Abstract
Proteins can be modified on lysines (K) with a single ubiquitin (Ub) or with polymers of Ub (polyUb). These different configurations and their respective topologies are primary factors for determining whether substrates are targeted to the proteasome for degradation or directed to nonproteolytic outcomes. We report here on the intrinsic ubiquitylation properties of UbcM2 (UBE2E3/UbcH9), a conserved Ub-conjugating enzyme linked to cell proliferation, development, and the cellular antioxidant defense system. Using a fully recombinant ubiquitylation assay, we show that UbcM2 is severely limited in its ability to synthesize polyUb chains with wild-type Ub. Restriction to monoubiquitylation is governed by multiple residues on the backside of the enzyme, far removed from its active site, and by lysine 48 of Ub. UbcM2 with mutated backside residues can synthesize K63-linked polyUb chains and to a lesser extent K6- and K48-linked chains. Additionally, we identified a single residue on the backside of the enzyme that promotes monoubiquitylation. Together, these findings reveal that a combination of noncatalytic residues within the Ubc catalytic core domain of UbcM2 as well as a lysine(s) within Ub can relegate a Ub-conjugating enzyme to monoubiquitylate its cognate targets despite having the latent capacity to construct polyUb chains. The two-fold mechanism for restricting activity to monoubiquitylation provides added insurance that UbcM2 will not build polyUb chains on its substrates, even under conditions of high local Ub concentrations.
The Ub system is a highly conserved enzymatic network for post-translationally modifying proteins. This conservation encompasses the functions of the system as well as many of its enzymatic components. The conjugation of Ub to substrates is mediated by a hierarchical enzyme cascade minimally composed of a Ub-activating enzyme (E1), a Ub-conjugating enzyme (E2), and a Ub protein ligase (E3). E1 activates Ub and transfers it to the active site cysteine of E2.1 Ub-charged E2s partner with specific E3s to transfer Ub onto substrates. E3s can be single proteins or multisubunit complexes, and their primary function is to recruit and facilitate the transfer of Ub to substrates. It is estimated that humans have at least two E1s, 38 E2s, and 600–1000 E3s (reviewed in ref (2)).
Target proteins can be modified on one or more lysine residues with either a single Ub (i.e., monoUb) or polymers of Ub (i.e., polyUb). Ub has seven lysines, and each can function as an acceptor during polyUb chain synthesis. As a result, polyUb chains can be homogeneous or heterogeneous. Homogeneous polyUb chains are synthesized utilizing a common acceptor lysine on the proximal Ub of the growing chain. For example, polyUb chains in which the Ub–Ub linkages are exclusively through K48 typically target substrates to the 26S proteasome for degradation.3−5 Alternatively, heterogeneous chains are composed of a composite of Ub–Ub linkages with different lysine residues serving as acceptor sites on the proximal Ub. Goldberg and colleagues have shown that two adjacent lysines on a single Ub can simultaneously serve as acceptors, giving rise to forked polyUb configurations.6,7 Similar findings have been reported for proteins isolated from growing yeast.8
Efforts to define the molecular mechanisms governing polyUb chain synthesis have provided insights into the fundamental relationships by which E2s and E3s cooperate (e.g., refs (6) and (9−17)). Recent insights have principally come from biochemical and structural studies that have elucidated how really interesting new gene (RING)/U-box E3 ligases stimulate Ub-charged E2s. Two primary mechanisms are now apparent. RING/U-box E3s function (1) to bring charged E2s into the proximity of the amino group of substrate lysine residues (e.g., ref (18)) and (2) to allosterically activate charged E2s by promoting catalytically favorable “closed” conformations in which Ub directly contacts helix 2 of the enzyme.19−21 This configuration optimally positions the thioester bond of the E2–Ub conjugate for nucleophilic attack. Additional characterization of E2–E3 component interactions using the ERAD E3 facilitator, Cue1p, and its partner E2, Ubc7p, has revealed that the Ubc7p binding region of Cue1p stimulates charging of the active site of Ubc7p by E1 as well as Ub discharge of the E2.22 The CUE domain of Cue1p further stimulates ERAD substrate turnover by binding to and stabilizing growing, K48-linked, polyUb chains.23
The type of Ub–Ub linkage synthesized during polyUb chain construction is governed both by the specific E2 involved and by whether this E2 partners with a RING/U-box E3 or with a homologous to E6-AP carboxyl-terminal (HECT) E3. When the pairing involves a RING/U-box E3, the E2 primarily dictates the specific Ub–Ub linkage. However, the E3 can impose limits on whether a given substrate is mono- or polyubiquitylated and on which type of Ub–Ub linkage within the given repertoire of an E2 is utilized.10 In contrast, a HECT E3 has an active site cysteine, accepts Ub from the E2 via transthiolation, and then transfers the Ub directly to the substrate.24 Thus, irrespective of the partnering E2, the HECT E3 primarily dictates the Ub–Ub linkage.6 Additional factors can also influence polyUb chain configuration. S5a/Rpn10 blocks the synthesis of forked chains that are produced by UbcH5 in cooperation with particular RING/U-box E3s. These forked chains are resistant to deubiquitylation and degradation by the proteasome, and by blocking their synthesis, S5a/Rpn10 effectively promotes the degradation of particular substrates.7
In this work, we investigated the molecular determinants governing the ubiquitylation capacity of UbcM2, a highly conserved metazoan enzyme of the class III E2s. UbcM2 possesses the conserved catalytic core domain characteristic of E2s (also known as the Ubc domain) as well as a unique 58-amino acid, N-terminal extension.25 Functionally, UbcM2 has been linked to cell cycle progression,26,27 development,28 turnover of damaged and misfolded proteins,25 and regulation of Nrf2, an antioxidant transcription factor,29 yet in vivo substrates remain to be identified; moreover, several in vitro studies have produced varied evidence with regard to whether UbcM2 is restricted to conjugating monoUb onto targets or can efficiently build polyUb chains. For example, mass spectrometry studies that aimed to analyze autoubiquitylation reactions and red fluorescent protein (RFP)–E3 fusion protein ubiquitylation showed that UbcM2 can synthesize polyUb chains composed of K11, K48, and K63 Ub–Ub linkages using wt Ub.10,30 Other studies, however, have shown that UbcM2 primarily attaches monoUb in reactions with E3 partners that function as proxy substrates in vitro (e.g., ref (9)). Consistent with this monoubiquitylating behavior, Day and co-workers recently reported that the N-terminal extensions of UbcM2 and two related enzymes (collectively known as the UBE2E enzymes or class III E2s) function to limit polyUb chain synthesis.31 Notably, all of these studies used different E3 ligase partners as proxy substrates, underscoring the fact that the capacity of UbcM2 to conjugate monoUb or polyUb may in part be dictated by its cognate E3 partner.
