Summary
We find that CsGAMYB1, a positive regulator of GA signalling, can regulate sex expression of cucumber. This provides a new insight into the mechanism of GA in sex determination.
Key words: CsGAMYB1, cucumber, ethylene, GAMYB, gibberellin, sex expression.
Abstract
Cucumber (Cucumis sativus L.) is a typical monoecious vegetable with individual male and female flowers, and has been used as a model plant for sex determination. It is well known that sex differentiation of cucumber can be regulated by phytohormones, such as gibberellic acid (GA) and ethylene. The molecular mechanism of female sex expression modulated by ethylene has been widely understood, but how GA controls male sex expression remains elusive. In hermaphroditic Arabidopsis and rice, GA can regulate stamen and anther development via the transcriptional regulation of GAMYB. Here we characterized a GAMYB homologue CsGAMYB1 in cucumber. We found that CsGAMYB1 is predominantly expressed in male flower buds, where its expression is upregulated by GA3 treatment. CsGAMYB1 protein is localized in the nucleus. CsGAMYB1 can partially rescue stamen development and fertility phenotypes of an Arabidopsis myb33 myb65 double mutant. However, constitutive overexpression of CsGAMYB1 in wild-type Arabidopsis resulted in male sterility, which mimics the effect of GA overdose in flower development. Knockdown of CsGAMYB1 in cucumber decreases the ratio of nodes with male and female flowers, and ethylene is not involved in this process. Our data suggest that CsGAMYB1 regulates sex expression of cucumber via an ethylene-independent pathway.
Introduction
Gibberellins (GAs), one kind of endogenous growth regulator, play an essential role in reproductive development of plants, especially in staminate development (Aya et al., 2009; Bai and Xu, 2013; King and Evans, 2003; Pharis and King, 1985; Plackett et al., 2011; Song et al., 2013). For example, GA application can promote development of male flowers in cucumber (Cucumis sativus L.) (Pike and Peterson, 1969; Wittwer and Bukovac, 1962). Additionally, GA-deficient mutants of Arabidopsis and tomato (Solanum lycopersicum) display abnormal stamen, and anther and pollen development leading to male sterility (Cheng et al., 2004; Goto and Pharis, 1999; Jacobsen and Olszewski, 1991; Koornneef and van der Veen, 1980; Nester and Zeevaart, 1988).
Recently, several studies have demonstrated that GA regulates staminate development via the GA signalling pathway (Aya et al., 2009; Cheng et al., 2004; Fleet and Sun, 2005; Sun, 2010; Sun, 2011). In this pathway, GA first binds the GID1 receptor and promotes binding of DELLA proteins (repressors of GA action, and plant growth and development) to GID1; this GA–GID1–DELLA complex then enables the rapid degradation of DELLA proteins by the proteasome, which releases the inhibitory effect of the DELLA proteins and allows GA action to occur (Fleet and Sun, 2005; Harberd et al., 2009; Murase et al., 2008; Plackett et al., 2014; Sun, 2010; Sun, 2011; Ueguchi-Tanaka et al., 2007). GAMYB, a positive regulator involved in the GA signalling pathway, has been known to act as an important downstream component in the degradation of DELLA proteins (Achard et al., 2004; Fleet and Sun, 2005; Olszewski et al., 2002). GAMYB was first identified in barley (Hordeum vulgare) aleurone cells, where its expression is upregulated by GA treatment (Gubler et al., 1995). HvGAMYB can bind specifically to GA-response elements in promoter regions of an α-amylase gene and other GA-regulated genes encoding hydrolytic enzymes, and constitutive expression of HvGAMYB mimics the positive effects of exogenous GA application on the transcriptional activation of these genes (Cercos et al., 1999; Gubler et al., 1999). GAMYB has also been demonstrated to play an important role in flower development, especially in anther development. For example, HvGAMYB shows high expression levels in barley anthers, where it is also upregulated by GA3. Overexpression of HvGAMYB in barley results in decreased anther length and male sterility (Murray et al., 2003), which phenocopies those plants treated with excessive exogenous GA (Colombo and Favret, 1996). In rice, loss-of-function mutations of GAMYB lead to abnormal anther and pollen development (Kaneko et al., 2004; Liu et al., 2010). In addition, OsGAMYB is also involved in GA-mediated programmed cell death (PCD) of tapetal cells, exine, and Ubisch body formation, and microarray analysis revealed that OsGAMYB can modulate most GA-regulated gene expression in rice anthers (Aya et al., 2009).
In Arabidopsis, there is a small family of GAMYB-like genes (Stracke et al., 2001), in which AtMYB33, AtMYB65, and AtMYB101 were identified to be able to substitute for barley and rice GAMYB in transactivating the α-amylase promoter (Gocal et al., 2001). Expression pattern analysis found that AtMYB33, AtMYB65, and AtMYB101 have a predominant expression in floral shoot apices and flowers, and the expression of AtMYB33 can be induced by exogenous GA (Achard et al., 2004; Gocal et al., 2001; Millar and Gubler, 2005). To further understand the function of AtGAMYBs, the deficient mutants for AtMYB33 and AtMYB65 were isolated, and the double mutant myb33 myb65 displays the phenotypes of shorter filaments, pollen abortion, and male sterility (Millar and Gubler, 2005). Furthermore, neither myb33 nor myb65 showed an abnormal phenotype compared with wild-type plants, suggesting that AtMYB33 and AtMYB65 are functionally redundant (Millar and Gubler, 2005). Taken together, these observations suggest that GAMYB is involved in GA-regulated stamen and anther development.
Cucumber is a typical monoecious vegetable with unisexual flowers, and has been served as a model plant for sex determination and differentiation (Malepszy and Niemirowicz-Szczytt, 1991). In young floral buds of cucumber, both stamen primordia and carpel primordia are initiated, and sex determination occurs just after the bisexual stage; subsequently, male or female flowers are formed and enlarged owing to the selective arrestment of carpel or stamen development, respectively (Bai et al., 2004; Malepszy and Niemirowicz-Szczytt, 1991). In this process, ethylene treatment can produce increased numbers of female flowers in cucumber (Iwahori et al., 1969; MacMurray and Miller, 1968), and the mechanism has been widely understood. Two major genes encoding ACC synthase (a key enzyme of ethylene biosynthesis), F (CsACS1G), and M (CsACS2) control female sex expression in cucumber; the F gene governs the development of female flowers (Knopf and Trebitsh, 2006; Mibus and Tatlioglu, 2004; Trebitsh et al., 1997), whereas the M gene inhibits stamen development in flower buds (Bie et al., 2013; Li et al., 2012; Saito et al., 2007; Wang et al., 2010; Yamasaki et al., 2001; Yamazaki et al., 2003). In addition, GA application can promote the male tendency (Pike and Peterson, 1969), but the molecular regulation remains elusive. Previous studies have confirmed that GA can modulate stamen and anther development via the transcriptional regulation of GAMYB in hermaphroditic plants, such as Arabidopsis and rice. However, in monoecious species cucumber whether GAMYB is involved in GA-regulated male tendency in the process of sex differentiation or not is still unknown. Therefore, in this study, a GAMYB orthologous gene in cucumber, designated as CsGAMYB1, was identified, and its spatial and temporal expression patterns were characterized. CsGAMYB1 is predominantly expressed in male flower buds, where its expression is upregulated by exogenous GA3 application, and CsGAMYB1 protein is localized in the nucleus. Ectopic expression of CsGAMYB1 can partially rescue the phenotypes of myb33 myb65 double mutant in Arabidopsis; however, constitutive overexpression of CsGAMYB1 in wild type resulted in male sterility. Furthermore, we generated CsGAMYB1-RNAi transgenic plants in cucumber and found that reduced transcript levels of CsGAMYB1 can result in decreased ratio of nodes with male and female flowers, but no effect on ethylene production and expression of F and M genes. Our results indicate that CsGAMYB1 can regulate ethylene-independent sex expression of cucumber.
Materials and methods
Plant materials and growth conditions
Monoecious cucumber (Cucumis sativus L.) line 3407 was used in this study. The seeds were germinated on wet filter paper in a Petri dish at 28 °C in dark overnight. Then the resulting seedlings were grown in a growth chamber under 16h/8h with 25 °C/18 °C in day/night. Upon the two true-leaf stage, plants were transferred to a greenhouse. The Arabidopsis mutant myb33 myb65 (Columbia background) was provided by Millar’s lab (Millar and Gubler, 2005), and Columbia (Col) was used as a wild-type control. Arabidopsis seeds were germinated on Murashige-Skoog (MS) medium, which contains 1% sucrose and 0.2% phytagar, at 4 °C for 3 d and then moved to 22 °C under a regime of 16h light/8h dark. Seedlings were transferred to soil 7–10 d after germination. For GA3 treatment, male flower buds of cucumber were sprayed with 200 μm GA3 (and mock-sprayed with 0.1% ethanol). Expression analyses were done after 4h of treatment.
