Abstract
Here we studied plasma metabolomic profiles as determinants of progression to ESRD in patients with Type 2 diabetes (T2D). This nested case-control study evaluated 40 cases who progressed to ESRD during 8-12 years of follow-up and 40 controls who remained alive without ESRD from the Joslin Kidney Study cohort. Controls were matched with cases for baseline clinical characteristics; although controls had slightly higher eGFR and lower levels of urinary albumin excretion than T2D cases. Plasma metabolites at baseline were measured by mass spectrometry-based global metabolomic profiling. Of the named metabolites in the library, 262 were detected in at least 80% of the study patients. The metabolomic platform recognized 78 metabolites previously reported to be elevated in ESRD (uremic solutes). Sixteen were already elevated in the baseline plasma of our cases years before ESRD developed. Other uremic solutes were either not different or not commonly detectable. Essential amino acids and their derivatives were significantly depleted in the cases, whereas certain amino acid-derived acylcarnitines were increased. All findings remained statistically significant after adjustment for differences between study groups in albumin excretion rate, eGFR or HbA1c. Uremic solute differences were confirmed by quantitative measurements. Thus, abnormal plasma concentrations of putative uremic solutes and essential amino acids either contribute to progression to ESRD or are a manifestation of an early stage(s) of the disease process that leads to ESRD in T2D.
INTRODUCTION
The incidence of End-Stage Renal Disease (ESRD) due to type 2 diabetes (T2D) increased over the last 20 years despite improving hyperglycemia control and increased renoprotective drugs use.(1) Clearly, a better understanding of the determinants responsible for progression to ESRD in T2D is urgently needed if this “epidemic” is to be contained.
Recently developed platforms for global metabolomic profiling are capable of examining hundreds of metabolites, so they are excellent tools to study complex metabolic alterations associated with progression of diabetic nephropathy.(2, 3) Reliable metabolomic data can be obtained with liquid or gas chromatography coupled with mass spectrometry (LC/GC-MS) or NMR spectroscopy. Among those, MS – based platforms are the most sensitive.(2, 4-6)
One of the hallmarks of progression to ESRD is plasma accumulation of certain metabolites, the so-called uremic solutes.(7-10) However, it is becoming apparent that increase in the levels of uremic solutes in blood may be more than a simple reflection of impaired kidney function.(11-13) The kidney is a key organ involved in the handling of major biochemical classes of metabolites. Kidney function includes filtration of metabolites via glomeruli, followed by their tubular secretion/reabsorption and synthesis/degradation in various components of the renal parenchyma. At present it is unclear whether elevated levels of uremic solutes precede or follow renal impairment. For example, elevated plasma concentration of uremic solutes may contribute to glomerular as well as tubular damage in diabetic nephropathy, and damage to those two components have been demonstrated in early nephropathy.(14, 15) Various alterations of certain biochemical classes of metabolites (amino acids, in particular) have been also reported in the associations with insulin resistance, type 2 diabetes or chronic kidney injury per se.(16-19)
To date, few metabolomic studies focusing on diabetic nephropathy have been performed in experimental models (20, 21) or in humans.(22-25) Nevertheless, the comparisons were either cross-sectional or focused on albuminuria progression rather than on the kidney failure, the ultimate outcome of the diabetic nephropathy.(22-25)
This study is the first that aims to survey the metabolomic profile of plasma in T2D subjects with normal or mildly impaired renal function at baseline who developed ESRD during the subsequent 8-12 years of follow-up. We aim to establish metabolomic profiles associated with subsequent progression to ESRD in T2D so we may hypothesize about the underlying mechanisms that initiate this progression.
RESULTS
Study groups and their characteristics
A cohort with T2D patients attending the Joslin Clinic was recruited into the Joslin Study of the Genetics of Kidney Complications. Of the 509 patients examined between 1992 and 1996. 410 were followed until the end 2004. During 8-12 years of follow-up 59 (14.4%) patients developed ESRD, 84 (20%) died without progressing to ESRD and 267 (65.1%) remained alive without progressing to ESRD. Details of the follow-up study were already published.(26)
For the present nested case-control study, we selected 40 patients who developed ESRD (cases of progressors to ESRD) and matched them with 40 patients who were alive as of 2004 without ESRD (controls for non-progressors). Of the 80 patients, 75 identified themselves as Caucasians of European origin. Baseline characteristics of the two selected study groups are summarized in Table 1. The groups were very similar with regard to most clinical characteristics. Progressors, however, had higher urinary albumin excretion and slightly lower eGFR. Despite the differences noted in median AER and mean eGFR, there was substantial overlap of the distributions in the two study groups. At baseline the majority of progressors and non-progressors were in CKD stage 2. CKD stage 3 was present in 7% of controls and 22% of cases, respectively. Overall the distribution of CKD stages was not statistically different between the study groups. 87% of non-progressors had annual eGFR decrease less than 3.5 ml/min/1.73m2. The median (25th, 75th percentile) decrease was −1.95 (−3.2, −0.8) ml/min/1.73m2 and the slope was determined based on the serial creatinine measurements over 7.6 (6.5-12.4) years. The study groups did not also differ regarding baseline plasma levels of parathormone.
Table 1.
Characteristics of subjects with T2D selected for nested case-controls study
| Baseline characteristic | Study groups: | ||
|---|---|---|---|
| Non-progressors | ESRD progressors | ||
| Controls (n=40) | Cases (n=40) | p value* | |
| Male (%) | 62 | 55 | n.s |
| Age (yr) | 56±11 | 59±7 | n.s |
| Duration of diabetes (yr) | 13±7 | 17±7 | n.s |
| Body mass index (kg/m2) | 29.7±5.9 | 31.5±7.1 | n.s |
| HbA1c (%) | 8.8±1.4 | 9.3±2.0 | n.s |
| Serum cholesterol (mg/dl) | 241±62 | 234±54 | n.s |
| Systolic blood pressure (mmHg) | 135±25 | 142±32 | n.s |
| Antihypertensive/renoprotective Treatment [%] | 74% | 62% | n.s |
| ACR (μg/g creatinine) | 308 (70,471) | 957 (382, 2265) <0.0001 | |
| eGFR (ml/min/1.73m2) | 87±23 | 75±19 | <0.01 |
| CKD stage 1 | 38% | 20% | |
| stage 2 | 55% | 58% | n.s. |
| stage 3 | 7% | 22% | |
| Parathormone (pg/ml) |
15 (9, 20) | 16 (10,31) | n.s. |
| Length of the follow up (years) | 10 (8, 12) | 7 (4, 9)† | 0.008 |
Proportion, mean ± standard deviation or median (25th, 75th percentile) are presented.
Abbreviations: T2D – type 2 diabetes, ESRD end-stage renal disease, ACR – albumin to creatinine ratio, eGFR – estimated glomerular filtration rate, HbA1c – hemoglobin A1c.
Bonferroni corrected
until ESRD development.
Results of global metabolomic analysis
Baseline plasma samples from study subjects were run on the Metabolon platform against a library containing mass spectra for 2400 chemically identified metabolites. A total of 445 named metabolites were detected. Of these, 183 belonged to drug-related metabolites or had low detectability and were excluded from further analysis. The remaining 262 were detected in at least 80% of the study subjects so we designate them as “common.” We repeated the assay in the follow-up plasma sample taken one to three years later for a subgroup of study patients to estimate intra-individual variation for each metabolite. The results by biochemical classes are summarized in Figure 1 and presented in details in supplemental Table 1. A rank correlation coefficient ≥0.4 between these paired samples was used to distinguish metabolites with persistent or transient concentrations. By this criterion, 119 of the common metabolites (45%) tracked well over time within individual and thus were considered persistent, defined here as “stable” and of particular interest, which remains in accordance with the systematic report by Floegel et al. (27)
Figure 1.
