 °C. Samples containing equal amounts of fresh weight of cells were incubated in the dark for 1min before a 300 μE m−2 s−1 light-emitting diode source was switched on. The contribution of mitochondrial respiration to the rate of oxygen production was subtracted when the same experiment was repeated with a fresh sample in the dark. The photosynthetic oxygen evolution rate was expressed as μM min–1 g–1 of fresh weight of cells and the percentage of photosynthetic activity was referred to control cells. All the rate measurements were performed in triplicate.
Protein oxidation analysis
The oxidative damage to the D1 protein (i.e. PsbA) of the PSII RC and general protein oxidation in ACSC were evaluated by western blot. Proteins were extracted from ~500mg of frozen cells as previously described (González-Pérez et al., 2011). Protein samples were subjected to 6M urea, 12% (w/v) SDS–PAGE and transferred overnight to nitrocellulose membranes. Nitrocellulose membranes were stained with Ponceau S solution (Applichem, Darmstadt, Germany) for 1min to visualize the protein transfer and then destained with water. The nitrocellulose membranes were blocked with 1% (w/v) bovine seum albumin (BSA) in 0.6% (w/v) Tween-20, 150mM NaCl, 50mM TRIS-HCl, pH 7.4 (T-TBS). For the immunodetection of PsbA, the nitrocellulose membranes were incubated for 1h at 37 °C with polyclonal antibodies against the C-terminal region of PsbA (Agrisera, Vännäs, Sweden) using a dilution of 1:5000 in T-TBS. After extensive washing in T-TBS, the immunocomplexed membranes were probed for 1h at 37 °C with an anti-rabbit, peroxidase-linked secondary antibody using a dilution of 1:2000 in T-TBS, and were washed with T-TBS once again. The immunodetection of PsbA was visualized with the 3,3’-diaminobenzidine tetrahydrochloride (DAB) substrate kit (Pierce Biotechnology, Rockford, IL, USA) or with luminol (Bio-Rad Laboratories, Hercules, CA, USA). To detect general protein oxidation, the extracted proteins were chemically modified with 2,4-dinitrophenylhydrazine (DNPH) and the corresponding carbonylated proteins were identified following the steps described in the OxyBlot protein oxidation detection kit from Millipore (Millipore, Billerica, MA, USA), and visualized with luminol (Bio-Rad Laboratories).
Indirect detection of 1O2
The production of 1O2 by RB was analysed in ACSC that were first broken in water or deuterium oxide (D2O), and then treated with RB at a concentration of 5 μM under continuous illumination at 150 μE m–2 s–1 or in the dark for 30min. After the treatment, the electrophoretic migration of the band of the D1/D2 heterodimer was inspected by western blot analysis, as described above, to evaluate 1O2 production qualitatively (Lupinkova and Komenda, 2004; Arellano et al., 2011).
Measuring cell death levels
After the (photo)oxidative treatments, dark-grown or light-grown cells were diluted 20 times into fresh culture medium and left in the dark for 24h to allow the progression of cell death-associated morphological changes. The vital stain fluorescein diacetate (FDA) was applied to ACSC to assess their cell viability. Cells were subsequently scored as alive, dead via necrosis, or dead via PCD using a UV-equipped light microscope according to the methods previously described (McCabe and Leaver, 2000; Reape et al., 2008; Doyle et al., 2010). Under UV radiation, a bright green fluorescence is observed in viable cells with an intact plasma membrane. Dead cells that displayed the hallmark PCD morphology consisting of a condensed protoplast, which had retracted away from the cell wall, were scored as dead via PCD. FDA-stained cells that did not exhibit bright green fluorescence or a retracted cell protoplast were scored as necrotic.
Target transcripts to evaluate early ROS-mediated responses
Target transcripts to monitor the responses of ACSC under the assayed oxidative stress conditions were selected from lists containing ROS markers specifically up-regulated by 1O2 and H2O2 (op den Camp et al., 2003; Gadjev et al., 2006; Laloi et al., 2007). The DEFENSIN-LIKE FAMILY (DEF) transcript named At2g43510 was chosen as a marker for general oxidative stress (Gadjev et al., 2006), whereas the PROFILIN FAMILY (PROF) transcript named At2g19760 was selected as a housekeeping marker for internal reference (Laloi et al., 2007). The primers designed to amplify the selected transcripts are given in Supplementary Table S1 available at JXB online.
RNA isolation and RT–PCR analysis
RNA isolation and relative quantification of mRNA expression were performed as previously described (Livak and Schmittgen, 2001; Chomczynski and Sacchi, 2006; González-Pérez et al., 2011). Other intermediate steps such as contaminating genomic DNA elimination, RNA reverse transcription, and reverse transcription–PCR (RT–PCR) runs are as previously described (González-Pérez et al., 2011). Expression levels were normalized using the housekeeping gene PROF (At2g19760).
Microarray experiments and data analysis
Transcriptional analyses were performed using Affymetrix GeneChip Arabidopsis genome ATH1 arrays (Affymetrix Inc., Santa Clara, CA, USA). The method to verify the quality of total, DNA-free RNA, and other technical details describing the microarray experiments are as previously described (González-Pérez et al., 2011). Experiments were performed using three biological replicates. Principal components analysis (PCA) was used to explore the variability between the replicates (Johnson and Wichern, 1998). Data from microarrays were standardized using quantile normalization and the robust multi-array average (RMA) method (Bolstad et al., 2003). Differential transcript expression was carried out using the Limma package (Smyth, 2004) available from Bioconductor (http://www.bioconductor.org/). Multiple testing adjustments of P-values were carried out according to the Benjamini and Hochberg methodology (Benjamini and Hochberg, 1995). A volcano plot was used to visualize transcripts with statistically significant differential expression (Cui and Churchill, 2003). Significantly over- or under-represented gene ontology (GO)/biological process terms were obtained using FatiScan from the Babelomics suite (http://babelomics.bioinfo.cipf.es/) as described previously (Al-Shahrour et al., 2004, 2007; Medina et al., 2010). Multiple testing adjustment of P-values was then carried out according to the false discovery rate (FDR) method (Benjamini and Hochberg, 1995; Benjamini and Yekutieli, 2001). GO terms were annotated from the Ensembl’s 56 (TAIR 10) release (http://www.ensembl.org).
Validation of microarray experiments
In order to validate the microarray experiments, several transcripts with statistically significant differential expression were selected from the list included in Supplementary Tables S2 and S3 at JXB online. The selected transcripts and their corresponding primers are given in Supplementary Table S4.
Co-regulation analysis
Data from the microarray experiments were clustered together with expression data from previously selected key treatments in Arabidopsis based on comparative analysis using Genevestigator (Hruz et al., 2008). Only transcripts presenting signal Log2 ratios ≥1 (induction) or ≤ –1 (repression) were considered for analysis. The hierarchical clustering analysis was then carried out using R statistical software, version 2.12.1 (http://www.r-project.org). Pearson’s correlation analysis was performed to evaluate the linear relationship between experimental treatments (Quinn and Keough, 2002). The raw microarray data have been deposited in the GEO database under the accession number GSE43551.
Results
Cellular location of the 1O2 photosensitizers in ACSC
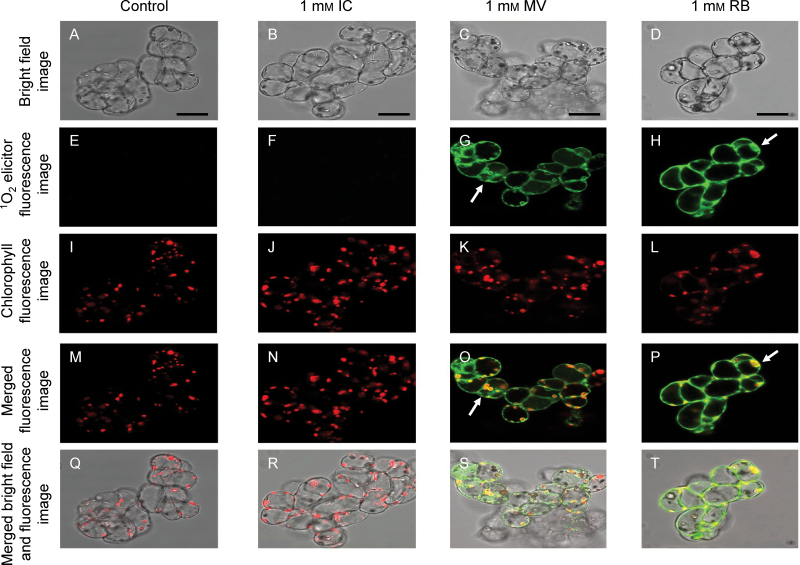
The cellular location of the 1O2 photosensitizers RB, MV, and IC was investigated on the basis of their fluorescence using a confocal microscope (Fig. 1). Additionally, red Chl autofluorescence was used to identify the chloroplasts inside the cells (Fig. 1I–L). The incubation with each photosensitizer was performed in the dark for only 10min to avoid unnecessary cellular damage. The Arabidopsis cells treated with 1mM IC did not exhibit fluorescence emission (Fig. 1F), suggesting that this dye barely permeates the cell or it was not retained within the cell after washing with 10mM PBS pH 7.4. In contrast to IC, the Arabidopsis cells treated with 1mM MV or 1mM RB showed a distinct green fluorescence around and inside the cells that clearly indicated that these photosensitizers were both able to cross the plasma membrane and to be partly retained on the cell boundary (i.e. cell wall and plasma membrane), and other cellular compartments (Fig. 1G, H). In spite of the ability of MV and RB to cross the cell boundary, a closer inspection of the fluorescence images revealed a significant difference between the dyes. MV accumulated in the chloroplast envelope and showed a ring-like fluorescence emission around these organelles, while the red Chl autofluorescence was still discernible (Fig. 1G, O), indicating that MV apparently did not reach the chloroplast interior. In contrast, the distinctive red Chl autofluorescence was blurred by the green fluorescence emission of RB (Fig. 1H, P), showing clearly that this dye could indeed cross the chloroplast envelope and accumulate inside this organelle.
