Abstract
Circadian clocks control a variety of neuronal, behavioral and physiological responses, via transcriptional regulation of a significant portion of the genome. We describe the complex communication network between the brain-specific central clock and the tissue-specific peripheral clocks that serve to synchronize the organism to both external and internal demands. In addition, we discuss and speculate how epigenetic processes are involved in creating transcriptional environments that are permissive to tissue-specific gene expression programs, which work in concert with the circadian machinery. Accumulating data shows that chromatin remodeling events may be critical for providing specificity and plasticity in circadian regulation, and metabolic cues may be involved in directing such epigenetic events. A detailed understanding of the communication cues between the central and peripheral clocks is crucial for a more complete understanding of the circadian system and the multiple levels of control that are implicated in maintaining biological timekeeping.
Circadian rhythms control the timing of numerous physiological processes over a 24-hour period, including sleep-wake cycles, thermoregulation, feeding, metabolic regulation and hormone production. These cyclic events are self-sustained and centrally controlled by the central pacemaker located within the suprachiasmatic nucleus (SCN)1. The SCN, located in the anterior hypothalamus, is a paired structure consisting of two nuclei, each of which contain approximately 10,000 neurons1. The central clock is receptive to environmental cues that ‘entrain’ or establish rhythmic periodicity of the circadian pacemaker. The most prominent entrainment cue is light, the photic input being transmitted to the SCN through the retino-hypothalamic tract2. Although the circadian clock operates as a strict timekeeping system, its responsiveness to entrainment cues endows it with remarkable plasticity which allows for the synchronization of the circadian pacemaker with the surrounding environment. Extensive disruption of circadian rhythms has been linked to numerous diseases including sleep disorders, depression, metabolic syndrome, cardiovascular disturbances and tumorigenesis3, 4.
At the heart of the molecular network that constitutes the circadian clock are the core transcription factors CLOCK and BMAL1 that heterodimerize and direct transcriptional activation of clock controlled genes (CCG), by binding to E-box sites on their promoters (Fig. 1). Among these CCGs, CLOCK and BMAL1 also direct transcription of their own repressors, period (PER) and cryptochrome (CRY) family members, creating a tightly self-regulated system1, 2, 5. During the day, transcription of PER and CRY is high, leading to protein translation of the circadian repressors, and resulting in formation of the inhibitory complex with CLOCK and BMAL1 that abolishes transcription of CCGs. The degradation of PER and CRY alleviates transcriptional repression and allows CLOCK:BMAL1 mediated transcription to again proceed, establishing an oscillatory rhythm in circadian gene expression. An additional level of circadian regulation exists with the orphan nuclear receptors RORα and REV-ERBα that activate and repress transcription of the Bmal1 gene, respectively1, 2. We refer the readers to some of the many review articles centered on the complex regulatory network of the circadian clock4, 6, 7.
Fig. 1. The circadian CLOCK network.
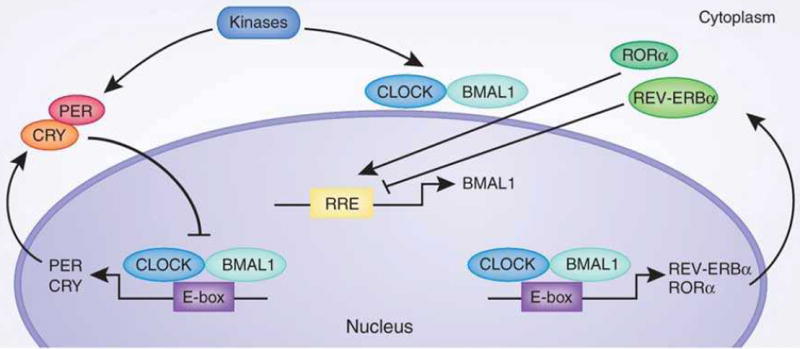
The core circadian transcription factors, CLOCK and BMAL1, direct E-box mediated transcription of clock controlled genes (CCGs), including activators and repressors of the circadian system. PER and CRY protein translation occurs at night and subsequently causes repression of the core CLOCK:BMAL1 transcriptional complex. Degradation of the PER/CRY repressors prompts a new circadian cycle whereby CLOCK:BMAL1 transcription is reinitiated. In addition to transcriptional regulation, post-translational modifications play a critical role in the modulation of circadian proteins. Here only phosphorylation is schematically presented. This can be elicited by a number of kinases including CKIε, CKIδ, CK2α, GSK3β and AMPK. Other post-translational modifications of clock proteins include acetylation, sumoylation and ubiquitination. RRE, REV-ERB/ROR response element; CK, Casein kinase; GSK3β, glycogen synthase kinase-3 beta; P, phosphorylation.