Cloning of CsGAMYB1
Total RNA was extracted from cucumber leaves using Promega’s SV Total RNA Isolation System, and cDNA was synthesized using MultiScribeTM reverse transcriptase (Applied Biosystems). The cDNA samples were amplified by PCR: 95 °C for 5min, 30 cycles of 95 °C for 30s, 52 °C for 30s, and 72 °C for 2.5min, and then 72 °C for 10min. The primers are listed in supplementary material Table S3 available at JXB online.
Sequence alignment and phylogenetic analysis
Through BLAST analysis in Phytozome (http://www.phytozome.net/search.php) or the Arabidopsis Information Resource (http://www.Arabidopsis.org) using the sequence information of CsGAMYB1 protein, the amino acid sequence of related GAMYB proteins in various species were obtained. The multiple sequence alignment of CsGAMYB1 and related GAMYB proteins was performed using ClustalW in the MEGA5 software package, and the boxes were drawn using the BoxShade web site (http://www.ch.embnet.org/software/BOX_form.html). The phylogenetic tree was constructed using the Neighbor–Joining (NJ) method (Saitou and Nei, 1987) with Poisson model and 1000 bootstrap replicates test through MEGA5 software.
Expression analysis by qRT-PCR
Total RNA was extracted using Promega’s SV Total RNA Isolation System, and cDNA was synthesized using MultiScribeTM reverse transcriptase (Applied Biosystems). Quantitative real-time RT-PCR (qRT-PCR) was performed using SYBR® Premix Ex Taq TM from TaKaRa (China) on an Applied Biosystems 7500 real-time PCR system. The cucumber α-TUBULIN (TUA) and Arabidopsis actin2 were used as internal controls in analysing gene expression in cucumber and Arabidopsis, respectively. And three biological replicates were performed for these experiments. The gene specific primers for qRT-PCR are listed in Supplementary Table S3 available at JXB online.
In situ hybridization
Shoot apex of 10-day-old seedlings, and male and female flower buds from 45-day-old cucumbers grown in the greenhouse were fixed, embedded, sectioned, and hybridized as described (Zhang et al., 2013). Digoxigenin-labelled sense and antisense RNA probes were generated using SP6 and T7 RNA polymerase (Roche) through PCR amplification respectively. The primer pairs are listed in Supplementary Table S3 available at JXB online.
Subcellular localization in onion epidermal cells
For transient expression in onion epidermal cells, the full-length coding region of CsGAMYB1 was cloned and fused to the upstream of the green fluorescent protein (GFP) between the EcoRI and BamHI sites in the pEZS-NL vector (http://deepgreen.stanford.edu) to generate 35S:GFP–CsGAMYB1; the empty pEZS-NL vector was used a control. The onion epidermal layers were prepared and bombarded, as previously described (Varagona et al., 1992), with gold particles containing the plasmid using a Bio-Rad PDS-1000/He particle delivery system. After bombardment, the onion epidermises were placed on MS medium and incubated in darkness at 22 °C for 24h. Fluorescence signals were detected using Olympus BX 51 fluorescence microscopy. The primer sequences used for vector construction are listed in Supplementary Table S3 available at JXB online.
Transformation of Arabidopsis
To make the CsGAMYB1 overexpression construct, full-length CsGAMYB1 cDNA was cloned and inserted into the pCAMBIA1305.1 vector between the BglII and SpeI sites. The construct was then introduced into Agrobacterium by electroporation and transformed into Col or myb33 myb65 plants as described (Clough and Bent, 1998). The transgenic plants were screened on MS medium with 25mg l–1 hygromycin. The primers for overexpression construct are listed in Supplementary Table S3 available at JXB online.
Transformation of cucumber
To generate the CsGAMYB1-RNAi transgenic plants of cucumber, two fragments of CsGAMYB1 were amplified using specific primers containing AscI (5’ end) and SwaI (3’ end) sites, and SpeI (5’ end) and BamHI (3’ end) sites. The two fragments were inversely inserted into the pFGC1008 vector, and the resulting CsGAMYB1-RNAi construct was then introduced into Agrobacterium by electroporation and transformed into the monoecious cucumber line 3407 using the cotyledon transformation method as described previously (Wang et al., 2014). The primers for RNAi construct are listed in Supplementary Table S3 available at JXB online.
Quantification of ethylene
To examine the ethylene production from cucumber, shoot apices were excised at the 4-leaf stage. The samples were enclosed in a 10ml vessel after weighing, and sealed with a rubber stopper. After incubation at 25 °C for 0.5h, 1ml of gas was extracted using a syringe and injected into a gas chromatograph (GC-9A, Shimadzu, Japan). Ethylene was quantified using an activated alumina column and hydrogen flame ionization detector (FID). Standard ethylene gas was used for calibrating the instrument.
Results
Identification of the CsGAMYB1 gene from cucumber
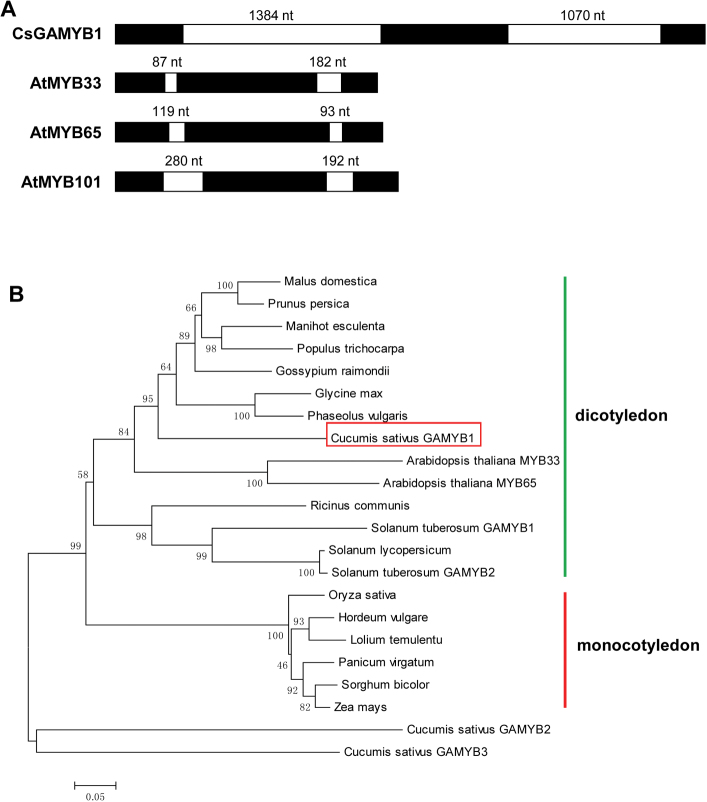
Through BLAST analysis in the Cucumber Genome Database (Huang et al., 2009), we discovered three GAMYB-like genes named as CsGAMYB1 (Csa009014), CsGAMYB2 (Csa019830), and CsGAMYB3 (Csa013555). The CsGAMYB1 gene shows the highest similarity compared with other GAMYB orthologues, so CsGAMYB1 was chosen and analysed in this study. CsGAMYB1 was cloned using cDNA derived from cucumber leaves. Consistent with three GAMYB orthologues in Arabidopsis (Achard et al., 2004; Gocal et al., 2001; Millar and Gubler, 2005), CsGAMYB1 also contains three exons and two introns (Fig. 1A), encoding 552 amino acids. The sequence alignment of the amino acid residues of CsGAMYB1 compared with other members of the GAMYB family was performed using ClustalW in the MEGA5 software package (Tamura et al., 2011). CsGAMYB1, HvGAMYB, AtMYB33, and AtMYB65 share an R2R3 repeat DNA-binding domain in their N-terminal regions (Kranz et al., 1998; Romero et al., 1998; Stracke et al., 2001); over this sequence, CsGAMYB1 shows high identity to HvGAMYB, AtMYB33, and AtMYB65, with 88.46%, 87.5%, and 87.5% identity, respectively. In addition, these proteins also contain three conserved motifs Box 1, Box 2, and Box 3, which are typical structures in the GAMYB family (Supplementary Fig. S1 available at JXB online) (Gocal et al., 2001).
Fig. 1.
Structural and phylogenetic analyses of GAMYB homologues in various species. (A) Structural analysis of GAMYB genes in cucumber and Arabidopsis. Exons and introns are shown in black and white, respectively. Cs, Cucumis sativus; At, Arabidopsis thaliana. (B) Phylogenetic analysis of GAMYB homologues in various species. This phylogenetic tree was constructed using the Neighbor–Joining (NJ) method through MEGA5 software. Eighteen species were used for this analysis and formed two main groups: dicotyledon group and monocotyledon group. A GAMYB homologue from cucumber is indicated in the box. Gene ID for each of the GAMYB protein used for this analysis is listed in the “accession numbers”. (This figure is available in colour at JXB online.)
