Abstract
The mechanisms for regulation of the Inhibitor of Apoptosis (IAP) Survivin in cells undergoing stress associated with tumor development and the tumor microenvironment are not well understood. The stress response transcription factors HIF-1α and Yin Yang 1 (YY1) were hypothesized to contribute to the upregulation of Survivin in tumor cells. As expected, U2OS cells overexpressing HIF-1α showed a 2- to 3-fold transactivation when transfected. Surprisingly, when YY1 was overexpressed in this survivin promoter reporter system, luciferase expression was repressed 30- to 40-fold. YY1 involvement in survivin promoter repression was confirmed using siRNA directed against YY1. These studies showed that knockdown of YY1 releases the survivin promoter from the observed repression and leads to a 3- to 5-fold increase in promoter activity above basal levels. A U2OS cell line containing a stable YY1 Tet-off system was used to determine whether a temporal increase in YY1 expression affects Survivin protein levels. A low to moderate decrease in Survivin protein was observed 24 h and 48 h after Tet removal. Studies also confirmed that YY1 is capable of directly binding to the survivin promoter. Collectively, these findings identify novel basal transcriptional requirements of survivin gene expression which are likely to play important roles in the development of cancer and resistance to its treatment.
Keywords: Survivin, YY1, Transcriptional repressor, HIF-1, IAP
1. Introduction
Survivin an unique mammalian inhibitor of apoptosis (IAP) protein, controls stress from the microenvironment through diverse functions within the cell including surveillance checkpoints, suppression of cell death, regulation of mitosis, and adaptation to unfavorable environments [1–3]. Epigenetic, genetic and post-translational mechanisms for survivin gene regulation have been described in many malignant cell types [4] with various transcription factors including Stat3 [5], HIF-1α [6], Rb-E2F1 [7], Dec1 [8], Sp1 [9], c-myc [10] and KLF5 [11] affecting its transcriptional upregulation. In addition, the tumor suppressor p53 and Rb-E2F2 have been shown to repress survivin transcription by direct binding to the survivin promoter in a lung adenocarcinoma cell line [12] and in normal human melanocytes [4]. An interesting polymorphism has also been described at a CDE/CHR repressor element in the survivin promoter that correlates with increased survivin mRNA and protein in cancer cell lines and not in normal cell line controls [13].
The transcription factor YY1 is known to have a fundamental role in normal biologic processes such as embryogenesis, differentiation, replication, and cellular proliferation [14]. YY1 exerts its effects on genes involved in these processes via its ability to initiate, activate, or repress transcription depending upon the context or recruited cofactors in which it binds [15,16]. One such family of cofactors are the histone deacetylases which have been shown to bind YY1 and repress transcription when targeted to promoters [17]. YY1 has been shown to interact with p300, PCAF and CBP, all which posses the histone acetyltransferase (HAT) activity [17]. YY1 may thus activate transcription by its recruitment of HAT proteins and repress transcription by recruiting HDACs.
Work by Affar et al. [18] previously showed that in a mouse YY1 knockdown model, survivin (BIRC5) levels were decreased. These findings lead to the hypothesis and impetus of our current work which was that YY1 overexpression would lead to survivin promoter control and Survivin protein regulation. The present study, therefore examined the transcriptional affect of YY1 on survivin in an osteosarcoma cell line derived from human bone osteosarcoma (U2OS). We found that when YY1 was overexpressed in U2OS cells, survivin mRNA and protein were repressed by YY1. By analyzing the survivin promoter activity, we further found that YY1 works as a transcriptional repressor of the survivin gene and we show for the first time that YY1 is capable of binding directly to the survivin core promoter thus acting as a transcription factor rather than a co-repressor.
2. Materials and methods
2.1. Antibodies and DNA vectors
All antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA) unless otherwise indicated. The plasmid expressing YY1 protein, pcDNA3/YY1 as well as the U6/yy1 siRNA and control U6/scrambled were kind gifts of Dr. Sui, Wake Forest and were described previously [19]. U6/yy1 siRNA vector was evaluated in our hands for effective knockdown (Supplemental Fig. 1). Survivin nested deletion constructs were previously described [9] and were a kind gift from Dr. Li, Roswell Park Memorial Institute.