Although the Ubc domains of UbcH5 family members (hereafter termed UbcH5) are 66% identical and 78% similar to the Ubc domain of UbcM2, UbcH5 processively constructs polyUb chains (e.g., refs (31) and (32)). This polyUb synthesizing behavior requires Ub-charged UbcH5 to self-associate via a noncovalent interaction between Ub and several residues on the so-called “backside” (i.e., opposite the catalytic pocket) of the enzyme.32 The high level of conservation between the Ubc domains of UbcM2 and UbcH5 but dramatically different Ub conjugating profiles offered the opportunity to identify residues within the Ubc domain that govern the intrinsic Ub transferring properties and linkage preferences of UbcM2. We report here that UbcM2 can synthesize K63-linked, and to a lesser extent K6- and K48-linked, polyUb chains but that this activity is largely suppressed, in the context of wt Ub. The suppression is mediated by multiple Ubc residues, antipodally situated from the active site of UbcM2, and by one or more lysines of Ub. Further, we have identified an additional residue distant from the catalytic pocket that is critical for the monoubiquitylating activity of UbcM2. Our data also reveal that the binding affinity of UbcM2 for its cognate E3 ligase can impact ubiquitylation capacity as tighter binding under particular reaction conditions can promote polyUb chain formation. Together, these findings highlight new mechanistic details by which E2 polyUb chain building activity can be mitigated to accommodate specialized roles in ubiquitylation cascades such as the attachment of a priming monoubiquitylation, which serves to limit “unintended” chain building on their substrates.
Experimental Procedures
Plasmids and Recombinant Protein Expression and Purification
Wild-type (wt) His6-T7-tagged UbcM2 and UbcH5b were cloned into pET28a (Novagen EMD Biosciences) for expression in BL21-star (DE3) Escherichia coli. UbcM2 point mutants were introduced using the QuikChange site-directed mutagenesis kit (Stratagene, Inc.) according to the manufacturer’s instructions. For recombinant expression, cultures were grown in Terrific Broth supplemented with 2% ethanol and 10 μg/mL kanamycin at 37 °C to an OD600 of ∼0.8, induced with 0.4 mM isopropyl β-d-1-thiogalactopyranoside (IPTG), and grown overnight at 22 °C. Bacteria were pelleted and lysed with an Emulsiflex C5 homogenizer (Avestin, Inc.), and lysates were rocked overnight with Ni-NTA His Bind Resin (Novagen EMD Biosciences) to purify the His6-T7-tagged E2s. The recombinant E2s were eluted from the resin with 200 mM imidazole, exchanged into 10 mM NaPO4, 70 mM KCl, and 1 mM MgCl2 (pH 7.2), aliquoted, and snap-frozen in liquid N2.
Ubiquitylation Assays
Fully recombinant ubiquitylation assays contained either AO7T or BD/BC 100/300 (residues 26–126 of BARD1 fused to residues 1–304 of BRCA1) synthesized in a coupled in vitro transcription/translation bacterial lysate system (PURExpress, New England Biolabs, Inc.) with or without 35S-labeled amino acids. These E3s/proxy substrates were then combined with recombinant E1 (62.5 nM), His6-T7-E2 (5 μM), an energy-regenerating system [100 mM Tris-HCl (pH 7.4), 0.4 mM MgATP, 1 mM MgCl2, 0.2 mM DTT, 2 mM phosphocreatine, 0.2% Tween 20, and 0.5 mg of creatine phosphokinase], and the indicated variant of Ub (50 μM). Reaction mixtures were incubated at 37 °C for 90 min (unless otherwise indicated) and reactions terminated by the addition of 1 volume of 2× concentrated Laemmli solubilizing buffer (2XSB). Reaction products were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE), and ubiquitylation of the 35S-labeled E3 ligases was visualized by fluorography. Reaction mixtures containing nonradiolabeled AO7T were analyzed by Western blotting with anti-K48-linked Ab (Cell Signaling Technology, Inc.; diluted 1:1000 in a 5% BSA/TBST mixture), anti-K63-linked Ab (Cell Signaling Technology, Inc.; diluted 1:1000 in a 5% BSA/TBST mixture), anti-Ub Ab (Santa Cruz Biotechnology; diluted 1:200 in a 5% BSA/TBST mixture), or anti-HA Ab (diluted 1:1000 in a 5% milk/TBST mixture). HRP-conjugated goat anti-rabbit secondary antibodies were diluted to a 1:2000 ratio to detect the K48 and K63 linkage-specific Abs, and HRP-conjugated goat anti-mouse secondary antibodies were diluted to a 1:10000 ratio to detect the anti-Ub and anti-HA Abs. In-house enhanced chemiluminescent reagents were used as substrates for the HRP. Blots were exposed to X-OMAT film or to an imager and figures prepared in Photoshop (version 8).
Lysine Reactivity Assays
In step 1 (E2 active site charging), recombinant E1 (200 nM), UbcM2 (2.2 μM), Ub (20 μM), an ATP-regenerating system, and buffer [25 mM NaPO4 and 150 mM NaCl (pH 7.0)] were incubated for 45 min at 37 °C. The charging reaction was terminated by the addition of 20 mM EDTA. In step 2 (lysine reactivity), reaction mixtures were supplemented with 0.5 M lysine, and at the indicated time points, aliquots were removed and solubilized with nonreducing buffer [50 mM Tris-HCl (pH 6.8), 4 M urea, 2% SDS, 10% glycerol, and 0.001% bromophenol blue] in preparation for SDS–PAGE and α-UbcM2 Western blotting to assess the amount of Ub-charged enzyme remaining. A second aliquot from the 15 min time point sample was solubilized with nonreducing buffer supplemented with β-mercaptoethanol and then heated to determine the level of autoubiquitylated enzyme.