To further understand the evolutionary relationship between CsGAMYB1 and other GAMYB homologues, phylogenetic analysis was performed using the Neighbor–Joining (NJ) method (Saitou and Nei, 1987) (Fig. 1B). CsGAMYB1 (shown in the box in Fig. 1B) is placed in the same clade as other GAMYB proteins, whereas CsGAMYB2 and CsGAMYB3 are distinct from this clade, suggesting that CsGAMYB1, but not CsGAMYB2 and CsGAMYB3, belongs to the GAMYB family in cucumber. A phylogenetic tree of the GAMYB family can be divided into two main groups: dicotyledon and monocotyledon. Within the dicotyledon group, GAMYB proteins in cucumber, which belongs to the cucurbitaceae family; Arabidopsis of the cruciferae family; kidney bean (Phaseolus vulgaris) and soybean (Glycine max) of the leguminosae family; and several woody plants such as apple (Malus domestica), cassava (Manihot esculenta), cotton (Gossypium raimondii), Prunus persica, and Populus trichocarpa all fall into the same clade, indicating that these plants may share a common origin. However, CsGAMYB1 is placed in a distinct group with GAMYB homologues in hermaphroditic species such as Arabidopsis, tomato, potato (Solanum tuberosum), kidney bean, and soybean, and it is also different from GAMYB groups in other monoecious plants such as cassava, Populus trichocarpa, and castor bean (Ricinus communis). These observations demonstrated that CsGAMYB1 is a GAMYB homologue in cucumber.
Expression pattern of CsGAMYB1 in cucumber
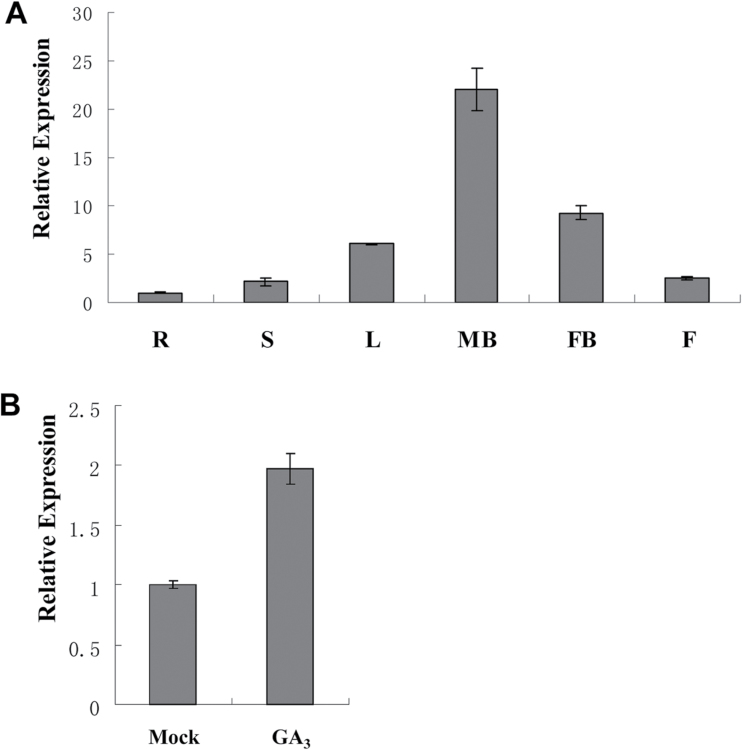
To obtain insights into the biological function of CsGAMYB1, we investigated its spatial and temporal expression patterns in cucumber. Quantitative real-time RT-PCR (qRT-PCR) was performed in various cucumber tissues including roots, stems, leaves, male flower buds, female flower buds, and fruits. CsGAMYB1 was expressed in all examined tissues, and the highest expression was detected in male flower buds (Fig. 2A), where its expression was increased almost two-fold by GA3 application (Fig. 2B). These data suggested that CsGAMYB1 may play an important role in cucumber male flower development.
Fig. 2.
qRT-PCR analyses of CsGAMYB1 in cucumber. (A) Tissue distribution of CsGAMYB1 in cucumber. R, root; S, stem; L, leaves; MB, male flower buds; FB, female flower buds; F, fruit. (B) Effect of GA3 on the expression of CsGAMYB1 in male flower buds. Male flower buds were mock-treated with 0.1% ethanol, or treated with 200 μM GA3 for 4h. The cucumber α-TUBULIN (TUA) was used as an internal control, and three biological replicates were performed for these experiments. Error bars indicate the standard errors. (This figure is available in colour at JXB online.)
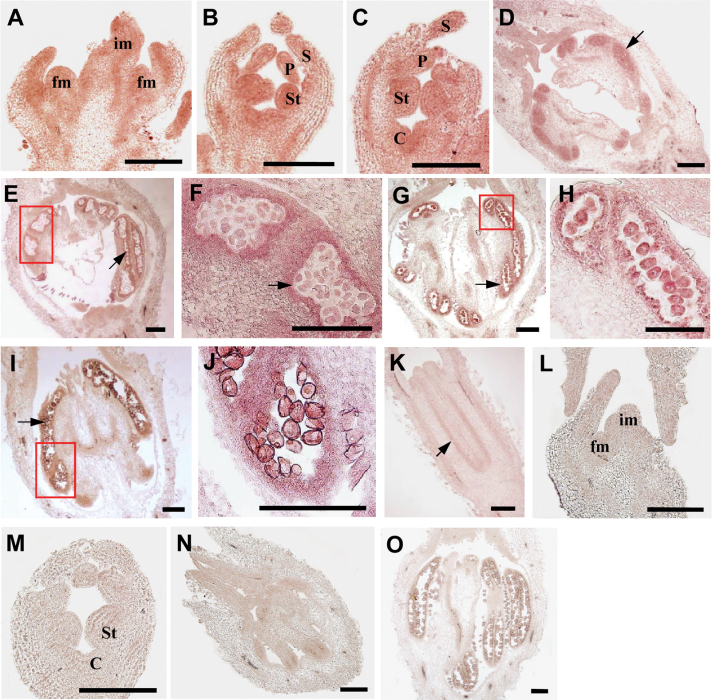
Further, we examined the detailed expression patterns of CsGAMYB1 during cucumber flower development by in situ hybridization (Fig. 3). CsGAMYB1 RNA was found throughout the inflorescence meristem (im) and floral meristem (fm) in stage 1 of cucumber flower development (Bai et al., 2004) (Fig. 3A). When flowers developed into the bisexual stages 4–5, the crucial periods that stamen primordia and carpel primordia initiate, higher expressions were detected in sepal primordia, petal primordia, stamen primordia, and carpel primordia (Fig. 3B, C). For male flowers, during the stages of microsporocytes (stage 9), meiosis (stage 10), uninuclear pollens (stage 11), and mature pollens (stage 12), CsGAMYB1 was predominately expressed in the microsporocytes (Fig. 3D), anther wall, and pollen grains (Fig. 3E–J). For female flowers, the expression of CsGAMYB1 was detected in the developing ovary of stage 8 (Fig. 3K), but the signal was weak. As negative controls, CsGAMYB1 sense probe hybridization showed no signals in the male flowers of stage 1, stage 5, stage 9, and stage 12 (Fig. 3L–O).
Fig. 3.
In situ hybridization of CsGAMYB1 during flower development in cucumber. Longitudinal sections of shoot apex (A and L, stage 1), male flower buds at stage 4 (B), stage 5 (C and M), stage 9 (D and N), stage 10 (E), stage 11 (G) and stage 12 (I and O), and female flower buds at stage 8 (K). The morphologies of tetrad in meiosis and pollens in the framed regions of E, G, and I are shown in F, H, and J, respectively. CsGAMYB1 sense probe hybridizations showed no signals in L–O. The arrows indicate the expression of CsGAMYB1 in microsporocytes, anther wall, pollen grains or ovary. im, inflorescence meristem; fm, floral meristem; S, sepal; P, petal; St, stamen; C, carpel. Bar=200 μm. (This figure is available in colour at JXB online.)
