Summary
PvSPX1 was found to be a positive regulator in the P signalling network of common bean, and is downstream of PvPHR1.
Key words: Bean, hairy roots, phosphate starvation, phosphorus homeostasis, root growth, SPX domain.
Abstract
Proteins containing the SPX domain are believed to play vital roles in the phosphorus (P) signalling network in plants. However, the functions of SPX proteins in legumes remain largely unknown. In this study, three SPX members, PvSPX1–PvSPX3 were cloned from common bean (Phaseolus vulgaris L.). It was found that the transcripts of all three PvSPX members were significantly enhanced in both bean leaves and roots by phosphate (Pi) starvation. Among them, the expression of nuclear localized PvSPX1 showed more sensitive and rapid responses to Pi starvation. Consistently, only overexpression of PvSPX1 resulted in increased root P concentration and modified morphology of transgenic bean hairy roots, such as inhibited root growth and an enlarged root hair zone. It was further demonstrated that PvSPX1 transcripts were up-regulated by overexpressing PvPHR1, and overexpressing PvSPX1 led to increased transcripts of 10 Pi starvation-responsive genes in transgenic bean hairy roots. Taken together, it is suggested that PvSPX1 is a positive regulator in the P signalling network of common bean, and is downstream of PvPHR1.
Introduction
Phosphorus (P) is an essential element for plant growth, and is easily fixed by soil particles due to its chemical properties. Therefore, low P availability adversely affects crop growth and production, especially on acid soils (Raghothama, 1999; Vance et al., 2003). To cope with low P stress, plants have developed a wide range of adaptive strategies, such as changes in root architecture and morphology (Liao et al., 2004; Zhou et al., 2008; Péret et al., 2011; Tian et al., 2012), increased exudation of protons and organic acids (Fox and Comerford, 1990; Ström et al., 2005; Taghipour and Jalali, 2012), and enhanced secreted or root-associated acid phosphatase activities (Del Pozo et al., 1999; Bozzo et al., 2002; Ligaba et al., 2004; C. Wang et al., 2009; Liang et al., 2010; Robinson et al., 2012; L.S. Wang et al., 2012). These adaptive strategies are tightly mediated by the P signalling network, which is composed of a wide array of regulators (Raghothama, 1999; Vance et al., 2003; Chiou and Lin, 2011).
Proteins containing the SPX domains have been demonstrated to play vital roles in the P signalling networks of yeast (Saccharomyces cerevisiae), Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), and rape (Brassica napus) (Ligaba et al., 2004; Duan et al., 2008; C. Wang et al., 2009, 2012; Z. Wang et al., 2009; Liu et al., 2010; Secco et al., 2012a, b; Yang et al., 2012). The SPX domain is named after SYG1/Pho81/XPR1 proteins, which contain a conserved domain in the N-terminal peptides of yeast SYG1 and PHO81, and human XPR1 proteins (Spain et al., 1995; Lenburg and O’Shea, 1996; Battini et al., 1999; Wang et al., 2004).
In yeast, several SPX domain-containing proteins involved in P acquisition and the signalling pathway have been identified (Secco et al., 2012b). PHO81 is a cyclin-dependent kinase (CDK) inhibitor (Lenburg and O’Shea, 1996). Under phosphate (Pi) starvation conditions, PHO81 inhibits the kinase activity of the PHO80–PHO85 complex against the Pho4 transcription factor, which subsequently regulates transcripts of several Pi starvation-responsive genes (Lenburg and O’Shea, 1996). Many yeast Pi transporters, such as Pho84, Pho87, Pho89, Pho90, and Pho91, also possess the SPX domain (Secco et al., 2012b). It is interesting that most SPX domain-harbouring proteins, including Vtc2, Vtc3, Vtc4, and Gde1, appear to play key regulatory roles in P homeostasis in yeast (Secco et al., 2012b).
In plants, four groups of proteins were also found to contain the SPX domain. Among them, three groups of proteins have the SPX domain in the N-terminus and other domains in the C-terminus, including an EXS (ERD1, XPR1, and SYG1), a major facility superfamily (MFS), or a RING-type zinc finger domain (Hamburger et al., 2002; Stefanovic et al., 2007; Lin et al., 2010; Secco et al., 2010; Kant et al., 2011; C. Wang et al., 2012). Similar to the functions of proteins containing the SPX domain in yeast, most of these plant members are involved in regulating P homeostasis in plants. Examples include OsSPX-MFS1 in rice (C. Wang et al., 2012), along with AtPHO1:1 and AtNLA in Arabidopsis (Stefanovic et al., 2007; Secco et al., 2010; Kant et al., 2011).
Recently, a specific group of proteins only containing the SPX domain have been characterized in plants, such as Arabidopsis and rice (Duan et al., 2008; C. Wang et al., 2009; Z. Wang et al., 2009; Liu et al., 2010; Yang et al., 2011). In Arabidopsis, four members only contain the SPX domain, namely AtSPX1, AtSPX2, AtSPX3, and AtSPX4 (Duan et al., 2008). Furthermore, expression patterns of several Pi starvation-responsive genes were positively and negatively regulated by AtSPX1 and AtSPX3, respectively (Duan et al., 2008). Similarly, the negative regulatory role of OsSPX1 was also suggested in rice, because transcription of several Pi starvation-responsive genes (e.g. OsPT2, OsPT6, and OsPAP10) was suppressed through OsSPX1 overexpression (C. Wang et al., 2009). Furthermore, it has recently been determined that OsSPX1 is downstream of OsPHR2 and OsPHO2 in the rice P signalling pathway (Liu et al., 2010).
Despite accumulated knowledge of the P signalling network in model plants (i.e. Arabidopsis and rice) (Chiou and Lin, 2011), information on Pi starvation-responsive pathways in other crops remains fragmentary. A group of Pi starvation-responsive genes (e.g. PvmiR399 and PvPS2:1) have been cloned and characterized in common bean (Phaseolus vulgaris L.), an important legume crop (Tian et al., 2007; Valdes-Lopez et al., 2008; Hernández et al., 2009; Liang et al., 2012a,b). This has facilitated elucidation of the P signalling network in bean, although this knowledge remains incomplete. Recently, essential roles for PvPHR1 and PvmiR399 have been suggested in P deficiency signalling (Valdes-Lopez et al., 2008). Nevertheless, other regulators are probably required as well. In a previous study, three expressed sequence tags (ESTs) with high homology to AtSPX1 were identified through screening a suppression subtractive hybridization library constructed from P-deficient bean (Tian et al., 2007). Among them, the full-length cDNA of PvIDS4-1 (i.e. PvSPX1) was cloned, and its expression levels were found to be up-regulated by Pi starvation in bean (Tian et al., 2007). However, the functions of PvSPX1 and other PvSPX genes in bean adaptation to P deficiency remain unknown. In this study, the full-length cDNA of the other two PvSPX genes (i.e. PvSPX2 and PvSPX3) was cloned. Subsequently, the expression patterns and functions of all three PvSPX gene family members as related to P availability were characterized in bean.
