Abstract
Translation control is a prevalent form of gene expression regulation in developmental and stem cell biology. A recent paper by Signer et al. measures protein synthesis rates in the mouse hematopoietic compartment and reveals the importance of diminished protein production for maintaining hematopoietic stem cell function and restraining oncogenic potential.
Translation control has long been known to serve as a major control point for gene regulation, not merely as an additional layer where “fine-tuning” occurs, but critical in directing key cellular processes central to stem cell biology (Thompson, 2007). This mode of gene regulation is likely to be of paramount importance in both germline and somatic stem cells that require tight control of gene expression to prevent unsolicited cell fate commitment. Linking the amount of protein synthesis to mRNA quality and translational accessibility in the cytoplasm provides unique advantages to long-lived, multipotent cells. Consistently, translational control has been shown to be important for the proliferation, growth, and differentiation of a diverse array of stem cells, such as Drosophila (Insco et al., 2012; Zhang et al., 2014) and C. elegans germ cells (Thompson, 2007), and mouse embryonic stem cells (Sampath et al., 2008). An interesting, recent study by Morrison and colleagues (Signer et al., 2014) extends the realm of protein synthesis to the adult hematopoietic stem cell compartment in mice. They provide a detailed quantitative analysis of protein synthesis within all major cell lineages in the hematopoietic compartment, revealing that protein production in hematopoietic stem cells (HSCs) and multipotent progenitors (MPPs) is markedly lower compared to other hematopoietic cell types. Moreover, tightly controlled protein synthesis appears to be critical for HSC function and serves as an important tumor suppressive mechanism.
Methods for precisely measuring protein synthesis rates in vivo have been difficult to achieve. A new fluorogenic assay using an alkyne analog of puromycin, O-propargyl-puromycin (OP-Puro), developed by the Salic lab (Liu et al., 2012), can be injected into mice where it is taken up by cells and incorporated into nascent polypeptides. This method can therefore quantify protein synthesis in individual cells. By employing OP-Puro, Signer et al. find that the amount of protein synthesized in HSCs in vivo is reduced compared to most other hematopoietic progenitors, including common myeloid progenitors (CMPs), granulocyte-macrophages (GMPs) and more differentiated cell types. A decrease in protein synthesis in HSCs is not entirely unexpected considering that more than 95% of these cells are in a quiescent state. Nevertheless, one of the most intriguing aspects of the Signer et al. study (Signer et al., 2014) is that reduction in the amount of proteins synthesized in HSCs appears to occur independently from cell cycle status, cell size, ribosomal RNA (rRNA), or total RNA content. Moreover, even forced entry into the cell cycle does not appear to revert this shutdown of protein synthesis. These studies however do not exclude the possibility that a shutdown in protein synthesis is simply a reflection of HSC quiescence compared to other progenitor lineages. Indeed, a dormant, quiescent phenotype may not be merely associated with a specific cell cycle status, but also with other key determinants, such as lower exposure to oxygen and different metabolic programs, which are unique to HSCs compared to more committed progenitors. By extension, a similar dramatic shutdown in protein synthesis occurs when cells undergo cellular senescence.
Morrison and colleagues next tested how reducing protein synthesis even further in HSCs would impact on their function. A ribosomal protein (RP) loss-of-function mouse mutant, Rpl24Bst/+, was previously shown to display an overall ~30% decrease in the rates of protein synthesis that, similar to Drosophila RP minute mutations, produces a smaller, yet largely physiologically normal adult mouse (Figure 1). Signer et al. (Signer et al., 2014) examine the potential of Rpl24Bst/+ HSCs to reconstitute the bone marrow compartment of a lethally irradiated mouse. Consistent with the observation that a delicate balance in protein production may be required for HSC function, an even further decrease in protein synthesis in Rpl24Bst/+ HSCs results in a cell autonomous reconstitution defect and impaired proliferative potential. Importantly, these effects are independent from p53 activation, known to be elicited downstream of some ribosomal defects.
Figure 1. Impact of protein synthesis in progenitor cells and downstream oncogenic pathways.
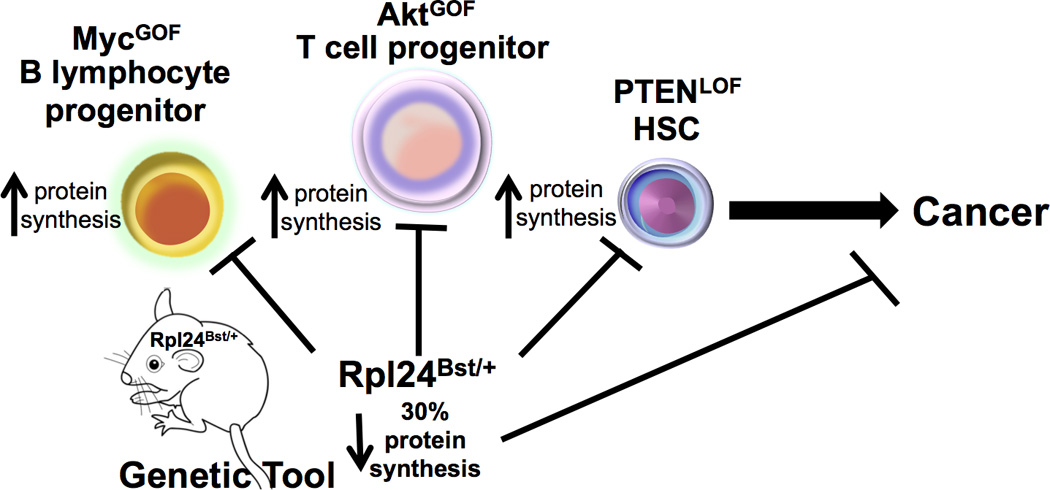
The Rpl24Bst/+ mouse is an important genetic tool to restore aberrant increases in protein synthesis downstream of oncogenic signaling in progenitor cells to normal levels. This reduction can suppress cancer formation downstream of Myc and PI3K-Akt-mTOR hyperactivation in B and T cell progenitors. Importantly, Signer et al. further show that restoring normal protein synthesis downstream loss of PTEN rescues HSC function and leukemogenesis. GOF: gain of function, LOF: loss of function.