2.2. Cell culture and transfection
The U2OS human osteosarcoma cell line was obtained from ATCC. U2OS cells with stable Tet-off YY1 were a kind gift from Dr. Sui, Wake Forest and were previously described [19]. Cell lines were maintained under an atmosphere of 5% CO2 at 37 °C in McCoys 5A media supplemented with 10% fetal bovine serum, 2 mmol/L of l-glutamine, and penicillin–streptomycin. The Tet-off cells were additionally maintained in G418, hygromycin B, and the tetracycline analogue doxycycline (50 ng/mL). YY1 expression was induced by transferring the cells to Tet-off medium, which is the same as control (Tet-on) medium except for the lack of doxycycline [19].
2.3. Transient transfection and reporter assays
U2OS cells were seeded in 12-well plates and grown to 60–80% confluence. A total 0.4 μg of the various survivin promoter-luciferase reporter plasmids were cotransfected with either 0.6 μg of pcDNA/YY1 or empty vector expression plasmids and 0.01 μg of pRL-tk using FuGENE 6 (Roche, Indianapolis, IN). Twenty-four hours after transfection, cells were lysed and assayed for luciferase activity by luminometer (Turner Design Systems, Sunnyvale, CA). Luciferase activity measurement was accomplished according to manufacturer’s instructions. The pLuc230 vector containing the CAT → GGG mutation was purchased from Origene, Rockville, MD.
2.4. Western blots
Cells were solubilized, proteins (20–40 μg) separated using 12 % Bis–Tris polyacrylamide gels, transferred onto polyvinylidene difluoride membranes (Millipore) and probed using the following antibodies: mouse monoclonal anti-YY1 (cat. # sc-7341; Santa Cruz Biotechnology, Inc., Santa Cruz, CA), and rabbit polyclonal anti-Survivin (cat. # NB500-201; Novus, Littleton, CO). Secondary antibodies (IR-Dye-conjugated) were goat anti-rabbit and goat anti-mouse immunoglobulin (LICOR, Lincoln, Nebraska). Immunoreactive bands were detected using the Odyssey imaging system (LICOR) and quantified using Image-Quant software.
2.5. Reverse transcriptase-PCR
Total RNA was extracted from cells at various time intervals using TRI-Reagent (Sigma, St. Louis, MO) and reverse-transcribed with SuperScript™ II RNase H− Reverse Transcriptase (Invitrogen™, Carlsbad, CA), as described by the manufacturer. One hundred nanograms of the resulting first-strand cDNA was used as template and amplified by PCR. Sequences of the oligonucleotide primer sets used for reverse transcription-PCR analysis are as follows: 5′-GCA TGG CTG CCC CGA CGT TG-3′ (sense) and 5′-GCT CCG GCC AGA GGC CTC AA-3′ (antisense) for survivin, 5′-GCT TCG AGG ATC AGA TTC TCA TCC-3′ (sense) and 5′-GAC TAC ATT GAA CAA ACG CTG GTC-3′ (antisense) for YY1, 5′-GCC AGA TCT CGG CGA AGT AAA-3′ (sense) and 5′-ATA TCC AGG CTG TGT CGA CTG-3′ (antisense) for HIF1, 5′-ATG ACT CGC GAT TTC AAA CCT-3′ (sense) and 5′-CTT TGA AGT CGA GAA TCC ATT-3′ (antisense) for p75/LEDGF, and, 5′-CTCATGACCACAGTCCATGC-3′ (sense) and 5′-TTACTCCTTGGAGGCCATGT-3′ (antisense) for beta actin. Products were visualized on ethidium bromide-stained agarose gels. Amplification of beta actin served as an internal control.