Materials and methods
Plant material and growth conditions
Seeds of common bean genotype G19833 were surface sterilized for 1min using 10% (v/v) H2O2 and then germinated in the dark on germination paper moistened with 1/4 strength modified nutrient solution as described previously (Yan et al., 2001). Five days after germination, seedlings were transferred to nutrient solution supplied with 5, 50, 100, or 500 μM KH2PO4 for a P dosage experiment. After 10 d, young leaves and roots were harvested. For time course experiments, seedlings were pre-treated in 1/4 strength nutrient solution for 7 d and then transplanted to nutrient solution containing 5 μM KH2PO4. Shoots and roots were each harvested at 0, 4, and 8 d after treatment for determination of fresh weight, total root length, and P content. Young leaves and roots were separately harvested for RNA extraction. Nutrient solution was well aerated and its pH was maintained between 5.8 and 6.0. Four biological replicates were included in all of the experiments.
Analysis of total root length and P content
Roots were scanned, and then the digital images were analysed using Win-Rhizo software (Régent Instruments, Canada) to measure total root length. Shoots and roots were kept separately at 75 °C until completely dry, and then were ground into powder for total P content analysis. P content was determined using the phosphorus–molybdate blue colour reaction as previously described (Murphy and Riley, 1962)
Cloning full length cDNAs of PvSPX2 and PvSPX3
Gene-specific primers were designed according to the EST sequences of PvSPX2 (EG594307) and PvSPX3 (EG594308) (Tian et al., 2007) (Supplementary Table S1 available at JXB online). Using the full-length cDNA library constructed from the roots of G19833 as a template, the 5’ and 3’ termini of each gene were amplified by the specific primers paired with T3 and T7 primers, respectively. The amplified DNA fragments were then cloned into the pMD18-T vector (TaKaRa, Japan) and sequenced. Sequences of PvSPX2 and PvSPX3 were analysed at the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/), and deposited in GenBank with accession numbers GU189405 for PvSPX2 and GU189406 for PvSPX3. Multiple sequence alignments were conducted using ClustalW 1.8. The phylogenetic tree was established using the Neighbor–Joining method of the MEGA 4.1 program.
RNA extraction and quantitative real-time PCR
Total RNA was isolated from young leaves and roots using RNAiso Plus reagent (TaKaRa) and treated with DNase I (TaKaRa). The first-strand cDNA was synthesized from total RNA using MMLV reverse transcriptase following the manual (Promega Inc., USA). The first-strand cDNA was then used for SYBR Green-monitored quantitative real-time PCR (qPCR) analysis, which was performed using a Rotor-Gene 3000 (Corbett Research, Australia). Expression levels of the tested genes were quantified relative to expression levels of the reference gene EF-1α (PvTC3216) using arbitrary units. The primer pairs used for qPCR analysis are shown in supplementary Table S1 at JXB online. All of the gene expression analyses had four biological replicates.
Subcellular localization analysis
The coding regions of PvSPX1 (EF191350), PvSPX2, and PvSPX3 without stop codons were separately cloned into the transient expression vector (pBEGFP), and fused with green fluorescent protein (GFP; Liang et al., 2010). For subcellular localization of PvSPXs in onion (Allium cepa) epidermal cells, the PvSPX–GFP fusion constructs and GFP empty vector control were separately transformed into onion epidermal cells using a helium-driven accelerator (PDS/1000, Bio-Rad). After the transformed cells were cultured on Murashige and Skoog (MS) medium for 16h, the GFP florescence was observed using a confocal scanning microscope system (TCS SP2, Leica, Germany) with 488nm excitation and 500–525nm emission filter wavelengths. For subcellular localization of PvSPXs in leaf epidermal cells of tobacco (Nicotiana tabacum), the PvSPX–GFP fusion constructs and GFP empty vector control were separately transformed into Agrobacterium tumefaciens strain GV3101, which were further used for transformation as previously described (Sparkes et al., 2006). After the transformation, plants were grown under normal conditions for 48h and the GFP florescence was observed using a fluorescence microscope (Leica DM5000B). The GFP fluorescence was imaged using a Leica DFC 480 camera.
Transformed genes in common bean hairy roots
The coding regions of PvSPX1, PvSPX2, PvSPX3, and PvPHR1 (EU500763) were inserted separately into the unique BamHΙ and MluΙ sites of the binary vector pYLRNAi as previously described (Liang et al., 2010). For PvSPX1 RNA interference (RNAi) construction, the same binary vector pYLRNAi was used by inserting the PvPSX1-specific fragment into the BamHΙ and HindIII, and the PstI and MluI sites, respectively. The overexpression, RNAi constructs, and the empty vector control (CK) were then separately transformed into Agrobacterium rhizogenes strain K599, which were further used for hairy root transformation. Transformed bean hairy roots were generated and maintained as described previously (Liang et al., 2012b). Briefly, sterilized bean seeds were germinated on half-strength MS medium. After 35h, the abaxial sides of cotyledons were wounded with a scalpel previously dipped into the overnight cultures of the transgenic A. rhizogenes strain K599. The wounded cotyledons were cultured in solid MS medium to develop hairy roots. The expression levels of the corresponding genes in hairy roots were verified through qPCR analysis. For P treatments, ~0.2g (fresh weight) of hairy roots was cultured in solid MS medium with or without the addition of 1.25mM KH2PO4. After 14 d growth, transgenic bean hairy roots were photographed using a microscope (Leica) and a Leica DFC 480 camera. The fresh weight and total P content of each transgenic line were determined as described above. The lateral root length was analysed using Win-Rhizo. Based on root hair density, lateral roots were separated into two zones, namely the root hair zone (i.e. the part of the root zone with >10 root hairs per 1mm root) and the non-root hair zone, and then the percentage of the root hair zones in the lateral roots was calculated. In total, 10 lateral roots were analysed for each replicate. For each treatment, four biological replicates were included.
To analyse the expression patterns of genes downstream of PvSPX1 in P signalling, total RNA was extracted from transgenic hairy roots grown under high P conditions. Subsequently, qPCR was conducted to analyse the expression of 11 genes downstream of PvSPX1: PvPT1 (TC27368), PvPHT2 (TC30856), Pv4 (CV536419), PvPAP1 (BAD05166), PvPAP2 (CAA04644), PvPAP3 (AC025293), PvPAP4 (AAF60317), PvPAP5 (ADK56125), PvPS2:1 (EF472460), PvLPR1-like (FE710903), and PvPDR2-like (TC44308). All qPCR primers (Supplementary Table S1 at JXB online) were designed according to the sequences downloaded from the Dana-Farber Cancer Institute (DFCI; http://compbio.dfci.harvard.edu/tgi/) for PvPHT2, PvLPR1-like, and PvPDR2-like, or from the GenBank database (http://www.ncbi.nlm.nih.gov/genbank) entries for PvPHR1, PvPT1, Pv4, PvPAP1, PvPAP2, PvPAP3, PvPAP4, PvPAP5, and PvPS2:1. To construct the PvSPX1 and PvSPX2 promoter fused with β-glucuronidase (GUS) vectors, the 5’-regulatory regions of 1.9kb for PvSPX1 and 1.6kb for PvSPX2 were each cloned, and inserted into the pCAMBIA 1391 vector. The constructs were transformed into the bean hairy roots as described above, and GUS activity was analysed as described before (Liang et al., 2012a).