The circadian network: From the SCN to the periphery
Whereas timekeeping is established by the central clock in the SCN, it must be maintained by other clock systems in surrounding regions of the brain, as well as in peripheral tissues. While entrainment to extrinsic cues modulates the master circadian clock, the biological pacemaker also requires responsiveness to intrinsic physiological cues to maintain synchrony within the circadian network. We briefly discuss some factors that synchronize SCN neurons to one another and establish rhythmicity within the central clock, and some examples of the humoral output factors of the SCN that transmit timekeeping to the peripheral clocks will be given.
The vasoactive intestinal polypeptide (VIP), in conjunction with its receptor (VPAC2R), is believed to be required for SCN synchronization. VIP is responsive to light and in VIP knockout mice, the normal rhythmic firing of SCN neurons is lost8. The SCN, in turn, also directs activity in other regions of the brain via secreted factors such as hormones and neurotransmitters. Vasopressin, for example, is considered an SCN neurotransmitter or a secreted humoral output factor. Vasopressin targets neurons within the dorsomedial hypothalamus (DMH) and the paraventricular nucleus (PVN) by directed projections emerging from the SCN9. Grafts of SCN in SCN-lesioned brains restored PVN circadian firing of neurons within the PVN region, while administration of vasopression similarly induced circadian rhythmicity in the hypothalamus 10. This data suggests that SCN projections to surrounding areas of the hypothalamus are critical, along with humoral factors, to direct and maintain circadian timekeeping in other regions of the brain.
Work from a decade ago showed that the central circadian pacemaker is not the only clock: most (if not all) peripheral tissues bear built-in circadian oscillators that are centrally controlled by the master clock. The fact that the central SCN clock dominates and directly controls the peripheral circadian oscillations was demonstrated by work using heterologous graft transplants of SCNs11, which resulted in dramatically dampened circadian oscillation in liver transcripts after SCN ablation12. Lesions within the SCN lead to altered cycles of hormonal synthesis, sleep patterns, drinking and locomotor activity1, 2, 11, presumably via a complex network of humoral factors that control behavior and physiology.
How does the SCN synchronize these peripheral clocks? Some intriguing results are beginning to provide some answers. Expression of the SCN secreted protein prokineticin 2 (PK2) is light-sensitive and the mRNA levels of PK2 oscillate in a circadian manner, suggesting a clock-controlled network. Levels of this protein are likely to regulate behavior and locomotor activity in mice, presumably through PK2 receptors (PKR2) found in surrounding regions of the brain, such as PVN and DMH13. Similarly, transforming growth factor alpha (TGFα) is another output signal of the SCN that has been implicated in sleep and locomotor activity by binding epidermal growth factor receptors found in the hypothalamic subparaventricular zone14. Glucocorticoid levels are similarly oscillatory, as monitored by ablation of the SCN and the resultant loss of rhythmic levels of plasma glucocorticoids and feeding patterns15. Of particular interest, glucocorticoids can also feedback and entrain the peripheral circadian clock, as dexamethasone treatment of rat-1 fibroblasts rapidly induced Per1 gene expression16. This effect is specific to peripheral clock systems, as SCN neurons are not entrained by dexamethasone treatment.
Overall, the central circadian clock system in the SCN can be entrained by multiple factors, which by signaling to peripheral clocks, demonstrates circadian plasticity and diversity in systemic timekeeping. It is unclear how such complex regulatory networks operate to maintain biological timekeeping. The extent of this systemic pacemaker network is not fully understood.
Clocks are everywhere, but is their clockwork the same?
It is clear that the interaction between the central and peripheral clocks is characterized by great heterogeneity. Furthermore, even though the SCN and the peripheral clocks are closely linked, their circadian functions and output are vastly divergent, begging the question on how the pacemakers intrinsic to these tissues may differ. For example, SCN neurons from neonatal rats displayed independent circadian rhythmicity in neuronal firing rate when cultured in vitro, while reversible disruption of action potential in these cells resulted in re-emergence of unaltered circadian behavior17. In contrast, cultured cells from peripheral tissues which have a circadian cycle, did not display sustained and synchronous oscillations18. Are there common features within the programs of circadian gene expression in the central versus the peripheral clocks? Also, among peripheral clocks, are there tissue-specific transcriptional regulators whose actions intersect with the clock machinery?
Several genome-wide array analyses have been centered on determining the proportion and specificity of cycling transcripts, to understand tissue-specific gene expression programs. The first remarkable finding indicated that ~10% of all expressed genes in any tissue are under circadian regulation12, 19. This unexpectedly high proportion of circadian transcripts suggests that the clock machinery may direct widespread events of cyclic chromatin remodeling and consequent transcriptional activation/repression. Furthermore, genome-wide studies comparing the central SCN pacemaker and peripheral tissues, such as the liver, revealed that between 5-10% of cycling genes where identical in both tissue types12, 19. A recent analysis covering 14 mouse tissues identified ~ 10,000 known genes showing circadian oscillations in at least one tissue. Not surprisingly, the number of common genes showing circadian oscillation in multiple tissues decreased drastically as the number of tissues included in the comparative analysis increased, with only 41 genes displaying circadian oscillation in at least 8 out of 14 tissues20. These findings underscore the presence of molecular interplays between the core clockwork, which can be assumed to be common to all tissues, and cell-specific transcriptional systems. Taking into consideration the view of the mammalian circadian clock as a transcriptional network21, 22, through which the oscillator acquires plasticity and robustness, it is reasonable to speculate that the clock network contributes to physiological responses by intersecting with cell-specific transcriptional pathways. We hypothesize that this complex protein interaction network implicates cell-specific chromatin remodeling events.