Fig. 1.
Representative confocal micrographs illustrating the location of IC, MV, and RB at a concentration of 1mM in ACSC. From top to bottom the following panels are represented: bright field images (A–D), 1O2 elicitor fluorescence images at 560–620nm (E–H), red Chl autofluorescence images at 680–700nm (I–L), merged Chl and 1O2 elicitor fluorescence images (M–P), and merged bright field and fluorescence images (Q–T). The 1O2 elicitor and red Chl fluorescence emissions were collected after excitation at 543nm and 633nm with an argon laser. Arrows in G, H, O, and P point to key features (see text for further details). Scale bars=50 μm.
Protein oxidation in ACSC as marker for photooxidative stress level
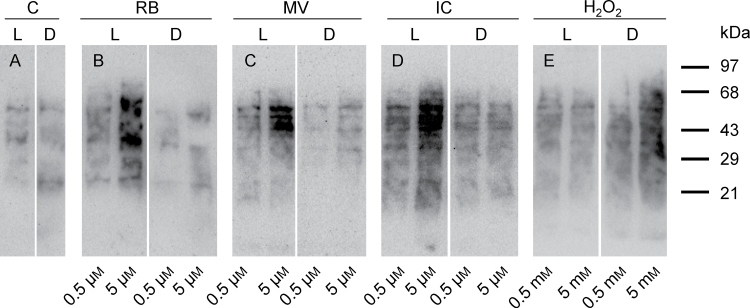
Experimental conditions were chosen to induce mild photooxidative stress in Arabidopsis cells in order to investigate defence responses that would be activated in a series of scenarios where the partitioning of 1O2 between the surrounding medium, the cells, and cellular organelles was imposed by the chemical nature of the 1O2 photosensitizers. Additionally, Arabidopsis cells were exposed to H2O2 for comparative purposes, and its concentration was adjusted to induce a level of oxidative stress similar to that caused by the three 1O2 elicitors. In order to avoid the superimposition of other signalling pathways that could become activated by severe damage, the lowest possible concentrations of RB, MV, IC, and H2O2 were chosen where oxidative damage was perceived in the first 30min of the light treatment. The level of carbonylated proteins was measured as a marker for oxidative stress in the cellular proteome of ACSC. Figure 2 shows the changes in the oxidation state of the cell proteome after the 30min treatment with two different concentrations for RB, MV, IC (i.e. 0.5 μM and 5 μM), and for H2O2 (i.e. 0.5mM and 5mM). A light treatment of ACSC with the photosensitizers at a concentration of 10 μM after 30min showed clear evidence of cellular lysis and a loss of cellular material after filtration, particularly in the case of RB. Additionally, homogenous light penetration in the culture medium was hampered at concentrations >10 μM due to the high molar absorptivity of the photosensitizers. Consequently, the concentration of 5 μM for each photosensitizer was considered as the upper limit for this photooxidative analysis. In the absence of the photosensitizers or H2O2, the OxyBlot experiments showed the immunodetection of a basal level of protein oxidation that did not differ appreciably between light and dark conditions after 30min (Fig. 2). This was not the case when the photosensitizers were added to the medium. The light treatments of ACSC in the presence of RB, MV, and IC at 5 μM revealed an evident increase in protein oxidation in their respective lanes that was not accompanied by similar changes in the dark treatments for the same concentration, showing a clear light-dependent effect on the protein oxidation of ACSC. The changes in protein oxidation of ACSC at 0.5 μM were subtle, but still discernible, for each photosensitizer when the respective treatments in light and dark conditions were compared. Therefore, 0.5 μM for each photosensitizer was considered high enough to induce mild photooxidative stress. When compared with control ACSC, the treatment of ACSC with H2O2 enhanced protein oxidation, but it was not light dependent. On the contrary, protein oxidation became more apparent in the dark, although this finding was not investigated further.
Fig. 2.
Protein oxidation analysis OxyBlot of ACSC under (photo)oxidative stress conditions. Control (A), RB (B), MV (C), IC (D), and H2O2 (E). ACSC were treated for 30min under continuous illumination at 150 μE m−2 s−1 (L) or in the dark (D) at two different concentrations for each photosensitizer (0.5 μM and 5 μM) and H2O2 (0.5mM and 5mM). Each lane was loaded with 30 μg of protein.
RT–PCR analysis of selected transcripts responding to photooxidative stress
Changes in the expression profile for some specific markers for 1O2 and H2O2 were investigated after a 30min treatment with RB, MV, and IC at 0.5 μM, and H2O2 at 500 μM. For the sake of clarity, the full name of the chosen specific markers and their respective locus identifiers are given in Supplementary Table S1 at JXB online. As shown in Supplementary Table S5, significant changes were observed in the expression profile of some selected markers after the treatments with RB. However, no significant changes in the expression profile of any of the selected ROS markers were observed after the treatments with IC and MV, a result that was further confirmed in the whole-genome transcriptional profiling analysis (see below). When ACSC were exposed to RB, the specific 1O2 markers NOD and SZF1 were significantly up-regulated, whereas the specific H2O2 marker SUBT was found to be down-regulated. The same ROS markers (i.e. NOD, SZF1, SUBT, and F9D12) did not show any significant change under dark conditions during the 30min treatment with 0.5 μM RB. No significant changes in the expression profile of the general oxidative stress marker (DEF) were observed in the presence of any of the 1O2 elicitors. In contrast to this, the 30min treatment with H2O2 resulted in slight up-regulation of DEF, but not of any of the selected H2O2 markers, confirming that 500 μM H2O2 also induced mild oxidative stress in ACSC under the experimental conditions used here.
Limma analyses of 1O2- and H2O2-mediated transcriptional responses of ACSC
The 1O2- and H2O2-mediated transcriptional responses of ACSC were further characterized using Affymetrix GeneChip Arabidopsis genome ATH1 arrays. The microarray experiments were designed and classified as follows: control (C1, C2, and C3), RB at 0.5 μM (RB1, RB2, and RB3), MV at 0.5 μM (MV1, MV2, and MV3), IC at 0.5 μM (IC1, IC2, and IC3), and H2O2 at 500 μM (HP1, HP2, and HP3); where the numbers 1–3 represent the number of biological replicates. After data normalization, the Limma package identified a total of 1705 transcripts which were differentially expressed in ACSC exposed to RB with an adjusted P-value <0.05 (Supplementary Table S2 at JXB online). Out of a total 1705 transcripts, 314 had a fold change >1 (Log2), whereas 171 exhibited a fold change < –1. Surprisingly, no transcripts with statistically significant differential expression (adjusted P-value <0.05) were found in cell cultures treated with either MV or IC. In comparison with the RB treatment, the treatment of ACSC with 500 μM H2O2 induced a mild transcriptional response characterized by the observation that both the number of transcripts differentially expressed (i.e. 568, adjusted P-value <0.05) and the number of transcripts with a fold change < –1 or > 1 (i.e. 270) were lower than in the RB treatment (Supplementary Table S3). Volcano plots are shown in Supplementary Fig. S1 to better summarize the experimental conditions that yielded statistically significant changes in transcript expression. It is worth noting that 32 out of the 314 up-regulated transcripts (and four out of the 171 down-regulated transcripts) with a statistically significant fold change |Log2|>1 after treatment with 0.5 μM RB were included in the lists of 296 up-regulated and 29 down-regulated transcripts described as specific markers for 1O2 according to the transcriptional profile analyses performed by Gadjev et al., (2006). Approximately 45% and 30% of the transcripts with up- and down-regulation after the treatment with 500 μM H2O2 were also present in the lists of up- and down-regulated transcripts after treatment with 0.5 μM RB. Surprisingly, the treatment with 500 μM H2O2 induced the expression (|Log2|>1) of only three transcripts defined as specific markers for H2O2, but 19 transcripts defined as specific markers for 1O2 (Gadjev et al., 2006) (Supplementary Table S6).
RT–PCR and Limma analyses were in line with respect to the treatments with 0.5 μM MV and 0.5 μM IC, and both confirmed that there were no statistically significant changes in the transcriptional profile of ACSC after these treatments. It was first hypothesized that the different cellular location of the three photosensitizers could explain the divergent changes in the transcriptional profile. However, it was also found reasonable that the differences were based on the inherent quantum yields of 1O2 for RB, MV, and IC, which are in a ratio of 0.75:0.37:0.095 (Kovacs et al., 2013). Although the most obvious thing to do was to adjust the MV and IC concentrations to reach 1O2 production levels similar to RB, several factors such as cellular lysis, non-homogeneous light penetration, and a direct cytotoxic effect by the photosensitizers discouraged the use of higher concentrations. Therefore, it could not be unambiguously determined whether cellular location was the sole reason why no significant changes in the transcriptional profile of ACSC were observed after the treatments with IC and MV, and, consequently, the study was centred on the treatments with RB and H2O2.
Microarray data validation by RT–PCR
The microarray data of the 30min treatments with 0.5 μM RB or 500 μM H2O2 were confirmed by RT–PCR. All the transcripts selected for validation (Supplementary Table S4 at JXB online) showed similar fold changes in expression when they were analysed by both techniques (Supplementary Fig. S2).