Subcellular localization of CsGAMYB1 protein
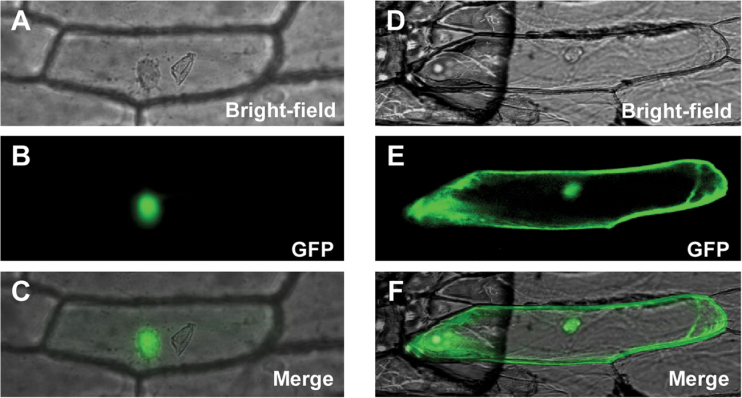
To further determine the subcellular localization of CsGAMYB1 protein, the GFP–CsGAMYB1 fusion protein was constructed under the control of the cauliflower mosaic virus (CaMV) 35S promoter and introduced into onion epidermal cells by particle bombardment. As shown in Fig. 4, the GFP–CsGAMYB1 fusion protein is localized in the nucleus (Fig. 4A–C). As a control, the signals of 35S:GFP are detected throughout the cell (Fig. 4D–F).
Fig. 4.
Subcellular localization of CsGAMYB1 protein in onion epidermal cells. 35S:GFP–CsGAMYB1 localized to the nucleus (A–C), whereas 35S:GFP localized throughout the cell (D–F). (This figure is available in colour at JXB online.)
CsGAMYB1 can partially rescue myb33 myb65 double mutant phenotypes in Arabidopsis
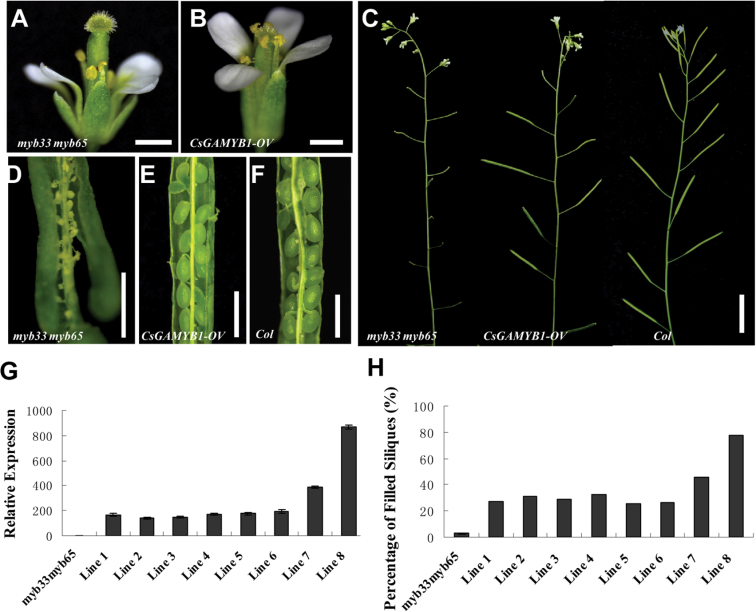
To investigate the biological role of CsGAMYB1, we ectopically expressed the full-length CsGAMYB1 cDNA under the control of a 35S promoter in an Arabidopsis myb33 myb65 double mutant, which displayed shorter filaments, pollen abortion, and male sterility (Millar and Gubler, 2005). A total of 23 independent transgenic lines were obtained, all of which could partially rescue the phenotypes of the myb33 myb65 double mutant, and all of which displayed similar phenotypes. As shown in Fig. 5, flowers in the transgenic plants had increased filaments length and pollen numbers as compared with those in the myb33 myb65 double mutant (Fig 5A, B). Consequently, fertility increased in the CsGAMYB1 transgenic plants (Fig 5C–F). For example, the myb33 myb65 plants displayed much smaller siliques (Fig 5C), which failed to set any seeds (Fig 5D), whereas ectopic expression of CsGAMYB1 resulted in normal siliques (Fig 5C), which set seeds with similar shape and numbers, compared with those in wild-type plants (Fig 5E, F). We chose eight transgenic lines to analyse expression of CsGAMYB1 and plant fertility. The fertility is defined as the percentage of siliques that set seeds per plant. The expression of CsGAMYB1 displayed different levels in these lines (Fig 5G), although the lines with the higher CsGAMYB1 mRNA levels showed the higher increase in fertility compared with the myb33 myb65 plants (Fig 5H). For example, the average fertility of myb33 myb65 plants was only 2.8%, whereas in the six lines (lines 1, 2, 3, 4, 5, and 6) which had lower CsGAMYB1 mRNA levels, the average fertility increased to 28.6%. However, in line 7, which had higher CsGAMYB1 mRNA expression, the fertility reached 46.0%, and the highest CsGAMYB1 mRNA level in line 8 resulted in 77.3% fertility, which was close to that in wild-type (92.2% fertility; Supplementary Tables S1 and S2 available at JXB online), suggesting that CsGAMYB1 can partially rescue the fertility of the myb33 myb65 double mutant, but the recovery relies on the levels of CsGAMYB1 expression. These results implied that the function of CsGAMYB1 in cucumber, which has unisexual flowers, is conserved with AtMYB33 and AtMYB65 in Arabidopsis, which has complete flowers with respect to stamen development and plant fertility.
Fig. 5.
Partial rescue of myb33 myb65 mutant by ectopic expression of CsGAMYB1 in Arabidopsis. (A, B) Flowers of myb33 myb65 (A) or CsGAMYB1 overexpression (B). (C) Inflorescences of myb33 myb65 (left), CsGAMYB1 overexpression (middle) or Col (right). (D–F) Opened siliques of myb33 myb65 (D), CsGAMYB1 overexpression (E) or Col (F). Bar=1mm, except C, in which the bar=1cm. (G) qRT-PCR analyses of CsGAMYB1 in eight selected transgenic lines in the myb33 myb65 background. The Arabidopsis actin2 was used as an internal control, and the experiments were repeated in three independent samples. Error bars indicate the standard errors. (H) Fertility in myb33 myb65 and 8 selected CsGAMYB1 transgenic lines. The fertility is defined as the percentage of siliques that set seeds per plant. Error bars indicate the standard errors. (This figure is available in colour at JXB online.)
Constitutive overexpression of CsGAMYB1 results in male sterility in Arabidopsis
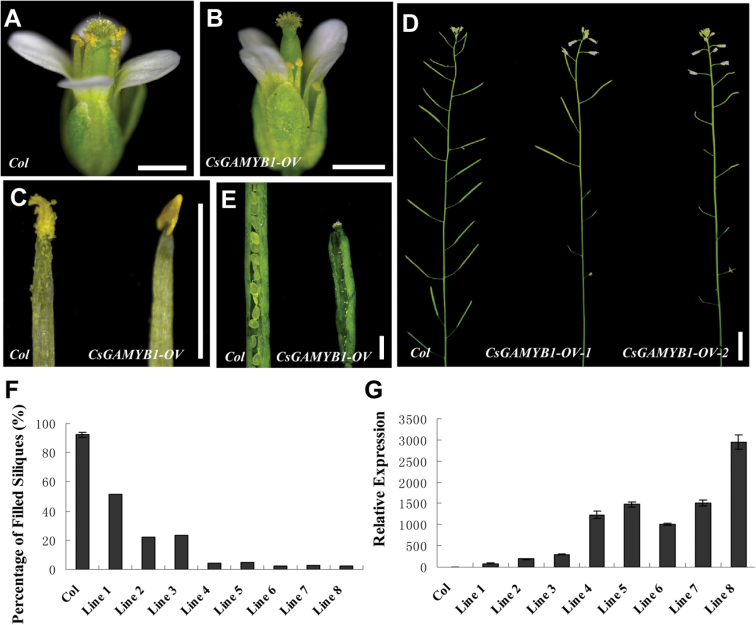
To further explore the function of CsGAMYB1, we also generated transgenic lines overexpressing CsGAMYB1 in Arabidopsis wild-type Columbia (Col); a total of 24 independent transgenic lines were obtained. In the transgenic plants, stamens were shorter than those in wild type and failed to fully extend to the pistil (Fig. 6A, B), and anthers were smaller than their wild-type counterparts and failed to generate pollen (Fig. 6C), leading to much smaller siliques, which failed to set any seeds (Fig. 6D, E).
Fig. 6.
Male sterility in CsGAMYB1 transgenic plants with high levels of CsGAMYB1 in Arabidopsis. (A and B) Flowers of Col (A) or CsGAMYB1 overexpression (B). (C) Stamens in Col (left) or CsGAIP overexpression (right). (D) Inflorescences of Col (left) or lines of CsGAMYB1 overexpression (middle and right). (E) Opened siliques of Col (left) or CsGAMYB1 overexpression (right). Bar=1mm, except D, in which the bar=1cm. (F) Fertility in Col and 8 selected CsGAMYB1 transgenic lines. The fertility is defined as the percentage of siliques that set seeds per plant. Error bars indicate the standard errors. (G) qRT-PCR analyses of CsGAMYB1 in eight selected transgenic lines in the Col background. The Arabidopsis actin2 was used as an internal control, and these experiments were repeated in three independent samples. Error bars indicate the standard errors. (This figure is available in colour at JXB online.)