Non-clock invitees to the circadian machinery
Neurons in the SCN express oscillating genes for the most part implicated in neuropeptide/neurotransmitter synthesis, cellular redox state and energy usage. Peripheral cells on the other hand express genes which are required for tissue-specific actions: hepatocytes, for example, display a unique profile, with oscillating transcripts primarily involved in metabolism19. It is hard to account for these fundamental differences solely based on the core clock machinery, as the core circadian proteins are conserved in the SCN and peripheral tissues. Instead, interactions between the core clock machinery and non-clock transcriptional networks may dictate tissue-specific circadian transcriptomes. In support of this idea, core clock components have been shown to interplay with other transcription factors, such as cAMP responsive element binding protein (CREB)23, the glucocorticoid receptor (GR)24 and the inhibitor of DNA binding 2 (Id2)25, suggesting that more extensive and definite interactions may contribute to the physiological specificity of individual peripheral oscillators. This concept could be further extended by simply considering that differences in gene expression profiles of peripheral circadian clock networks could be caused by the preferential recruitment of specialized transcription factors or co-regulatory proteins that dictate tissue-specific transcriptomes. In this respect it is notable that transcription factors such as myc associated factor X (Max), MAX interactor 1 (Mxi1), HNF1 homeobox B (Hnf1b), upstream transcription factor 2 (Usf2) and the CCAAT-box nuclear factor I/X (Nfix) were rhythmic exclusively in the liver, and not in SCN neurons19. This data implies that the transcriptional ‘environment’, including tissue-specific transcription factors, may supplement the core circadian machinery in peripheral clock systems (Fig. 2).
Fig. 2. What underlies the different genomic responses of central versus peripheral clocks?
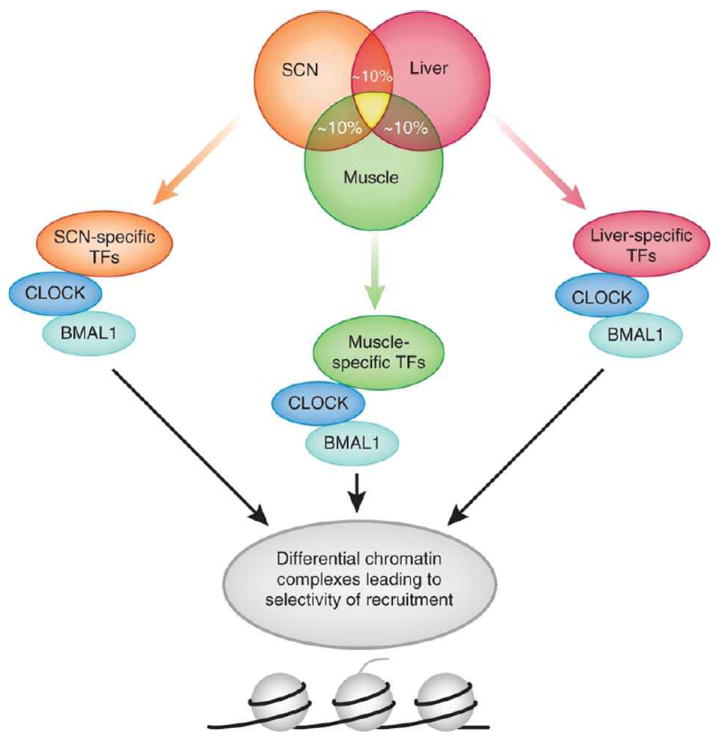
Approximately 10% of transcripts in a given tissue display circadian expression. Of all oscillatory transcripts in each tissue (here schematically represented as a circle for SCN, liver and muscle), only a fraction of ~5-10% is common between two given tissues, and that fraction decreases drastically when intersecting more than two tissues (the yellow area in the middle of the circles)12, 19-22. These differences can be accounted by the contribution of tissue-specific transcription factors (TFs) which interplay with the circadian machinery (here simplistically represented by the CLOCK:BMAL1 complex). The differential composition of these complexes in different tissues and circadian times might bestow selectivity of recruitment to chromatin loci corresponding to promoters of CCGs.