GST Pull-Down Assay
35S-labeled AO7T and BD/BC 100/300 were expressed in a coupled in vitro transcription/translation bacterial lysate system and then combined with 10 μM GST-UbcM2 or 20 μM GST and glutathione (GSH)-Sepharose beads. Reaction mixtures were incubated in a 4 °C thermomixer at 900 rpm for 90 min in 1× binding buffer [10 mM HEPES-KOH (pH 7.4), 55 mM potassium acetate, 1 mM magnesium acetate, 0.1 mM EGTA, 0.25% Tween 20, and 150 mM NaCl], after which the bead-associated proteins (bound) were pelleted for 1 min at 13000 rpm. The unbound fraction was collected and solubilized in 2XSB. The bound fraction was washed three times with ice-cold 1× binding buffer and then solubilized in 2XSB; 75% of the bound and 37.5% of the unbound proteins were resolved by SDS–PAGE. GST-bound proteins were visualized by Coomassie Brilliant Blue (CBB) staining and the 35S-labeled proteins by fluorography.
Results
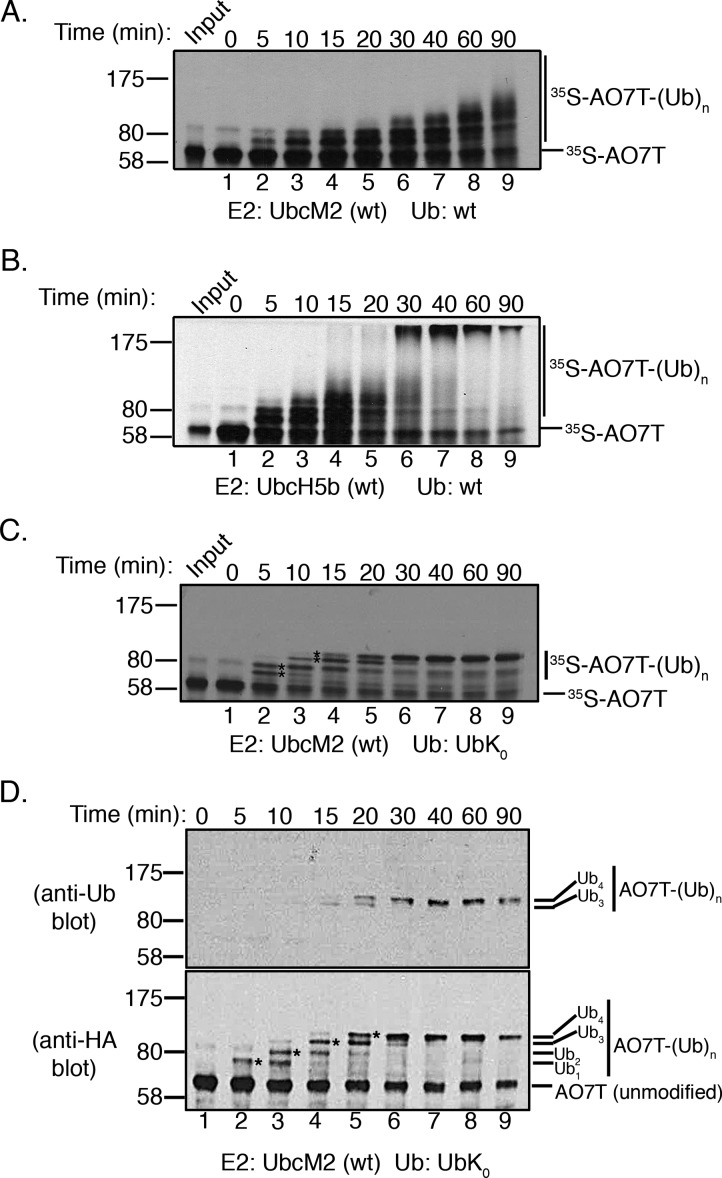
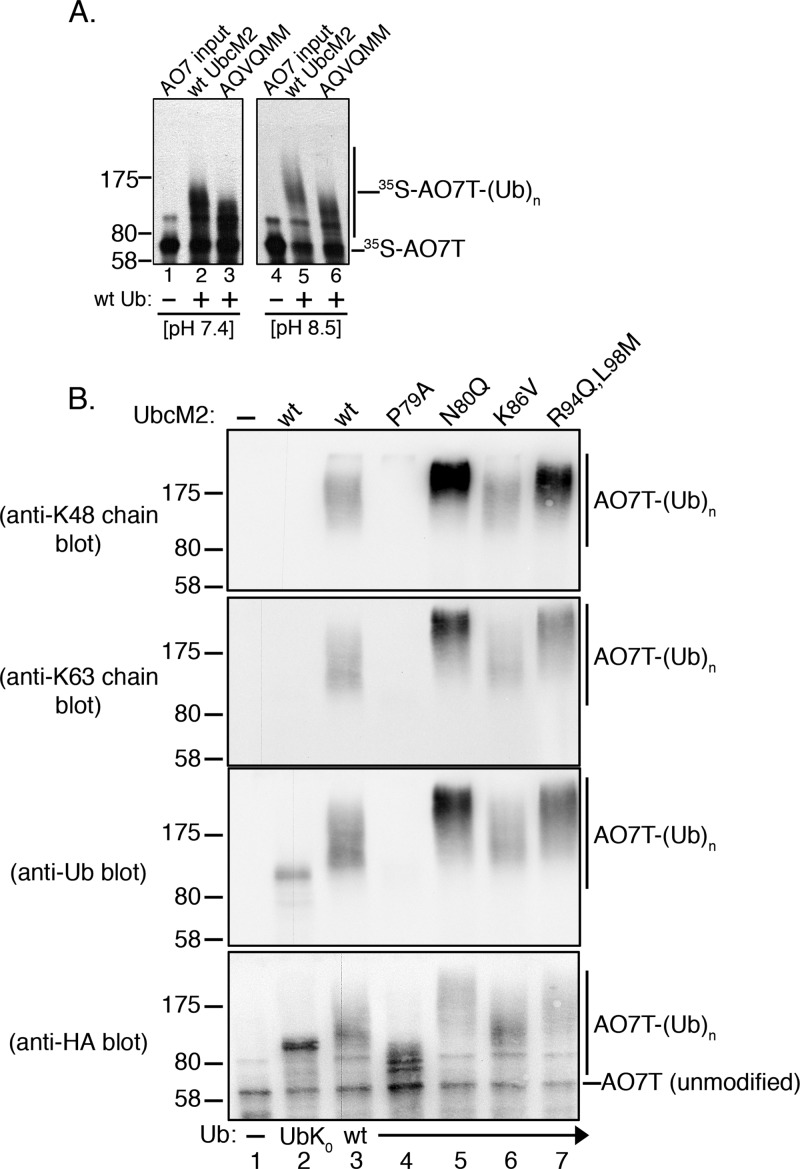
To characterize the Ub conjugating properties of UbcM2, we implemented a fully recombinant assay to compare the E3-dependent ubiquitylation kinetics of UbcM2 and UbcH5b, a UbcH5 family member. In this assay, the E3 ligase serves as a proxy substrate. Because all reaction components are expressed in bacteria or bacterial lysates, changes in migration of the proxy substrate during SDS–PAGE can be attributed specifically to the attachment of Ub, ruling out all other post-translational modifications. The ligase used in these assays is AO7 (RNF25), a RING finger domain-containing E3 that binds both UbcM2 and UbcH5.33 Because of the limited expression and solubility of full length AO7, we used AO7T, a truncation mutant spanning residues 1–360 and containing the RING finger domain.33 Hemaglutinin (HA)-tagged, 35S-labeled AO7T (HA-AO7T) was synthesized in a coupled in vitro transcription/translation bacterial lysate system and combined with recombinant E1, His6-T7-tagged UbcM2 (H6T-UbcM2) or H6T-UbcH5b, an energy-regenerating system, and wt Ub. Reactions were conducted at pH 7.4, and [35S]AO7T ubiquitylation was followed over time by SDS–PAGE and fluorography. UbcM2 conjugated Ub to AO7T such that distinct band shifts were produced, consistent with the attachment of monoUb(s) (Figure 1A, lanes 2–7). At the 60 and 90 min time points, a small amount of smeared density at the high-molecular weight end of the pattern is observed, consistent with polyUb chain synthesis on AO7T (Figure 1A, lanes 8 and 9). This limited polyUb chain synthesis occurred nonprocessively; i.e., reaction products appear in stepwise order and subsequently disappear in conjunction with the appearance of the next slower-migrating band (Figure 1A, lanes 8 and 9). By contrast, UbcH5b constructed polyUb chains processively on AO7T, i.e., following initial priming reactions in which monoUb(s) is added (Figure 1B, lanes 2–4), products with distinct chain lengths are not observed, and only high-molecular weight end products are detected (Figure 1B, lanes 6–9).