Among the 24 transgenic lines, 8 lines were chosen for further analysis. Of these lines, five lines (lines 4, 5, 6, 7 and 8) were sterile and three other lines (lines 1, 2 and 3) exhibited partial fertility. For example, in these five sterile lines, few siliques that set seeds were present, from which few seeds were obtained, and the average fertility was only 3.1%. However, there were some normal siliques in lines 1, 2, and 3, characterized by the similar seeds number per silique compared with that of wild-type, resulting in partial fertility, with 51.2%, 21.7%, and 23.4%, respectively (Fig. 6F, Supplementary Table S2 available at JXB online). The expression of CsGAMYB1 in these eight transgenic plants was also analysed (Fig. 6G). We found that CsGAMYB1 mRNA expressions in the five sterile lines were more than 3.4-fold higher than those in the three partial sterile plants. Line 8, which had only 1.9% fertility, displayed the highest level of CsGAMYB1 mRNA. In line 8, CsGAMYB1 mRNA level was more than 35-fold higher than that in line 1, in which the fertility reached 51.2%. This correlation between high levels of CsGAMYB1 mRNA and sterility in multiple transgenic lines suggests that constitutive overexpression of CsGAMYB1 results in male sterility in Arabidopsis in a dose-dependent manner.
CsGAMYB1 can regulate sex expression of cucumber via an ethylene-independent pathway
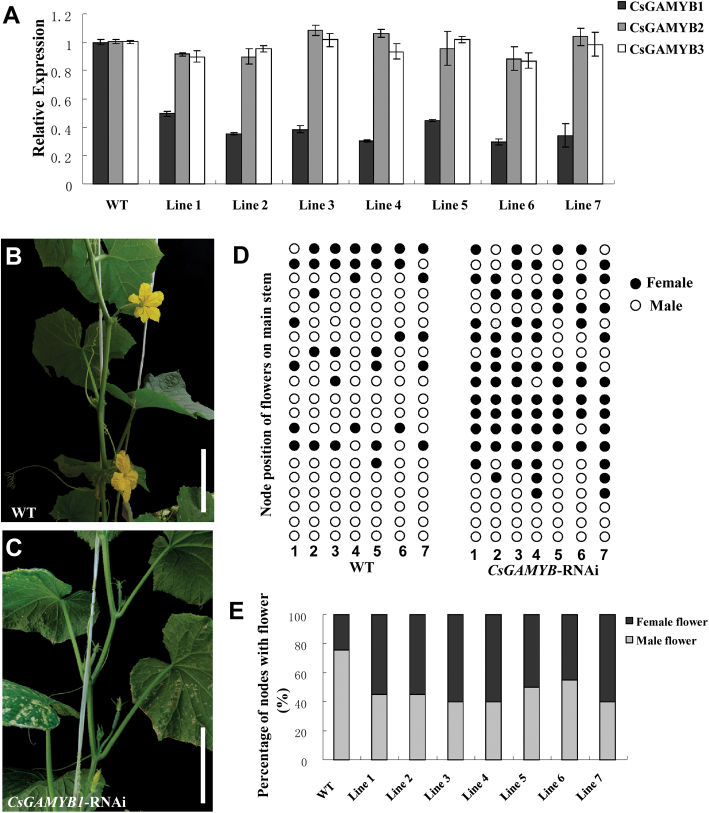
To further determine the biological function of CsGAMYB1 in cucumber, a double-strand RNAi construct containing the specific sequence of CsGAMYB1 under the control of 35S promoter was introduced into the monoecious cucumber plants. A total of seven independent RNAi lines were obtained. In these RNAi lines, the transcript levels of CsGAMYB1 were significantly reduced, whereas the expressions of the other two GAMYB-like genes, CsGAMYB2 and CsGAMYB3, had no change as compared with those in the wild-type plants (Fig. 7A). This suggests that CsGAMYB1 expression was effectively knocked down by RNAi, and this process had no effect on the expression of other GAMYB-like genes that show high sequence similarity with CsGAMYB1.
Fig. 7.
Sex expression of the flowers in the CsGAMYB1-RNAi plants of cucumber. (A) qRT-PCR analyses of CsGAMYB1, CsGAMYB2, and CsGAMYB3 in wild-type (WT) plants and transgenic RNAi lines. The cucumber α-TUBULIN (TUA) was used as an internal control, and the experiments were repeated in three times. Error bars indicate the standard errors. (B, C) Phenotypes of WT (B) or transgenic RNAi plants (C) which are both 60 d old. (D) Diagrammatic data of sex expression of the flowers on the first 20 nodes of the main stems in WT and transgenic RNAi lines. The black and white circles represent female and male flowers, respectively. The numbers indicate the various transgenic lines. (E) The percentage of the nodes with male or female flowers up to the 20th node on the main stem of WT and transgenic RNAi lines. (This figure is available in colour at JXB online.)
When CsGAMYB1-RNAi plants grew until anthesis of flowers on node 20, the sex of the flowers on each node of the main stem was recorded (Fig. 7B–E). The percentage of nodes with male flowers was 75.7% in wild type, and 45% in the CsGAMYB1-RNAi plants, whereas the proportion of nodes with female flowers was 24.3% and 55%, respectively. So, the ratio of nodes with male and female flowers in the transgenic RNAi plants decreased by almost 4-fold in comparison with that in wild-type, indicating that CsGAMYB1 may promote male tendency or inhibit female tendency, leading to a significant change in the sex expression of flowers in cucumber.
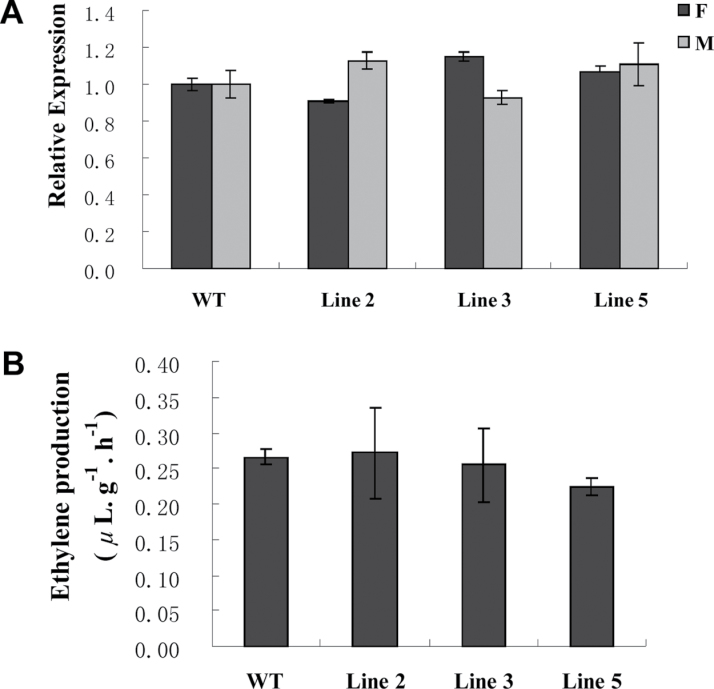
It has been reported that accumulation of F (CsACS1G) and M (CsACS2) mRNA and evolution of ethylene can control the sex determination of cucumber. To test whether there is a correlation between CsGAMYB1-regulated and ethylene-modulated sex expression of cucumber, we measured the expression of F and M genes and ethylene production in the shoot apices of CsGAMYB1-RNAi plants at the 4-leaf stage. As shown in Fig. 8, in transgenic lines 2, 3, and 5, neither F and M mRNA levels nor ethylene production had any significant changes as compared with wild type (Fig. 8A and B), suggesting that ethylene may not be involved in the pathway of CsGAMYB1-regulated sex expression of cucumber.
Fig. 8.
Ethylene is not involved in CsGAMYB1-regulated sex expression of cucumber. (A) Expression of F (CsACS1G) and M (CsACS2) by qRT-PCR in WT and transgenic RNAi lines. The cucumber α-TUBULIN (TUA) was used as an internal control, and the experiments were repeated in triplicate independent samples. Error bars indicate the standard errors. (B) Quantification of ethylene released from shoot apices in WT and transgenic RNAi lines at 4-leaf stage. Amount of ethylene was measured per 1g fresh weight and per h. Vertical bars indicate the standard errors of the mean for triplicate samples. (This figure is available in colour at JXB online.)