Results
Plant growth was affected by P availability
P deficiency significantly inhibited bean growth, as reflected by decreases in plant fresh weight, total root length, and total P content (Supplementary Table S2 at JXB online). With an increased duration of P deficiency, plant fresh weight and total P content gradually decreased. After 8 d of Pi starvation, the total P contents of bean shoots and roots were reduced by 32% and 30%, respectively, as compared with high P conditions (Supplementary Table S2). Similarly, total root length was also significantly decreased by P deficiency. Total root length at 4 d and 8 d of P deficiency was reduced by 39% and 55%, respectively, as compared with under high P conditions (Supplementary Table S2).
Identification and bioinformatics analysis of PvSPX2 and PvSPX3
Based on the reported EST sequences of PvSPX2 and PvSPX3, the full-length cDNAs of both PvSPX2 and PvSPX3 were cloned from a full-length cDNA library of G19833 subjected to P deficiency. The coding regions of PvSPX2 and PvSPX3 were 861bp and 756bp in length, respectively. Alignment analysis showed that PvSPX2 and PvSPX3 exhibited 75% and 50% similarity to PvSPX1, respectively.
Phylogenetic analysis showed that plant proteins containing the SPX domain could be divided into four groups, namely SPX, SPX-EXS, SPX-MFS, and SPX-RING (Fig. 1). Furthermore, SPX proteins could be further subdivided into three groups. Among them, PvSPX1, PvSPX2, and PvSPX3 belong to group I, which includes AtSPX1 and AtSPX2 in Arabidopsis, as well as OsSPX1 and OsSPX2 in rice (Fig. 1).
Fig. 1.
Phylogenetic analysis of SPX proteins in plants. The first two letters of each protein label represent the abbreviated species name: At, Arabidopsis thaliana; Os, Oryza sativa; Pv, Phaseolus vulgaris. AtSPX1 (At5g20150), AtSPX2 (At2g26660), AtSPX3 (At2g45130), AtSPX4 (At5g15330), AtPHO1 (AT3G23430), AtPHO1: H1 (At1g68740), AtPHO1: H2 (At2g03260), AtPHO1: H3 (At1g14040), AtPHO1: H4 (At4g25350), AtPHO1: H5 (At2g03240), AtPHO1: H6 (At2g03250), AtPHO1: H7 (At1g26730), AtPHO1: H8 (At1g35350), AtPHO1: H9 (At3g29060), AtPHO1: H10 (At1g69480), OsSPX1 (Os03g0343400), OsSPX2 (Os02g10780), OsSPX3 (Os10g25310), OsSPX4 (Os03g61200), OsSPX5 (Os03g29250), OsSPX6 (Os07g42330), OsNLA (Os02g0673200), OsSPX-MFS1 (Os04g0573000), OsSPX-MFS2 (Os02g0678200), PvSPX1 (EF191350), PvSPX2 (EG594307), PvSPX3 (EG594308).
Temporal expression patterns of PvSPX genes in response to Pi starvation
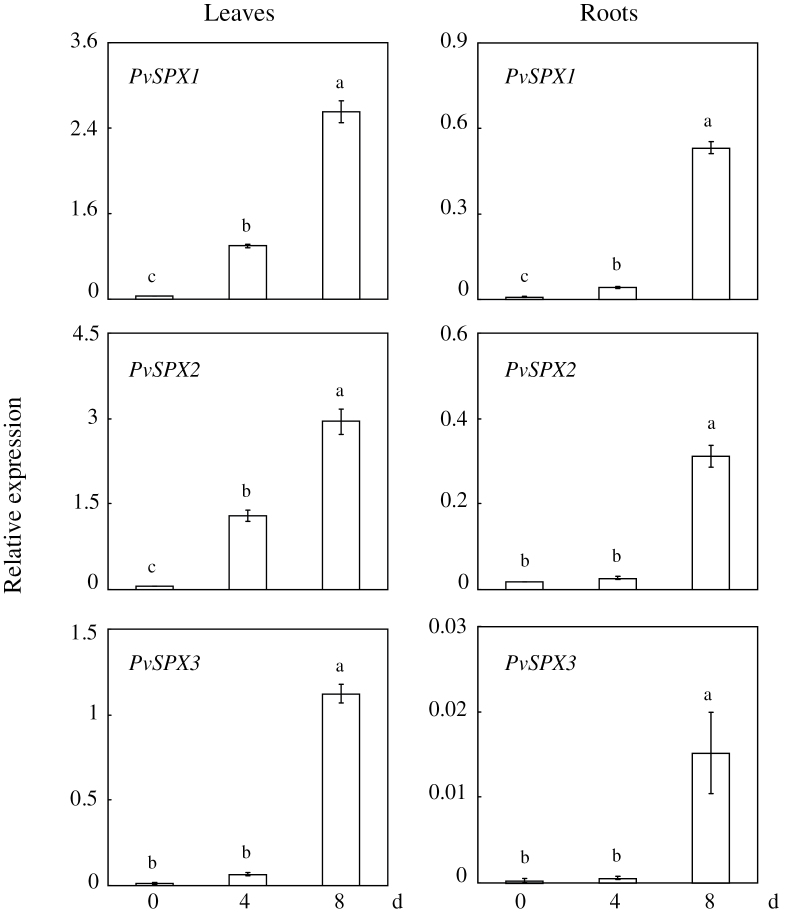
The temporal expression patterns of the three PvSPX genes in bean leaves and roots were analysed by qPCR. As shown in Fig. 2, their expression levels were significantly increased over time and reached their highest levels after 8 d of low P treatment (Fig. 2). However, their expression patterns varied in leaves and roots at 4 d of P deficiency (Fig. 2). After 4 d of P deficiency, significantly increased transcription was observed for PvSPX1 and PvSPX2 in leaves, while for PvSPX3 transcription was not increased either in leaves or in roots (Fig. 2). This suggests that PvSPX1 and PvSPX2 respond to Pi starvation earlier than PvSPX3 in bean.
Fig. 2.
Temporal expression patterns of PvSPX genes in response to Pi starvation in leaves and roots of bean. Seedlings were hydroponically grown under normal conditions for 7 d, and then subjected to P deficiency for 0, 4, and 8 d. Total RNA isolated from leaves and roots of plants was used for qPCR analysis. Expression levels of the tested genes were quantified relative to expression levels of the reference gene EF-1α (PvTC3216) using arbitrary units. Each bar is the mean of four replicates with the standard error. Different letters represent significant differences at the 0.05 level.