Stability of the common metabolites within individual in subjects with type 2 diabetes in plasma samples taken 1-2 years apart.
Footnote: Spearman rank correlation coefficients ( r ) are presented per individual measurements. Blue line and number represents median value per a specific class.
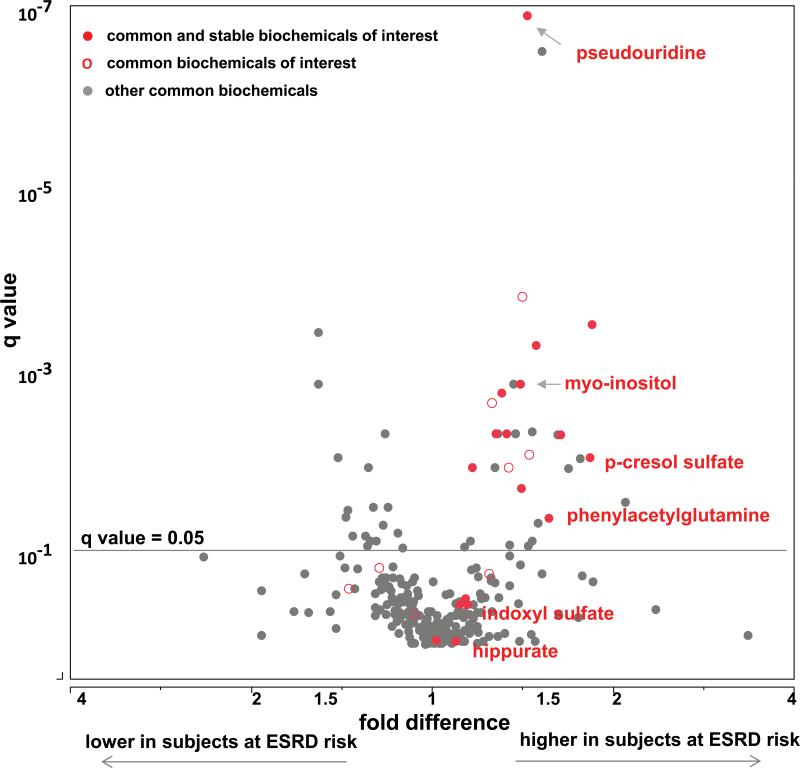
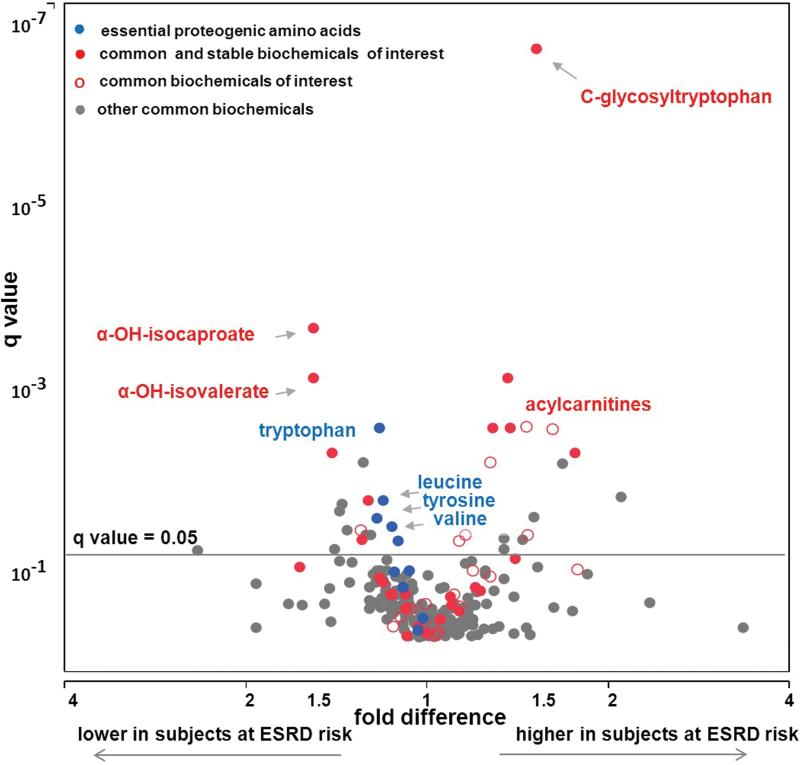
Fold differences between the mean plasma concentration in progressors to ESRD and non-progressors were analyzed for the 262 common metabolites (Table 2). In the analysis of fold difference, 49 (19%) were significant after adjusting for multiple comparisons (q value <0.05). Among the subset of 119 metabolites stable over time, 28 (23%) differed significantly from 1.0. These results are summarized in Table 2 according to biochemical class and whether the metabolite has been identified as a uremic solute. The proportion of metabolites associated with ESRD progression was similar regardless of whether the metabolite was stable over time. Nevertheless, we focused further analysis on the stable ones because their involvement in a prolonged process such as progression to ESRD is more plausible. Among the biochemical classes, the proportion associated with ESRD was high for amino acids and their derivatives, carbohydrates, and modified nucleotides (40%, 42%, and 57%, respectively), intermediate (16%) for other metabolites, and low (4%) for lipids (see Supplemental Figure 1A-1D).
Table 2.
Summary of global metabolomic analysis: frequency of significant fold differences between plasma concentrations in cases (who subsequently progressed to ESRD) and controls (did not progress) according to type of metabolite and its recognition as a uremic solute
| Biochemical class: | All common metabolites | Uremic solutes | ||||
|---|---|---|---|---|---|---|
| Total | Associated with ESRD* | Total | Associated with ESRD* | |||
| n | n | (%) | n | n | (%) | |
| Lipids: | ||||||
| Common metabolites† | 126 | 4 | (3%) | 0 | 0 | (0%) |
| Stable metabolites ‡ | 51 | 2 | (4%) | 0 | 0 | (0%) |
| Amino acid and derivatives: | ||||||
| Common metabolites† | 67 | 25 | (37%) | 10 | 3 | (30%) |
| Stable metabolites ‡ | 35 | 14 | (40%) | 6 | 2 | (33%) |
| Carbohydrates: | ||||||
| Common metabolites† | 20 | 9 | (45%) | 6 | 6 | (100%) |
| Stable metabolites ‡ | 14 | 6 | (42%) | 5 | 5 | (100%) |
| Nucleotides: | ||||||
| Common metabolites† | 12 | 7 | (58%) | 10 | 7 | (70%) |
| Stable metabolites ‡ | 7 | 4 | (57%) | 5 | 4 | (80%) |
| Other metabolites: | ||||||
| Common metabolites† | 37 | 4 | (11%) | 1 | 0 | (0%) |
| Stable metabolites ‡ | 12 | 2 | (16%) | 0 | 0 | (0%) |
| TOTAL: | ||||||
| Common metabolites† | 262 | 49 | (19%) | 27 | 16 | (55%) |
| Stable metabolites over time ‡ | 119 | 28 | (23%) | 18 | 12 | (67%) |
Values of fold difference were significantly different from 1.0 at a q-value <0.05. See also Figure 1 A & B.
Detectable in plasma of ≥80% of patients
Common and stable over time, i.e. Spearman correlation coefficient ≥0.4 between measurements taken 1-2 years apart from the same individual. Out of 29 stable metabolites associated with ESRD, five were not examined further.