Discussion
CsGAMYB1 may be the homologue for both AtMYB33 and AtMYB65
In Arabidopsis, the GAMYB family has three members: MYB33, MYB65, and MYB101 (Gocal et al., 2001), in which MYB33 and MYB65 are functionally redundant in anther development (Millar and Gubler, 2005). Although in cucumber, there are also three putative GAMYB-like genes: CsGAMYB1, CsGAMYB2, and CsGAMYB3. CsGAMYB1 is closely related to AtMYB33 and AtMYB65, whereas CsGAMYB2 and CsGAMYB3 may not belong to the GAMYB family (Fig. 1B). Similar to AtMYB33 and AtMYB65, CsGAMYB1 also has an R2R3 repeat DNA-binding domain and three conserved motifs containing Box 1, Box 2, and Box 3 (Supplementary Fig. S1 available at JXB online). In addition, CsGAMYB1 can partially rescue the phenotypes of myb33 myb65 double mutant in Arabidopsis, with respect to stamen development and plant fertility (Fig. 5). These observations suggested that CsGAMYB1 acts as the homologue for both AtMYB33 and AtMYB65 in cucumber.
CsGAMYB1 has both conserved and divergent functions with its hermaphroditic homologues
In hermaphroditic plants, GAMYB has been shown to play an important role in stamen and anther development. For example, HvGAMYB in barley, OsGAMYB in rice, and AtMYB33 and AtMYB65 in Arabidopsis were strongly expressed in floral organs, especially in stamens and anthers, but weakly in mature pollen grains, and the expression was upregulated by exogenous GA3 (Aya et al., 2009; Gocal et al., 2001; Kaneko et al., 2004; Murray et al., 2003; Tsuji et al., 2006). Loss of function of OsGAMYB in rice, and AtMYB33 and AtMYB65 in Arabidopsis resulted in abnormal staminate specific organs, such as shorter filaments and pollen abortion caused by endless expansion of the tapetum, leading to male sterility (Alonso-Peral et al., 2010; Aya et al., 2009; Kaneko et al., 2004; Liu et al., 2010; Millar and Gubler, 2005), whereas overexpression of HvGAMYB in barley also led to decreased anther length and male sterility (Murray et al., 2003). This suggests that GAMYB homologues have a conserved role in staminate development, but the regulatory mechanism may be different in various species. In our study, CsGAMYB1 was highly expressed in male flower buds, where its expression was also up-regulated by GA3 application (Fig. 2), and CsGAMYB1 rescued the phenotypes of Arabidopsis double mutant myb33 myb65 in stamen development and plant fertility (Fig. 5, Supplementary Table S1 available at JXB online), indicating that CsGAMYB1 may also function as a positive regulator for staminate development as those of Arabidopsis MYB33 and MYB65. Meanwhile, ectopic expression of CsGAMYB1 in Arabidopsis wild type resulted in reduced filaments, aborted pollen, and male sterility (Fig. 6, Supplementary Table S2 available at JXB online), similar to overexpression of HvGAMYB in barley, supporting the hypothesis that CsGAMYB1 has a conserved role in staminate development.
However, unlike the weak expression of HvGAMYB, OsGAMYB, AtMYB33, and AtMYB65 in mature pollen grains, CsGAMYB1 is expressed throughout male flower development containing the mature pollen grains in cucumber (Fig. 3), but there are no significant difference in anthers and pollen development of male flowers between CsGAMYB1RNAi and wild-type plants in cucumber (data not shown), which is divergent with the phenotypes of the GAMYBdeficient mutants in hermaphroditic species. Moreover, GAMYB genes in Arabidopsis might mediate GA signalling in flowering responses by activating the expression of a floral meristem identity gene, LEAFY (Gocal et al., 2001). Also, in the grass Lolium temulentum, the content of GAs increases in the leaves and SAM (shoot apical meristem) with exposure to long-day which is sufficient to induce flowering (King et al., 2001), and the increase in GAs level is followed by increased expression of LtGAMYB in the shoot apex, suggesting that LtGAMYB plays an important role in GA-regulated flower initiation (Gocal et al., 1999). But in cucumber, even though high expression was detected in the inflorescence meristem (im) and floral meristem (fm) (Fig. 3A), flowering time of either male or female flowers seemed to be undisturbed upon partial loss of function of CsGAMYB1 in the transgenic RNAi plants, indicating that CsGAMYB1 may not be involved in floral initiation of cucumber. These observations verified that monoecious CsGAMYB1 displays conserved as well as divergent functions with its hermaphroditic homologues.
The possible roles of CsGAMYB1 in Arabidopsis
Ectopic expression of CsGAMYB1 in Arabidopsis leads to reduced filament length, aborted pollen, and male sterility (Fig. 6), and it is probably caused by a number of events. For example, CsGAMYB1 may interfere with some MYB transcription factors of Arabidopsis that have been shown to be needed in GA-regulated stamen development (Cheng et al., 2009; Millar and Gubler, 2005). Alternatively, CsGAMYB1 possibly up-regulates the target genes of these MYB transcription factors and there may be feedback in this process. In addition, it could result from an accumulated level of GA signalling, which mimics the effect of GA overdose in flower development and male fertility (Colombo and Favret, 1996; Jacobsen and Olszewski, 1993). Interestingly, this developmental phenomenon owing to GA signalling is also observed in other plants. For instance, constitutive overexpression of HvGAMYB in barley causes male sterility (Murray et al., 2003), whereas loss of function of RGA and GAI, repressors of GA signalling and plant growth and development, results in shorter stamen filaments and reduced pollen levels as well as fertility in Arabidopsis (Dill and Sun, 2001). Overall, despite ectopic expression of CsGAMYB1 in Arabidopsis having an important role in stamen development, the regulatory mechanism is unclear, and further analysis is needed.
Moreover, CsGAMYB1 can partially rescue the fertility of a myb33 myb65 double mutant, and the recovery levels rely on the levels of CsGAMYB1 expression (Fig. 5), whereas constitutive overexpression of CsGAMYB1 results in male sterility in Arabidopsis (Fig. 6), suggesting that although CsGAMYB1 is important for stamen development, the effect is dose dependent. And in the transgenic plants overexpressing CsGAMYB1 in Arabidopsis, the expression of CsGAMYB1 in line 8 is higher than that in line 4, 5, 6, and 7, but they are all male sterile (Fig. 6F and G, Supplementary Table S2 available at JXB online), indicating that the level of expression of CsGAMYB1 in line 4, 5, 6, and 7 is sufficient to lead to sterility.
GA and ethylene may regulate sex expression of cucumber via two parallel pathways
The sex determination of cucumber occurs owing to the selective arrestment of either carpel or stamen development at the bisexual stage (Bai et al., 2004; Malepszy and Niemirowicz-Szczytt, 1991). In the pathway of ethylene-regulated female sex expression, F gene governs female flowers formation (Knopf and Trebitsh, 2006; Mibus and Tatlioglu, 2004; Trebitsh et al., 1997), whereas M gene inhibits stamen development in flower buds (Yamasaki et al., 2001; Yamazaki et al., 2003). CsGAMYB1 can enhance the ratio of nodes with male and female flowers (Fig. 7), probably owing to either promotion in male tendency or repression of female sex expression. However, even though CsGAMYB1 is highly expressed in both stamen primordia and carpel primordia at the bisexual stage, its expression remains high in the male specific organs, but weak in female flowers during later stages (Figs 2 and 3), suggesting that CsGAMYB1 may promote the development of male flowers and inhibit the development of female flowers. In addition, in the CsGAMYB1-RNAi plants, the proportion of nodes with male flowers and female flowers is reduced and increased, respectively (Fig. 7), and no bisexual flowers are observed (data not shown). So, we conclude that CsGAMYB1 may modulate the sex expression of cucumber by promoting male tendency and inhibiting the formation of female flowers at the same time, which potentially is an overlapping role with both F and M genes, even though the specific functions are opposite. Moreover, in the process of male flower development, CsGAMYB1 expresses ubiquitously in the male flower, including sepal, petal, and stamen (Fig. 3), but has no effect on the male floral patterning when CsGAMYB1 is knocked down by RNAi (data not shown), which possibly results from a number of events, one of which may be partial functional redundancy of three CsGAMYBs of cucumber. Although phylogenetic analysis has suggested that CsGAMYB2 and CsGAMYB3 may not belong to the GAMYB family (Fig. 1B), they show high similarity compared with CsGAMYB1 (data not shown). Given that Arabidopsis GAMYBs have specific as well as partially overlapping roles, we speculate that CsGAMYB1 may regulate male sex expression of cucumber specifically, whereas the three CsGAMYBs are likely to be functionally redundant in the development of male floral organs. However, for elucidating the functional similarities and differences among these three CsGAMYBs, the functional analysis of CsGAMYB2 and CsGAMYB3 in cucumber is the best way to elucidate this in future studies.