2.6. Electrophoretic mobility shift assay
Nuclear extracts were prepared as previously described [20], with the only modification that N-N-(L-3-trans-carboxyoxirane-2-carbonyl)-l-leucyl-agmatine (E64) and 4-(2-aminoethyl)-benzolsulfonyl fluoride (“Pefabloc SC”) were included as protease inhibitors in concentrations suggested by the manufacturer (Boehringer, Mannheim, Germany). Protein concentration in nuclear extracts was determined using the BCA assay (Pierce) according to the manufacturers instructions. Oligos used were as follows: Two YY1 sites (YY1 sites underlined): 5′-GC GCT CCC GAC ATG CCC CGC GGC GCG CCA TTA ACC GCC A-3′; YY1 Site 1 5′-TG CGC TCC CGA CAT GCC CCG CG-3′; YY1 Site 2 CGC GGC GCG CCA TTA ACC GCC A-3′ YY1 Consensus 5′-CGC TCC CCG GCC ATC TTG GCG GCT GGT-3′. All oligos were annealed by incubating at 95 °C for 2 min, then ramp cooling to room temperature. The DNA–protein binding reaction was performed in 20 μl reaction mixtures including 10% glycerol, 12 mM HEPES pH 7.9, 4 mM Tris HCl pH 8.0, 1 mM EDTA, and 3 μg BSA. Binding reactions were incubated at room temperature for 30 min, then for an addition 60 min at 4 °C with anti-YY1 antibody (cat. # sc-281; Santa Cruz Biotechnology, Santa Cruz, CA) added to the appropriate reactions. The DNA–protein complexes were resolved on 5.5% non-denaturing polyacrylamide gel (29:1 cross-linking ratio) run in Tris–Borate buffer, dried and exposed using the Storm 860 Phosphoimager (Amersham Biosciences). Competitions in EMSA were performed as above, except that 25 ng of poly (dI–dC)·(dI–dC) per reaction were used.
2.7. Statistical analysis
All data in reporter assays and semiquantitative PCR are presented as means ± standard deviation.
3. Results
3.1. HIF-1α and YY1 transcriptionally regulate surviving
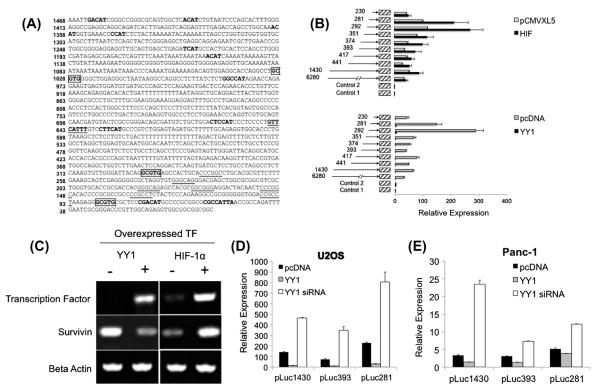
HIF-1α has previously been shown to be a transcriptional regulator of survivin [6,21,22]. However, it is currently unknown if YY1 plays a role in survivin transcription. To determine possible bindings sites for YY1, using a computer-based approach, the survivin promoter was scanned for putative HIF-1α, and YY1 binding sites using the online tools TFSearch (http://www.cbrc.jp/research/db/TFSEARCH.html), Promo Program (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3), and previously published consensus sequences [23]. Fig. 1A shows the locations of all identified HIF-1α and YY1 consensus sites in the first 6280 bp of the survivin promoter. Using survivin promoter nested deletions in a luciferase reporter system [24], the ability of YY1 and HIF-1 to activate or repress survivin promoter activity was tested (Fig. 1B). The survivin promoter nested deletions were utilized to assist in identification of regions of the promoter that are essential to regulation of the survivin promoter by each transcription factor tested. Therefore constructs ranging from 230 bp upstream of the survivin start site up to 6280 bp upstream of the start site were utilized. When HIF-1α was overexpressed in U2OS cells a 2- to 3-fold induction was seen in all constructs tested except +230 bp and +6280 bp. However, when YY1 was overexpressed, contrary to our initial hypothesis, there was a 30- to 40-fold repression of survivin promoter activity in all constructs tested. To further examine these findings, we evaluated endogenous survivin transcript levels after overexpression of HIF-1α and YY1 in U2OS cells (Fig. 1C). The results were consistent with survivin transcriptional upregulation by HIF-1α as seen in the previous reporter experiments, and down-regulation of survivin after YY1 overexpression.
Fig. 1.
Effect of HIF-1, and YY1 overexpression on survivin promoter activity and transcript levels. (A) Proximal survivin promoter schematic. Analysis revealed multiple putative YY1 binding sites (bolding), HIF-1 binding sites (boxed segments), and heat shock elements (GAA). For reference, putative SP1 sites are denoted as underlined segments. (B) Luciferase reporter assays were performed using survivin promoter constructs ranging in length from +6280 bp to +230 bp. Error bars represent the standard deviation of duplicate luminescence measurement. Results are representative of two repeat experiments. (C) RT-PCR analysis of survivin expression following overexpression of YY1, and HIF-1. Cells were transfected with the corresponding empty vector for each transcription factor. (D) YY1 siRNA relieves the survivin promoter from transcriptional repression. Luciferase reporter assays were performed after YY1 overexpression and siRNA knockdown in (D) U2OS and (E) Panc-1 cells. Three survivin promoter reporter constructs were tested (pluc1430, pLuc 393, and pLuc 281). Relative expression indicates promoter activity relative to luciferase activity in the presence of empty vector and background activity. Error bars represent the standard deviation of duplicate luminescence measurements. Results are representative of two repeated experiments.