The above notions provoke some considerations: if tissue-specific transcriptional networks regulate the circadian transcriptome in peripheral pacemakers, what is the contribution of the CLOCK:BMAL1 core machinery? Specifically, given the dominance of the SCN in the control of systemic rhythms, is the modulation of peripheral circadian profiles exerted on the CLOCK:BMAL1 heterodimer complex or on tissue-specific transcription factors? It is known that CLOCK:BMAL1 expression and function are required in peripheral tissues for rhythmic circadian gene expression26, yet the tissue-specific localization of co-regulatory proteins may be essential. It is important to stress that at least some of the tissue-specific transcription factors interacting with the core clock machinery could be encoded by CCGs, and their respective expression modulated directly by CLOCK:BMAL1. Interestingly, a number of genes encoding tissue-specific transcription factors have E-box motifs in their promoters, and the expression of these clock target genes do oscillate in a circadian manner. These clock-controlled transcription factors include TEA domain family member 4 (Tead4), upstream transcription factor 1 (Usf1), pre B-cell leukemia transcription factor 2 (Pbx2), v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (Maf-g), to name a select few, that are involved in skeletal muscle, metabolic tissue, blood cells, and skeletal muscle/heart, respectively27. These notions suggest that peripheral tissues can contribute to rhythmic gene expression, though these transcription factors are clock-controlled, implicating a hierarchy within the clock system. Also, though the master pacemaker still dictates timekeeping, there could be a secondary contribution from peripheral clock transcription networks to determine tissue-specificity and gene expression. Finally, as previously mentioned, the significant proportion of circadian transcripts indicates a potential presence of extensive chromatin remodeling events. The difference between central and peripheral oscillators may extend also to this level. Indeed, accumulating evidence indicates the presence of cell-specific chromatin remodeling mechanisms.
The circadian epigenome
Since transcription of peripheral clock genes has to be modulated by the specific transcriptional environment, how are these factors integrated in regulating tissue-specific circadian gene expression? Epigenetic control has been described on a number of levels, including DNA methylation, histone modifications, histone variants, small RNAs and/or a combination of all these events, some of which may be inherited, that participate in modulating gene expression state. While our understanding of the mechanisms that direct circadian epigenetic control is still limited, emerging evidence implicates histone modifications and some chromatin remodeling proteins (Fig. 3) as particularly important in directing circadian epigenetic control. In turn, it is important to elucidate how these modifications themselves are regulated.
Fig. 3. Chromatin remodeling and the circadian clock.
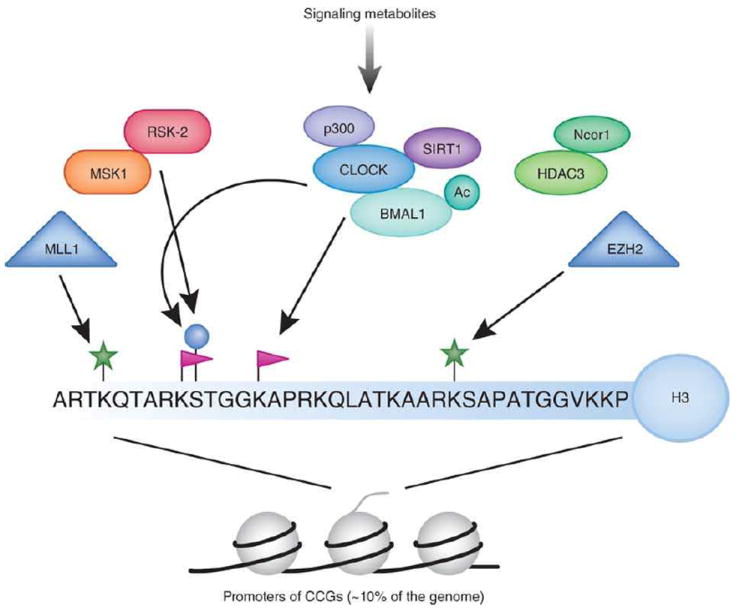
Directed chromatin-modifying events responsible for rhythmic CCG transcription, comprise a major portion of the circadian epigenome. Our current understanding of post-translational modifications of the H3 tail suggests that phosphorylation (serine 10), acetylation (lysine 9/14) and methylation (lysine 4/27) are associated to circadian transcription. Some chromatin modifiers may be directly or indirectly modulated by the circadian system. Acetylation of non-histone proteins can also occur in a clock-dependent manner, this is the case for BMAL138 and the GR24. The involvement of the NAD+-dependent deacetylase SIRT1 in circadian control and its physical interaction with CLOCK revealed a link between circadian clock and cellular metabolism30, 39. The metabolic state of the cell has a robust response on epigenetic control, and some metabolic cues have been found to oscillate. The extent to which the circadian clock regulates the metabolic state of the cell is only beginning to emerge, and the full elucidation of these concepts is awaited.