Progression to ESRD according to plasma concentration of uremic solutes
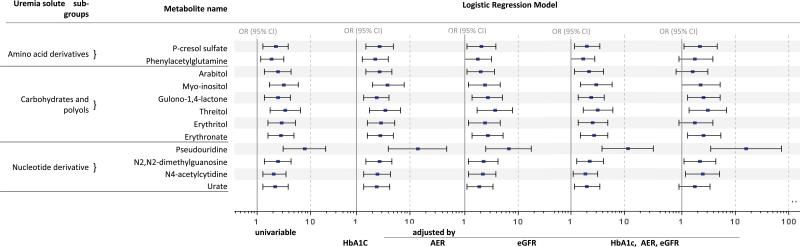
The fold difference between progressors and non-progressors and its q value for each of these metabolites are plotted in Figure 2A according to whether the metabolite has been identified as a uremic solute. Among 119 metabolites that were common and stable over time within individual, 18 are known as uremic solutes and, of these, the fold difference was significantly increased above 1.0 for 12 (67%). To obtain a descriptive measure of their effects on the risk of progression to ESRD, we turned to logistic regression analysis so we can express the associations in terms of odds ratios for the outcome, ESRD. The results are grouped by biochemical class in Figure 3, which also shows the odds ratios after adjustment for the clinical covariates, AER, eGFR and HbA1c. The fact that the associations remained after adjustment for AER, eGFR and HbA1c suggests that the effects of these uremic solutes are potentially independent from the clinical characteristics.
Figure 2.
Multivariate analysis (volcano plot) of all common metabolites measured on the Metabolon platform and their association with progression to ESRD are demonstrated as a fold difference (x axis) and significance adjusted for multiple comparisons and presented as q values (y axis). Uremic solutes comprise metabolites of interests in Figure 2A, amino acids are metabolites of interests in Figure 2B. Uremic solutes are not displayed in Figure 2B. Common and stable metabolites of interest are represented as red circles ( ), common, but not stable over time as red empty circles (
), common, but not stable over time as red empty circles ( ), all other common as grey circles (
), all other common as grey circles ( ). Blue circles represent essential amino acids (
). Blue circles represent essential amino acids ( ).
).
Figure 3.
Logistic regression analysis of the effect of the plasma concentration of metabolites identified as uremic solutes on the risk of progression to ESRD in patients with T2D.
Footnote: Data are odds ratios and 95% confidence intervals (OR, 95% CI) estimated for an effect of one standard deviation change of the metabolite. Abbreviations: ESRD – end stage renal disease, T2D – type 2 diabetes, HbA1c – hemoglobin A1c, AER – albumin excretion rate, eGFR – estimated glomerular filtration rate.
Among the amino acid-derived uremic solutes associated with progression to ESRD were two, p-cresol sulfate and phenylacetylglutamine, produced by the gut microbiome. Their effects on the risk of progression to ESRD were strong. For example, the odds ratio for progression to ESRD for a one standard deviation increase in plasma p-cresol sulfate concentration was 2.3 (95%CI; 1.3, 3.9) in univariable analysis. The effect of phenylacetylglutamine was similar, but slightly less than that of p-cresol sulfate. Of the six polyol derived uremic solutes significantly associated with the risk of progression to ESRD, myo-inositol was the one most strongly associated, odds ratio: 3.2 (95% CI; 1.7, 5.9). Of the four nucleotide-derived uremic solutes significantly associated with the risk of progression to ESRD, three are derived from degradation of RNA and the fourth (urate or uric acid) is derived from degradation of DNA. The strongest association with progression to ESRD was for pseudouridine, odds ratio 7.8 (95% CI; 3.1, 19).
Progression to ESRD according to plasma concentration of amino acids and their derivatives
Thirty-nine metabolites representing amino acids or their derivatives were common and stable over time and 14 of them (29%) were associated with risk of progression to ESRD. The fold difference between progressors and non-progressors and its q-value for each of these metabolites are plotted in Figure 2B along with other common and stable metabolites after removal of the 16 uremic solutes (including 2 amino acid derivatives).
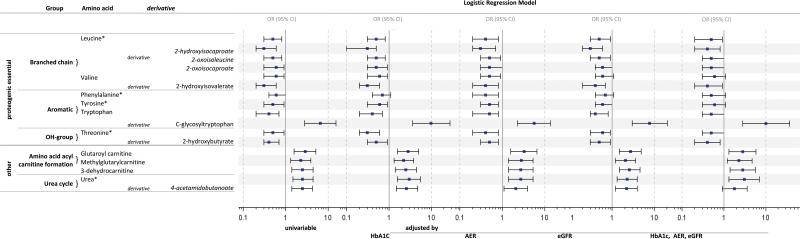
The effects of the remaining 12 amino acids on risk of ESRD were estimated with logistic regression Figure 4. In addition, five essential amino acids are present, although they were not stable over time. As in Figure 3, the odds ratios after adjustment for the group differences in AER, eGFR and HbA1c are also shown. It is important to note that none of the associations in Figure 4 were diminished by the adjustments, therefore they seem to be independent of these clinical covariates. In contrast to the associations with uremic solutes, concentrations of many of these metabolites were higher in the non-progressors than the progressors. For example, low concentrations of six essential proteogenic amino acids were associated with progression to ESRD. The odds ratio for progression to ESRD for a one standard deviation increase in the plasma concentration of leucine was 0.5 (95% CI; 0.3, 0.8) and odds ratios for the remaining 5 amino acids were similar Figure 4. In addition, five amino acid derivatives were negatively associated with risk of progression to ESRD. The odds ratio for a one standard deviation increase in plasma concentration of 2-hydroxyisocaproate (leucine derivative) was 0.3 (95%CI; 0.2, 0.6), and the odds ratios for the remaining five derivatives were similar.
Figure 4.
Logistic regression analysis of the effect of the plasma concentration of proteogenic amino acids and amino acid derivatives on the risk of progression to ESRD in subjects with T2D.
Footnote: Data are odds ratios and 95% confidence intervals (OR, 95% CI) estimated for an effect of one standard deviation change of the metabolite. * Metabolite was not stable over time but is shown for its biological relevance. Abbreviations: ESRD – end stage renal disease, T2D – type 2 diabetes,HbA1c – hemoglobin A1c, AER – albumin excretion rate, eGFR – estimated glomerular filtration rate, OR – odds ratio.
A few amino acid derivatives were positively associated with progression to ESRD. C-glycosyltryptophan was elevated and the most significantly different between progressors and non-progressors among the metabolites shown in Figure 2B. The odds ratio for a one standard deviation increase in its plasma concentration was 6.6 (95%CI; 2.8, 15). The five remaining metabolites, which are derivatives related to acylcarnitines and the urea cycle, were positively associated with progression to ESRD with smaller odds ratios.
Among other metabolites associated with ESRD risk, there were two lipids, dihomo-linolenate (20:3n3) and docosapentaenoate (n3 DPA; 22:5n3). Both are ω6 fatty acids and were negatively associated with risk of progression to ESRD. Gluconate and two metabolites involved in vitamin B metabolism: pantothenate and N1-methyl-2-pyridone-5-carboxamide were positively associated with progression to ESRD. Data for these five metabolites were not shown.
Reduction of redundant data
Among metabolites significantly associated with progression to ESRD, some may reflect on shared underlying biology. To evaluate the potential redundancy among these metabolites, we created a Spearman rank correlation matrix. Approximately two-thirds of the metabolites were strongly correlated (data not shown).