3.2. YY1 affects basal levels of survivin
To provide further evidence for the ability of YY1 to affect basal survivin promoter activity, YY1 knockdown was performed (Fig. 1D and E). Because previous experiments showed survivin promoter activity repression across all constructs tested, pLuc1430, 393, and 281 were chosen for this experiment. In U2OS (Fig. 1D) and Panc-1 (Fig. 1E) cells, siRNA knockdown of YY1 relieved the survivin promoter of basal promoter activity repression, indicated by an increased in luciferase reporter activity of approximately 3- to 4-fold in each construct tested. The overexpression of YY1 again repressed promoter activity to nearly undetectable levels, a finding consistent with our previously described experiments.
3.3. YY1 overexpression modulates Survivin protein levels
To investigate whether YY1 expression can affect Survivin expression at the protein level, Western blot analysis was done using a U2OS YY1 tet-off cell line [19]. Twenty-four hours after tet removal, a significant YY1 overexpression was recorded (Fig. 2). Survivin protein levels diminished at 24 h. However, after 48 h of incubation in tet-free media, a measurable reduction in Survivin protein expression was seen (Fig. 2).
Fig. 2.
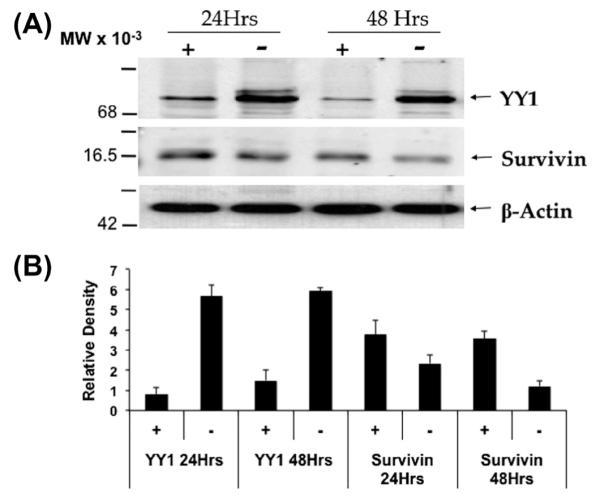
Survivin expression decreases after 48 h of YY1 overexpression. Western blot analysis of Survivin expression after YY1 overexpression via tet-off U2OS cells was analyzed. (A) U2OS cells that stably express a YY1 overexpression vector under the control of a tetracycline responsive promoter were lysed and protein was extracted for western blot analysis. +/− indicates the presence or absence, respectively, of tet in the culture media. Data is representative of 3 independent experiments. (B) Densitometric analysis of Western blot bands. Bars represent density of YY1 or Survivin normalized to the actin band density.
3.4. Putative YY1 bindings sites found in the survivin promoter are employed in survivin’s regulation
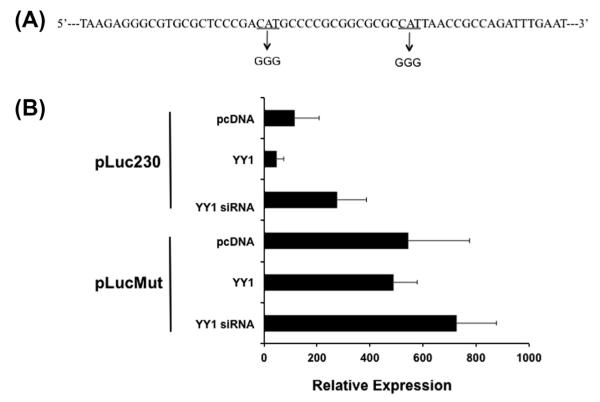
Repression of survivin promoter activity in our luciferase reporter system was seen in all constructs tested, including the shortest construct containing 230 bp of the promoter, which has previously been shown to include the core promoter for survivin. Fig. 1 illustrates two putative YY1 binding sites within the first 230 bp of the survivin promoter, and we therefore designed to investigate the involvement of these two sites as key areas involved in repression of basal survivin transcription by YY1. Site-directed mutagenesis was employed to define the role of these two sites in survivin transcription. Fig. 3A illustrates the mutation of each CAT region of the putative YY1 sites to GGG. This region was chosen for mutation based on previous studies indicating that the core sequence preferred by YY1 is CCAT or ACAT [23]. When both putative YY1 binding sites were mutated, neither YY1 overexpression nor knockdown altered the basal survivin promoter activity (Fig. 3B). Furthermore, the basal survivin promoter activity was increased relative to non-mutated promoter activity. These data support a role for these putative YY1 binding sites in basal survivin transcription.