Sex differentiation of cucumber exists plasticity, and male and female expression can be changed by GA and ethylene, respectively (Iwahori et al., 1969; MacMurray and Miller, 1968; Pike and Peterson, 1969; Wittwer and Bukovac, 1962); however, whether there is a crosstalk between these two pathways is unknown. In our study, we certified that GA can modulate sex expression of cucumber via the transcriptional regulation of CsGAMYB1, which acts a positive factor of GA signalling pathway (Figs 2B and 7), and this process has no effect on ethylene biosynthesis and production (Fig. 8), indicating that GA-CsGAMYB1-regulated male sex expression and ethylene-modulated female sex expression of cucumber might take two independent pathways. Our study on GA-CsGAMYB1 reveals a new model for sex expression, which enhances our understanding of sex determination in cucumber and provides the basis for molecular flower induction and high-yield cultural practices. Besides, given that the signalling pathway GA–GID1–DELLA complex play important roles in plant growth and development, particularly in staminate development (Cheng et al., 2004; Dill and Sun, 2001; Fleet and Sun, 2005; Griffiths et al., 2006; Hou et al., 2008; Sun, 2010; Sun, 2011; Tyler et al., 2004), and GAMYB acts as an important downstream component of the DELLA proteins in Arabidopsis (Achard et al., 2004; Fleet and Sun, 2005; Olszewski et al., 2002), identifying the position of CsGAMYB1 in GA response pathways and the relationship between CsGAMYB1 and the GA–GID1–DELLA complex will shed light on the molecular mechanism of male sex expression of cucumber regulated by GA signalling.
Accession numbers
Sequence data of GAMYB proteins in this study can be found in the Cucumber Genome DataBase, Arabidopsis Genome Initiative, Phytozome or GenBank/EMBL/Swiss-Prot databases under the following accession numbers: CsGAMYB1 (Csa009014), CsGAMYB2 (Csa019830), CsGAMYB3 (Csa013555), AtMYB33 (AT5G06100), AtMYB65 (AT3G11440), Malus domestica (MDP0000147309), Prunus persica (ppa003628m.g), Populus trichocarpa (Potri.003G189700), Manihot esculenta (cassava4.1_004606m.g), Gossypium raimondii (Gorai.009G301100), Glycine max (Glyma13g25716), Phaseolus vulgaris (Phvul.011G191300), Ricinus communis (29686.t000037), Solanum tuberosum GAMYB2 (PGSC0003DMG400005918), Solanum tuberosum GAMYB1 (PGSC0003DMG400022689), Solanum lycopersicum (Solyc06g073640.2), Oryza sativa (Os01g59660), Sorghum bicolor (Sb03g037680), Zea mays (GRMZM2G139688), Panicum virgatum (Pavirv00069350m.g), Hordeum vulgare (AAG22863), Lolium temulentu (AAD31395).
Supplementary data
Supplementary data are available at JXB online
Fig. S1. Sequence alignment of amino acid residues of CsGAMYB1 with other GAMYB proteins.
Table S1. CsGAMYB1 can partially rescue the fertility of myb33myb65 in Arabidopsis.
Table S2. Overexpression of CsGAMYB1 resulted in partial sterility in Arabidopsis
Table S3. List of primers and their uses.
Acknowledgments
This work was supported by National Public Service Sectors (Agriculture) Project of China (201203003), National High Technology Research and Development Program (863) of China (2012AA100103), National Science and Technology Projects of China (2013BAD20B01), Innovational Team Program of Beijing Industrial Technology System for Fruit-vegetables and Scientific and Technological Research Project of Beijing (D131100000713001) to HR. We thank Anthony A. Millar for providing the myb33 myb65 seeds, Chunjiang Zhou (Cornell University) for revising the manuscript, and members of the Ren lab for helpful discussions and technical assistance. YZ and HR designed the experiments, YZ performed most of the experiments and wrote the paper along with XZ and XL (Xingwang Liu), BL and WW helped with the in situ hybridization, CC helped with the qRT-PCR, XL (Xiaofeng Liu) helped the subcellular localization, and SY helped with the measurement of ethylene production.
References
- Achard P, Herr A, Baulcombe DC, Harberd NP. 2004. Modulation of floral development by a gibberellin-regulated microRNA. Development 131, 3357–3365 [DOI] [PubMed] [Google Scholar]
- Alonso-Peral MM, Li J, Li Y, Allen RS, Schnippenkoetter W, Ohms S, White RG, Millar AA. 2010. The microRNA159-regulated GAMYB-like genes inhibit growth and promote programmed cell death in Arabidopsis . Plant Physiology 154, 757–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aya K, Ueguchi-Tanaka M, Kondo M, Hamada K, Yano K, Nishimura M, Matsuoka M. 2009. Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB. Plant Cell 21, 1453–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai SL, Peng YB, Cui JX, Gu HT, Xu LY, Li YQ, Xu ZH, Bai SN. 2004. Developmental analyses reveal early arrests of the spore-bearing parts of reproductive organs in unisexual flowers of cucumber (Cucumis sativus L.). Planta 220, 230–240 [DOI] [PubMed] [Google Scholar]
- Bai SN, Xu ZH. 2013. Cucumber unisexual flowers, sex and sex differentiation. International Review of Cell and Molecular Biology 304, 1–56 [DOI] [PubMed] [Google Scholar]
- Bie BB, Pan JS, He HL, Yang XQ, Zhao JL, Cai R. 2013. Molecular cloning and expression analysis of the ethylene insensitive3 (EIN3) gene in cucumber (Cucumis sativus). Genetics and Molecular Research 12, 4179–4191 [DOI] [PubMed] [Google Scholar]
- Cercos M, Gomez-Cadenas A, Ho THD. 1999. Hormonal regulation of a cysteine proteinase gene, EPB-1, in barley aleurone layers: cis- and trans-acting elements involved in the co-ordinated gene expression regulated by gibberellins and abscisic acid. Plant Journal 19, 107–118 [DOI] [PubMed] [Google Scholar]
- Cheng H, Qin LJ, Lee SC, Fu XD, Richards DE, Cao DN, Luo D, Harberd NP, Peng JR. 2004. Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131, 1055–1064 [DOI] [PubMed] [Google Scholar]
- Cheng H, Song S, Xiao L, Soo HM, Cheng Z, Xie D, Peng J. 2009. Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis . PLoS Genetics 5, e1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . Plant Journal 16, 735–743 [DOI] [PubMed] [Google Scholar]
- Colombo N, Favret EA. 1996. The effect of gibberellic acid on male fertility in bread wheat. Euphytica 91, 297–303 [Google Scholar]
- Dill A, Sun TP. 2001. Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana . Genetics 159, 777–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleet CM, Sun TP. 2005. A DELLAcate balance: the role of gibberellin in plant morphogenesis. Current Opinion in Plant Biology 8, 77–85 [DOI] [PubMed] [Google Scholar]
- Gocal GF, Poole AT, Gubler F, Watts RJ, Blundell C, King RW. 1999. Long-day up-regulation of a GAMYB gene during Lolium temulentum inflorescence formation. Plant Physiology 119, 1271–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocal GF, Sheldon CC, Gubler F, et al. 2001. GAMYB-like genes, flowering, and gibberellin signaling in Arabidopsis . Plant Physiology 127, 1682–1693 [PMC free article] [PubMed] [Google Scholar]
- Goto N, Pharis RP. 1999. Role of gibberellin in the development of floral organs of the gibberellin-deficient mutant, ga1-1, of Arabidopsis thaliana . Canadian Journal of Botany-revue Canadienne De Botanique 77, 944–954 [Google Scholar]
- Griffiths J, Murase K, Rieu I, et al. 2006. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis . Plant Cell 18, 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Kalla R, Roberts JK, Jacobsen JV. 1995. Gibberellin-regulated expression of a myb gene in barley aleurone cells: Evidence for Myb transactivation of a high-pI α-amylase gene promoter. Plant Cell 7, 1879–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler F, Raventos D, Keys M, Watts R, Mundy J, Jacobsen JV. 1999. Target genes and regulatory domains of the GAMYB transcriptional activator in cereal aleurone. Plant Journal 17, 1–9 [DOI] [PubMed] [Google Scholar]
- Harberd NP, Belfield E, Yasumura Y. 2009. The angiosperm Gibberellin-GID1-DELLA growth regulatory mechanism: how an “inhibitor of an inhibitor” enables flexible response to fluctuating environments. Plant Cell 21, 1328–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Hu WW, Shen L, Lee LY, Tao Z, Han JH, Yu H. 2008. Global identification of DELLA target genes during Arabidopsis flower development. Plant Physiology 147, 1126–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Li R, Zhang Z, et al. 2009. The genome of the cucumber, Cucumis sativus L. Nature Genetics 41, 1275–1281 [DOI] [PubMed] [Google Scholar]