A cluster analysis revealed clusters (see Figure 5) mirroring the patterns of our grouping based on the biological relevance presented in Figures 3 and 4, respectively.(28, 29) Clusters 1 and 2 comprised uremic solutes and C-glycosyltryptophan. Cluster 3 included carnitine derivatives, urate and urea. Cluster 4 comprised essential amino acids, cluster 5 their keto- and cluster 6 their hydroxylderivatives, respectively. In a logistic regression model including the leading metabolites from each cluster, erythritol, glutaryl carnitine and alphahydroxyisovalerate (from clusters 2, 3 and 6) remained significant. Odds ratios for an effect of one standard deviation difference in those metabolites considered together were: for erythritol 2.1 (95% CI 1.0, 4.5), for glutaroyl carnitine, 2.6 (95% CI 1.3, 5.4) and for 2-hydroxyisovalerate, 0.4 (95% CI 0.2, 0.9), respectively. Discrimination ability was c=0.89 for this model, while it was 0.74 to 0.75 for models including the significant metabolites separately. The p value for the difference tested by integrated discrimination improvement (IDI) test was p<0.001 for each single model in comparison with the model including all four metabolites. When either pseudouridine or C-glycosyltryptophan (the most significantly different metabolites between the progressors and non-progressors present in Cluster 2) were added to the logistic model with the representative of the other clusters, the effect of the remaining metabolites became borderline or non-significant.
Figure 5.
Hierarchical cluster analysis (Ward's method) of the metabolites significantly associated with progression to ESRD. Separate clusters are marked in different colors. Distance scale is shown. Footnote: C1-C6 represent respective clusters.
Targeted quantitative metabolites measurements
To validate our findings, we performed targeted quantitative measurements of nine common and stable metabolites over time identified on the Metabolon global biochemical profiling platform. Five out of nine metabolites were the ones strongly associated with progression to ESRD. Among the remaining four, two metabolites, indoxyl sulfate and hippurate were uremic solutes reported in the literature, but not significantly associated with progression in the global profiling study. The remaining two measured were allantoin and uracil, linked in the metabolic pathways related to urate and pseudouridine, respectively. Allantoin was not sufficiently detectable by our quantitative methods (data not shown). For each of the other metabolites, association with progression to ESRD was consistent with the result obtained via global profiling. Also the association remained after adjustment for AER, eGFR and HbA1c (Table 3). Odds ratio for ESRD progression was 1.7 (95%CI; 1.0, 2.8) per one standard deviation of the logarithmically transformed metabolite, p-cresol sulfate and phenylacetylglutamine and higher than 2.5 for every other significant metabolite (pseudouridine, p-cresol sulfate, myoinositol and urate). We confirmed that hippurate and indoxyl sulfate were not significantly associated with ESRD in our study. Logistic regression of the pseudouridine/uracil ratio did not improve the discrimination ability of the model (data not shown).
Table 3.
Logistic regression analysis of the effect of plasma concentration of uremic solutes measured by targeted quantitative metabolomics on the risk of progression to ESRD in subjects with T2D.
| Uremic solute name | mean ± std | Logistic Regression Model univariable | |
|---|---|---|---|
| μmol/L | OR (95% CI) | p | |
| Significant in the global profiling analysis | |||
| P-cresol sulfate | 33±30 | 1.7 (1.0, 2.8) | 0.050 |
| Phenylacetylglutamine | 4±5 | 1.7 (1.0, 2.9) | 0.041 |
| Myo-inositol | 52±35 | 2.8 (1.5, 5.5) | 0.002 |
| Pseudouridine | 1.5±0.8 | 2.4 (1.3, 4.3) | 0.004 |
| Urate | 275±75* | 2.5 (1.3, 4.8) | 0.007 |
| Non significant in the global profiling analysis | |||
| Hippurate | 8.0 ±8.5 | 0.9 (0.6, 1.5) | 0.798 |
| Indoxyl sulphate | 8.0±9.3 | 1.3 (0.8, 2.1) | 0.280 |
| Uracil | 1.9±6.8 | 1.3 (0.8, 2.0) | 0.355 |
Data are odds ratios (95% confidence interval) OR (95% CI) estimated as the effect per one standard deviation of the logarithmically transformed metabolite. Abbreviations: ESRD – end stage renal disease, T2D – type 2 diabetes, std – standard deviation
In order to convert urate (uric acid) μmol/L concentrations to mg/dl, please divide the values by 59.5. Concentration of the uric acid 275 μmol/L is equal to 4.6 mg/dl.
DISCUSSION
This study was performed in subjects with T2D, the majority of whom had normal renal function at baseline. Half progressed to ESRD and half did not during a decade of follow-up. In baseline plasma, progressors could be distinguished from non-progressors by high concentrations of metabolites referred to as uremic solutes and low concentrations of certain amino acids and their derivatives. This is the first demonstration that abnormal plasma concentrations of certain metabolites are associated with risk of progression to ESRD at a very early stage of diabetic nephropathy.
Uremic solutes, as catalogued by EUTox group,(7, 9) comprise compounds of different biochemical classes: amino acid derivatives, certain alcohol/polyols and modified nucleosides among them. Of the 18 known uremic solutes that were detected as common and stable metabolites with the Metabolon platform, 12 were elevated in subjects who progressed to ESRD. On the basis of results of multiple cross-sectional studies (7-10) the immediate interpretation of our findings might be that the increased concentration of uremic solutes was due to significant impairment of renal function in subjects who progressed to ESRD during 8-12 years of follow-up. However, the majority of progressors at baseline were in CKD stages 1 and 2 and had only slightly lower baseline eGFR than non-progressors (75±19 ml/min vs. 87 ±23ml/min). In multivariable analyses, the odds ratios of progression to ESRD for specific uremic solutes were only minimally affected by adjustment for eGFR, AER and HbA1c. In the following we discuss each sub-group of uremic solutes associated with risk of ESRD.
Phenyl compounds, such as p-cresol sulfate and phenylacetylglutamine are the most extensively studied solutes known to increase in the uremic state.(8, 11, 13) These solutes can be toxic to endothelial cells and can contribute to increased risk of cardiovascular complications in patients with renal impairment.(30, 31) In humans these metabolites are exogenous and are produced by intestinal bacterial flora before they are absorbed into plasma and excreted through the kidney.(8, 11, 13, 32) Evidence confirming the microbiome as a source for these solutes was recently provided in a study of ESRD subjects with and without a colon.(11) In our study, high plasma concentrations of these solutes were associated with progression to ESRD. It would be valuable to determine how different or similar those concentrations are in comparison with subjects at competing risk of subsequent cardiovascular complications.(33)
Three additional solute derivatives of amino acids that are synthesized in the gut, phenol sulfate, indoleacetate and 3-indoxyl sulfate, were elevated in plasma of progressors compared to non-progressors, however, the differences did not reach statistical significance (see supplemental Table 2). All these findings support the notion that the gut microbiome (11, 13, 32) might control plasma levels of amino acid-derived uremic solutes, and their high levels increase the risk of progression to ESRD in subjects with T2D. The increase in circulating levels of these solutes also may be attributed to dietary factors.(34)
Elevated plasma concentrations of myo-inositol and other polyols were strongly associated with progression to ESRD in our study. These metabolites accumulate in plasma in the uremia state and during acute kidney injury.(9, 35, 36) The kidney is the most important organ for whole body metabolism of myo-inositol, as both the synthetic and degradative enzymes, L-myo-inositol-1-phosphate synthetase and myo-inositol oxygenase (MIOX), are highly expressed in the renal parenchyma.(37, 38) Myo-inositol can be obtained also from dietary sources. Regulation of its plasma levels is complex, involving glomerular filtration, reabsorption in proximal tubules in competition with glucose transport, apparent conversion to chiro-inositol in tissues and, finally, catabolic degradation of myo-inositol by MIOX, a protein that is upregulated by hyperglycemia.(36, 38-44) The high concentration gradients of myo-inositol in certain tissues such as kidney and the vascular endothelium are maintained by the osmoregulatory sodium/myo-inositol cotransporter 1 (SMIT1)(39, 40) Glucose/myo-inositol imbalance was demonstrated to induce proliferative and pro-fibrotic response in the proximal tubules in vitro and to alter the immune responses in leukocytes.(45, 46) Increased urinary excretion of myo-inositol and inositol imbalance in muscle tissue (high myo/chiro-inositol ratios) were reported in subjects with T2D.(42-44) In our study chiroinositol levels were hardly detectable (data not shown).