Figure 1.
UbcM2 has a severely limited capacity to synthesize polyUb chains on AO7T with wt Ub. (A) Fully recombinant in vitro ubiquitylation assays containing recombinant E1, H6T-UbcM2, wt Ub, energy, and [35S]AO7T (produced in a bacterial TNT expression system) were incubated at 37 °C for the indicated times. Reaction mixtures were solubilized and processed for SDS–PAGE and fluorography. (B) Same as panel A except testing H6T-UbcH5b. For panels A and B, [35S]AO7T input was loaded to the left of lane 1 and the migration of molecular weight markers is denoted on the left. Unmodified [35S]AO7T is marked on the right of each fluorograph with a hashmark and Ub-modified [35S]AO7T [[35S]AO7T-(Ub)n] with a vertical line. (C) Assay similar to that in panel A except wt Ub has been replaced with lysine-less Ub (UbK0). Distinct bands representing the attachment of one, two, three, or four monoUbK0 molecules to AO7T are denoted with asterisks between lanes 2 and 3 and lanes 3 and 4. (D) Assay similar to that in panel C except that HA-AO7T is not radiolabeled with [35S]Met/Cys but rather reaction products are visualized by either anti-Ub (top) or anti-HA Western blotting (bottom). The number of UbK0 molecules conjugated to HA-AO7T is indicated to the right of each blot. The sensitivity of the anti-Ub antibody is such that only HA-AO7T modified with either three or four UbK0 molecules is detected. All experiments were repeated at least three independent times.
To corroborate the monoubiquitylation pattern observed with UbcM2 using wt Ub, we repeated the time course experiments with lysine-less Ub (UbK0), a variant that can be conjugated to a substrate but cannot support polyUb chain extension. The results from these assays define four bands corresponding to four monoubiquitylation events (Figure 1C, asterisks between lanes 2 and 3 and lanes 3 and 4). By the 40 min time point, fully monoubiquitylated AO7T accumulates (Figure 1C, lane 7). To further validate that the slower-migrating AO7T bands represent ubiquitylated forms of the proxy substrate, we repeated the UbK0 time course using nonradiolabeled HA-AO7T and performed Western blot analyses using an anti-Ub antibody and an anti-HA antibody. These data clearly illustrate the four monoubiquitylations (Figure 1D, bottom blot, asterisks between lanes 2 and 3, lanes 3 and 4, lanes 4 and 5, and lanes 5 and 6) and stepwise, nonprocessive product formation. Furthermore, the anti-Ub blot revealed that a minimum of three UbK0 molecules on AO7T was required for detection by the pan Ub antibody (Figure 1D, top blot, lanes 4 and 5).