GO process terms associated with response to stimuli after (photo)oxidative treatments
In an attempt to identify GO/biological process terms that became over-represented after the treatments with RB and H2O2, functional enrichment was carried out using FatiScan from the Babelomics suite (http://babelomics.bioinfo.cipf.es). Briefly, this tool makes use of the complete list of genes available in the ATH1 arrays (i.e. ~24 000) and detects significantly up- or down-regulated blocks of functionally related transcripts ordered by differential expression after the treatment, no matter whether the differential transcriptional expression is found to be statistically significant or not. The treatment with 0.5 μM RB exhibited an extensive list of over-represented biological processes (42 GO terms) that were associated with responses to (i) stimuli (i.e. 25 out of 42; defence and immune responses, responses to abiotic, biotic, organic substance, etc.) and (ii) metabolic, cellular, and developmental processes (i.e. 17 out of 42; nitrogen, nucleic acid, and protein metabolic processes, transcription, protein folding and modification, cell cycle, PCD, apoptosis, etc.) (Supplementary Table S7 at JXB online). In the presence of 500 μM H2O2, 46 GO terms were over-represented with statistical significance and they could be divided into GO terms ascribed to responses to (i) stimuli (i.e. 20 out of 46) and (ii) metabolic, cellular, and developmental processes (i.e. 27 out of 46) (Supplementary Table S8). Sixteen out of the 20 over-represented defence processes for the treatment with H2O2 were common with the RB treatment (Supplementary Tables S7 and S8). Other GO/defence process terms for the H2O2 treatment were associated with metal ion responses.
RB and H2O2 inhibit the photosynthetic activity of ACSC
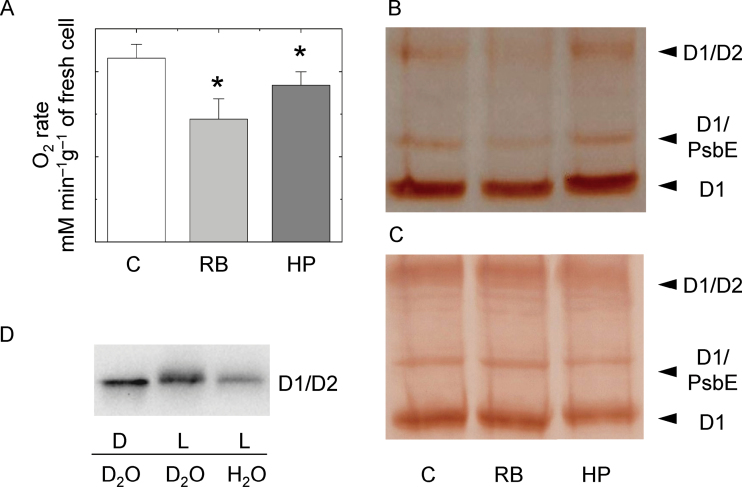
The photosynthetic oxygen evolution rate of ACSC was examined in the presence of 0.5 μM RB and 500 μM H2O2 (Fig. 3A). It decreased to ~65% of the initial value under control conditions when RB was present, suggesting that the 1O2 produced by RB inside chloroplasts could inhibit the photosynthetic activity of ACSC. The treatment with 500 μM H2O2 also brought a slight inhibition of the photosynthetic activity of ACSC, indicating that exogenous H2O2 easily permeated the plasma membrane and entered chloroplasts.
Fig. 3.
(A) Oxygen evolution rates of ACSC under control conditions (C) and after the 30min treatments with 0.5 μM RB and 500 μM H2O2, *P-value <0.05. (B and C) Western blot analysis of the D1 protein of PSII after the 30min treatment with 0.5 μM RB and 500 μM H2O2 (HP) under continuous illumination (B) and in the dark (C). Lane 1, control (C); lane 2, RB; lane 3, HP. Each lane was loaded with 70 μg of protein. The upper, middle, and lower band show the D1/D2 heterodimer of PSII, the adduct between the monomeric D1 protein and the α-subunit of the cytochrome b559 (D1/PsbE), and the monomeric D1 (D1), respectively. (D) Western blot analysis of the D2/D1 heterodimer of PSII after the 30min treatment of broken ACSC with 5 μM RB under continuous illumination at 150 μE m−2 s−1 (L) or in the dark (D) in water or deuterium oxide (D2O). Each lane was loaded with 30 μg of protein.
In order to establish whether the presence of 1O2 and H2O2 could damage the D1 protein of PSII in ACSC, cultured cells were subjected to a western blot analysis using an antibody raised against the D1 protein (Fig. 3B). In control conditions, three bands were distinguished; the lower and more intense band corresponded to the monomeric D1 protein of the PSII RC, whereas the upper band corresponded to the D1/D2 heterodimer of the PSII RC. The intermediate band, which is often observed to react against the D1 antibody, was a faint band that corresponded to an adduct between the D1 protein and the α-subunit of cytochrome b 559 of PSII (Arellano et al., 2011, and references therein). The intensity of the band ascribed to the monomeric D1 protein was lower in the presence of 0.5 μM RB, but it does not change in the treatment with 500 μM H2O2 (Fig. 3B). As a control experiment, the incubation with RB and H2O2 was also performed in the dark, and the results showed no damage to the D1 protein (Fig. 3C). Neither significant damage to the D1 protein nor a substantial decrease in the photosynthetic oxygen evolution rate were to be expected in the presence of 500 μM H2O2 on the basis of the study by Miyao et al. (1995), who reported a more visible and progressive damage to the D1 protein in the range of millimolar concentrations of H2O2. However, this does not exclude that other photosynthetic complexes (i.e. PSI or Rubisco) could be more sensitive to H2O2 inhibition (Nakano et al., 2006).
To demonstrate qualitatively that RB produced 1O2 under the present experimental conditions, broken ACSC were treated with this photosensitizer at a concentration of 5 μM in both water and D2O, where the lifetime of singlet oxygen notably varies from ~4 μs to ~70 μs. It is known that the D1/D2 heterodimer exhibits a shift in its electrophoretic migration due to changes in protein conformation after oxidation of amino acid residues prone to react with 1O2 (Lupinkova and Komenda, 2004; Arellano et al., 2011). Figure 3D showed that 5 μM RB induced a shift of the immunodetected band of the D1/D2 heterodimer toward the cathodic end of the gel together with a slight broadening after the light treatment. The shift was more prominent when the broken ACSC were exposed to the 30min light treatment in D2O, where the lifetime for 1O2 was longer and, consequently, the changes in the conformation of the D1/D2 heterodimer induced by 1O2 were more acute.
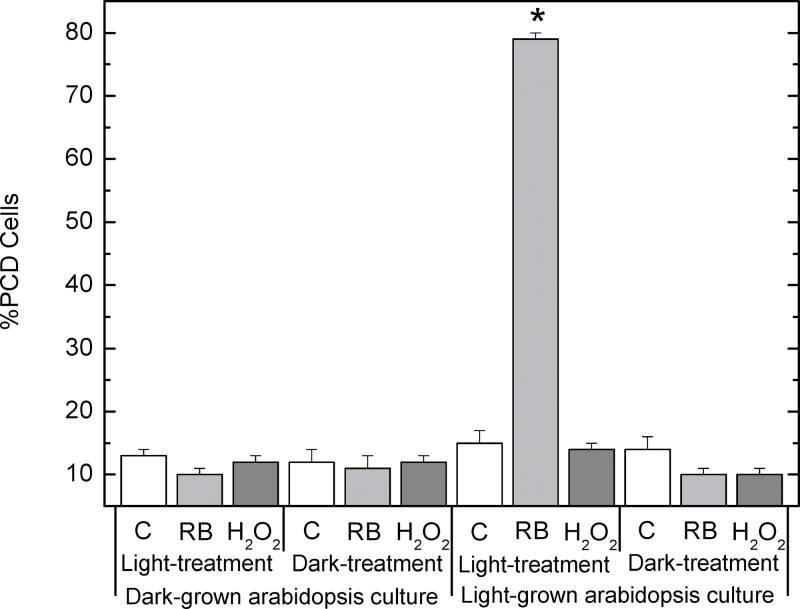
RB, but not H2O2, affects PCD rates in light-grown ACSC
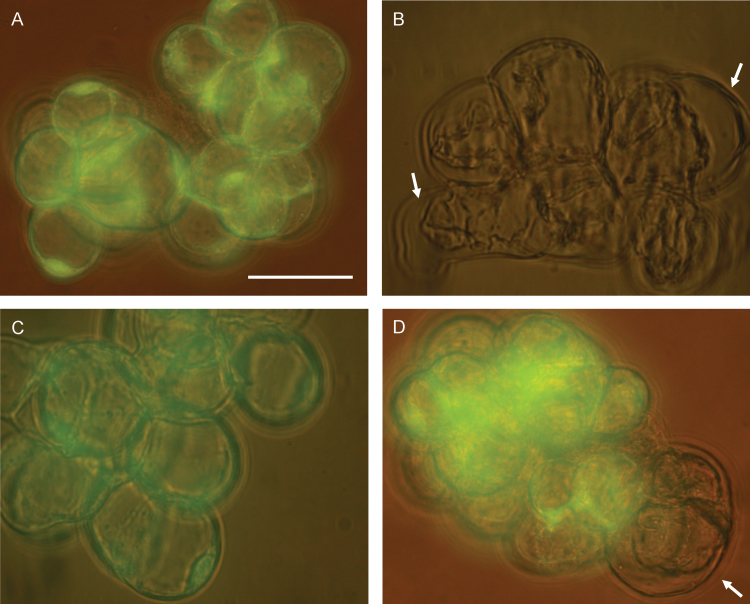
The production of ROS (and particularly 1O2) in chloroplasts has been demonstrated to play a key role in PCD regulation (Doyle et al., 2010; Kim et al., 2012). Therefore, the two treatments with RB or H2O2—both compounds able to enter chloroplasts—could affect the viability of ACSC and induce PCD. In order to determine if this was the case, visible morphological PCD changes, together with FDA fluorescence, were investigated. Figure 4 shows four representative bright field images of light-grown cells under control conditions, after treatment with 0.5 μM RB or 500 μM H2O2 for 30min under continuous illumination at 150 μE m−2 s−1, and after exposure to HL (i.e. 1800 μE m−2 s−1) for 90min. Under control conditions, light-grown cells exhibited the standard morphology (i.e. excessive hydration in cellular cultures) and were largely viable (12–15% PCD; Fig. 5), as demonstrated by the FDA fluorescence. In contrast to this, a clear gap between the cell wall and the plasma membrane was discernible in many of the light-grown cells treated with 0.5 μM RB, and bright green fluorescence could barely be detected in the cells stained with FDA (Fig. 4), indicating that PCD was greatly induced in light-grown ACSC after the RB treatment (79%; Fig. 5). The condensation of the cell protoplast could not be observed when light-grown cells were subjected to 500 μM H2O2, and bright green fluorescence was detected in the presence of FDA (Fig. 4). PCD was 14% (Fig. 5), indicating that the assayed concentration of H2O2 did not induce PCD significantly under these experimental conditions and that higher H2O2 concentrations are required for the activation of PCD (Desikan et al., 1998, 2001). Likewise, cell viability of light-grown cells exposed to HL was examined to explore whether 1O2 production by PSII was capable of activating PCD. HL treatment at 1800 μE m−2 s−1 for 30min did not affect cell viability (González-Pérez et al., 2011) and only HL treatments for (or beyond) 90min resulted in morphological changes associated with PCD (Fig. 4). No significant effects were observed on PCD rates when light-grown ACSC were subjected to chemical treatments with 0.5 μM RB or 500 μM H2O2 for 30min in the dark (Fig. 5).