Plasma concentrations of several nucleotide derivatives that are considered to be uremic solutes were also strongly associated with progression to ESRD in our study. Among these derivatives, elevated concentration of pseudouridine in plasma was the strongest and most statistically significant predictor of progression. Pseudouridine belongs to the group of modified nucleosides that are regarded as indicators of whole-body RNA turnover.(47) These metabolites are increased in patients with malignancies,(48) and with uremia.(8, 49-51) Pseudouridine is synthesized from uracil(52, 53) and constitutes an end product, as it is not catabolized in humans.(54) Trace study in humans with radiolabeled pseudouridine showed that the kidney handling includes glomerular filtration and also tubular reabsorption.(55) High plasma levels of two other pyrimidine derivatives that are closely correlated with plasma levels of pseudouridine also increased the risk of ESRD progression (N2, N2-dimethylguanosine, and N4-acetylcytidine). These three nucleotides increase in plasma as urinary excretion decreases and accumulate significantly in the uremic state.(51)
Urate (uric acid) is a metabolite of purine metabolism. In our study, increased plasma concentrations were associated with progression to ESRD. Urate is another compound known to accumulate in the uremic state.(8, 9) Its increase, however, is disproportionally small due to compensatory mechanisms including increased enteric excretion, decreased production(56) and possibly altered tubular handling.(57) In our recent study, elevated plasma level of urate was a strong predictor of early renal function decline during follow-up of a large cohort of subjects with T1D.(58)
What are the mechanisms through which the elevated plasma levels of uremic solutes identified in our study could increase risk of progression to ESRD? The common feature of these organic solutes is that all are handled by the tubular compartment. Uremic toxins (including indoxyl sulfate and uric acid) have been reported to cause proximal tubular injury via different mechanisms, tubulointerstitial hypoxia, endoplasmic reticulum stress or via organic anion transporters (OAT)-mediated mechanisms (59, 60 61). Increase in the urinary excretion of indoxyl sulfate predicted albuminuria development in Pima Indian subjects with T2D and normal renal function at baseline.(63) The SLC22 family of transporters comprises OATs, urate transporters and organic carnitine transporters. Numerous family members are expressed in the proximal tubules on either basolateral or apical side. Their function was extensively reviewed recently.(61, 62) OAT knockout animals accumulate uremic solutes in plasma(62), whereas decreased expression of certain basolateral organic transporters has been identified in chronic kidney failure.(63, 64) Hippurate is an interesting example of a uremic solute that is not a uremic toxin. It is one of the most highly accumulated metabolites in ESRD, the concentrations are up to 100 times that in normal subjects.(9-11) However, in vitro, hippurate does not demonstrate an OAT-dependent toxic effect.(65) Our study groups had preserved renal function and baseline hippurate levels did not differ between cases and controls.
Kidney protein turnover, as compared with muscle and splanchnic turnover, is characterized by the highest rates of protein synthesis and amino acid oxidation, mainly in the tubulointerstitium.(17, 66) Depletion of the circulating pools of branched chain amino acids and tryptophan are known phenomena accompanying advanced chronic kidney disease.(10, 16, 17) On the other hand, increase in those amino acids was shown to predict development of T2D. (19) Interestingly, our study revealed that not only branched and aromatic, but also all other essential amino acids and their derivatives were lower in subjects who progressed to ESRD than in those who were non-progressors. In an experimental model of acute kidney injury, one of the strongest metabolic responses to nephrotoxins was massive excretion of all essential amino acids.(35, 67) It needs to be determined whether impaired tubular reabsorption contributes to the decreased levels of amino acids in early diabetic nephropathy. Amino acids that were decreased in subjects at risk of ESRD in our study all have a neutral charge. They are handled by the B0AT1 cotransporter responsible for the luminal influx, the heterodimeric exchanger and the facilitated diffusion transporter TAT1 in charge of the basolateral efflux and possibly by other transporters.(68, 69)
Among amino-acid derivatives, one C-glycosyltryptophan showed a different pattern of association than the others. Its plasma concentration was the highest in progressors when compared with non-progressors. After pseudouridine it had the second highest fold difference between the study groups. Plasma concentrations of both were very highly correlated. Interestingly, they both carry a C-glycosylation linkage, a rare type of posttranslational protein modification.(70) The biological meaning of this mutual characteristic remains unknown. Increased expression of the proteins containing certain forms of C-glycosylated tryptophan in the aortic vessels have been reported in the diabetic rats.(71) C-glycosylated tryptophan also correlates with eGFR.(72)
Analysis of acylcarnitines revealed that the increased concentrations were independently associated with risk of progression to ESRD. Acylcarnitines are filtered through the kidney and about 75% are excreted into urine.(73) Serum acylcarnitines deriving from lipid and amino acids are inversely correlated with GFR in individuals with normal as well as with impaired renal function.(18, 74, 75) Acylcarnitines transport is regulated by organic carnitine transporters in the kidney.(61) It is interesting also that in our study amino acid- deriving (but not lipid-deriving) acylcarnitines were increased in the subjects at risk. They are generated via beta-oxidation of the branched chain amino acids. Those amino acids and their intermediate keto acid derivatives were also depleted in our study (2-oxoisoleucine, 2-oxoisocaproate), which may suggest an enhanced mitochondrial amino acids beta-oxidation.
Strengths and limitations of our study should be considered. In contrast to multiple case-control studies previously reported, ours is the first follow-up study in which progressors and non-progressors were studied at the time when the majority had normal renal function. We used the most comprehensive metabolomics platform with the broadest spectrum of the metabolites examined. We acknowledge that the study groups were small and that, although the prevailing majority was of homogenous ancestry origin, not all the study subjects were of the same ancestry. Therefore our findings, which are exploratory in nature, warrant replication study. Nevertheless, the differences between the study groups were statistically significant when individual metabolites or grouped-metabolites were compared. Traditional pathway analysis was not performed because more than half of the common metabolites observed in this study are not yet among those with universal pathway database annotations (52, 53).
STUDY GROUPS AND METHODS
Study group
Between 1991 and 1995, a cohort with T2D was recruited into the Joslin Study of the Genetics of Type 2 Diabetes and Kidney Complications (half with normoalbuminuria and half with microalbuminuria or proteinuria). The cohort was followed-up until 2004 for the occurrence of ESRD or death unrelated to ESRD. Details of the recruitment, examination, and follow-up of this cohort were already published elsewhere.(26) Among 410 individuals, 59 developed ESRD and 84 died without ESRD as ascertained by United States Renal Data System(1), National Death Index(76) and medical records review.
For this nested case-control study, 40 out of 56 incident cases of ESRD who had sufficient stored plasma samples at baseline available were selected. A group of 40 subjects from among those who survived and were without ESRD as of the end of follow-up were selected as controls. They were grouped-matched with cases with regard to gender, age, and baseline eGFR. For comparison of cases and controls we used baseline clinical characteristics as reported previously.(26)
As a small reproducibility study, ten study subjects (balanced by caseness status) had plasma samples selected 2.2+0.8 years after baseline. All plasma samples were stored at −70°C until analysis. The study protocol and informed consent procedures were approved by the Joslin Diabetes Center Institutional Review Board.