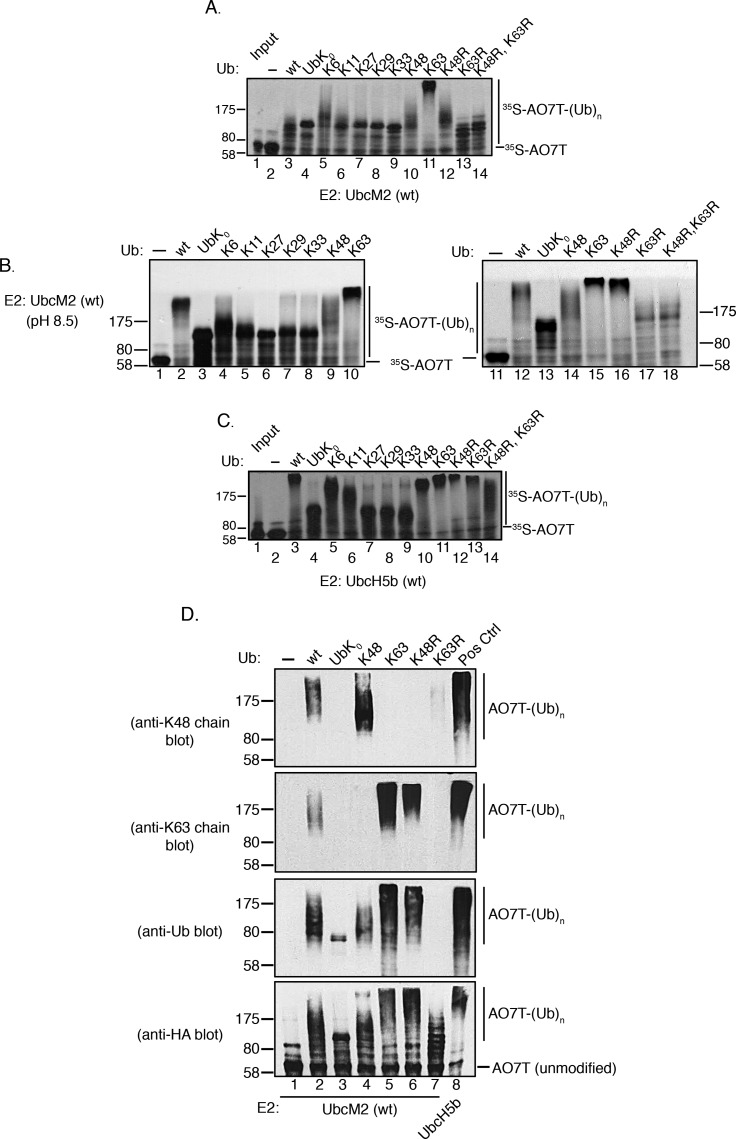
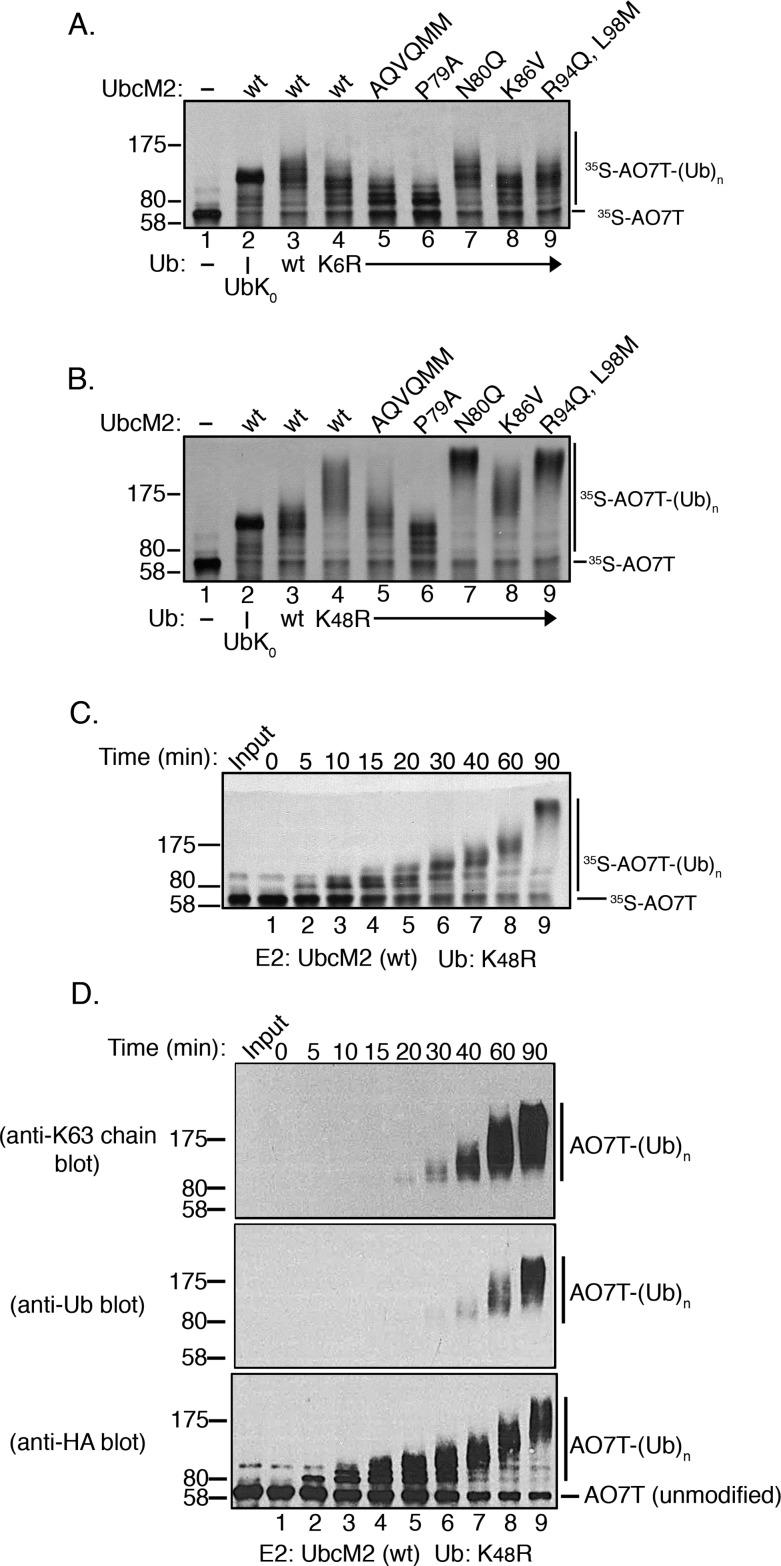
The limited polyUb chain building capacity of UbcM2 prompted us to investigate the Ub–Ub linkage preference of the enzyme. UbcM2 was incubated with either wt Ub, UbK0, or variants having only a single intact lysine, and identical reactions were conducted with UbcH5b for comparison. With regard to the nomenclature used to designate these Ub mutants, variants containing only a single intact lysine with the other six lysines mutated are named to indicate the intact lysine. For example, the mutant in which only K48 is present but the remaining six lysines have been mutated to arginines is termed “K48 Ub” in figures, whereas the mutant in which only K6 is present but the other six lysines have been mutated to arginines is termed “K6 Ub”. Likewise, Ub mutants in which a specific lysine(s) has been replaced with an arginine are given the name and number of the substituted lysine. For example, “K48R Ub” has a single substitution of K48 with arginine, but the six remaining lysines (K6, K11, K27, K29, K33, and K63) are present. Interestingly, these experiments showed that the polyUb chain building capacity of UbcM2 was enhanced by particular Ub variants. Specifically, polyUb chain synthesis was markedly augmented in reaction mixtures exclusively containing K63 Ub (Figure 2A, lane 11) and to a lesser extent in reaction mixtures using K6 Ub and K48 Ub (Figure 2A, lanes 5 and 10, respectively). In contrast, the products formed with the Ub variants containing only a single Lys residue (K11, K27, K29, or K33) appeared to be identical to the products formed with UbK0, confirming these as monoubiquitylated species. The preference for synthesizing K63-linked chains was corroborated by a failure of the enzyme to construct polyUb chains with K63R Ub (Figure 2A, lane 13). Notably, UbcM2 displayed modest chain building capacity with K48R Ub (Figure 2A, lane 12), implying that, in the context of wt Ub, K48 suppresses chain synthesis by the enzyme. Chain extension was more readily visualized at pH 8.5, especially with wt Ub, presumably because of the increased nucleophilicity of the ε-amino group on the attacking lysine of Ub (Figure 2B, lane 2). By comparison, UbcH5b extensively polyubiquitylated AO7T with wt Ub, K6 Ub, K11 Ub, K48 Ub, and K63 Ub at pH 7.4 (Figure 2C, lanes 3, 5, 6, 10, and 11, respectively) but did not synthesize polyUb chains with K27 Ub, K29 Ub, and K33 Ub (Figure 2C, lanes 7–9, respectively).
Figure 2.