Fig. 4.
Representative field images showing changes in the morphology of light-grown Arabidopsis cells under control conditions (A), after the 30min treatment with 0.5 μM RB (B) and 500 μM H2O2 (C), and after the 90min treatment at 1800 μE m−2 s−1 (D). Bright green fluorescence of fluorescein diacetate following excitation with UV radiation in A, C, and D indicates that Arabidopsis cells remain viable after the stress treatment. Arrows in B and D illustrate some of the gaps between the cell wall and the plasma membrane after the induction of PCD. Scale bars=50 μm.
Fig. 5.
PCD in light-grown and dark-grown Arabidopsis cells subjected to 30min chemical treatments at 150 μE m−2 s−1 or in the dark. Black bar, 0.5 μM RB; grey bar, 500 μM H2O2; white bar, control samples. *P-value <0.05.
Further experiments were performed with dark-grown cells containing proplastids instead of functional chloroplasts (Doyle et al., 2010), and cells were subjected to chemical treatments with 0.5 μM RB or 500 μM H2O2 for 30min under continuous illumination at 150 μE m–2 s–1 or in the dark. The results showed that the viability of the chemically treated cells was similar to that of control cells for dark-grown cells; there was no effect whether the chemical treatment was with 0.5 μM RB or 500 μM H2O2, or whether the cells were exposed to light or remained in the dark during the RB or H2O2 treatments (Fig. 5). Considered together, these results showed that the presence of developed chloroplasts and illumination were essential for the activation of PCD in ACSC treated with 0.5 μM RB.
Co-regulation of 1O2-producing mutants and HL treatments with RB-treated ACSC
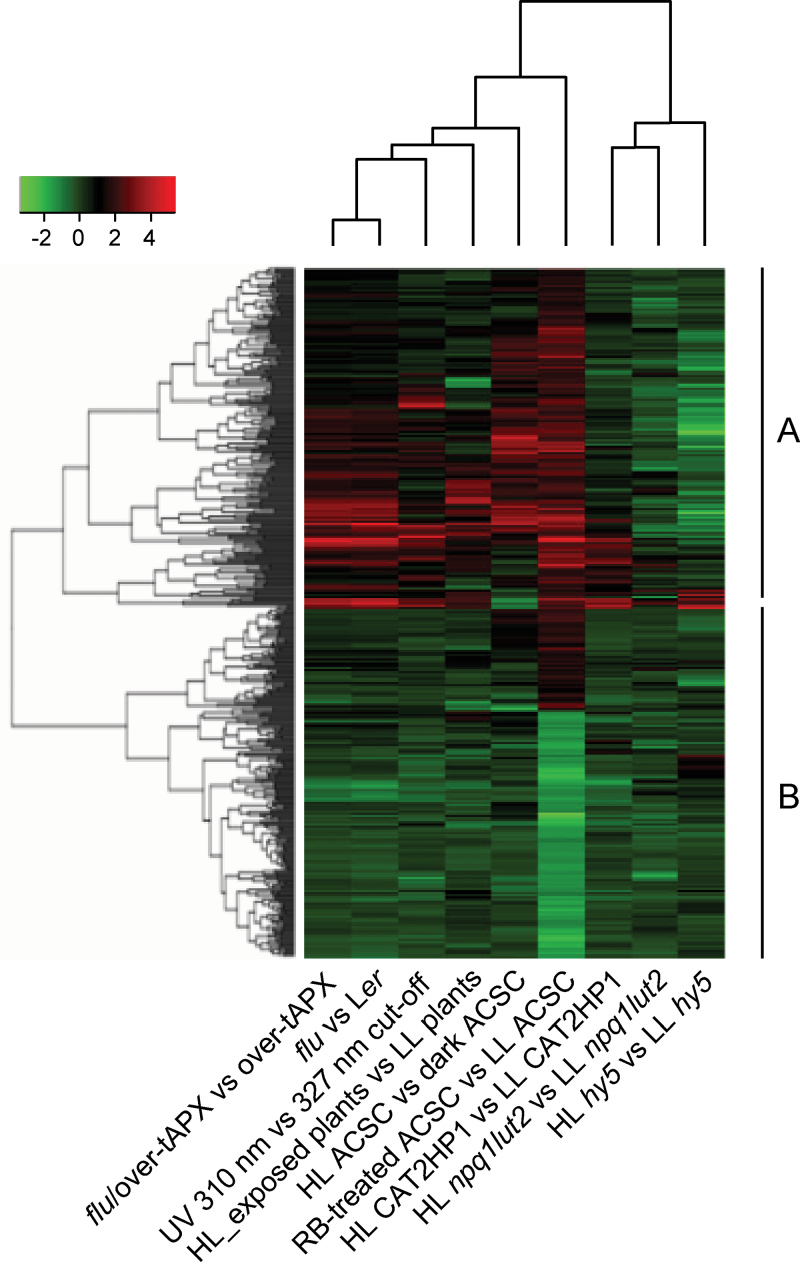
Over the last 14 years, analysis of the transcriptional profile of the flu mutant family has shed much light on 1O2-mediated defence responses in plants and has also provided lists of specific transcriptional markers and cis-regulatory elements (Meskauskiene et al., 2001; op den Camp et al., 2003; Gadjev et al., 2006; Petrov et al., 2012). The flu mutant family is characterized by the high production of 1O2 after the dark–light shift in response to the accumulation of Pchlide in thylakoids. In contrast to the flu mutant, high levels of 1O2 are produced in response to excess excitation energy in PSII of wild-type plants when photosynthetic activity is inhibited (Mullineaux and Baker, 2010). It is well established that 1O2 is the major ROS generated under HL conditions (Triantaphylidès et al., 2008; Triantaphylidès and Havaux, 2009) and, consequently, the activation of defence responses under HL conditions ought to share a set of 1O2-responsive transcripts with the flu mutant family. GENEVESTIGATOR can be used to perform comparative genome-wide analyses under different experimental conditions (Hruz et al., 2008), and here it has been used to identify light treatments or Arabidopsis mutants that exhibit a transcriptional expression pattern similar to that of ACSC treated with RB. Six experimental conditions are described in GENEVESTIGATOR as HL studies, although the light irradiance used in those studies ranged from 400 μE m–2 s–1 to 1800 μE m–2 s–1 (Vandenabeele et al., 2004; Walters et al., 2004; Vanderauwera et al., 2005; Kleine et al., 2007; Rossel et al., 2007; Caldana et al., 2011; González-Pérez et al., 2011). Additionally, the GENEVESTIGATOR database contains the transcriptional profiling of Arabidopsis after perturbation with UV radiation, a ‘light’ perturbation also known to lead to the production of 1O2 and other ROS at high doses (Brosche et al., 2002; Ulm et al., 2004; Hideg et al., 2013). Among the Arabidopsis mutants described to produce 1O2, the database of GENEVESTIGATOR only includes the flu mutant family (Laloi et al., 2007), but not others. The CEL files of the double mutant npq1lut2 were kindly provided by Dr Alboresi and co-workers (Alboresi et al., 2011) and were included in the genome-wide analysis. The study by Ramel and co-workers (2012b , 2013) could not be included in the co-regulation analysis because these authors used CATMAv5 instead of Affymetrix microarrays (Ramel et al., 2012a , b ), and only a manual inspection was performed with their microarray data (see the Discussion). Using a list of 400 transcripts of the 0.5 μM RB treatment with |Log2|>1, it was found that the biclustering tool of GENEVESTIGATOR identified a group of transcripts exhibiting an expression pattern similar to that described in the flu mutant family. Additionally, other treatments such as HL irradiance beyond 1400 μE m–2 s–1 or UV radiation also proved to have a similar expression pattern. In contrast, HL treatments below 1400 μE m–2 s–1 showed a poor correlation. In a more robust analysis, the total number of 485 transcripts with significant differential expression (with |Log2|>1) in the treatment with 0.5 μM RB was subjected to hierarchical clustering in order to identify both experimental conditions with similar differential transcript expression and clusters of transcripts that could be rich in specific markers for 1O2.