Fig. 3.
Mutation of two putative YY1 binding sites in the proximal survivin promoter alters promoter activity (A) The two putative YY1 binding sites (contained within pLuc230 construct) were mutated from the core YY1 recognition site CAT to GGG. (B) Luciferase reporter assay. U2OS cells were transfected with either (1) pLuc230, the standard pGL3 vector containing 230 unmutated bp of the survivin promoter, or (2) pLucMut in which the two putative YY1 binding sites were mutated from CAT to GGG. Each vector was cotransfected with either empty pcDNA, YY1, or YY1 siRNA as well as pRL-tk for transfection efficiency internal control. Error bars represent standard deviation of duplicate luminescence measurement, and results are representative of multiple experiments.
3.5. YY1 interacts with the core survivin promoter
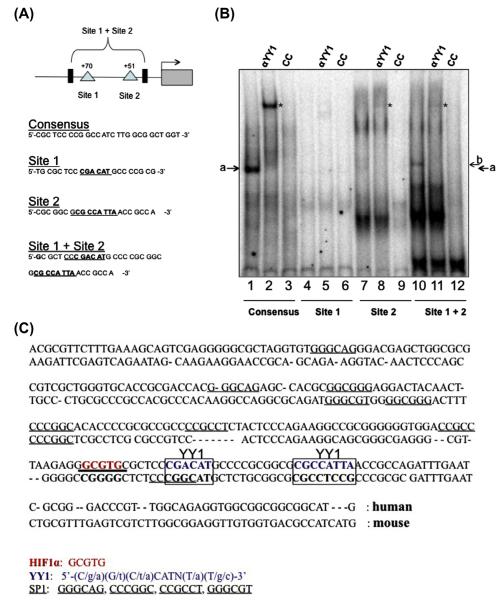
YY1 can exert transcriptional changes via direct DNA binding or through protein–protein interaction. To determine if survivin repression seen in reporter assays, RT-PCR, and Western blotting is the result of YY1’s direct binding to the survivin promoter at locations identified in Fig. 1, electrophoretic mobility shift assays (EMSA) were performed. Two putative YY1 binding sites located in the survivin core promoter (Fig. 4A) were studied. To validate the study, a YY1 consensus sequence was used (Santa Cruz Biotechnology, Santa Cruz, CA). Strong YY1 binding to the consensus sequence was observed (Fig. 4B; lane 1, arrow) providing a baseline for experimental comparison analysis. Supershift (lane 2, asterisk) and cold competition (lane 3) confirmed the identity and specificity of the YY1 binding. When a probe for Site 1 was used, no YY1 binding or supershift was seen (lanes 4 and 5), indicating that it may not be the primary site of binding at least under basal conditions. When a probe for Site 2 was used, a possible double band was seen at the appropriate location, possibly representing a more specific binding site for YY1. Importantly, these bands were super-shifted with the addition of YY1 antibody, and competed using cold competition, confirming the specificity of the results. Binding was enhanced by use of a probe containing both putative YY1 binding sites (lane 10). Supershift and cold competition was again used to confirm the specificity of the binding (lanes 11 and 12). These results indicate that the most proximal YY1 consensus site located on the survivin promoter is a target of YY1 binding and regulation, but binding to this site is increased with the inclusion of the second YY1 recognition site.
Fig. 4.