The Rpl24Bst/+ mouse was previously established as an important genetic tool to restore aberrant increases in protein synthesis downstream of oncogenic signaling to normal levels and was shown to suppress cancer formation downstream of Myc and PI3K-Akt-mTOR hyperactivation in B and T cell progenitors (Barna et al., 2008; Hsieh et al., 2010) (Figure 1). These oncogenic pathways stimulate increases in protein synthesis by directly regulating the expression and activity of ribosome components and translation factors. Signer et al. similarly show that Pten deletion, a critical negative regulator of PI3K-AktmTOR signaling, within HSCs leads to hyperactivation of protein synthesis, which is restored to relatively normal levels in the Rpl24Bst/+ background (Figure 1). Deletion of Pten has been shown to deplete HSCs and promote leukemogenesis. Strikingly, blocking increases in protein synthesis in a Pten-deficient background restores, to a large extent, normal HSC function and suppresses leukemogenesis. These findings suggest that a delicate balance in protein synthesis serves a vital role for HSC function and tumor suppression.
Overall, the Signer et al. study reveals an exquisite layer of control in protein synthesis within the hematopoietic stem cell compartment. However, the intrinsic or extrinsic mechanisms responsible for low rates of protein production in HSCs as well as MPPs remain undetermined. Deletion of a critical component of the mTORC1 complex, Raptor, which controls the phosphorylation of key translation initiation components such as 4EBP1 (a negative regulator of eIF4E) and RPS6K, is known to play a critical role in HSC regeneration and leukemogenesis (Kalaitzidis et al., 2012). In the current study, Signer et al. show that HSCs and MPPs display a reduction in 4EBP1 phosphorylation compared to other hematopoietic cells. Together, these findings hint at the possibility that regulation of cap-dependent translation initiation itself may be the critical mode for shutting down protein production in HSCs.
Importantly, it is not clear why HSCs require lower rates of protein synthesis for proper function. Imperative to this question is whether dampened protein synthesis simply reflects the general shutdown of gene expression at many levels associated with a quiescent program. For example, the differentiation of ES cells into more committed progenitors induces an anabolic switch with global increases in transcript abundance as well as protein synthesis and increased ribosomal loading of transcripts (Sampath et al., 2008). How might the findings of Signer et al. more broadly extend to other stem cell compartments where translation control is known to be important? Intriguingly, Drosophila ovarian stem cells show increased, higher rates of rRNA synthesis relative to their immediate progeny (Zhang et al., 2014). This is linked to highly specific changes in the expression of a subset of proteins, such as those in the BMP pathway, which affect proliferation and cell fate decisions. Thus, perceived global changes in protein synthesis or ribosome biogenesis may have more select outcomes on stem cell lineages and translation of key mRNAs. Unraveling whether select transcripts are similarly influenced by global changes in the protein synthesis rates in HSCs quantified by Signer et al. will be an exciting future direction. The answers to these questions will also be critical to advance our understanding of human disorders known as “ribosomopathies” (McCann and Baserga, 2013), where mutations in components of the ribosome are associated with hematopoietic dysfunction, including perturbations in HSC function, and increased cancer susceptibility.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Barna M, Pusic A, Zollo O, Costa M, Kondrashov N, Rego E, Rao PH, Ruggero D. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature. 2008;456:971–975. doi: 10.1038/nature07449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh AC, Costa M, Zollo O, Davis C, Feldman ME, Testa JR, Meyuhas O, Shokat KM, Ruggero D. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cancer Cell. 2010;17:249–261. doi: 10.1016/j.ccr.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insco ML, Bailey AS, Kim J, Olivares GH, Wapinski OL, Tam CH, Fuller MT. A self-limiting switch based on translational control regulates the transition from proliferation to differentiation in an adult stem cell lineage. Cell Stem Cell. 2012;11:689–700. doi: 10.1016/j.stem.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaitzidis D, Sykes SM, Wang Z, Punt N, Tang Y, Ragu C, Sinha AU, Lane SW, Souza AL, Clish CB, et al. mTOR complex 1 plays critical roles in hematopoiesis and Pten-loss-evoked leukemogenesis. Cell Stem Cell. 2012;11:429–439. doi: 10.1016/j.stem.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Xu Y, Stoleru D, Salic A. Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc Natl Acad Sci U S A. 2012;109:413–418. doi: 10.1073/pnas.1111561108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann KL, Baserga SJ. Genetics. Mysterious ribosomopathies. Science. 2013;341:849–850. doi: 10.1126/science.1244156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath P, Pritchard DK, Pabon L, Reinecke H, Schwartz SM, Morris DR, Murry CE. A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2008;2:448–460. doi: 10.1016/j.stem.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Signer RA, Magee J, Salic A, Morrison SJ. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature. 2014 doi: 10.1038/nature13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B, Wickens M, Kimble J. Translational control in development. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 507–544. [Google Scholar]
- Zhang Q, Shalaby NA, Buszczak M. Changes in rRNA transcription influence proliferation and cell fate within a stem cell lineage. Science. 2014;343:298–301. doi: 10.1126/science.1246384. [DOI] [PMC free article] [PubMed] [Google Scholar]