In the first instance, it is worth noting that the hy5 (long hypocotyl5 transcription factor mutant of Arabidopsis) and npq1lut2 mutants were subjected to light irradiance of 1000 μE m–2 s–1 (Kleine et al., 2007; Alboresi et al., 2011) and exhibited a poor correlation with the other selected experimental systems (Supplementary Table S9 at JXB online); they clustered together with catalase-deficient (CAT2HP1) plants (Fig. 6). CAT2HP1 plants in HL accumulate H2O2 (Vanderauwera et al., 2005) and exhibit a distinctive transcriptional profile when compared with other experimental systems where 1O2, ozone (O3), or superoxide radical (O2 –) accumulates (Gadjev et al., 2006). The other set of experiments clustered and, in particular, the transcriptional profile of the treatment with 0.5 μM RB showed the highest correlation with the flu family mutants (Supplementary Table S9).
Fig. 6.
Overall picture of the hierarchical clustering analysis on the 485 transcripts with statistically differential expression and fold change |Log2|>1 in ACSC after the treatment with 0.5 μM RB. Data were clustered together with available expression microarray data from HL experiments and Arabidopsis mutants included in the GENEVESTIGATOR database: HL hy5 vs LL hy5, long hypocotyl5 transcription factor mutant of A. thaliana (Col-0) seedlings exposed to HL irradiance (1000 μE m–2 s–1) for 3h versus low light (LL) irradiance (100 μE m–2 s–1) (Kleine et al., 2009); HL npq1lut2 vs LL npq1lut2, double mutant of 6-week-old plants of A. thaliana (Col-0) lacking violaxanthin de-epoxidase and lycopene-ε-cyclase activities exposed to HL irradiance (1000 μE m–2 s–1) for 2h versus LL irradiance (25 μE m–2 s–1) (Alboresi et al., 2011); HL CAT2HP1 vs LL CAT2HP1, 6-week-old catalase-deficient plants of A. thaliana (Col-0) exposed to HL irradiance (1600–1800 μE m–2 s–1) for 8h versus LL irradiance (100–140 μE m–2 s–1) (Vanderauwera et al., 2005); RB-treated ACSC vs LL ACSC, this study; HL ACSC vs dark ACSC, Arabidopsis cell suspension cultures exposed to HL irradiance (1000 μE m–2 s–1) for 0.5h versus 1h dark conditions (González-Pérez et al., 2011); HL-exposed plants vs LL plants; mature wild-type plants of A. thaliana (Col-0) exposed to HL irradiance (1400–1600 μE m–2 s–1) for 1h versus LL irradiance (40–70 μE m–2 s–1) (Rossel et al., 2007); UV 310nm vs 327nm cut-off; seedlings of A. thaliana (Wassilewskija) exposed for 15min at midday to UV-B from Philips TL 40W/12 UV fluorescent tubes unfiltered through a 3mm quartz plate versus filtered through a 3mm transmission WG327cut-off filter (Ulm et al., 2004); flu vs Ler, the flu mutant of A. thaliana (Ler) exposed to light irradiance (90 μE m–2 s–1) for 2h after incubation in the dark for 8h versus dark conditions (Laloi et al., 2007); flu/over-tAPX vs over-tAPX, the flu mutant of A. thaliana (Ler) overexpressing the thylakoid-bound ascorbate peroxidase (tAPX) exposed to light irradiance (90 μE m–2 s–1) for 2h after incubation in the dark for 8h versus dark conditions (Laloi et al., 2007). See the text for a detailed description of the main clusters A and B.
In the second instance, two main clusters of transcripts were observed and named A and B (Fig. 6). Cluster A consisted of 242 transcripts and was characterized by the presence of the largest number of specific markers for 1O2 (i.e. 27 out of 36). The value of 3.42 for the odds ratio [i.e. 27×(242–9)/9×(243–27)] indicated that the enrichment of specific markers for 1O2 in cluster A was statistically significant (P-value of 1.5×10–3). Cluster B with 243 transcripts included all the down-regulated transcripts with a fold change <1 in the treatment with 0.5 μM RB and it did not show any significant enrichment in specific markers for any type of ROS. Table 1 summarizes the list of specific markers for ROS identified in cluster A. It is worth noting that six out of the 27 specific markers for 1O2 identified in cluster A were also found in the list of the 229 up-regulated transcripts with a fold change >1 in the treatment with 500 μM H2O2 (Supplementary Table S6 at JXB online).
Table 1.
1O2 markers with statistically significant enrichment (P<0.05) in cluster A of the hierarchical clustering analysis of the 485 transcripts with fold change |Log2|>1 in ACSC after the 30min treatment with 0.5 μM RB
| Element | Locus ID | Annotation |
|---|---|---|
| 248799_at | At5g47230 | ERF5, ethylene responsive element binding factor 5 |
| 266821_at | At2g44840 | ERF13, ethylene responsive element-binding factor 13 |
| 248448_at | At5g51190 | AP2 (Apetala2) domain-containing transcription factor, putative |
| 262211_at | At1g74930 | Member of the dehydration response element-binding (DREB) subfamily A-5 of ERF/AP2 transcription factor family |
| 259992_at | At1g67970 | HSFA8, heat shock transcription factor A8 |
| 247707_at | At5g59450 | SCL11, Scarecrow-like transcription factor 11 |
| 252679_at | At3g44260 | CCR4-NOT transcription complex protein, putative |
| 257511_at | At1g43000 | Zinc-binding family protein |
| 252474_at | At3g46620 | Zinc finger (C3HC4-type RING finger) family protein |
| 258436_at | At3g16720 | Zinc ion binding/protein binding |
| 256306_at | At1g30370 | Lipase class 3 family protein |
| 246600_at | At5g14930 | SAG101, senescence-associated gene 101; triacylglycerol lipase |
| 245777_at | At1g73540 | NUDT21, Nudix hydrolase homologue 21 |
| 248793_at | At5g47240 | NUDT8, Nudix hydrolase homologue 8 |
| 252592_at | At3g45640 | MPK3, mitogen-activated protein (MAP) kinase 3 |
| 257751_at | At3g18690 | MKS1, MAP kinase substrate 1 |
| 255872_at | At2g30360 | CIPK11, Cystathionine β-lyase-interacting protein kinase 11 |
| 251683_at | At3g57120a | Protein kinase family protein |
| 252470_at | At3g46930a | Protein kinase family protein |
| 263320_at | At2g47180 | GOLS1, galactinol synthase 1 |
| 247811_at | At5g58430 | EXO70B1, exocyst subunit EXO70 family protein B1 |
| 247240_at | At5g64660a | U-box domain-containing protein |
| 256999_at | At3g14200a | DNAJ heat shock N-terminal domain-containing protein |
| 246108_at | At5g28630 | Glycine-rich protein |
| 266259_at | At2g27830 | Similar to pentatricopeptide (PPR) repeat-containing protein |
| 246270_at | At4g36500a | Similar to unknown protein (TAIR:At2g18210.1) |
| 245840_at | At1g58420 | Similar to unknown protein (TAIR:At1g10140.1) |
a Specific markers for 1O2 that are also up-regulated in ACSC after the treatment with 500 μM H2O2.
Discussion
Chloroplast-dependent 1O2-mediated PCD in ACSC treated with RB
RB, which accumulates in chloroplasts of Arabidopsis cells, caused a surge in 1O2 production that activated transcriptional defence responses leading to the physiological induction of PCD. However, treatment with MV or IC, which do not enter chloroplasts, did not activate any statistical change in the transcriptional profile of ACSC. In spite of this, it could not be unambiguously concluded that the divergent subcellular location of the three 1O2 elicitors was the sole cause of this observation because the 1O2 quantum yields of the three elicitors are inherently different and, in particular, lower for MV and IC (Kovacs et al., 2013). Therefore, this physiological defence response was investigated in more detail in both dark-grown and light-grown Arabidopsis cells in order to establish whether its activation was initiated by 1O2 production inside the chloroplasts or instead was independent of cellular location. It was determined that PCD was not activated by 1O2 produced by RB in dark-grown cells that had not developed chloroplasts; a result that was in full agreement with the recent study by Kim and co-workers (2012), who observed that the 1O2-mediated cell death in seedlings of the leaf-variegated mutant variegated2 was only induced in cells of the green leaf sector of this mutant containing fully developed chloroplasts, but not in the white sector where undifferentiated plastids were present. Chloroplasts are known both to be an important source of defence signalling molecules (i.e. ROS and precursors of defence hormones) and to have a central role in defence responses and the hypersensitive response (HR), a form of PCD localized at the site of pathogen attack that requires light for its development in many cases (Karpinski et al., 2003; Coll et al., 2011; Kacprzyk et al., 2011). Likewise, the involvement of chloroplasts and ROS in PCD was recently demonstrated in ACSC (Doyle et al., 2010), supporting the view that chloroplasts play a significant role in PCD regulation. In some studies carried out with C. reinhardtii, it has been proposed that 1O2 can escape from the chloroplasts and interact with an 1O2 sensor located in the cytoplasm or even reach the nucleus (Fischer et al., 2007). However, on the basis of the present results with RB, which does not activate PCD in dark-grown cells, it is difficult to reconcile the results with a role for 1O2 itself as a signalling molecule outside chloroplasts unless extremely, non-physiological HL conditions or very high concentrations of 1O2 elicitors come into play. Hence it is proposed that 1O2-mediated PCD in ACSC is initiated only via developed chloroplasts.
Under the experimental conditions used here, 500 μM H2O2 did not activate PCD in cells grown in dark or light conditions. Low PCD rates in ACSC subjected to H2O2 were shown here; however, higher H2O2 concentrations or longer incubation times than those described in this study were required to increase the PCD levels (Desikan et al., 1998, 2001).