YY1 directly interacts with the survivin promoter. (A) Schematic of survivin promoter representing regions investigated for YY1 binding. (B) Electrophoretic mobility shift assay. Nuclear extract was prepared from U2OS cells. 32p labelled probes were incubated with nuclear extracts either alone (Lanes 1, 4, 7 and 10), with anti-YY1 antibody (lanes 2, 5, 8 and 11) or cold competitor probes in 100× excess (lanes 3, 6, 9 and 12). Arrow indicates YY1 bound to probe. * indicates supershift. (C) Comparison of human and mouse survivin promoter sequences. Boxed segments represent the 2 putative YY1 binding segments of the survivin promoter contained within the pLuc230 construct that were mutated in previous experiments. There is lack of homology between mouse and human at both putative YY1 binding sites found in the first 230 bp of the survivin promoter.
4. Discussion
Interestingly, compared with mouse model knockdown work by Affar et al. [18] which showed decreased survivin (BIRC5) levels when YY1 was knocked down, human survivin increased with YY1 knockdown. Human & mouse survivin core promoter sequence alignment showed a lack of homology existing at both putative YY1 sites investigated (Fig. 4C). This may in part explain why YY1 appears to negatively regulate survivin transcription in our cell culture model, whereas in mice YY1 may contribute to survivin’s positive regulation.
We provide several lines of evidence that YY1 represses survivin promoter activity in U2OS cells. YY1 regulates targets genes through a host of mechanisms including protein–protein interactions that allow it to act as a coactivator or corepressor and by direct DNA binding. In the present study, we identified a survivin core promoter sequence containing two putative YY1 binding sites and showed that YY1 binds directly to both sites. Binding affinity for the survivin promoter was lower than for a consensus sequence known to efficiently bind YY1 [25], which may represent a technical limitation owed to the extremely high GC content (70–80%) of the survivin promoter. Because of the highly complicated nature of transcriptional regulatory networks, it is also possible that a proper scaffold is required for optimal binding. Although YY1 binding is strongest at the most proximal site on the survivin promoter (Site 2), binding is improved by inclusion of a second putative YY1 binding site (Site 1) that by itself appears to facilitate no YY1 binding.
The transcriptional and post-transcriptional networks regulating survivin expression are complex [26], and it is therefore possible that downstream of YY1’s downregulation of survivin transcription other factors are significantly involved in determining the ultimate expression of survivin and the clinical sequelae that result. It is also important to note that while our work demonstrates robust survivin promoter activity reduction via reporter assay, the resulting reduction in mRNA and protein is modest. The extent to which YY1-mediated reduction of survivin expression results in increased apoptosis, alterations in cell cycle progression, or modulation of other hallmarks of cancer progression is currently under investigation in our laboratory.
It was previously believed that targeting of transcription factors for cancer therapy was not practical owing to the complexity of transcriptional networks. However, it is increasingly believed that drug or small molecule inhibitor-mediated interruption of transcription factor binding represents an important approach to cancer therapeutics. The small molecule inhibitor YM155 is currently in phase II clinical trials for several types of cancer including diffuse large B-cell lymphoma [27], prostate cancer [28], melanoma [29], and non-small cell lung cancer [30] due to it’s previously observed ability to induced apoptosis and reduce tumor bulk in various in vitro and in vivo models [31]. Reduction in survivin transcription after YM155 treatment has also been reported [32] and is believed to be a key mechanism for the apparent sensitization of tumors to cell death that has been observed.
Our discovery of a novel transcriptional repressor of survivin may provide new ways of understanding survivin expression in the context of cellular stress resulting from chemo- and radiotherapy. We also provide evidence for a possible positive role in YY1 overexpression in human cancer. The clinical significance of this finding across different cancer types has yet to be determined.
Supplementary Material
Acknowledgments
Grant Support: This research was supported by the NIGMS of the NIH under award # 2 R25 GM060507 and by the NIHDMH of the NIH under award numbers P20MD006988 and 5P20MD001632 (project 3 NRW). The content is the responsibility of the authors and does not necessarily represent the official views of the NIH. Funding was also obtained from a National Merit Test Bed (NMTB) award sponsored by the Department of the Army under Cooperative Agreement Number DAMD17-97-2-7016 (NRW).
Footnotes
Conflict of interest The authors declare no conflict of interest.
Appendix A. Supplementary data Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.bbrc.2014.01.169.