Dosage responses of PvSPX genes to P availability
Expression patterns of the three PvSPX members studied here were tightly dependent on P availability in the medium (Fig. 3). Their highest transcript levels were observed in both leaves and roots supplied with 5 μM P, and were decreased with increased P availability (Fig. 3). When the applied P concentration was increased to 500 μM, transcription of each PvSPX gene was negligible (Fig. 3). However, slight differences existed among their expression patterns as related to P availability. Transcript levels of PvSPX1 and PvSPX2 in both leaves and roots declined significantly when the applied P concentration was increased from 100 μM to 500 μM, but that of PvSPX3 did not (Fig. 3), suggesting that expression of PvSPX1 and PvSPX2 might be more sensitive to P availability than that of PvSPX3.
Fig. 3.
Dosage response of PvSPX genes to P deficiency. Seedlings were grown in nutrient solution supplied with 5, 50, 100, or 500 μM KH2PO4. After 10 d, total RNA was isolated from leaves and roots for qPCR analysis. Expression levels of the tested genes were quantified relative to expression levels of the reference gene EF-1α (PvTC3216) using arbitrary units. Each bar is the mean of four replicates with the standard error. Different letters represent significant differences at the 0.05 level.
Subcellular localization of PvSPX proteins
To determine the subcellular localization, the coding regions of the three PvSPX genes were fused with the GFP reporter gene and transiently expressed in onion and tobacco epidermal cells. Subcellular localization was visualized by detecting GFP signal in the transformed onion and tobacco epidermal cells. The empty vector containing 35S:GFP was used as a control. The results showed that the three PvSPX members were found in various subcellular localizations (Fig. 4). Signals of GFP fusion with PvSPX1 and PvSPX2 were only detected in the nuclei of onion and tobacco epidermal cells (Fig. 4). However, GFP fusion with PvSPX3 was observed in many areas in onion and tobacco epidermal cells, suggesting that PvSPX3 might be localized in the cytoplasm and nuclei (Fig. 4).
Fig. 4.
Subcellular localization of PvSPXs. (A) Transient expression of the pBEGFP construct and PvSPX–GFP fusion in onion epidermal cells. Scale bars=50 μm. (B) Transient expression of the pBEGFP construct and PvSPX–GFP fusion in tobacco epidermal cells. Scale bars=20 μm. The first row shows the empty vector control, followed by PvSPX1–GFP, PvSPX2–GFP, and PvSPX3–GFP constructs. Cells were observed by the green fluorescence of GFP and the PvSPX–GFP proteins.
Functional analysis of PvSPX genes in transgenic hairy roots
The functions of PvSPX genes were further analysed in bean transgenic hairy roots by overexpressing PvSPX1, PvSPX2, and PvSPX3. Significantly increased transcripts of the three PvSPX genes in the transgenic bean hairy roots were verified through qPCR analysis (Supplementary Fig. S1 at JXB online). Subsequently, the transgenic hairy roots were grown in MS medium with or without P application for 14 d. The results showed that only overexpressing PvSPX1 could inhibit hairy root growth, as reflected by reduced fresh weight of hairy roots under both P conditions (Fig. 5A, B). Compared with the control lines, the fresh weight of the PvSPX1 overexpression line was reduced by ~60% in high P and 40% in low P (Fig. 5B). Furthermore, the P concentration in the PvSPX1 overexpression line was higher than that in the control line by ~45% in high P and 30% in low P (Fig. 5C). In contrast, the fresh weight and P concentration of both PvSPX2 and PvSPX3 overexpression lines were similar to those in the control line at the two P levels (Fig. 5B, C). Similarly, suppressed PvSPX1 did not affect hairy root fresh weight and P concentration, compared with those in the empty vector (CK) controls (Supplementary Figs S2, S3).
Fig. 5.
Growth and P concentration of bean hairy roots at two P levels. (A) Photograph of bean hairy roots grown at two P levels. Scale bars=1cm. (B) Fresh weight of bean hairy roots at two P levels. (C) P concentration in bean hairy roots. Bean hairy roots were grown in media containing 0 μM (–P) or 1.25mM (+P) KH2PO4 for 14 d. THe fresh weight and P concentration were measured. Each bar is the mean of four replicates with the standard error. Asterisks represent significant differences between overexpressing PvSPX and CK for the same trait in t-tests. *0.01<P≤0.05; **P≤0.01. OX1 and OX2 indicate two transgenic bean hairy root lines overexpressing PvSPX1, PvSPX2, or PvSPX3. CK1 and CK2 indicate the two transgenic lines transformed with the empty vector.
Root morphology was further investigated in all hairy root lines at the two P levels through determination of the percentage of the root hair zones (i.e. the part of root zone with >10 root hairs per 1mm root) in bean hairy roots. The percentage of the root hair zones in all hairy root lines was ~80% without P application (Fig. 6). With P application, the percentage of the root hair zones of CK, and PvSPX2 and PvSPX3 overexpression lines was decreased by >50% (Fig. 6). However, for the PvSPX1 overexpression line, applied P did not affect the percentage of the root hair zone (Fig. 6). Interestingly, similar results were also observed in PvSPX1 RNAi lines, in which the percentage of the root hair zone was not affected by P application (Supplementary Fig. S4 at JXB online). The results suggest that expression of PvSPX1 might regulate enlargement of the root hair zones at a high P level.
Fig. 6.
Root morphology and the percentage of the root hair zones in bean hairy roots at two P levels. (A) Photographs of roots in bean hairy roots at two P levels. (B) Percentage of the root hair zone in bean hairy roots at two P levels. Bean hairy roots were grown in media containing 0 μM (–P) or 1.25mM (+P) KH2PO4 for 14 d. Ten lateral roots were selected from each replicate for further analysis. Photographs were taken of three zones of the lateral roots, namely the root tip, root middle, and root base. Scale bars=1mm. OX1 and OX2 indicate two transgenic bean hairy root lines overexpressing PvSPX1, PvSPX2, or PvSPX3. CK1 and CK2 indicate the two transgenic lines transformed with the empty vector. Asterisks represent significant differences between two P treatments for the same trait at the 0.05 level.