The functional enrichment analysis of the transcriptional profiling of light-grown ACSC also provided lines of evidence for the activation of PCD after the treatment with 0.5 μM RB, but not with 500 μM H2O2. A direct comparison between the 30min, 2-fold up-regulated transcripts in ACSC treated with 0.5 μM RB and the 30min early-induced, 2-fold up-regulated transcripts in the flu mutant (op den Camp et al., 2003) showed that >40% of the up-regulated transcripts in ACSC (i.e. 138 of out 314) were included in the above list of up-regulated transcripts in the flu mutant. Likewise, the list of the 30min, 2-fold, up-regulated transcripts in ACSC, treated with 0.5 μM RB, contained 32 specific markers for 1O2 (Supplementary Table S6 at JXB online), seven specific markers for H2O2, and no specific markers for O2 – when it was compared with lists of specific markers for ROS provided by Gadjev and co-workers (2006). This indicated that 1O2 produced by RB in ACSC induced the expression of a list of transcripts that resembled the list observed in the flu mutant and confirmed that the transcriptional defence responses observed were mediated by 1O2 production inside chloroplasts. However, they did not seem to be associated with the EX1/EX2- and EDS1-dependent signalling pathway (see below). In addition, the analysis of the over-represented GO/biological process terms in Arabidopsis cells treated with 0.5 μM RB showed that most of them corresponded to responses to pathogens, several biotic and abiotic stimuli, and PCD, which were also observed in the flu mutant (op den Camp et al., 2003). Unexpectedly, the list of the 2-fold up-regulated transcripts in ACSC after the treatment with 500 μM H2O2 also exhibited some overlap (<30%) with the list of the 30min early-induced, 2-fold up-regulated transcripts in the flu mutant and 19 specific markers for 1O2 (Supplementary Table S6), and only three specific markers for H2O2 were identified. The fact that the treatment with 500 μM H2O2 inhibited the expression of several chloroplast transcripts (Log2 <1) particularly associated with photosynthesis (Supplementary Table S3, locus identifier in a cyan background) suggested that H2O2 clearly entered chloroplasts and activated transcriptional defence responses that had common characteristics with those described for the 0.5 μM RB treatment and the flu mutant. This is confirmed in Supplementary Tables S7 and S8, where a significant number of over-represented GO/biological process terms after the treatments with 0.5 μM RB and 500 μM H2O2 appeared in both lists. However, a closer inspection of both lists disclosed that GO terms such as resistance response to pathogen (GO:0009814 and GO:009627), protein ubiquitination (GO:0016567), and PCD (GO:0012501 and GO:0006915) were only present in the treatment with 0.5 μM RB. The list of the 30min, up-regulated transcripts in ACSC after the treatment with 0.5 μM RB contained key transcripts associated with the above GO terms that included: resistance (R) transcripts encoding TIR-NB-LRR-type resistance and CC-NB-LRR proteins (At5g41750; At5g41740; At1g58807; At1g59124; At1g59620), MITOGEN-ACTIVATED PROTEIN KINASES (MPK3, At3g45640; MPKK4, At1g51660), the ubiquitin ligase PLANT U-BOX17 (PUB17, At1g29340), a NUDIX HYDROLASE HOMOLOG (NUDT7, At4g12720), BCL-2-ASSOCIATED ATHANOGENE (BAG3, At5g07220), the lipase-like protein PHYTOALEXIN DEFICIENT 4 (PAD4, At3g52430), the ELICITOR PEPTIDE 2 PRECURSOR (PROPEP2, At5g64890), the transcription factor WRKY70 (At3g56400), and the ANKYRIN REPEAT-CONTAINING PROTEIN 2A (AKR2A; At4g35450); the underlined transcripts of which were also present in the early-induced, 2-fold up-regulated transcripts of the flu mutant. In contrast, the list of up-regulated transcripts in ACSC treated with 500 μM H2O2 contained only four transcripts associated with PCD and resistance response to pathogens (BAG3, At5g07220; PROPEP2, At5g64890; AKR2A, At4g35450; and MYB30, At3g28910), but none of them was included in the list of the early-induced, 2-fold up-regulated transcripts in the flu mutant.
It has been proposed that ethylene (ET) and salicylic acid (SA) signalling pathways function additionally during the 1O2-mediated PCD and that the oxylipins 12-oxophytodienoic acid (OPDA) and dinor-OPDA antagonize the cell death-inducing activity of jasmonic acid (JA) in the flu mutant (Danon et al., 2005). In a search for key transcripts associated with the biosynthesis of the above enzymes in ACSC treated with 0.5 μM RB, it was observed that 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID (ACC) SYNTHASE 6 (ACC6, At4g11280) was present together with another eight ethylene-responsive element binding factors (ERFs) that included all those early-induced ERFs listed by Danon and co-workers (2005). In addition, transcripts encoding enzymes associated with the biosynthesis of OPDA in chloroplasts such as ALLENE OXIDE SYNTHASE (AOS, At5g42650), the conjugation of OPDA with glutathione such as GLUTATHIONE S-TRANSFERASE 5 (GST5, At2g29450), and the biosynthesis of JA in peroxisomes such as 3-oxo-2-[2’(Z)-pentenyl]-cyclopentane-1-octanoic (OPC-8:0) CoA LIGASE 1 (OPCL1, At1g20510) were also present. However, there was no evidence for the up-regulation of EDS1 (At3g48090) known to be required for the biosynthetic activation of SA and the modulation of the 1O2-mediated PCD in the flu mutant (Ochsenbein et al., 2006). The essential regulation role of EDS1 is mediated by its interaction with PAD4, and also by the product of the SENESCENCE-ASSOCIATED GENE 101 (SAG101, At5g14930), two proteins that have defence regulatory functions beyond stabilizing EDS1 in TIR-NB-LRR-type R gene-triggered resistance as demonstrated by silencing EDS1 with a double-standed DNA (dsRNA) interference construct (Feys et al., 2005). The evidence that key transcripts associated with the biosynthesis pathway of ET and JA, together with PAD4 and SAG101, are up-regulated in ACSC with 0.5 μM RB suggests that they could be responsible for the induction of PCD through a signalling pathway that does not depend on EDS1. This agrees with the recent study by Ramel et al. (2013), who established that 1O2 activated cell death in the ch1 mutant under HL stress, although no evidence for the activation of the EX1/EX2- and EDS1-dependent signalling pathway was found. In contrast, JA was described to have a key role in the 1O2-mediated cell death, finding significant differences in the activation or repression of transcripts associated with the biosynthesis of this hormone under HL or acclimatory conditions. It is worth noting that an acclimatory response, but not PCD, was activated in ACSC subjected to a 30min treatment with HL (1800 μE m−2 s−1) (González-Pérez et al., 2011; Gutiérrez et al., 2011) and that no evidence was found for the up-regulation of EDS1, PAD4 and SAG101 nor for transcripts encoding enzymes associated with the biosynthesis of JA in peroxisomes.
Similarly, a search for key early-induced transcripts associated with the biosynthesis and signalling cascade of ET, JA, and SA (listed in the study by Danon and co-workers, 2005) in ACSC treated with 500 μM H2O2 did not yield any match except for a transcript encoding the transcription factor ERF6 (At4g17490), supporting the view that PCD was not activated either. Of interest is the observation that ACSC treated with 500 μM H2O2 exhibited several up-regulated transcripts encoding UDP-glycosyl transferases (UGTs) and GST, some of them known to be early-induced by H2O2, such as UGT73B3, UGT73B5, and GST6 and with an important role in plant defence responses (Chen and Singh, 1999; Langlois-Meurinne et al., 2005).
Correlation between the flu family mutants and ACSC treated with RB
The hierarchical clustering analysis of 485 co-regulated transcripts between ACSC treated with 0.5 μM RB, 1O2-producing mutants, and wild-type plants subjected to HL showed that ACSC treated with 0.5 μM RB had higher Pearson’s correlation with the flu family mutants than with the npq1lut2 mutant. ACSC treated with 0.5 μM RB had 27 specific markers for 1O2 in common with the flu mutant (Table 1), but only two out of the 27 were up-regulated in the npq1lut2 mutant exposed to HL for 2h (At5g47240 and At3g14200), where an acclimatory response was reported (Alboresi et al., 2011). A manual inspection of the up-regulated transcripts (with Log2 ratios ≥1) in ch1 (Ramel et al., 2013) showed that four (At2g29450, At2g30360, At2g44840, and At3g44260) were listed among the 27 specific markers for 1O2 in cluster A when this mutant was subjected to HL for 2 d, but none when ch1 was first acclimated to HL. Interestingly, the former up-regulated transcripts are involved in response to ET and JA stimuli. A transcriptional analysis of defence responses mediated by carotenoid oxidation products (i.e. β-cyclocitral) with origin in the reaction between 1O2 and the β-carotene molecules (housed in the PSII RC) also proved that 1O2 production by PSII RC induced an acclimatory rather than a PCD response (Ramel et al., 2012a , b ). A manual inspection of the up-regulated transcripts in the aforementioned study also indicated that only two out of the 27 specific markers for 1O2 in cluster A (At3g46620 and At3g14200) were up-regulated after the treatment with β-cyclocitral. In the study by Rossel and co-workers (2007), leaves were directly exposed to HL for 30min. The list of up-regulated transcripts contained several specific markers for 1O2, eight of which were included in cluster A (At1g74930, At2g30360, At2g35710, At2g44840, At2g47180, At3g44260, At3g46930, At5g47240, see Table 1 for their description). Despite the fact that this HL treatment induced a significant number of specific markers for 1O2, the underlined transcripts of which were involved in response to ET and JA stimuli, the authors described the activation of a (systemic acquired) acclimatory response. The production of 1O2 and other ROS has also been demonstrated in plants exposed to high doses of UV, and their effects on the expression of transcripts encoding enzymes with an antioxidant role and with a UV-dependent expression have been recently investigated (Hideg et al., 2013). However, the overlap between transcripts encoding several antioxidative enzymes in the flu mutant and the UV-induced transcripts was very limited. In fact, a single match with the transcript encoding GLUTAREDOXIN (GRX480, At1g28480)—also induced in other chemical treatments—was detected. A similar conclusion was reached when the 14 UV-induced transcripts were compared with the 2-fold, up-regulated transcripts in ACSC treated with 0.5 μM RB, as only GRX480 was also up-regulated. Although the hierarchical clustering analysis shows certain correlation between experiments where 1O2 is produced, the genome-wide expression profiling and, consequently, the 1O2-mediated defence responses (i.e. acclimation or PCD) vary significantly between experimental conditions, where the EX1/EX2- and EDS1-dependent signalling pathway is not activated or overlaps with other 1O2-mediated signalling pathways belonging to a more complex signalling network.