UbcM2 can synthesize K63-linked polyUb chains. (A) H6T-UbcM2 in vitro ubiquitylation assays conducted at pH 7.4 with either no Ub (lane 2), wt Ub (lane 3), lysine-less Ub (UbK0, lane 4), Ub variants in which only the indicated lysine is intact and the six other lysines have been mutated to arginine (lanes 5–11), or variants in which only the indicated lysine has been mutated to arginine (lanes 12–14). Unmodified [35S]AO7T is marked on the right with a hashmark, and Ub-modified [35S]AO7 [[35S]AO7T-(Ub)n] is marked with a vertical line. The migration of molecular weight markers is denoted at the left. (B) Same as panel A except reactions were conducted at pH 8.5 and samples were run in parallel on two gels. (C) Same as panel A except testing H6T-UbcH5b. (D) Recombinant ubiquitylation assays using nonradiolabeled HA-AO7T were incubated for 90 min with the indicated variants of Ub. Reaction products were analyzed by Western blotting with the antibody listed to the left of each blot. As a positive control for the synthesis of K48- and K63-linked polyUb chains, UbcM2 was replaced with UbcH5b (lane 8, Pos Ctrl). All experiments were repeated a minimum of three independent times.
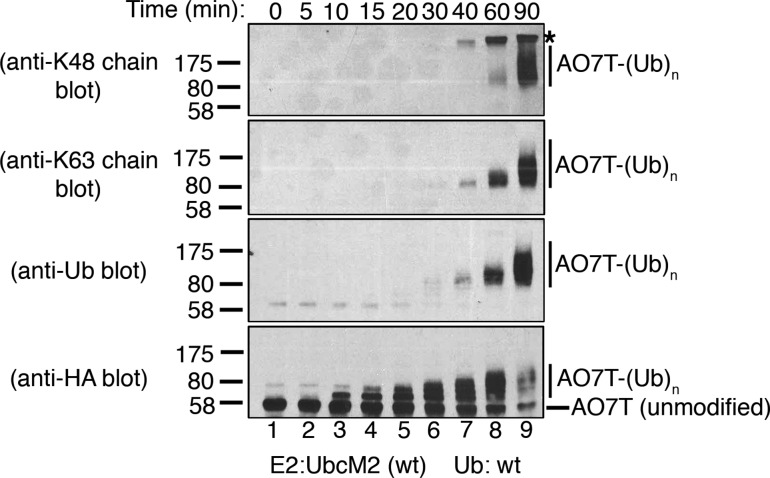
To firmly establish that the UbcM2-mediated changes in AO7T migration correspond to the attachment of polyUb chains, we conducted recombinant assays with nonradiolabeled AO7T. Reaction products were resolved by SDS–PAGE and subjected to Western blotting with antibodies specific for (1) K48-linked polyUb chains, (2) K63-linked polyUb chains, (3) pan Ub, and (4) the HA tag appended to the amino terminus of AO7T. These experiments validated that UbcM2 can modestly synthesize K48-linked chains using K48 Ub and robustly synthesize K63-linked chains using K63 Ub (Figure 2D, lane 4, anti-K48 chain blot, and lane 5, anti-K63 chain blot, respectively). The generation of K63-linked chains in reaction mixtures containing K48R Ub was also confirmed (Figure 2D, lane 6, anti-K63 chain blot), consistent with the preference of UbcM2 for synthesizing K63 linkages and for doing so with an increased capacity in the absence of Ub Lys48. These data also highlight the preference of UbcM2 for synthesizing K63-linked polyUb chains by showing that Ub immunoreactivity was not detected in reaction mixtures containing K63R Ub despite the accumulation of monoubiquitylated forms of AO7T (Figure 2D, lane 7, anti-Ub and anti-HA blots, respectively). Notably, fully monoubiquitylated AO7T from the UbK0 reaction could be detected with the Ub antibody but, as expected, not the linkage-specific antibodies (Figure 2D, lane 3, anti-Ub blot). Similarly, in reaction mixtures containing wt Ub, the Ub antibody detected a signal at and above the migration of fully monoubiquitylated AO7T (Figure 2D, lane 2, anti-Ub blot) but did not detect a signal corresponding to the partially monoubiquitylated species that migrated below the 80 kDa marker. In complementary time course experiments with wt Ub, we first detected polyUb chain synthesis at 40 min and the K48- and K63-linked chains were synthesized more robustly by the 60 and 90 min time points (Figure 3, lanes 7–9, anti-K48 and anti-K63 chain blots).