PvSPX participates in the P signalling network in bean
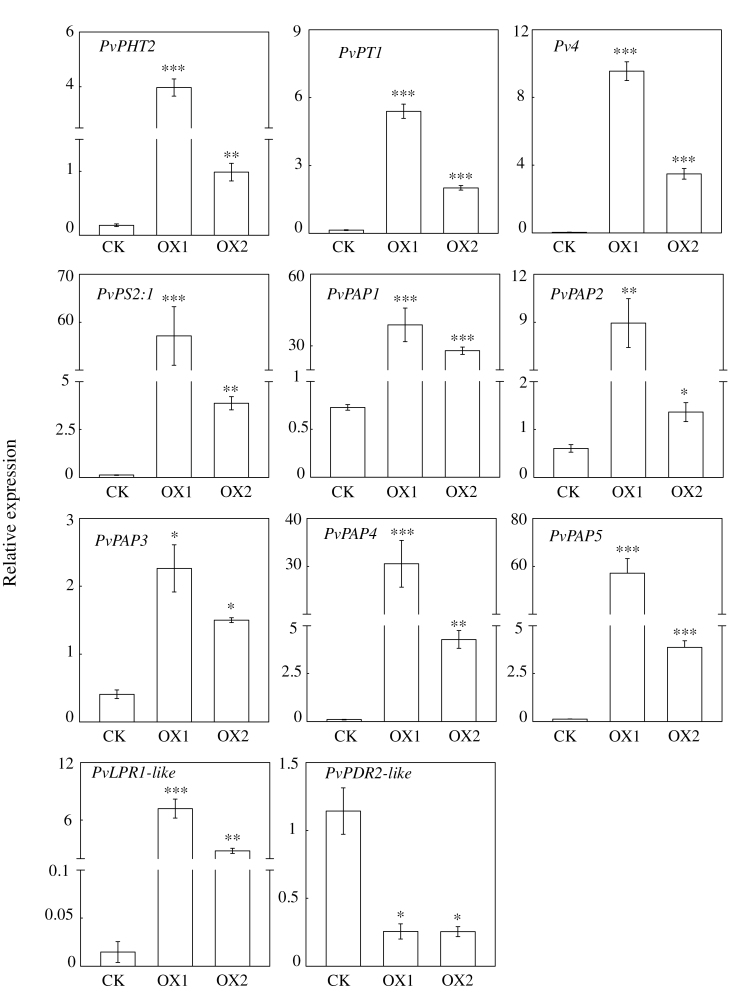
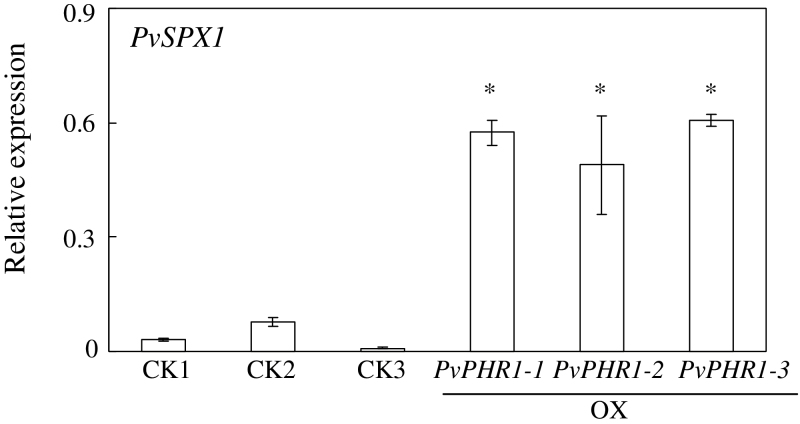
The expression patterns of 11 genes were investigated in the transgenic hairy roots overexpressing PvSPX1 in order to illustrate the regulatory role of PvSPX1 in the P signalling network in bean. Among them, nine genes were previously characterized as Pi starvation-responsive genes, namely two Pi transporters (PvPT1 and PvPHT2), five purple acid phosphatases (PvPAP1–PvPAP5), Pv4, and PvPS2:1. The other two genes (PvLPR1-like and PvPDR2-like) exhibit high homology with AtLPR1 and AtPDR2, respectively, which both regulate root growth in Arabidopsis. The qPCR analysis showed that overexpressing PvSPX1 led to significantly increased transcription of 10 genes compared with the control line, namely PvPT1, PvPHT2, Pv4, PvPAP1–PvPAP5, PvPS2:1, and PvLPR1-like (Fig. 7). Consistently, suppressed transcripts of PvSPX1 resulted in lower expression patterns of several genes—PvPHT2, PvPAP3, PvPS2:1, and PvLPR1-like (Supplementary Fig. S5 at JXB online). The results suggest that expression of these genes is positively regulated by PvSPX1. However, expression levels of PvPDR2-like were inhibited in the PvSPX1 overexpression lines and increased in the PvSPX1 RNAi lines, compared with those in the control line (Fig. 7; Supplementary Fig. S7), suggesting that PvPDR2-like is negatively regulated by PvSPX1 in bean. Similarly, PvSPX2 overexpression resulted in increased transcripts of several genes downstream of PvSPX1, except PvPDR2-like (Supplementary Fig. S6), suggesting that PvSPX2 might have a similar regulatory role to PvSPX1. However, overexpression of PvSPX3 did not affect expression patterns of genes downstream of PvSPX1 (Supplementary Fig. S6). Furthermore, significantly increased transcription of PvSPX1 was obviously observed in the transgenic bean hairy roots with overexpression of PvPHR1 (Fig. 8), suggesting that PvSPX1 lies downstream of PvPHR1.
Fig. 7.
Transcription levels of downstream genes of PvSPX1 in CK and PvSPX1-overexpressing bean hairy roots. Expression patterns of downstream genes were determined by qPCR in CK and two PvSPX1 overexpression hairy root lines grown in MS medium containing 1.25mM P. Expression levels of the tested genes were quantified relative to expression levels of the reference gene EF-1α (PvTC3216) using arbitrary units. OX1 and OX2 indicate two transgenic bean hairy root lines overexpressing PvSPX1. CK indicates the transgenic line transformed with the empty vector. Asterisks represent significant differences of downstream gene expression levels between PvSPX1-overexpressing and CK in t-tests. *0.01<P≤0.05; **0.001<P≤0.01; ***P≤0.001.
Fig. 8.
PvSPX1 transcripts in CK and PvPHR1-overexpressing transgenic bean hairy roots. Expression levels of PvSPX1 were determined in CK and three PvPHR1-overexpressing hairy root lines grown in MS medium containing 1.25mM P by qPCR. Expression levels of the tested genes were quantified relative to the expression levels of the reference gene EF-1α (PvTC3216) using arbitrary units. PvPHR1-1, PvPHR1-2, and PvPHR1-3 indicate three transgenic bean hairy root lines overexpressing PvPHR1. CK1, CK2, and CK3 indicate three transgenic lines transformed with the empty vector. An asterisk indicates a significant difference in PvSPX1 expression between PvPHR1-overexpressing and CK lines at the 0.05 level.