Conclusion
In brief, PCD has been activated in Arabidopsis cell cultures treated with submicromolar concentrations of RB, a potent, artificial 1O2 photosensitizer that accumulates inside chloroplasts. The fact that this 1O2-mediated physiological response occurs in light-grown cultures, but not in dark-grown cultures, suggests that its activation requires functional chloroplasts. Additionally, a co-regulation analysis using the early-induced transcripts with a statistically significant fold change |Log2|>1 in ACSC treated with RB showed that this treatment had high correlation with the flu family mutants (although there was no evidence for the up-regulation of EDS1), but low correlation with other 1O2-producing mutants or HL treatments in which an acclimatory response was activated instead of cell death.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Volcano plots of the microarray experiments:
Figure S2. Validation of the microarray experiments in ACSC after the 30min treatments with 0.5 μM RB and 500 μM H2O2.
Table S1. Transcripts and their corresponding primers selected for monitoring early ROS-mediated responses in ACSC under (photo)oxidative stress.
Table S2. Set of transcripts down- and up-regulated in ACSC after the 30min treatment with 0.5 μM RB (adjusted P-value <0.05).
Table S3. Set of transcripts down- and up-regulated in ACSC after the 30min treatment with 500 μM H2O2 (adjusted P-value <0.05).
Table S4. Transcripts and their corresponding primers selected for validation of the microarray experiments in ACSC under (photo)oxidative stress.
Table S5. Fold changes (Log2) in the expression of selected transcripts responding to ROS production in ACSC after the 30min treatment with RB, MV, and IC at 0.5 μM, and H2O2 at 500 μM.
Table S6. Up-regulated specific markers for 1O2 in ACSC after several chemical treatments.
Table S7. Tree view of over-represented GO/biological process terms in Arabidopsis cell suspension culture after the 30min treatment with 0.5 μM RB (adjusted P-value <0.05).
Table S8. Tree view of the over-represented GO/biological process terms in Arabidopsis cell suspension culture after the 30min treatment with 500 μM H2O2 (adjusted P-value <0.05).
Table S9. Pearson’s correlation between different experimental conditions and Arabidopsis mutants.
Acknowledgements
This work is funded by the Junta de Castilla y León (ref: CSI002A10-2). CTD was funded by the Irish Research Council for Science, Engineering and Technology. The group is very grateful to the ‘Institut de Biochimie & Physiologie Moléculaire des Plantes’ (INIA), Montpellier, France for providing the cell culture. We also thank Mr José Javier Martín for technical assistance.
Glossary
Abbreviations:
- ACSC
Arabidopsis thaliana cell suspension cultures
- Chl
chlorophyll
- DAB
3,3′-diaminobenzidine tetrahydrochloride
- DNPH
2,4-dinitrophenylhydrazine
- ET
ethylene
- EX
EXECUTER
- FDA
fluorescein diacetate
- HL
high light
- IC
indigo carmine
- JA
jasmonic acid
- LL
low light
- MV
Methylene Violet
- OPDA
12-oxophytodienoic acid
- PCD
programmed cell death
- Pchlide
protochlorophyllide
- RB
Rose Bengal
- ROS
reactive oxygen species
- SA
salicylic acid.
References
- Alboresi A, Dall’Osto L, Aprile A, Carillo P, Roncaglia E, Cattivelli L, Bassi R. 2011. Reactive oxygen species and transcript analysis upon excess light treatment in wild-type Arabidopsis thaliana vs a photosensitive mutant lacking zeaxanthin and lutein. BMC Plant Biology 11, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shahrour F, Arbiza L, Dopazo H, Huerta-Cepas J, Mínguez P, Montaner D, Dopazo J. 2007. From genes to functional classes in the study of biological systems. BMC Bioinformatics 8, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shahrour F, Díaz-Uriarte R, Dopazo J. 2004. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics 20, 578–580 [DOI] [PubMed] [Google Scholar]
- Arellano JB, Li H, González-Pérez S, Gutiérrez J, Melø TB, Vacha F, Naqvi KR. 2011. Trolox, a water-soluble analogue of α-tocopherol, photoprotects the surface-exposed regions of the photosystem II reaction center in vitro. Is this physiologically relevant? Biochemistry 50, 8291–8301 [DOI] [PubMed] [Google Scholar]
- Axelos M, Curie C, Mazzolini L, Bardet C, Lescure B. 1992. A protocol for transient gene expression in Arabidopsis thaliana protoplasts isolated from cell suspension cultures. Plant Physiology and Biochemistry 30, 123–128 [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 57, 289–300 [Google Scholar]
- Benjamini Y, Yekutieli D. 2001. The control of the false discovery rate in multiple testing under dependency. Annals of Statistics 29, 1165–1188 [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193 [DOI] [PubMed] [Google Scholar]
- Brosche M, Schuler MA, Kalbina I, Connor L, Strid A. 2002. Gene regulation by low level UV-B radiation: identification by DNA array analysis. Photochemical and Photobiological Sciences 1, 656–664 [DOI] [PubMed] [Google Scholar]
- Caldana C, Degenkolbe T, Cuadros-Inostroza A, Klie S, Sulpice R, Leisse A, Steinhauser D, Fernie AR, Willmitzer L, Hannah MA. 2011. High-density kinetic analysis of the metabolomic and transcriptomic response of Arabidopsis to eight environmental conditions. The Plant Journal 67, 869–884 [DOI] [PubMed] [Google Scholar]
- Chang CC, Yang YT, Yang JC, Wu HD, Tsai T. 2008. Absorption and emission spectral shifts of rose bengal associated with DMPC liposomes. Dyes and Pigments 79, 170–175 [Google Scholar]
- Chen WQ, Singh KB. 1999. The auxin, hydrogen peroxide and salicylic acid induced expression of the Arabidopsis GST6 promoter is mediated in part by an ocs element. Plant Journal 19, 667–677 [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. 2006. The single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction: twenty-something years on. Nature Protocols 1, 581–585 [DOI] [PubMed] [Google Scholar]
- Coll NS, Epple P, Dangl JL. 2011. Programmed cell death in the plant immune system. Cell Death and Differentiation 18, 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui XQ, Churchill GA. 2003. Statistical tests for differential expression in cDNA microarray experiments. Genome Biology 4, 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danon A, Miersch O, Felix G, op den Camp RG, Apel K. 2005. Concurrent activation of cell death-regulating signaling pathways by singlet oxygen in Arabidopsis thaliana . The Plant Journal 41, 68–80 [DOI] [PubMed] [Google Scholar]
- Desikan R, Burnett EC, Hancock JT, Neill SJ. 1998. Harpin and hydrogen peroxide induce the expression of a homologue of gp91-phox in Arabidopsis thaliana suspension cultures. Journal of Experimental Botany 49, 1767–1771 [Google Scholar]
- Desikan R, Mackerness SAH, Hancock JT, Neill SJ. 2001. Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiology 127, 159–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle SM, Diamond M, McCabe PF. 2010. Chloroplast and reactive oxygen species involvement in apoptotic-like programmed cell death in Arabidopsis suspension cultures. Journal of Experimental Botany 61, 473–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys BJ, Wiermer M, Bhat RA, Moisan LJ, Medina-Escobar N, Neu C, Cabral A, Parker JE. 2005. Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. The Plant Cell 17, 2601–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer BB, Hideg E, Krieger-Liszkay A. 2013. Production, detection, and signaling of singlet oxygen in photosynthetic organisms. Antioxidant and Redox Signaling 18, 2145–2162 [DOI] [PubMed] [Google Scholar]
- Fischer BB, Krieger-Liszkay A, Hideg E, Snyrychova I, Wiesendanger M, Eggen RIL. 2007. Role of singlet oxygen in chloroplast to nucleus retrograde signaling in Chlamydomonas reinhardtii . FEBS Letters 581, 5555–5560 [DOI] [PubMed] [Google Scholar]
- Gadjev I, Vanderauwera S, Gechev TS, Laloi C, Minkov IN, Shulaev V, Apel K, Inze D, Mittler R, van Breusegem F. 2006. Transcriptomic footprints disclose specificity of reactive oxygen species signaling in arabidopsis. Plant Physiology 141, 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gálvez-Valdivieso G, Mullineaux PM. 2010. The role of reactive oxygen species in signalling from chloroplasts to the nucleus. Physiologia Plantarum 138, 430–439 [DOI] [PubMed] [Google Scholar]
- González-Pérez S, Gutiérrez J, García-García F, Osuna D, Dopazo J, Lorenzo Ó, Revuelta JL, Arellano JB. 2011. Early transcriptional defense responses in arabidopsis cell suspension culture under high light conditions. Plant Physiology 156, 1439–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez J, González-Pérez S, García-García F, Lorenzo Ó, Arellano JB. 2011. Does singlet oxygen activate cell death in Arabidopsis cell suspension cultures? Analysis of the early transcriptional defense responses to high light stress. Plant Signaling and Behavior 6, 1937–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hideg E, Ayaydin F, Tandori J, Kalai T, Kovacs L. 2011. Effects of singlet oxygen elicitors on plant leaves. Are these all caused by ROS? European Biophysics Journal 40, 176–176 [Google Scholar]
- Hideg E, Jansen MAK, Strid Å. 2013. UV-B exposure, ROS, and stress: inseparable companions or loosely linked associates? Trends in Plant Science 18, 107–115 [DOI] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmerman P. 2008. Genevestigator V3: a reference expression database for the meta-analysis of transcriptiones. Advances in Bioinformatics 2008, 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RA, Wichern DW. 1998. Applied multivariate statistical analysis, 2nd edn Upper Saddle River, NJ: Prentice Hall [Google Scholar]