Studies on the effect of light on the central pacemaker provided the first clue that chromatin remodeling would be implicated in circadian control28. Light is a major factor responsible for entraining the circadian clock, acting through the retina and signaling to the SCN2. Light cues were found to induce phosphorylation of histone H3 at serine 10 (H3S10) specifically in SCN neurons, resulting in the induction of immediate early gene expression (c-fos) and circadian gene expression (Per1)28. Subsequently, H3 acetylation at K9/K14 was found to be rhythmic at Per1, Per2 and Cry1 promoters in the liver29, 30. Although it is still unclear whether H3S10 phosphorylation contributes to other histone modifications in SCN neurons, H3S10 phosphorylation has been shown to facilitate an increase in K9/K14 H3 acetylation in fibroblasts31. Is this regulation occurring in an identical manner in both SCN and peripheral clocks, and which chromatin modifiers are responsible for these events? In this respect it is worth noting that chromatin modifiers, such as kinases and histone acetyltransferases (HATs), coexist in nuclear complexes so to facilitate combinatorial histone modifications.
Another level of complexity may be provided by the multitude of specific modifiers for specific histone sites. For example, H3S10 phosphorylation is elicited by various kinases, but the kinase involved in H3S10 phosphorylation in SCN neurons has not been identified. Yet, multiple kinases have been involved in modulating the circadian machinery7. Specificity may be elicited by selected kinases in various circadian contexts and tissues. For example, kinases acting as chromatin modifiers may have a role in circadian ‘memory’ and timekeeping. Others could be instrumental as endpoints of signaling pathways involved in ‘triggering’ circadian response or inducing entrainment. Whereas much more needs to be uncovered at this level, we speculate that chromatin remodeling in the SCN may involve activation of signaling pathways by specific neuropeptides, neurotransmitters or brain-specific metabolites. Potential SCN or even neuronal-specific signaling cues could dictate actions of ubiquitously expressed kinases (and other histone-modifying enzymes) to act in a tissue-specific manner in the brain. For instance, gamma amino butyric acid (GABA) is a neurotransmitter critically involved in the SCN in both inhibitory and excitatory neuronal responses. GABA neurotransmitter binding to its receptors has been shown to activate the extracellular signal-regulated protein kinases 1/2 (ERK1/2) in a GABA receptor subtype dependent manner32. While these events may be linked to H3 acetylation, we can only speculate on how these epigenetic signaling events may be tightly self-regulated within the central circadian clock system. Neurotransmitters acting to remodel chromatin in the brain, may be selective in kinase selection and recruitment to facilitate tissue-specific events pertinent only in the SCN.
Acetylation is a histone modification that has been directly associated to gene activation. The search for HATs responsible for cyclic K9/K14 H3 acetylation revealed that CREB-binding protein (CBP) and p300 were implicated in CLOCK:BMAL1 dependent transcription of circadian gene expression4. In a manner analogous to kinase activation by brain-specific factors, potential hints exist regarding the pathways through which HATs are recruited to the central circadian clock system and how these could be different in peripheral tissues. N-methyl-D-aspartic acid (NMDA) has been implicated in synaptic plasticity and selectively bound its receptors (NMDAR) on SCN neurons, which are believed to play a critical role in entraining circadian clocks33. Interestingly, NMDA activated transcription of CREB-responsive genes and resulted CBP recruitment to the c-Fos promoter34. Similarly, dopamine activated signaling through D2 receptors potentiated circadian transcription by increasing CBP phosphorylation and recruitment to CLOCK:BMAL135. Since both dopamine and NMDA are involved in modulating circadian rhythms, this could be one mechanism by which the central circadian clock machinery can dictate SCN-specific circadian events. Further studies will be needed to confirm this possibility.
An additional relevant finding demonstrated that the circadian transcription factor CLOCK has intrinsic HAT activity, targeting K9/K14 H3 in a cyclic manner36. CLOCK and BMAL1 are recruited to target promoters in a rhythmic manner30,37, and are hypothesized to generate a permissive chromatin state for transcription in concert with other chromatin remodeling regulators. In addition to histone targets, CLOCK also acetylated non-histone proteins in a circadian manner, such as BMAL138 and GR24, suggesting that additional targets may exist, which may be different in various circadian tissues. Interesting, BMAL1 acetylation was found be to critical for circadian function since it regulated its interaction with the repressor CRY1 to effectively repress transcription of circadian genes38.
How chromatin remodelers, and particularly histone deacetylases (HDACs), elicit their action in a specific manner is still unclear. The search for HDACs with specific circadian function has lead to the identification of various enzymes. The class III of HDACs includes the sirtuins, a group of deacetylases whose activity is modulated by the coenzyme nicotinamide adenine dinucleotide (NAD+). Thus, changes in the intracellular levels of NAD+ directly link chromatin remodeling to metabolism (see below). SIRT1 was found to interact with CLOCK and to be responsible for deacetylation of BMAL130 and PER239, as well as rhythmic alterations in acetylation state of histone H3 lysine 9/1430. We hypothesize the presence of a CLOCK chromatin complex (CCC), in which CLOCK associates with BMAL1, SIRT1 and other chromatin remodelers. It is also tempting to speculate that the composition and recruitment of the CCC may be dynamic and circadian.