Discussion
Proteins containing the SPX domain have been well documented to be involved in the P signalling pathway of yeast and model plants, including Arabidopsis and rice (Lenburg and O’Shea, 1996; Duan et al., 2008; C. Wang et al., 2009; Z. Wang et al., 2009; Lin et al., 2010; Secco et al., 2012a). However, involvement of SPX proteins in P signalling remains largely unknown in legumes. In this study, three PvSPX genes were cloned and comparatively characterized as related to Pi starvation in bean. The results demonstrated that PvSPX1 is an important regulator in the P signalling network of common bean, which shows several novel functions in regulating root growth, P homeostasis, and downstream gene transcription.
Since the transcription of several Pi starvation-responsive genes was noticeably increased and decreased in the PvSPX1 overexpression and RNAi transgenic bean hairy roots, respectively, PvSPX1 appears to be a positive regulator in the bean P signalling network (Fig. 7; Supplementary Fig. S7 at JXB online). Furthermore, PvSPX1 appears to be a downstream gene of PvPHR1, because overexpressing PvPHR1 led to increased transcription of PvSPX1 in bean hairy roots (Fig. 8). Similarly, it has been demonstrated that AtSPX1 and OsSPX1 were downstream genes of AtPHR1 and OsPHR2 in the P signalling pathways of Arabidopsis and rice, respectively (Duan et al., 2008; C. Wang et al., 2009; Liu et al., 2010). However, regulatory roles of PvSPX1, AtSPX1, and OsSPX1 in the P signalling pathways seemed to vary among species despite them showing several similar properties, such as nuclear localization and Pi starvation-induced expression patterns (Figs 2–4). In rice, OsSPX1 has been considered as a negative regulator in the P signalling network because overexpressing OsSPX1 significantly suppressed the expression levels of 10 Pi starvation-induced genes (C. Wang et al., 2009). Also, OsSPX1 suppression resulted in increased transcripts of OsPT2 and OsPT8 in rice (C. Wang et al., 2009; Liu et al., 2010). However, AtSPX1 was considered as a positive regulator in the Arabidopsis P signalling pathway because overexpressing AtSPX1 led to increased transcription of several genes increased by Pi starvation, such as AtACP5 and AtRNS1 (Duan et al., 2008). Therefore, it seems that the regulatory roles of SPX1 in dicots might differ from those in monocot plants, which needs to be further studied.
Consistent with the enhanced expression levels of two Pi transporter genes (PvPHT2 and PvPT1), a significantly increased P concentration was observed in bean hairy roots overexpressing PvSPX1, especially under high P conditions (Fig. 5C). This suggests that PvSPX1 is involved in regulating P homeostasis in bean roots. Similarly, it has been documented that suppressed expression of OsSPX1 led to more P accumulation in both leaves and roots in rice under high P conditions (C. Wang et al., 2009; Liu et al., 2010). Taken together, these results suggest that SPX might control P homeostasis in plants through regulating expression of Pi transporter (PT) genes. However, the molecular mechanisms underlying SPX regulation of PT transcription remain largely unknown. Since PvSPX1 has regulatory roles which appear to contrast with those of OsSPX1 and AtSPX3 in P signalling pathways, it is plausible that SPX might not directly control downstream gene expression. It will be important to clarify the functions of SPX through identification of other P signalling regulators interacting with SPX in plants.
Another novel feature of PvSPX1 is its involvement in regulating root growth and root morphology in bean roots. Changes in root morphology, such as inhibition of root elongation and stimulation of root hair growth, are well accepted as typical responses of plant roots to Pi starvation (Péret et al., 2011). It was found here that overexpressing PvSPX1 significantly inhibited hairy root growth at two P levels (Fig. 5B), but led to enlarged root hair zones (Fig. 6). This suggests that overexpression of PvSPX1 could enhance root morphological modifications in adaptation to P deficiency.
In bean, it has been documented that the P-efficient genotype G19833 has greater root hair density and longer root hair length than the P-inefficient genotype DOR364 in low P conditions (Yan et al., 2004). In this study, the Pi starvation-induced PvSPX1 was originally cloned from G19833. Furthermore, higher PvSPX1 expression levels were found in G19833 than in DOR364 (data not shown) at low P, suggesting positive contributions of PvSPX1 to superior P efficiency in G19833 through regulation of root morphology. Subsequently, two genes regulating root growth in transgenic bean hairy roots, PvLPR1-like and PvPDR2-like, were cloned and their transcription was investigated Overexpression of PvSPX1 led to increased expression of PvLPR1-like and reduced expression of PvPDR2-like (Fig. 7). Since it has been documented that AtLPR1 and AtPDR2 are two critical components regulating root growth in opposite ways (Ticconi et al., 2004, 2009; Reymond et al., 2006; Svistoonoff et al., 2007; Wang et al., 2010; Miura et al., 2011), it is conceivable that changes in root morphology in bean result from up-regulation of PvSPX1, with consequent effects on transcripts of PvLPR1-like and PvPDR2-like.
Although the three PvSPX proteins studied here exhibit high homology, and belong to the same subgroup in phylogenetic tree analysis (Fig. 1), diverse properties and functions of PvSPXs were observed in response to Pi starvation, as reflected by different expression patterns, variations in subcellular localization, and dissimilar growth of transgenic bean hairy roots. In response to Pi starvation, it seems that PvSPX1 and PvSPX2 might be earlier responsive genes which are more sensitive to Pi starvation than PvSPX3 in bean leaves. At 4 d of Pi starvation, transcripts of both PvSPX1 and PvSPX2 in bean leaves were significantly increased, while PvSPX3 remained unchanged (Fig. 2). Also, with an increase in available Pi from 100 μM to 500 μM in the medium, significantly decreased transcription was observed for PvSPX1 and PvSPX2, but not for PvSPX3 (Fig. 3). Interestingly, through their promoter-fused GUS activity analysis in bean hairy roots, it was found that PvSPX1 and PvSPX2 exhibited similar spatial expression patterns (Supplementary Fig. S7 at JXB online), and similar responses to P deficiency as well as subcellular localization (Figs 2, 4), but only overexpression of PvSPX1 resulted in inhibited root growth, increased root P concentration, and changes of morphological traits in transgenic bean hairy roots (Figs 5, 6), strongly suggesting diverse functions of PvSPX members, and PvSPX requirement of other P signalling regulators to regulate P homeostasis and root growth in bean.
Similarly, diverse functions of SPX members have been demonstrated in Arabidopsis and rice (Duan et al., 2008; C. Wang et al., 2009). In Arabidopsis, suppressed AtSPX3 led to an increased P concentration in shoots, and aggregative responses to Pi starvation (Duan et al., 2008). However, knock-down of AtSPX1, AtSPX2, or AtSPX4 did not alter the phenotypes of Arabidopsis at two P levels (Duan et al., 2008). In rice, suppressed plant growth was observed through overexpression of OsSPX1 and OsSPX3, as well as suppression of OsSPX1 (C. Wang et al., 2009; Z. Wang et al., 2009). However, functions of other SPX members as related to P deficiency in rice still remain unknown.