- Jouanneau JP, Péaud-Lenoël C. 1967. Growth and synthesis of proteins in cell suspensions of a kinetin dependent tobacco. Physiologia Plantarum 20, 834–850 [Google Scholar]
- Kacprzyk J, Daly CT, McCabe PF. 2011. The botanical dance of death: programmed cell death in plants. In: Kader JC, Delseny M, eds. Advances in botanical research. Amsterdam: Elsevier Ltd, 169–261 [Google Scholar]
- Karpinski S, Gabrys H, Mateo A, Karpinska B, Mullineaux PM. 2003. Light perception in plant disease defence signalling. Current Opinion in Plant Biology 6, 390–396 [DOI] [PubMed] [Google Scholar]
- Kauss D, Bischof S, Steiner S, Apel K, Meskauskiene R. 2012. FLU, a negative feedback regulator of tetrapyrrole biosynthesis, is physically linked to the final steps of the Mg++-branch of this pathway. FEBS Letters 586, 211–216 [DOI] [PubMed] [Google Scholar]
- Kim C, Apel K. 2013. Singlet oxygen-mediated signalling in plants: moving from flu to wild type reveals an increasing complexity. Photosynthesis Research 116, 455–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CH, Meskauskiene R, Apel K, Laloi C. 2008. No single way to understand singlet oxygen signalling in plants. EMBO Reports 9, 435–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Meskauskiene R, Zhang SR, Lee KP, Ashok ML, Blajecka K, Herrfurth C, Feussner I, Apel K. 2012. Chloroplasts of arabidopsis are the source and a primary target of a plant-specific programmed cell death signaling pathway. The Plant Cell 24, 3026–3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T, Kindgren P, Benedict C, Hendrickson L, Strand A. 2007. Genome-wide gene expression analysis reveals a critical role for CRYPTOCHROME1 in the response of arabidopsis to high irradiance. Plant Physiology 144, 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T, Voigt C, Leister D. 2009. Plastid signalling to the nucleus: messengers still lost in the mists? Trends in Genetics 25, 185–190 [DOI] [PubMed] [Google Scholar]
- Kóvacs L, Ayaydin F, Kálai T, Tandori J, Kós PB, Hideg E. 2013. Assessing the applicability of singlet oxygen photosensitizers in leaf studies. Photochemistry and Photobiology (in press) [DOI] [PubMed] [Google Scholar]
- Laloi C, Stachowiak M, Pers-Kamczyc E, Warzych E, Murgia I, Apel K. 2007. Cross-talk between singlet oxygen- and hydrogen peroxide-dependent signaling of stress responses in Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 104, 672–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlois-Meurinne M, Gachon CMM, Saindrenan P. 2005. Pathogen-responsive expression of glycosyltransferase genes UGT73B3 and UGT73B5 is necessary for resistance to Pseudomonas syringae pv tomato in Arabidopsis. Plant Physiology 139, 1890–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledford HK, Chin BL, Niyogi KK. 2007. Acclimation to singlet oxygen stress in Chlamydomonas reinhardtii . Eukaryotic Cell 6, 919–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KP, Kim C, Landgraf F, Apel K. 2007. EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 104, 10270–10275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leisinger U, Rufenacht K, Fischer B, Pesaro M, Spengler A, Zehnder AJB, Eggen RIL. 2001. The glutathione peroxidase homologous gene from Chlamydomonas reinhardtii is transcriptionally up-regulated by singlet oxygen. Plant Molecular Biology 46, 395–408 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- Lupinkova L, Komenda J. 2004. Oxidative modifications of the photosystem II D1 protein by reactive oxygen species: from isolated protein to cyanobacterial cells. Photochemistry and Photobiology 79, 152–162 [DOI] [PubMed] [Google Scholar]
- McCabe PF, Leaver CJ. 2000. Programmed cell death in cell cultures. Plant Molecular Biology 44, 359–368 [DOI] [PubMed] [Google Scholar]
- Medina I, Carbonell J, Pulido L, et al. 2010. Babelomics: an integrative platform for the analysis of transcriptomics, proteomics and genomic data with advanced functional profiling. Nucleic Acids Research 38, W210–W213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meskauskiene R, Nater M, Goslings D, Kessler F, op den Camp RG, Apel K. 2001. FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana . Proceedings of the National Academy of Sciences, USA 98, 12826–12831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyao M, Ikeuchi M, Yamamoto N, Ono T. 1995. Specific degradation of the D1 protein of photosystem II by treatment with hydrogen peroxide in darkness: implications for the mechanism of degradation of the D1 protein under illumination. Biochemistry 34, 10019–10026 [DOI] [PubMed] [Google Scholar]
- Morrison H, Mohammad T, Kurukulasuriya R. 1997. Photobiological properties of methylene violet. Photochemistry and Photobiology 66, 245–252 [DOI] [PubMed] [Google Scholar]
- Mullineaux PM, Baker NR. 2010. Oxidative stress: antagonistic signaling for acclimation or cell death? Plant Physiology 154, 521–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano R, Ishida H, Makino A, Mae T. 2006. In vivo fragmentation of the large subunit of ribulose-1,5-bisphosphate carboxylase by reactive oxygen species in an intact leaf of cucumber under chilling-light conditions. Plant and Cell Physiology 47, 270–276 [DOI] [PubMed] [Google Scholar]
- Ochsenbein C, Przybyla D, Danon A, Landgraf F, Gobel C, Imboden A, Feussner I, Apel K. 2006. The role of EDS1 (enhanced disease susceptibility) during singlet oxygen-mediated stress responses of Arabidopsis. The Plant Journal 47, 445–456 [DOI] [PubMed] [Google Scholar]
- op den Camp RG, Przybyla D, Ochsenbein C, et al. 2003. Rapid induction of distinct stress responses after the release of singlet oxygen in arabidopsis. The Plant Cell 15, 2320–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov V, Vermeirssen V, De Clercq I, Van Breusegem F, Minkov I, Vandepoele K, Gechev TS. 2012. Identification of cis-regulatory elements specific for different types of reactive oxygen species in Arabidopsis thaliana . Gene 499, 52–60 [DOI] [PubMed] [Google Scholar]
- Quinn GP, Keough MJ. 2002. Experimental design and data analysis for biologists. Cambridge: Cambridge University Press [Google Scholar]
- Ramel F, Birtic S, Cuine S, Triantaphylidès C, Ravanat J-L, Havaux M. 2012a. Chemical quenching of singlet oxygen by carotenoids in plants. Plant Physiology 158, 1267–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel F, Birtic S, Ginies C, Soubigou-Taconnat L, Triantaphylides C, Havaux M. 2012b. Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proceedings of the National Academy of Sciences, USA 109, 5535–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel F, Ksas B, Akkari E, Mialoundama AS, Monnet F, Krieger-Liszkay A, Ravanat JL, Mueller MJ, Bouvier F, Havaux M. 2013. Light-induced acclimation of the arabidopsis chlorina1 mutant to singlet oxygen. The Plant Cell 25, 1445–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel F, Sulmon C, Serra AA, Gouesbet G, Couee I. 2012. Xenobiotic sensing and signalling in higher plants. Journal of Experimental Botany 63, 3999–4014 [DOI] [PubMed] [Google Scholar]
- Reape TJ, Molony EM, McCabe PF. 2008. Programmed cell death in plants: distinguishing between different modes. Journal of Experimental Botany 59, 435–444 [DOI] [PubMed] [Google Scholar]
- Roberts EL, Burguieres S, Warner IM. 1998. Spectroscopic studies of indigo carmine dye in organized media. Applied Spectroscopy 52, 1305–1313 [Google Scholar]
- Rossel JB, Wilson PB, Hussain D, Woo NS, Gordon MJ, Mewett OP, Howell KA, Whelan J, Kazan K, Pogson BJ. 2007. Systemic and intracellular responses to photooxidative stress in Arabidopsis . The Plant Cell 19, 4091–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology 3, article 3. [DOI] [PubMed] [Google Scholar]
- Triantaphylidès C, Havaux M. 2009. Singlet oxygen in plants: production, detoxification and signaling. Trends in Plant Science 14, 219–228 [DOI] [PubMed] [Google Scholar]
- Triantaphylidès C, Krischke M, Hoeberichts FA, Ksas B, Gresser G, Havaux M, Van Breusegem F, Mueller MJ. 2008. Singlet oxygen is the major reactive oxygen species involved in photooxidative damage to plants. Plant Physiology 148, 960–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm R, Baumann A, Oravecz A, Mate Z, Adam E, Oakeley EJ, Schafer E, Nagy F. 2004. Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis . Proceedings of the National Academy of Sciences, USA 101, 1397–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenabeele S, Vanderauwera S, Vuylsteke M, Rombauts S, Langebartels C, Seidlitz HK, Zabeau M, Van Montagu M, Inze D, Van Breusegem F. 2004. Catalase deficiency drastically affects gene expression induced by high light in Arabidopsis thaliana . The Plant Journal 39, 45–58 [DOI] [PubMed] [Google Scholar]
- Vanderauwera S, Zimmermann P, Rombauts S, Vandenabeele S, Langebartels C, Gruissem W, Inze D, van Breusegem F. 2005. Genome-wide analysis of hydrogen peroxide-regulated gene expression in Arabidopsis reveals a high light-induced transcriptional cluster involved in anthocyanin biosynthesis. Plant Physiology 139, 806–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters RG, Ibrahim DG, Horton P, Kruger NJ. 2004. A mutant of Arabidopsis lacking the triose-phosphate/phosphate translocator reveals metabolic regulation of starch breakdown in the light. Plant Physiology 135, 891–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.