Global metabolomics profiling
All plasma samples (80 baseline and 10 from the early follow up timepoint) were subjected to global metabolomic profiling (Metabolon, Inc, Durham, NC). (5, 6) The detailed protocols of global metabolomics profiling are provided in the Supplementary information.
Uremic solutes
The European Uremic Toxins (EUTox) Work Group, initiated in 1999, consists of 24 European Research Institutes and provides the most comprehensive encyclopedic list of systematically and critically reviewed uremic solutes/toxins.(7, 9) Metabolites measured with the global profiling were classified as uremic solutes/toxins based on the EUTox list prepared in 2003, revisited in 2012 as well as based on selected relevant other publications.(7-9, 11, 13, 32) Seventy eight uremic solutes are available in the Metabolon library. For detailed information of the detectable uremic solutes in our study, please see Supplemental Table 2.
Targeted quantitative measurements of metabolites
Method description of the quantitative methods is provided in the Supplementary information.
Data analysis
Differences in clinical characteristics between the two study groups were tested by analysis of variance for continuous variables and chi-square test for categorical variables. Preliminary data cleaning included investigations of detectability, batch effects and outliers at the metabolite- and individual-levels (heatmap, principal component procedures; data not shown).(29, 77) Volcano plots were generated for common metabolites based on the fold difference between the outcome groups and the p-value obtained in a general linear model. Adjustment for multiple comparison was performed with a positive false discovery rate (pFDR) q value <0.05 for significance.(78) The effect of a metabolite was estimated using logistic regression. After transformation of the metabolite concentration to normally distributed ranks, its effect on risk of progression was expressed as the odds ratio for a one standard deviation difference.(26) Clinical covariates and metabolites measured quantitatively were transformed to their (base 10) logarithms for the logistic analysis. Non-common metabolites were analyzed as categorical variables by chi-square, but this analysis did not result in identifying additional significant metabolites (data not shown).
Correlations between the continuous variables were examined with Spearman rank correlation. Data reduction was carried out with hierarchical cluster analysis using the Ward method based on values transformed to normally distributed ranks. Top metabolites from the multivariate volcano plots analysis were included. More than a half of the detected metabolites lacked pathway identifiers, preventing us from a comprehensive canonical pathway analysis. Data analysis was performed with SAS 9.3 and JMP Pro 9.0.0 softwares (Cary, NC).
Supplementary Material
Acknowledgments
This study was supported by grants from the National Institutes of Health (NIH): Diabetes Research Center - Joslin, Pilot and Feasibility Grant, P30DK036836 to Dr. M. Niewczas; DK41526 and DK67638 to A.S. Krolewski; DK39773 and DK 72381 to JV Bonventre; DK94292, DK89503 and DK097153 to S. Pennathur; DK84439 and DK80123 to TW Meyer. T. Sirich was supported by the Mitsubishi Tanabe Pharma Corporation NKF Fellowship for the Study of Uremia, and J. Skupien by the JDRF fellowship grant 3-2009-397.
Footnotes
Supplementary information
Supplementary information is available at Kidney International's website.
Disclosure: R.P.Mohney, E.D.Karoly and E.M.Kensicki are employees of Metabolon, Inc. and, as such, have affiliations with or financial involvement with Metabolon, Inc. The remaining authors have nothing to disclose.
Reference List
- 1.U.S.Renal Data System . USRDS 2010 Annual Data Report: Atlas of Chronic Kidney Disease and End-Stage Renal Disease in the United States. National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases; Bethesda, MD: 2010. 2010. [Google Scholar]
- 2.Wang JH, Byun J, Pennathur S. Analytical approaches to metabolomics and applications to systems biology. Semin Nephrol. 2010;30:500–11. doi: 10.1016/j.semnephrol.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Portilla D, Schnackenberg L, Beger RD. Metabolomics as an extension of proteomic analysis: study of acute kidney injury. Semin Nephrol. 2007;27:609–20. doi: 10.1016/j.semnephrol.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dehaven CD, Evans AM, Dai H, Lawton KA. Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J Cheminform. 2010;2:9. doi: 10.1186/1758-2946-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans AM, Dehaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small- molecule complement of biological systems. Anal Chem. 2009;81:6656–67. doi: 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- 6.Ohta T, Masutomi N, Tsutsui N, et al. Untargeted metabolomic profiling as an evaluative tool of fenofibrate-induced toxicology in Fischer 344 male rats. Toxicol Pathol. 2009;37:521–35. doi: 10.1177/0192623309336152. [DOI] [PubMed] [Google Scholar]
- 7.Duranton F, Cohen G, De Smet R, et al. Normal and pathologic concentrations of uremic toxins. J Am Soc Nephrol. 2012;23:1258–70. doi: 10.1681/ASN.2011121175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyer TW, Hostetter TH. Uremia. N Engl J Med. 2007;357:1316–25. doi: 10.1056/NEJMra071313. [DOI] [PubMed] [Google Scholar]
- 9.Vanholder R, De Smet R, Glorieux G, et al. Review on uremic toxins: classification, concentration, and interindividual variability. Kidney Int. 2003;63:1934–43. doi: 10.1046/j.1523-1755.2003.00924.x. [DOI] [PubMed] [Google Scholar]
- 10.Rhee EP, Souza A, Farrell L, et al. Metabolite profiling identifies markers of uremia. J Am Soc Nephrol. 2010;21:1041–51. doi: 10.1681/ASN.2009111132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aronov PA, Luo FJ, Plummer NS, et al. Colonic contribution to uremic solutes. J Am Soc Nephrol. 2011;22:1769–76. doi: 10.1681/ASN.2010121220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eloot S, Schepers E, Barreto DV, et al. Estimated glomerular filtration rate is a poor predictor of concentration for a broad range of uremic toxins. Clin J Am Soc Nephrol. 2011;6:1266–73. doi: 10.2215/CJN.09981110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyer TW, Hostetter TH. Uremic solutes from colon microbes. Kidney Int. 2012;81:949–54. doi: 10.1038/ki.2011.504. [DOI] [PubMed] [Google Scholar]
- 14.Mogensen CE. Microalbuminuria, blood pressure and diabetic renal disease: origin and development of ideas. Diabetologia. 1999;42:263–85. doi: 10.1007/s001250051151. [DOI] [PubMed] [Google Scholar]
- 15.Vaidya VS, Niewczas MA, Ficociello LH, et al. Regression of microalbuminuria in type 1 diabetes is associated with lower levels of urinary tubular injury biomarkers, kidney injury molecule-1, and N-acetyl-beta-D-glucosaminidase. Kidney Int. 2011;79:464–70. doi: 10.1038/ki.2010.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cano NJ, Fouque D, Leverve XM. Application of branched-chain amino acids in human pathological states: renal failure. J Nutr. 2006;136:299S–307S. doi: 10.1093/jn/136.1.299S. [DOI] [PubMed] [Google Scholar]