Taken together, the results demonstrate that PvSPX1 is a positive regulator in the P signalling network of common bean, and is downstream of PvPHR1 (Fig. 9). Increased transcription of PvSPX1 led to significantly coordinated expressions of a group of Pi starvation-responsive genes, which could dramatically regulate changes of root morphology, Pi acquisition and mobilization, as well as P homeostasis in bean roots (Fig. 9).
Fig. 9.
A suggested model for PvSPX1 involvement in the P signalling network of bean. Arrowheads show the presence of positive regulation. Flat-ended lines show negative regulation. Dotted lines represent the putative regulatory pathway. The question mark indicates uncertainy in the network.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Expression of PvSPX genes in transgenic bean hairy roots.
Figure S2. Expression of PvSPX1 in PvSPX1 RNAi transgenic bean hairy roots.
Figure S3. Growth and P concentration of bean hairy roots in CK and PvSPX1 RNAi transgenic lines at two P levels.
Figure S4. Percentage of root hair zone in bean hairy roots with suppressed PvSPX1 at two P levels.
Figure S5. Transcription levels of downstream genes of PvSPX1 in CK and PvSPX1 RNAi transgenic lines.
Figure S6. Transcription levels of downstream genes of PvSPX1 in CK and overexpression transgenic lines of PvSPX2 or PvSPX3.
Figure S7. Expression patterns of PvSPX1 and PvSPX2 through their promoter:GUS analysis.
Table S1. List of primers used in the study.
Table S2. Effects of phosphorus availability on bean growth.
Acknowledgements
This work was supported by grants from the National Key Basic Research Special Funds of China [2011CB100301]; the National Natural Science Foundation of China [31101593 and 31025022]; the Project of Science and Technology New Star in Zhujiang Guangzhou city [2011J2200055]; and the Special Fund for Science and Technology of Guangdong Province in 2012 for Construction of State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources. The authors acknowledge Dr Thomas Walk for critical reading of the manuscript.
References
- Battini JL, Rasko JE, Miller AD. 1999. A human cell-surface receptor for xenotropic and polytropic murine leukemia viruses: possible role in G protein-coupled signal transduction. Proceedings of the National Academy of Sciences, USA 96, 1385–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozzo GG, Raghothama KG, Plaxton WC. 2002. Purification and characterization of two secreted purple acid phosphatase isozymes from phosphate-starved tomato (Lycopersicon esculentum) cell cultures. European Journal of Biochemistry 269, 6278–6286 [DOI] [PubMed] [Google Scholar]
- Chiou TJ, Lin S. 2011. Signaling network in sensing phosphate availability in plants. Annual Review of Plant Biology 62, 185–206 [DOI] [PubMed] [Google Scholar]
- Del Pozo JC, Allona I, Rubio V, Leyva A, De La Peña A, Aragoncillo C, Paz-Ares J. 1999. A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilising/oxidative stress conditions. The Plant Journal 19, 579–589 [DOI] [PubMed] [Google Scholar]
- Duan K, Yi K, Dang L, Huang H, Wu W, Wu P. 2008. Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. The Plant Journal 54, 965–975 [DOI] [PubMed] [Google Scholar]
- Fox TR, Comerford NB. 1990. Low-molecular-weight organic acids in selected forest soils of the southeastern USA. Soil Science Society of America Journal 54, 139–144 [Google Scholar]
- Hamburger D, Rezzonico E, MacDonald-Comber PJ, Somerville C, Poirier Y. 2002. Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. The Plant Cell 14, 889–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández G, Valdes-Lopez O, Ramirez M, et al. 2009. Global changes in the transcript and metabolic profiles during symbiotic nitrogen fixation in phosphorus-stressed common bean plants. Plant Physiology 151, 1221–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S, Peng M, Rothstein SJ. 2011. Genetic regulation by NLA and microRNA 827 for maintaining nitrate-dependent phosphate homeostasis in Arabidopsis . PLoS Genetics 7, e1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenburg ME, O’Shea EK. 1996. Signaling phosphate starvation. Trends in Biochemical Sciences 21, 383–387 [PubMed] [Google Scholar]
- Liang C, Chen Z, Yao Z, Tian J, Liao H. 2012a. Characterization of two putative protein phosphatase genes and their involvement in phosphorus efficiency in Phaseolus vulgaris . Journal of Integrative Plant Biology 54, 400–411 [DOI] [PubMed] [Google Scholar]
- Liang C, Sun L, Yao Z, Liao H, Tian J. 2012b. Comparative analysis of PvPAP gene family and their functions in response to phosphorus deficiency in common bean. PLoS One 7, e38106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Tian J, Lam HM, Lim BL, Yan X, Liao H. 2010. Biochemical and molecular characterization of PvPAP3, a novel purple acid phosphatase isolated from common bean enhancing extracellular ATP utilization. Plant Physiology 152, 854–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H, Yan X, Rubio G, Beebe SE, Blair MW, Lynch JP. 2004. Genetic mapping of basal root gravitropism and phosphorus acquisition efficiency in common bean. Functional Plant Biology 31, 959–970 [DOI] [PubMed] [Google Scholar]
- Ligaba A, Yamaguchi M, Shen H, Sasaki T, Yamamoto Y, Matsumoto H. 2004. Phosphorus deficiency enhances plasma membrane H+-ATPase activity and citrate exudation in greater purple lupin (Lupinus pilosus). Functional Plant Biology 31, 1075–1083 [DOI] [PubMed] [Google Scholar]
- Lin SI, Santi C, Jobet E, et al. 2010. Complex regulation of two target genes encoding SPX-MFS proteins by rice miR827 in response to phosphate starvation. Plant and Cell Physiology 51, 2119–2131 [DOI] [PubMed] [Google Scholar]
- Liu F, Wang Z, Ren H, Shen C, Li Y, Ling HQ, Wu C, Lian X, Wu P. 2010. OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice. The Plant Journal 62, 508–517 [DOI] [PubMed] [Google Scholar]
- Miura K, Lee J, Gong Q, Ma S, Jin JB, Yoo CY, Miura T, Sato A, Bohnert HJ, Hasegawa PM. 2011. SIZ1 regulation of phosphate starvation-induced root architecture remodeling involves the control of auxin accumulation. Plant Physiology 155, 1000–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy J, Riley J. 1962. A modified single solution method for the determination of phosphate in natural water. Analytica Chimica Acta 27, 31–35 [Google Scholar]
- Péret B, Clément M, Nussaume L, Desnos T. 2011. Root developmental adaptation to phosphate starvation: better safe than sorry. Trends in Plant Science 16, 442–450 [DOI] [PubMed] [Google Scholar]