References
- [1].Altieri DC. Survivin, versatile modulation of cell division and apoptosis in cancer. Oncogene. 2003;22(53):8581–8589. doi: 10.1038/sj.onc.1207113. [DOI] [PubMed] [Google Scholar]
- [2].Li F, Ambrosini G, Chu EY, et al. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396(6711):580–584. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- [3].Altieri DC. The case for survivin as a regulator of microtubule dynamics and cell-death decisions. Curr. Opin. Cell Biol. 2006;18(6):609–615. doi: 10.1016/j.ceb.2006.08.015. [DOI] [PubMed] [Google Scholar]
- [4].Raj D, Liu T, Samadashwily G, Li F, Grossman D. Survivin repression by p53, Rb and E2F2 in normal human melanocytes. Carcinogenesis. 2008;29(1):194–201. doi: 10.1093/carcin/bgm219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gritsko T, Williams A, Turkson J, et al. Persistent activation of stat3 signaling induces survivin gene expression and confers resistance to apoptosis in human breast cancer cells. Clin Cancer Res. 2006;12(1):11–19. doi: 10.1158/1078-0432.CCR-04-1752. [DOI] [PubMed] [Google Scholar]
- [6].Peng XH, Karna P, Cao Z, Jiang BH, Zhou M, Yang L. Cross-talk between epidermal growth factor receptor and hypoxia-inducible factor-1alpha signal pathways increases resistance to apoptosis by up-regulating survivin gene expression. J. Biol. Chem. 2006;281(36):25903–25914. doi: 10.1074/jbc.M603414200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Dasgupta P, Kinkade R, Joshi B, Decook C, Haura E, Chellappan S. Nicotine inhibits apoptosis induced by chemotherapeutic drugs by up-regulating XIAP and survivin. Proc. Natl. Acad. Sci. U.S.A. 2006;103(16):6332–6337. doi: 10.1073/pnas.0509313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Li Y, Xie M, Yang J, et al. The expression of antiapoptotic protein survivin is transcriptionally upregulated by DEC1 primarily through multiple sp1 binding sites in the proximal promoter. Oncogene. 2006;25(23):3296–3306. doi: 10.1038/sj.onc.1209363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Li F, Altieri DC. Transcriptional analysis of human survivin gene expression. Biochem. J. 1999;344(Pt 2):305–311. doi: 10.1042/0264-6021:3440305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cosgrave N, Hill AD, Young LS. Growth factor-dependent regulation of survivin by c-myc in human breast cancer. J. Mol. Endocrinol. 2006;37(3):377–390. doi: 10.1677/jme.1.02118. [DOI] [PubMed] [Google Scholar]
- [11].Zhu N, Gu L, Findley HW, et al. KLF5 interacts with p53 in regulating survivin expression in acute lymphoblastic leukemia. J. Biol. Chem. 2006;281(21):14711–14718. doi: 10.1074/jbc.M513810200. [DOI] [PubMed] [Google Scholar]
- [12].Hoffman WH, Biade S, Zilfou JT, Chen J, Murphy M. Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J. Biol. Chem. 2002;277(5):3247–3257. doi: 10.1074/jbc.M106643200. [DOI] [PubMed] [Google Scholar]
- [13].Xu Y, Fang F, Ludewig G, Jones G, Jones D. A mutation found in the promoter region of the human survivin gene is correlated to overexpression of survivin in cancer cells. DNA Cell Biol. 2004;23(7):419–429. doi: 10.1089/1044549041474788. [DOI] [PubMed] [Google Scholar]
- [14].Shi Y, Lee JS, Galvin KM. Everything you have ever wanted to know about Yin Yang 1. Biochim. Biophys. Acta. 1997;1332(2):F49–F66. doi: 10.1016/s0304-419x(96)00044-3. [DOI] [PubMed] [Google Scholar]
- [15].Gordon S, Akopyan G, Garban H, Bonavida B. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene. 2006;25(8):1125–1142. doi: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- [16].Deng Z, Wan M, Cao P, Rao A, Cramer SD, Sui G. Yin Yang 1 regulates the transcriptional activity of androgen receptor. Oncogene. 2009;10:3746–3757. doi: 10.1038/onc.2009.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wang X, Feng Y, Xu L, et al. YY1 restrained cell senescence through repressing the transcription of p16. Biochim. Biophys. Acta. 2008;1783(10):1876–1883. doi: 10.1016/j.bbamcr.2008.05.015. [DOI] [PubMed] [Google Scholar]