- 17.Garibotto G, Sofia A, Saffioti S, Bonanni A, Mannucci I, Verzola D. Amino acid and protein metabolism in the human kidney and in patients with chronic kidney disease. Clin Nutr. 2010;29:424–33. doi: 10.1016/j.clnu.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 18.Goek ON, Doring A, Gieger C, et al. Serum metabolite concentrations and decreased GFR in the general population. Am J Kidney Dis. 2012;60:197–206. doi: 10.1053/j.ajkd.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 19.Wang TJ, Larson MG, Vasan RS, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–53. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H, Saha J, Byun J, et al. Rosiglitazone reduces renal and plasma markers of oxidative injury and reverses urinary metabolite abnormalities in the amelioration of diabetic nephropathy. Am J Physiol Renal Physiol. 2008;295:F1071–F1081. doi: 10.1152/ajprenal.90208.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao T, Zhang H, Zhao T, et al. Intrarenal metabolomics reveals the association of local organic toxins with the progression of diabetic kidney disease. J Pharm Biomed Anal. 2012;60:32–43. doi: 10.1016/j.jpba.2011.11.010. [DOI] [PubMed] [Google Scholar]
- 22.Makinen VP, Soininen P, Forsblom C, et al. Diagnosing diabetic nephropathy by 1H NMR metabonomics of serum. MAGMA. 2006;19:281–96. doi: 10.1007/s10334-006-0054-y. [DOI] [PubMed] [Google Scholar]
- 23.Makinen VP, Tynkkynen T, Soininen P, et al. Metabolic diversity of progressive kidney disease in 325 patients with type 1 diabetes (the FinnDiane Study). J Proteome Res. 2012;11:1782–90. doi: 10.1021/pr201036j. [DOI] [PubMed] [Google Scholar]
- 24.van der Kloet FM, Tempels FW, Ismail N, et al. Discovery of early-stage biomarkers for diabetic kidney disease using ms-based metabolomics (FinnDiane study). Metabolomics. 2012;8:109–19. doi: 10.1007/s11306-011-0291-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Yan L, Chen W, et al. Metabonomics research of diabetic nephropathy and type 2 diabetes mellitus based on UPLC-oaTOF-MS system. Anal Chim Acta. 2009;650:16–22. doi: 10.1016/j.aca.2009.02.027. [DOI] [PubMed] [Google Scholar]
- 26.Niewczas MA, Gohda T, Skupien J, et al. Circulating TNF Receptors 1 and 2 Predict ESRD in Type 2 Diabetes. J Am Soc Nephrol. 2012;23(3):507–515. doi: 10.1681/ASN.2011060627. J Am Soc Nephrol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Floegel A, Drogan D, Wang-Sattler R, et al. Reliability of serum metabolite concentrations over a 4-month period using a targeted metabolomic approach. PLoS One. 2011;6:e21103. doi: 10.1371/journal.pone.0021103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milligan GV. A Review Of Monte Carlo Tests Of Cluster Analysis. Multivariate Behavioral Research. 1981;16(3):379–407. doi: 10.1207/s15327906mbr1603_7. [DOI] [PubMed] [Google Scholar]
- 29.Xia J, Wishart DS. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat Protoc. 2011;6:743–60. doi: 10.1038/nprot.2011.319. [DOI] [PubMed] [Google Scholar]
- 30.Itoh Y, Ezawa A, Kikuchi K, Tsuruta Y, Niwa T. Protein-bound uremic toxins in hemodialysis patients measured by liquid chromatography/tandem mass spectrometry and their effects on endothelial ROS production. Anal Bioanal Chem. 2012;403:1841–50. doi: 10.1007/s00216-012-5929-3. [DOI] [PubMed] [Google Scholar]
- 31.Meijers BK, Claes K, Bammens B, et al. p-Cresol and cardiovascular risk in mild-to- moderate kidney disease. Clin J Am Soc Nephrol. 2010;5:1182–9. doi: 10.2215/CJN.07971109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicholson JK, Holmes E, Kinross J, et al. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–7. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 33.Go AS, Chertow GM, Fan D, McCulloch CE, Hsu CY. Chronic kidney disease and the risks of death, cardiovascular events, and hospitalization. N Engl J Med. 2004;351:1296–305. doi: 10.1056/NEJMoa041031. [DOI] [PubMed] [Google Scholar]
- 34.Boudonck KJ, Mitchell MW, Nemet L, et al. Discovery of metabolomics biomarkers for early detection of nephrotoxicity. Toxicol Pathol. 2009;37:280–92. doi: 10.1177/0192623309332992. [DOI] [PubMed] [Google Scholar]
- 35.Clements RS, Jr., DeJesus PV, Jr., Winegrad AI. Raised plasma-myoinositol levels in uraemia and experimental neuropathy. Lancet. 1973;1:1137–41. doi: 10.1016/s0140-6736(73)91143-4. [DOI] [PubMed] [Google Scholar]
- 36.Arner RJ, Prabhu KS, Thompson JT, Hildenbrandt GR, Liken AD, Reddy CC. myo-Inositol oxygenase: molecular cloning and expression of a unique enzyme that oxidizes myo-inositol and D-chiro-inositol. Biochem J. 2001;360:313–20. doi: 10.1042/0264-6021:3600313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nayak B, Kondeti VK, Xie P, et al. Transcriptional and post-translational modulation of myo-inositol oxygenase by high glucose and related pathobiological stresses. J Biol Chem. 2011;286:27594–611. doi: 10.1074/jbc.M110.217141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berry GT, Mallee JJ, Kwon HM, et al. The human osmoregulatory Na+/myo-inositol cotransporter gene (SLC5A3): molecular cloning and localization to chromosome 21. Genomics. 1995;25:507–13. doi: 10.1016/0888-7543(95)80052-n. [DOI] [PubMed] [Google Scholar]
- 39.Mallee JJ, Atta MG, Lorica V, et al. The structural organization of the human Na+/myo inositol cotransporter (SLC5A3) gene and characterization of the promoter. Genomics. 1997;46:459–65. doi: 10.1006/geno.1997.5055. [DOI] [PubMed] [Google Scholar]
- 40.Wang S, Mitu GM, Hirschberg R. Osmotic polyuria: an overlooked mechanism in diabetic nephropathy. Nephrol Dial Transplant. 2008;23:2167–72. doi: 10.1093/ndt/gfn115. [DOI] [PubMed] [Google Scholar]
- 41.Kennington AS, Hill CR, Craig J, et al. Low urinary chiro-inositol excretion in non- insulin-dependent diabetes mellitus. N Engl J Med. 1990;323:373–8. doi: 10.1056/NEJM199008093230603. [DOI] [PubMed] [Google Scholar]
- 42.Larner J, Brautigan DL, Thorner MO. D-chiro-inositol glycans in insulin signaling and insulin resistance. Mol Med. 2010;16:543–52. doi: 10.2119/molmed.2010.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daughaday WH, Larner J. The renal excretion of inositol in normal and diabetic human beings. J Clin Invest. 1954;33:326–32. doi: 10.1172/JCI102901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ziyadeh FN, Simmons DA, Snipes ER, Goldfarb S. Effect of myo-inositol on cell proliferation and collagen transcription and secretion in proximal tubule cells cultured in elevated glucose. J Am Soc Nephrol. 1991;1:1220–9. doi: 10.1681/ASN.V1111220. [DOI] [PubMed] [Google Scholar]
- 45.Bartnicki P, Zbrog Z, Baj Z, Tchorzewski H, Luciak M. Myoinositol may be a factor in uremic immune deficiency. Clin Nephrol. 1997;47:197–201. [PubMed] [Google Scholar]
- 46.Charette M, Gray MW. Pseudouridine in RNA: what, where, how, and why. IUBMB Life. 2000;49:341–51. doi: 10.1080/152165400410182. [DOI] [PubMed] [Google Scholar]
- 47.Seidel A, Brunner S, Seidel P, Fritz GI, Herbarth O. Modified nucleosides: an accurate tumour marker for clinical diagnosis of cancer, early detection and therapy control. Br J Cancer. 2006;94:1726–33. doi: 10.1038/sj.bjc.6603164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Daniewska-Michalska D, Motyl T, Gellert R, et al. Efficiency of hemodialysis of pyrimidine compounds in patients with chronic renal failure. Nephron. 1993;64:193–7. doi: 10.1159/000187313. [DOI] [PubMed] [Google Scholar]
- 49.Gerrits GP, Monnens LA, De Abreu RA, Schroder CH, Trijbels JM, Gabreels FJ. Disturbances of cerebral purine and pyrimidine metabolism in young children with chronic renal failure. Nephron. 1991;58:310–4. doi: 10.1159/000186442. [DOI] [PubMed] [Google Scholar]
- 50.Niwa T, Takeda N, Yoshizumi H. RNA metabolism in uremic patients: accumulation of modified ribonucleosides in uremic serum. Technical note. Kidney Int. 1998;53:1801–6. doi: 10.1046/j.1523-1755.1998.00944.x. [DOI] [PubMed] [Google Scholar]
- 51. [April 15, 2013]; http://www.genome.jp/kegg/pathway.html.