- Raghothama KG. 1999. Phosphate acquisition. Annual Review of Plant Physiology and Plant Molecular Biology 50, 665–693 [DOI] [PubMed] [Google Scholar]
- Reymond M, Svistoonoff S, Loudet O, Nussaume L, Desnos T. 2006. Identification of QTL controlling root growth response to phosphate starvation in Arabidopsis thaliana . Plant, Cell and Environment 29, 115–125 [DOI] [PubMed] [Google Scholar]
- Robinson WD, Park J, Tran HT, Del Vecchio HA, Ying S, Zins JL, Patel K, McKnight TD, Plaxton WC. 2012. The secreted purple acid phosphatase isozymes AtPAP12 and AtPAP26 play a pivotal role in extracellular phosphate-scavenging by Arabidopsis thaliana . Journal of Experimental Botany 63, 6531–6542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secco D, Baumann A, Poirier Y. 2010. Characterization of the rice PHO1 gene family reveals a key role for OsPHO1;2 in phosphate homeostasis and the evolution of a distinct clade in dicotyledons. Plant Physiology 152, 1693–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secco D, Wang C, Arpat BA, Wang ZY, Whelan J. 2012a. The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytologist 193, 842–851 [DOI] [PubMed] [Google Scholar]
- Secco D, Wang C, Shou H, Whelan J. 2012b. Phosphate homeostasis in the yeast Saccharomyces cerevisiae, the key role of the SPX domain-containing proteins. FEBS Letters 586, 289–295 [DOI] [PubMed] [Google Scholar]
- Spain BH, Koo D, Ramakrishnan M, Dzudzor B, Colicelli J. 1995. Truncated forms of a novel yeast protein suppress the lethality of a G protein alpha subunit deficiency by interacting with the beta subunit. Journal of Biological Chemistry 270, 25435–25444 [DOI] [PubMed] [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C. 2006. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nature Protocols 1, 2019–2025 [DOI] [PubMed] [Google Scholar]
- Stefanovic A, Ribot C, Rouached H, Wang Y, Chong J, Belbahri L, Delessert S, Poirier Y. 2007. Members of the PHO1 gene family show limited functional redundancy in phosphate transfer to the shoot, and are regulated by phosphate deficiency via distinct pathways. The Plant Journal 50, 982–994 [DOI] [PubMed] [Google Scholar]
- Ström L, Owen AG, Godbold DL, Jones DL. 2005. Organic acid behaviour in a calcareous soil implications for rhizosphere nutrient cycling. Soil Biology and Biochemistry 37, 2046–2054 [Google Scholar]
- Svistoonoff S, Creff A, Reymond M, Sigoillot-Claude C, Ricaud L, Blanchet A, Nussaume L, Desnos T. 2007. Root tip contact with low-phosphate media reprograms plant root architecture. Nature Genetic 39, 792–796 [DOI] [PubMed] [Google Scholar]
- Taghipour M, Jalali M. 2012. Effect of low-molecular-weight organic acids on kinetics, release and fractionation of phosphorus in some calcareous soils of western Iran. Environmental Monitoring and Assessment 185, 5471–5482 [DOI] [PubMed] [Google Scholar]
- Tian J, Venkatachalam P, Liao H, Yan X, Raghothama K. 2007. Molecular cloning and characterization of phosphorus starvation responsive genes in common bean (Phaseolus vulgaris L.). Planta 227, 151–165 [DOI] [PubMed] [Google Scholar]
- Tian J, Wang X, Tong Y, Chen X, Liao H. 2012. Bioengineering and management for efficient phosphorus utilization in crops and pastures. Current Opinion in Biotechnology 23, 866–871 [DOI] [PubMed] [Google Scholar]
- Ticconi CA, Delatorre CA, Lahner B, Salt DE, Abel S. 2004. Arabidopsis pdr2 reveals a phosphate-sensitive checkpoint in root development. The Plant Journal 37, 801–814 [DOI] [PubMed] [Google Scholar]
- Ticconi CA, Lucero RD, Sakhonwasee S, Adamson AW, Creff A, Nussaume L, Desnos T, Abel S. 2009. ER-resident proteins PDR2 and LPR1 mediate the developmental response of root meristems to phosphate availability. Proceedings of the National Academy of Sciences, USA 106, 14174–14179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdes-Lopez O, Arenas-Huertero C, Ramirez M, Girard L, Sanchez F, Vance CP, Luis RJ, Hernández G. 2008. Essential role of MYB transcription factor: PvPHR1 and microRNA: PvmiR399 in phosphorus-deficiency signalling in common bean roots. Plant, Cell and Environment 31, 1834–1843 [DOI] [PubMed] [Google Scholar]
- Vance CP, Uhde-Stone C, Allan DL. 2003. Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytologist 157, 423–447 [DOI] [PubMed] [Google Scholar]
- Wang C, Huang W, Ying Y, Li S, Secco D, Tyerman S, Whelan J, Shou H. 2012. Functional characterization of the rice SPX-MFS family reveals a key role of OsSPX-MFS1 in controlling phosphate homeostasis in leaves. New Phytologist 196, 139–148 [DOI] [PubMed] [Google Scholar]
- Wang C, Ying S, Huang H, Li K, Wu P, Shou H. 2009. Involvement of OsSPX1 in phosphate homeostasis in rice. The Plant Journal 57, 895–904 [DOI] [PubMed] [Google Scholar]
- Wang LS, Dong JS, Gao ZY, Liu D. 2012. The Arabidopsis gene HYPERSENSITIVE TO PHOSPHATE STARVATION 3 encodes ETHYLENE OVERPRODUCTION 1. Plant and Cell Physiology 56, 1093–1105 [DOI] [PubMed] [Google Scholar]
- Wang X, Wang Y, Tian J, Lim BL, Yan X, Liao H. 2009. Overexpressing AtPAP15 enhances phosphorus efficiency in soybean. Plant Physiology 151, 233–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ribot C, Rezzonico E, Poirier Y. 2004. Structure and expression profile of the Arabidopsis PHO1 gene family indicates a broad role in inorganic phosphate homeostasis. Plant Physiology 135, 400–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Hu H, Huang H, Duan K, Wu Z, Wu P. 2009. Regulation of OsSPX1 and OsSPX3 on expression of OsSPX domain genes and Pi-starvation signaling in rice. Journal of Integrative Plant Biology 51, 663–674 [DOI] [PubMed] [Google Scholar]
- Yan X, Liao H, Beebe SE, Blair MW, Lynch JP. 2004. QTL mapping of root hair and acid exudation traits and their relationship to phosphorus uptake in common bean. Plant and Soil 265, 17–29 [Google Scholar]
- Yan X, Liao H, Trull MC, Beebe SE, Lynch JP. 2001. Induction of a major leaf acid phosphates does not confer adaptation to low P availability in common bean. Plant Physiology 125, 1901–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang GZ, Ding GD, Shi L, Cai HM, Xu FS. 2012. Characterization of phosphorus starvation-induced gene BnSPX3 in Brassica napus . Plant and Soil 350, 339–351 [Google Scholar]
- Zhou J, Jiao F, Wu Z, Li Y, Wang X, He X, Zhong W, Wu P. 2008. OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiology 146, 1673–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.