Additional studies indicate that SIRT1 is not the only HDAC involved in circadian regulation. Indeed, delineating the roles of chromatin modifiers in the SCN versus peripheral clock systems may help address and further clarify contributions of circadian epigenome family members. Microarray studies have revealed that the expression of HDAC1 and HDAC2 oscillated in a circadian manner in rat-1 fibroblasts27. In addition, HDAC3 when complexed with nuclear receptor corepressor 1 (Ncor1) was needed for peripheral circadian gene expression, including metabolic genes in the liver40. These notions suggest that a number of factors may coexist within the circadian epigenome, possibly in nuclear complexes with a certain degree of redundancy.
It is reasonable to speculate that circadian gene expression is dictated by various epigenetic factors, depending on dynamic assembly of chromatin remodeling complexes in diverse tissues. Intriguingly, HATs and HDACs have often been found in the same complexes, indicating that their action may be balanced by a number of regulatory events. These large nuclear complexes also contain histone methyltransferases (HMTs). Histone methylation plays important roles in modulating chromatin states and subsequent gene expression profiles, being involved in both activation (specifically methylation at H3K4) and silencing. In terms of the central circadian system, little is known about chromatin methylation marks in the SCN. Recent findings have identified a HMT, mixed lineage leukemia 1 (MLL1), as critically involved in oscillatory methylation of H3K4 at circadian gene promoters, a mark which subsequently enhanced H3K9/14 acetylation and gene expression41. It has been reported that methylation of histone H3K27 (a silencing mark) was mediated by the HMT enhancer of zeste (EZH2) at Per1 and Per2 gene promoters in the liver, and was implicated in transcriptional repression42. The question remains whether EZH2 is involved only in the peripheral system as a repressor of the circadian system, or if EZH2 plays a role in neurons of the SCN or in other areas of the brain.
Given the chromatin remodeling functions of a number of kinases, HATs, HDACs and HMTs implicated in the brain, the question is what effect this has on neuronal function? Long-term memory is thought to be a transcription/translation event, requiring a number of transcription factors and accompanying molecules to mediate the effects of chromatin remodeling on memory acquisition. Glucocorticoids (which are known to have circadian expression) in the hippocampus and the insular cortex were found to rely on GR, CREB and CBP to induce H3K14 acetylation and subsequently enhanced object memory recognition43. Mitogen and stress-activated protein kinase 1 (MSK1, a downstream target of ERK/MAPK) deficient mice displayed inhibited fear conditioning and spatial learning, as a result of decreased histone H3S10 phosphorylation and H3K14 acetylation in the hippocampus44. Interestingly, SIRT1, whose role in circadian control has been discussed30, 39, was recently connected to cognitive function. Indeed, loss of SIRT1 disrupted synaptic plasticity and memory function through a microRNA (miR-134)-dependent mechanism that post-transcriptionally targeted expression of CREB and brain-derived neurotrophic factor (BDNF)45. It would be fascinating to determine whether the interplay between circadian control and memory formation involves SIRT1 and its chromatin remodeling function.
Linking the circadian epigenome & cellular metabolic state
The molecular and physiological overlap between circadian rhythms and metabolic control is extensive6. As previously discussed, the HDAC SIRT1 is implicated in the circadian epigenome, by counteracting the acetylation events mediated by CLOCK and other HATs. As NAD+ levels dictate the deacetylase activity of SIRT1, the recent finding demonstrating that the levels of NAD+ oscillate in a circadian manner has profound implications. CLOCK:BMAL1 were found to regulate directly the expression of a critical enzyme within the NAD+ salvage pathway, nicotinamide phosphoribosyltransferase (NAMPT)46, 47. This finding is a critical link between the metabolic state of the cell and chromatin remodeling, and suggests the circadian clock ultimately can control and fine tune cofactor levels that are needed to regulate the timekeeping system. This data also opens up more questions regarding central versus peripheral pacemakers. Are the oscillatory levels of NAD+ found only in peripheral clock systems, or does NAD+ oscillate in the SCN via an identical mechanism? We also can speculate on a system where NAD+ levels are regulated in a tissue-specific manner, suggesting a multi-tiered network of control. Another relevant question relates to other NAD+-dependent enzymes: could the oscillation of this metabolite lead to a circadian regulation of other chromatin remodeling enzymes, such as the poly-ADP ribose polymerases (PARPs)? Finally, the interplay between SIRT1 and the AMP-activated kinase (AMPK) is intriguing, since AMPK regulates energy homeostasis and has been shown to regulate the circadian machinery by controlling the stability of the CRY proteins48.
Similar to NAD+ levels, the cofactor adenosine triphosphate (ATP) has been shown to oscillate in the SCN in vivo in a robust manner, following the typical circadian light/dark cycle49. Again, this data suggests that the circadian clock functions as regulator of metabolites, in this case, specifically in the SCN. ATP is required for numerous enzymes within the body, including kinases and various chromatin remodeling enzymes. Therefore, if levels of ATP oscillate, this has implications on many biological processes within the cell. As an example, the ATP-dependent enzyme involved in chromatin topography and ultimately transcription machinery accessibility to DNA is the SWI/SNF complex. Importantly, there is clear rhythmic oscillation in multiple components of the SWI/SNF complex in both SCN and liver as indicated by the expression database compiled from microarray studies19. Interestingly, ATP-dependent chromatin remodeling has been already linked to circadian control in Neurospora50. While it is established that the circadian clock regulates an exceptional variety of cellular functions, the extent to which it may control metabolite levels still needs to be properly explored.