- 52. [April 15, 2013]; http://www.hmdb.ca.
- 53.Huang S, Mahanta N, Begley TP, Ealick SE. Pseudouridine monophosphate glycosidase: a new glycosidase mechanism. Biochemistry. 2012;51:9245–55. doi: 10.1021/bi3006829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bernert JT, Jr., Bell CJ, Guntupalli J, Hannon WH. Pseudouridine is unsuitable as an endogenous renal clearance marker. Clin Chem. 1988;34:1011–7. [PubMed] [Google Scholar]
- 55.Vaziri ND, Freel RW, Hatch M. Effect of chronic experimental renal insufficiency on urate metabolism. J Am Soc Nephrol. 1995;6:1313–7. doi: 10.1681/ASN.V641313. [DOI] [PubMed] [Google Scholar]
- 56.Anzai N, Kanai Y, Endou H. New insights into renal transport of urate. Curr Opin Rheumatol. 2007;19:151–7. doi: 10.1097/BOR.0b013e328032781a. [DOI] [PubMed] [Google Scholar]
- 57.Ficociello LH, Rosolowsky ET, Niewczas MA, et al. High-Normal Serum Uric Acid Increases Risk of Early Declining Renal Function In Type 1 Diabetes: Results of 6-year Follow-up. Diabetes Care. 2010;33(6):1337–1343. doi: 10.2337/dc10-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chiang CK, Tanaka T, Nangaku M. Dysregulated oxygen metabolism of the kidney by uremic toxins: review. J Ren Nutr. 2012;22:77–80. doi: 10.1053/j.jrn.2011.10.028. [DOI] [PubMed] [Google Scholar]
- 59.Kawakami T, Inagi R, Wada T, Tanaka T, Fujita T, Nangaku M. Indoxyl sulfate inhibits proliferation of human proximal tubular cells via endoplasmic reticulum stress. Am J Physiol Renal Physiol. 2010;299:F568–F576. doi: 10.1152/ajprenal.00659.2009. [DOI] [PubMed] [Google Scholar]
- 60.Ahn SY, Bhatnagar V. Update on the molecular physiology of organic anion transporters. Curr Opin Nephrol Hypertens. 2008;17:499–505. doi: 10.1097/MNH.0b013e32830b5d5d. [DOI] [PubMed] [Google Scholar]
- 61.Wang L, Sweet DH. Renal Organic Anion Transporters (SLC22 Family): Expression, Regulation, Roles in Toxicity, and Impact on Injury and Disease. AAPS J. 2012 doi: 10.1208/s12248-012-9413-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Monica TA, Mac LM, Muller A, Brandoni A, Anzai N, Endou H. Altered renal elimination of organic anions in rats with chronic renal failure. Biochim Biophys Acta. 2005;1740:29–37. doi: 10.1016/j.bbadis.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 63.Naud J, Michaud J, Beauchemin S, et al. Effects of chronic renal failure on kidney drug transporters and cytochrome P450 in rats. Drug Metab Dispos. 2011;39:1363–9. doi: 10.1124/dmd.111.039115. [DOI] [PubMed] [Google Scholar]
- 64.Motojima M, Hosokawa A, Yamato H, Muraki T, Yoshioka T. Uremic toxins of organic anions up-regulate PAI-1 expression by induction of NF-kappaB and free radical in proximal tubular cells. Kidney Int. 2003;63:1671–80. doi: 10.1046/j.1523-1755.2003.00906.x. [DOI] [PubMed] [Google Scholar]
- 65.Broer S. Amino acid transport across mammalian intestinal and renal epithelia. Physiol Rev. 2008;88:249–86. doi: 10.1152/physrev.00018.2006. [DOI] [PubMed] [Google Scholar]
- 66.Williams RE, Major H, Lock EA, Lenz EM, Wilson ID. D-Serine-induced nephrotoxicity: a HPLC-TOF/MS-based metabonomics approach. Toxicology. 2005;207:179–90. doi: 10.1016/j.tox.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 67.Verrey F, Ristic Z, Romeo E, et al. Novel renal amino acid transporters. Annu Rev Physiol. 2005;67:557–72. doi: 10.1146/annurev.physiol.67.031103.153949. [DOI] [PubMed] [Google Scholar]
- 68.Verrey F, Singer D, Ramadan T, Vuille-dit-Bille RN, Mariotta L, Camargo SM. Kidney amino acid transport. Pflugers Arch. 2009;458:53–60. doi: 10.1007/s00424-009-0638-2. [DOI] [PubMed] [Google Scholar]
- 69.Furmanek A, Hofsteenge J. Protein C-mannosylation: facts and questions. Acta Biochim Pol. 2000;47:781–9. [PubMed] [Google Scholar]
- 70.Ihara Y, Manabe S, Kanda M, et al. Increased expression of protein C-mannosylation in the aortic vessels of diabetic Zucker rats. Glycobiology. 2005;15:383–92. doi: 10.1093/glycob/cwi012. [DOI] [PubMed] [Google Scholar]
- 71.Yonemura K, Takahira R, Yonekawa O, Wada N, Hishida A. The diagnostic value of serum concentrations of 2-(alpha-mannopyranosyl)-L-tryptophan for normal renal function. Kidney Int. 2004;65:1395–9. doi: 10.1111/j.1523-1755.2004.00521.x. [DOI] [PubMed] [Google Scholar]
- 72.Reuter SE, Evans AM. Carnitine and acylcarnitines: pharmacokinetic, pharmacological and clinical aspects. Clin Pharmacokinet. 2012;51:553–72. doi: 10.1007/BF03261931. [DOI] [PubMed] [Google Scholar]
- 73.Fouque D, Holt S, Guebre-Egziabher F, et al. Relationship between serum carnitine, acylcarnitines, and renal function in patients with chronic renal disease. J Ren Nutr. 2006;16:125–31. doi: 10.1053/j.jrn.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 74.Sun J, Shannon M, Ando Y, et al. Serum metabolomic profiles from patients with acute kidney injury: a pilot study. J Chromatogr B Analyt Technol Biomed Life Sci. 2012;893-894:107–13. doi: 10.1016/j.jchromb.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.K/DOQI clinical practice guidelines for chronic kidney disease: evaluation, classification, and stratification. Am J Kidney Dis. 2002;39:S1–266. [PubMed] [Google Scholar]
- 76.CDC/National Center for Health Statistics.Data Access - National Death Index. Queried on October 10, 2010 [Google Scholar]
- 77.van den Berg RA, Rubingh CM, Westerhuis JA, van der Werf MJ, Smilde AK. Metabolomics data exploration guided by prior knowledge. Anal Chim Acta. 2009;651:173–81. doi: 10.1016/j.aca.2009.08.029. [DOI] [PubMed] [Google Scholar]
- 78.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.