Figure 3.
UbcM2 can synthesize K48- and K63-linked polyUb chains on AO7T. Recombinant ubiquitylation assays using nonradiolabeled HA-AO7T and wt Ub were incubated for the indicated times and solubilized, and the reaction products were analyzed by Western blotting. Unmodified AO7T is marked to the right of the anti-HA blot with a hashmark, and Ub-modified AO7T [AO7T-(Ub)n] is marked with a vertical line to the right of each blot. The migration of molecular weight markers and the antibodies used are denoted at the left. The asterisk denotes a nonspecific band detected by the anti-K48 linkage antibody. This band is derived from either a protein present in the energy-regenerating system or the PUREXPRESS bacterial lysate used to synthesize HA-AO7T. This band is detected at the 40, 60, and 90 min time points when AO7T is absent (i.e., unprogrammed PUREXPRESS bacterial lysate is used in the ubiquitylation reaction) (data not shown). Experiments were repeated a minimum of three independent times.
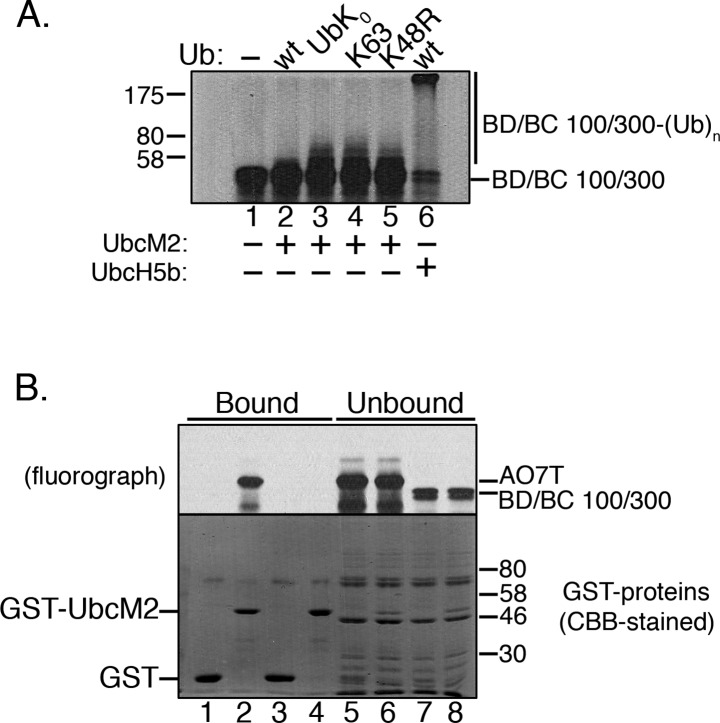
We next tested the capacity of UbcM2 to polyubiquitylate BD/BC 100/300, an established proxy substrate32 consisting of residues 26–126 of BARD1 and residues 1–304 of BRCA1, the heterodimeric E3 ligase (Figure 4A, lanes 2 and 3). UbcM2 only weakly monoubiquitylated BD/BC 100/300 and, furthermore, failed to construct K63-linked polyUb chains or to display enhanced ubiquitylating activity with K48R Ub (Figure 4A, lanes 4 and 5, respectively). As previously reported,32 UbcH5b synthesized polyUb chains on BD/BC 100/300 with wt Ub (Figure 4A, lane 6). The failure of UbcM2 to synthesize polyUb chains on BD/BC 100/300 correlated with a failure of the E2 to coprecipitate the proxy substrate. GST-UbcM2 did not precipitate 35S-labeled BD/BC 100/300, whereas GST-UbcM2 stably associated with 35S-labeled AO7T (Figure 4B, lane 4 vs lane 2).
Figure 4.
Affinity of UbcM2 for a proxy substrate that correlates with the capacity to attach polyUb chains. (A) In vitro ubiquitylation assay with H6T-UbcM2 and a 35S-labeled fusion of BARD1/BRCA1 100/300 (BD/BC 100/300) consisting of residues 26–126 of BARD1 fused to residues 1–304 of BRCA1. The Ub variant used in each reaction is indicated at the top of the fluorograph. In lane 6, H6T-UbcH5b was used in place of H6T-UbcM2. The migrations of unmodified BD/BC 100/300 and the Ub-modified fusion protein [BD/BC 100/300-(Ub)n] are indicated at the right, and the migration of molecular weight markers is denoted at the left. (B) Parallel GST fusion protein pull downs of 35S-labeled AO7T or 35S-labeled BD/BC 100/300. Fractions of bead-bound (75% of total) and unbound (37.5% of total) proteins were resolved by SDS–PAGE. 35S-labeled proteins were visualized by fluorography (top) and GST proteins by CBB staining (bottom). Assays were repeated a minimum of three independent times.