In conclusion, circadian pacemakers are required for numerous biological processes that regulate our daily behavior and metabolism. This is achieved through a network of internal timekeepers that constitutes the circadian clock, a finely tuned system that contributes to homeostasis by governing the timing and synchronization of physiological events. The central circadian clock found in the SCN directs a tightly controlled circadian network that transmits cues to peripheral clock system and thus maintains systemic biological rhythms. We have discussed how this balance is determined through a complex and highly refined program of gene expression. The circadian epigenome is an extensive network of transcription factors and regulatory proteins that establish a permissive state for the cyclic opening of specific chromatin loci. How these regulatory events may contribute to what could be defined as ‘circadian memory’, the anticipation of an upcoming circadian cycle, is still not fully understood. Likewise, the contribution of the epigenome to the plasticity of the circadian system to adapt to changing rhythms also needs further exploration. In addition, the involvement of microRNA pathways in circadian control is emerging, and this may be where tissue-specificity of the SCN versus peripheral clocks lies. Finally, the role of DNA methylation in circadian biology is tantalizing since this is a process by which cellular memory might be established and maintained. The circadian epigenome appears to constitute the ideal interface where signaling, metabolism and neuronal transmission may converge, leading to the plasticity that characterizes the circadian clock. It is also likely that it is within the epigenome that lie some of the key features that differentiate the central versus peripheral pacemakers.
Acknowledgments
We thank all members of our laboratory for discussions and support. Work in the laboratory is supported by the National Institute of Health, by the Inserm (Institut National de la Sante et la Recherche Medicale), France, and Sirtris Pharmaceutical Inc., a GSK Company.
References
- 1.Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol. 2001;63:647–76. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- 2.Welsh DK, Takahashi JS, Kay SA. Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol. 2010;72:551–77. doi: 10.1146/annurev-physiol-021909-135919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bechtold DA, Gibbs JE, Loudon AS. Circadian dysfunction in disease. Trends Pharmacol Sci. 2010;31:191–8. doi: 10.1016/j.tips.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Sahar S, Sassone-Corsi P. Metabolism and cancer: the circadian clock connection. Nat Rev Cancer. 2009;9:886–96. doi: 10.1038/nrc2747. [DOI] [PubMed] [Google Scholar]
- 5.Thresher RJ, et al. Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science. 1998;282:1490–4. doi: 10.1126/science.282.5393.1490. [DOI] [PubMed] [Google Scholar]
- 6.Eckel-Mahan K, Sassone-Corsi P. Metabolism control by the circadian clock and vice versa. Nat Struct Mol Biol. 2009;16:462–7. doi: 10.1038/nsmb.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–48. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- 8.Aton SJ, Colwell CS, Harmar AJ, Waschek J, Herzog ED. Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat Neurosci. 2005;8:476–83. doi: 10.1038/nn1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalsbeek A, Fliers E, Hofman MA, Swaab DF, Buijs RM. Vasopressin and the output of the hypothalamic biological clock. J Neuroendocrinol. 22:362–72. doi: 10.1111/j.1365-2826.2010.01956.x. [DOI] [PubMed] [Google Scholar]
- 10.Tousson E, Meissl H. Suprachiasmatic nuclei grafts restore the circadian rhythm in the paraventricular nucleus of the hypothalamus. J Neurosci. 2004;24:2983–8. doi: 10.1523/JNEUROSCI.5044-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ralph MR, Foster RG, Davis FC, Menaker M. Transplanted suprachiasmatic nucleus determines circadian period. Science. 1990;247:975–8. doi: 10.1126/science.2305266. [DOI] [PubMed] [Google Scholar]
- 12.Akhtar RA, et al. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol. 2002;12:540–50. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- 13.Cheng MY, et al. Prokineticin 2 transmits the behavioural circadian rhythm of the suprachiasmatic nucleus. Nature. 2002;417:405–10. doi: 10.1038/417405a. [DOI] [PubMed] [Google Scholar]
- 14.Kramer A, et al. Regulation of daily locomotor activity and sleep by hypothalamic EGF receptor signaling. Science. 2001;294:2511–5. doi: 10.1126/science.1067716. [DOI] [PubMed] [Google Scholar]
- 15.Abe K, Kroning J, Greer MA, Critchlow V. Effects of destruction of the suprachiasmatic nuclei on the circadian rhythms in plasma corticosterone, body temperature, feeding and plasma thyrotropin. Neuroendocrinology. 1979;29:119–31. doi: 10.1159/000122913. [DOI] [PubMed] [Google Scholar]