- [18].Affar EB, Gay F, Shi Y, et al. Essential dosage-dependent functions of the transcription factor Yin Yang 1 in late embryonic development and cell cycle progression. Mol. Cell Biol. 2006;26(9):3565–3581. doi: 10.1128/MCB.26.9.3565-3581.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sui G, Affar el B, Shi Y, et al. Yin Yang 1 is a negative regulator of p53. Cell. 2004;117(7):859–872. doi: 10.1016/j.cell.2004.06.004. [DOI] [PubMed] [Google Scholar]
- [20].Schreiber E, Matthias P, Muller MM, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17(15):6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wei H, Wang C, Chen L. Proliferating cell nuclear antigen, survivin, and CD34 expressions in pancreatic cancer and their correlation with hypoxia-inducible factor 1alpha. Pancreas. 2006;32(2):159–163. doi: 10.1097/01.mpa.0000202961.71600.9b. [DOI] [PubMed] [Google Scholar]
- [22].Bai H, Ge S, Lu J, Qian G, Xu R. Hypoxia inducible factor-1alpha-mediated activation of survivin in cervical cancer cells. J. Obstet. Gynaecol. Res. 2013;39(2):555–563. doi: 10.1111/j.1447-0756.2012.01995.x. [DOI] [PubMed] [Google Scholar]
- [23].Hyde-DeRuyscher RP, Jennings E, Shenk T. DNA binding sites for the transcriptional activator/repressor YY1. Nucleic Acids Res. 1995;23(21):4457–4465. doi: 10.1093/nar/23.21.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Li F, Altieri DC. The cancer antiapoptosis mouse survivin gene: characterization of locus and transcriptional requirements of basal and cell cycle-dependent expression. Cancer Res. 1999;59(13):3143–3151. [PubMed] [Google Scholar]
- [25].Chaput N, Flament C, Viaud S, et al. Dendritic cell derived-exosomes: biology and clinical implementations. J. Leukocyte Biol. 2006;80(3):471–478. doi: 10.1189/jlb.0206094. [DOI] [PubMed] [Google Scholar]
- [26].Zhang M, Yang J, Li F. Transcriptional and post-transcriptional controls of survivin in cancer cells: novel approaches for cancer treatment. J. Exp. Clin. Cancer Res. 2006;25(3):391–402. [PMC free article] [PubMed] [Google Scholar]
- [27].Cheson BD, Bartlett NL, Vose JM, et al. A phase II study of the survivin suppressant YM155 in patients with refractory diffuse large B-cell lymphoma. Cancer. 2012;118(12):3128–3134. doi: 10.1002/cncr.26510. [DOI] [PubMed] [Google Scholar]
- [28].Tolcher AW, Quinn DI, Ferrari A, et al. A phase II study of YM155, a novel small-molecule suppressor of survivin, in castration-resistant taxanepretreated prostate cancer. Ann. Oncol. 2012;23(4):968–973. doi: 10.1093/annonc/mdr353. [DOI] [PubMed] [Google Scholar]
- [29].Lewis KD, Samlowski W, Ward J, et al. A multi-center phase II evaluation of the small molecule survivin suppressor YM155 in patients with unresectable stage III or IV melanoma. Invest. New Drugs. 2011;29(1):161–166. doi: 10.1007/s10637-009-9333-6. [DOI] [PubMed] [Google Scholar]
- [30].Giaccone G, Zatloukal P, Roubec J, et al. Multicenter phase II trial of YM155, a small-molecule suppressor of survivin, in patients with advanced, refractory, non-small-cell lung cancer. J. Clin. Oncol. 2009;27(27):4481–4486. doi: 10.1200/JCO.2008.21.1862. [DOI] [PubMed] [Google Scholar]
- [31].Nakahara T, Kita A, Yamanaka K, et al. YM155, a novel small-molecule survivin suppressant, induces regression of established human hormone-refractory prostate tumor xenografts. Cancer Res. 2007;67(17):8014–8021. doi: 10.1158/0008-5472.CAN-07-1343. [DOI] [PubMed] [Google Scholar]
- [32].Tao YF, Lu J, Du XJ, et al. Survivin selective inhibitor YM155 induce apoptosis in SK-NEP-1 Wilms tumor cells. BMC Cancer. 2012;12:619. doi: 10.1186/1471-2407-12-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.