- Iwahori S, Lyons JM, William LS. 1969. Induced femaleness in cucumber by 2-chloroethanephosphonic acid. Nature 222, 271–272 [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE. 1991. Characterization of the arrest in anther development associated with gibberellin deficiency of the gib-1 mutant of tomato. Plant Physiology 97, 409–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE. 1993. Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 5, 887–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Inukai Y, Ueguchi-Tanaka M, et al. 2004. Loss-of-function mutations of the rice GAMYB gene impair alpha-amylase expression in aleurone and flower development. Plant Cell 16, 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Evans LT. 2003. Gibberellins and flowering of grasses and cereals: prizing open the lid of the “florigen” black box. Annual Review of Plant Biology 54, 307–328 [DOI] [PubMed] [Google Scholar]
- King RW, Moritz T, Evans LT, Junttila O, Herlt AJ. 2001. Long-day induction of flowering in Lolium temulentum involves sequential increases in specific gibberellins at the shoot apex. Plant Physiology 127, 624–632 [PMC free article] [PubMed] [Google Scholar]
- Knopf RR, Trebitsh T. 2006. The female-specific Cs-ACS1G gene of cucumber. A case of gene duplication and recombination between the non-sex-specific 1-aminocyclopropane-1-carboxylate synthase gene and a branched-chain amino acid transaminase gene. Plant and Cell Physiology 47, 1217–1228 [DOI] [PubMed] [Google Scholar]
- Koornneef M, van der Veen JH. 1980. Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) heynh. Theoretical and Applied Genetics 58, 257–263 [DOI] [PubMed] [Google Scholar]
- Kranz HD, Denekamp M, Greco R, et al. 1998. Towards functional characterization of the members of the R2R3-MYB gene family from Arabidopsis thaliana . Plant Journal 16, 263–276 [DOI] [PubMed] [Google Scholar]
- Li Z, Wang S, Tao Q, Pan J, Si L, Gong Z, Cai R. 2012. A putative positive feedback regulation mechanism in CsACS2 expression suggests a modified model for sex determination in cucumber (Cucumis sativus L.). Journal of Experimental Botany 63, 4475–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Bao W, Liang W, Yin J, Zhang D. 2010. Identification of gamyb-4 and analysis of the regulatory role of GAMYB in rice anther development. Journal of Integrative Plant Biology 52, 670–678 [DOI] [PubMed] [Google Scholar]
- MacMurray AL, Miller CM. 1968. Cucumber sex expression modified by 2-chloroethanephosphonic acid. Science 162, 1397–1398 [DOI] [PubMed] [Google Scholar]
- Malepszy S, Niemirowicz-Szczytt K. 1991. Sex determination in cucumber (Cucumis sativus) as a model system for molecular biology. Plant Science 80, 39–47 [Google Scholar]
- Mibus H, Tatlioglu T. 2004. Molecular characterization and isolation of the F/f gene for femaleness in cucumber (Cucumis sativus L.). Theoretical and Applied Genetics 109, 1669–1676 [DOI] [PubMed] [Google Scholar]
- Millar AA, Gubler F. 2005. The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17, 705–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase K, Hirano Y, Sun TP, Hakoshima T. 2008. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456, 459–463 [DOI] [PubMed] [Google Scholar]
- Murray F, Kalla R, Jacobsen J, Gubler F. 2003. A role for HvGAMYB in anther development. Plant Journal 33, 481–491 [DOI] [PubMed] [Google Scholar]
- Nester JE, Zeevaart JAD. 1988. Flower development in normal tomato and a gibberellin-deficient (ga-2) mutant. American Journal of Botany 75, 45–55 [Google Scholar]
- Olszewski N, Sun T-P, Gubler F. 2002. Gibberellin signaling: biosynthesis, catabolism, and response pathways. The Plant Cell 14 (suppl.), S61–S80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pharis RP, King RW. 1985. Gibberellins and reproductive development in seed plants. Annual Review of Plant Physiology 36, 517–568 [Google Scholar]
- Pike LM, Peterson CE. 1969. Gibberellin A4/A7, for induction of staminate flowers on the gynoecious cucumber (Cucumis sativus L.). Euphytica 18, 106–109 [Google Scholar]
- Plackett AR, Ferguson AC, Powers SJ, Wanchoo-Kohli A, Phillips AL, Wilson ZA, Hedden P, Thomas SG. 2014. DELLA activity is required for successful pollen development in the Columbia ecotype of Arabidopsis . New Phytologist 201, 825–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plackett AR, Thomas SG, Wilson ZA, Hedden P. 2011. Gibberellin control of stamen development: a fertile field. Trends in Plant Science 16, 568–578 [DOI] [PubMed] [Google Scholar]
- Romero I, Fuertes A, Benito MJ, Malpica JM, Leyva A, Paz-Ares J. 1998. More than 80 R2R3-MYB regulatory genes in the genome of Arabidopsis thaliana . Plant Journal 14, 273–284 [DOI] [PubMed] [Google Scholar]
- Saito S, Fujii N, Miyazawa Y, Yamasaki S, Matsuura S, Mizusawa H, Fujita Y, Takahashi H. 2007. Correlation between development of female flower buds and expression of the CS-ACS2 gene in cucumber plants. Journal of Experimental Botany 58, 2897–2907 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4, 406–425 [DOI] [PubMed] [Google Scholar]
- Song S, Qi T, Huang H, Xie D. 2013. Regulation of stamen development by coordinated actions of jasmonate, auxin, and gibberellin in Arabidopsis . Molecular Plant 6, 1065–1073 [DOI] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B. 2001. The R2R3-MYB gene family in Arabidopsis thaliana . Current Opinion in Plant Biology 4, 447–456 [DOI] [PubMed] [Google Scholar]
- Sun TP. 2010. Gibberellin-GID1-DELLA: a pivotal regulatory module for plant growth and development. Plant Physiology 154, 567–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP. 2011. The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Current Biology 21, R338–R345 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28, 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebitsh T, Staub JE, O’Neill SD. 1997. Identification of a 1-aminocyclopropane-1-carboxylic acid synthase gene linked to the female (F) locus that enhances female sex expression in cucumber. Plant Physiology 113, 987–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji H, Aya K, Ueguchi-Tanaka M, et al. 2006. GAMYB controls different sets of genes and is differentially regulated by microRNA in aleurone cells and anthers. Plant Journal 47, 427–444 [DOI] [PubMed] [Google Scholar]
- Tyler L, Thomas SG, Hu JH, Dill A, Alonso JM, Ecker JR, Sun TP. 2004. DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis . Plant Physiology 135, 1008–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Nakajima M, Motoyuki A, Matsuoka M. 2007. Gibberellin receptor and its role in gibberellin signaling in plants. Annual Review of Plant Biology 58, 183–198 [DOI] [PubMed] [Google Scholar]
- Varagona MJ, Schmidt RJ, Raikhel NV. 1992. Nuclear localization signal(s) required for nuclear targeting of the maize regulatory protein Opaque-2. Plant Cell 4, 1213–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DH, Li F, Duan QH, Han T, Xu ZH, Bai SN. 2010. Ethylene perception is involved in female cucumber flower development. Plant Journal 61, 862–872 [DOI] [PubMed] [Google Scholar]
- Wang H, Sui X, Guo J, Wang Z, Cheng J, Ma S, Li X, Zhang Z. 2014. Antisense suppression of cucumber (Cucumis sativus L.) sucrose synthase 3 (CsSUS3) reduces hypoxic stress tolerance. Plant, Cell and Environment 37, 795–810 [DOI] [PubMed] [Google Scholar]
- Wittwer SH, Bukovac MI. 1962. Staminate flower formation on gynoecious cucumber as influenced by the various gibberellins. Naturwissenshaften 49, 305–306 [Google Scholar]
- Yamasaki S, Fujii N, Matsuura S, Mizusawa H, Takahashi H. 2001. The M locus and ethylene-controlled sex determination in andromonoecious cucumber plants. Plant and Cell Physiology 42, 608–619 [DOI] [PubMed] [Google Scholar]
- Yamazaki Y, Fujii N, Takahashi H. 2003. Characterization of ethylene effects on sex determination in cucumber plants. Sexual Plant Reproduction 16, 103–111 [Google Scholar]
- Zhang X, Zhou Y, Ding L, Wu Z, Liu R, Meyerowitz EM. 2013. Transcription repressor HANABA TARANU controls flower development by integrating the actions of multiple hormones, floral organ specification genes, and GATA3 family genes in Arabidopsis . Plant Cell 25, 83–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.