- 16.Balsalobre A, et al. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–7. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- 17.Welsh DK, Logothetis DE, Meister M, Reppert SM. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron. 1995;14:697–706. doi: 10.1016/0896-6273(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 18.Yamazaki S, et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–5. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- 19.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–20. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 20.Yan J, Wang H, Liu Y, Shao C. Analysis of gene regulatory networks in the mammalian circadian rhythm. PLoS Comput Biol. 2008;4:e1000193. doi: 10.1371/journal.pcbi.1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baggs JE, et al. Network features of the mammalian circadian clock. PLoS Biol. 2009;7:e52. doi: 10.1371/journal.pbio.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ueda HR, et al. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005;37:187–92. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 23.Travnickova-Bendova Z, Cermakian N, Reppert SM, Sassone-Corsi P. Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc Natl Acad Sci U S A. 2002;99:7728–33. doi: 10.1073/pnas.102075599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nader N, Chrousos GP, Kino T. Circadian rhythm transcription factor CLOCK regulates the transcriptional activity of the glucocorticoid receptor by acetylating its hinge region lysine cluster: potential physiological implications. FASEB J. 2009;23:1572–83. doi: 10.1096/fj.08-117697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duffield GE, et al. A role for Id2 in regulating photic entrainment of the mammalian circadian system. Curr Biol. 2009;19:297–304. doi: 10.1016/j.cub.2008.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duffield GE, et al. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr Biol. 2002;12:551–7. doi: 10.1016/s0960-9822(02)00765-0. [DOI] [PubMed] [Google Scholar]
- 28.Crosio C, Cermakian N, Allis CD, Sassone-Corsi P. Light induces chromatin modification in cells of the mammalian circadian clock. Nat Neurosci. 2000;3:1241–7. doi: 10.1038/81767. [DOI] [PubMed] [Google Scholar]
- 29.Etchegaray JP, Lee C, Wade PA, Reppert SM. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421:177–82. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- 30.Nakahata Y, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–40. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheung P, et al. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell. 2000;5:905–15. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- 32.Tu H, et al. Dominant role of GABAB2 and Gbetagamma for GABAB receptor-mediated-ERK1/2/CREB pathway in cerebellar neurons. Cell Signal. 2007;19:1996–2002. doi: 10.1016/j.cellsig.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 33.Ding JM, et al. Resetting the biological clock: mediation of nocturnal circadian shifts by glutamate and NO. Science. 1994;266:1713–7. doi: 10.1126/science.7527589. [DOI] [PubMed] [Google Scholar]
- 34.Impey S, et al. Phosphorylation of CBP mediates transcriptional activation by neural activity and CaM kinase IV. Neuron. 2002;34:235–44. doi: 10.1016/s0896-6273(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 35.Yujnovsky I, Hirayama J, Doi M, Borrelli E, Sassone-Corsi P. Signaling mediated by the dopamine D2 receptor potentiates circadian regulation by CLOCK:BMAL1. Proc Natl Acad Sci U S A. 2006;103:6386–91. doi: 10.1073/pnas.0510691103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 37.Ripperger JA, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet. 2006;38:369–74. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- 38.Hirayama J, et al. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007;450:1086–90. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- 39.Asher G, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–28. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 40.Alenghat T, et al. Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature. 2008;456:997–1000. doi: 10.1038/nature07541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katada S, Sassone-Corsi P. The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nat Struct Mol Biol. 2010 doi: 10.1038/nsmb.1961. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Etchegaray JP, et al. The polycomb group protein EZH2 is required for mammalian circadian clock function. J Biol Chem. 2006;281:21209–15. doi: 10.1074/jbc.M603722200. [DOI] [PubMed] [Google Scholar]
- 43.Roozendaal B, et al. Membrane-associated glucocorticoid activity is necessary for modulation of long-term memory via chromatin modification. J Neurosci. 30:5037–46. doi: 10.1523/JNEUROSCI.5717-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chwang WB, Arthur JS, Schumacher A, Sweatt JD. The nuclear kinase mitogen- and stress-activated protein kinase 1 regulates hippocampal chromatin remodeling in memory formation. J Neurosci. 2007;27:12732–42. doi: 10.1523/JNEUROSCI.2522-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao J, et al. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 466:1105–9. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–7. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramsey KM, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–4. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lamia KA, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–40. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Womac AD, Burkeen JF, Neuendorff N, Earnest DJ, Zoran MJ. Circadian rhythms of extracellular ATP accumulation in suprachiasmatic nucleus cells and cultured astrocytes. Eur J Neurosci. 2009;30:869–76. doi: 10.1111/j.1460-9568.2009.06874.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Belden WJ, Loros JJ, Dunlap JC. Execution of the circadian negative feedback loop in Neurospora requires the ATP-dependent chromatin-remodeling enzyme CLOCKSWITCH. Mol Cell. 2007;25:587–600. doi: 10.1016/j.molcel.2007.01.010. [DOI] [PubMed] [Google Scholar]