Together, the results shown in Figures 1–4 provide five important insights into UbcM2 function. First, UbcM2 has a restricted ability to synthesize polyUb chains and is largely limited to monoubiquitylating AO7T. Second, UbcM2 has a latent capacity to generate polyUb chains, with a preference for K63-linked chains. Third, the polyubiquitylating activity requires extended reaction times, even at 37 °C, and is therefore unlikely to be physiologically relevant under most conditions. Fourth, the (latent) polyubiquitylating activity is partially suppressed by Ub residue Lys48, as chain building activity is partially recovered in reactions using K48R Ub. Fifth, tight binding to its cognate E3 can facilitate polyUb chain synthesis by UbcM2.
To determine if UbcM2 preferentially synthesizes polyUb chains with K63 Ub and K48R Ub because of an enhanced capacity to be loaded with or to discharge these Ub variants, we performed lysine reactivity assays. The premise of this assay is that some E2s, such as UbcM2, can transfer Ub from their active sites to free lysine in the absence of an E3 ligase.34,35 In step 1 of the assay, the E2 is charged with Ub. The charging reaction is then terminated by adding EDTA. For step 2, reaction mixtures are supplemented with free lysine, and at different time points, aliquots are removed, solubilized, and analyzed by nonreducing SDS–PAGE and anti-UbcM2 Western blotting. Nonreducing SDS–PAGE allows tracking of the UbcM2–Ub species as a function of time exposed to free lysine. These experiments showed that UbcM2 is comparably charged with each Ub variant (Figure 1A,B of the Supporting Information, lanes 2 and 5) and that the extent and kinetics of Ub discharge by free lysine are comparable among wt Ub, K63 Ub, and K48R Ub (Figure 1A,B of the Supporting Information). To see if the observed chain building activity with K63 Ub or K48R Ub is due to the Ub variants being better acceptor Ubs, we also monitored the discharge of UbK0 from the active site of UbcM2 in the presence of a 5-fold molar excess of either Ub variant. No differences were detected (data not shown). These findings indicate that the differences in polyUb chain synthesis observed with the variant forms of Ub are not due to differences in the intrinsic aminolysis activity of the UbcM2–Ub conjugates or to differences in the properties of the donor Ubs. Consistent with this conclusion, nuclear magnetic resonance (NMR) experiments demonstrated that neither the Ubc domain nor full length UbcM2 displayed preferential binding affinity for K63 Ub (M. Cook, E. Duncan, and R. Klevit, unpublished results).
The disparity in polyUb capacity between UbcM2 and UbcH5b suggests that residues differing between the two enzymes govern the Ub conjugating behavior of UbcM2. We initially tested whether the unique 58-residue N-terminal extension of UbcM2 plays a role, as reported for reactions of UbcM2 with several RING E3s (e.g., CIAP2, Mdm2, and CARP2).31 A His6-T7-tagged mutant of UbcM2 lacking the N-terminal extension (i.e., ΔN-UbcM2) was analyzed side by side with the wt enzyme. In all cases, the ubiquitylating activity (mono and poly) and linkage preference of ΔN-UbcM2 were indistinguishable from those of the wt enzyme, indicating that the N-terminal extension neither represses UbcM2 polyUb chain synthesis nor dictates the types of chains built when UbcM2 partners with AO7T (Figure 5).
Figure 5.
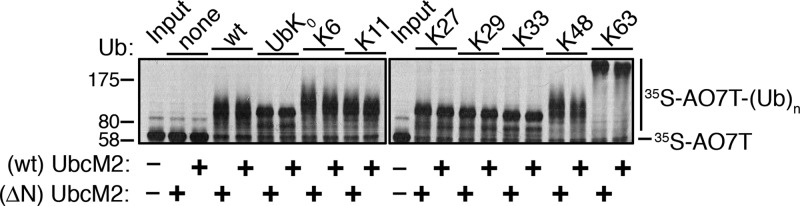
Unique amino-terminal extension of UbcM2 does not dictate the in vitro Ub conjugating behavior of the enzyme when it is partnered with AO7T. In vitro ubiquitylation assays comparing the polyUb chain building capacity of wt and ΔN-UbcM2, a mutant lacking the N-terminal 58-amino acid extension. Ub variants included in each 90 min reaction are indicated above the panels. The migrations of unmodified ([35S]AO7T) and ubiquitylated AO7T [[35S]AO7T-(Ub)n] are marked at the right and molecular weight markers at the left. The assay was repeated a minimum of three independent times.
We next analyzed the contribution of a patch of residues on the backside of UbcM2 (i.e., antipodal from the active site) where the corresponding residues of UbcH5b form a noncovalent interaction with Ub. This backside binding to Ub is essential for UbcH5 family members to processively synthesize polyUb chains.32 Only six of the 12 residues comprising this patch are conserved in UbcM2. We simultaneously mutated the six nonconserved residues to the corresponding UbcH5b residues. Using the residue numbering for UbcM2, the substitutions were P79A, N80Q, K86V, R94Q, L98M, and T207M, and we refer to this mutant as “AQVQMM”. We predicted that this mutant might mimic UbcH5 with respect to processive chain building. Instead, AQVQMM was modestly impaired in its ability to ubiquitylate [35S]AO7T (Figure 6A, lane 2 vs lane 3), and this difference was exacerbated at pH 8.5 because of the increased polyUb synthesizing capacity of the wt enzyme at the higher pH (Figure 6A, lane 5 vs lane 6). To further study these backside mutations, a panel of single and double mutations were analyzed by Western blotting with the linkage-specific and pan Ub antibodies. This analysis revealed that several backside residues differentially affect UbcM2 ubiquitylation capacity. Notably, the P79A mutation reduced the efficiency of AO7T monoubiquitylation by UbcM2 (Figure 6B, anti-Ub and anti-HA blots, lane 4 vs the wt enzyme in lane 3). This decrease in activity was not due to a deficiency in being charged with Ub by E1 or to discharge capacity, as loading of the mutant enzyme and transfer of Ub to an acceptor lysine occurred at levels comparable to those seen for wt UbcM2 (Figure 1C of the Supporting Information). Surprisingly, P79A UbcM2 largely retained its capacity to polyubiquitylate AO7T using K63 Ub (Figure 2A of the Supporting Information), indicating that the mutant does not exhibit gross conformational defects or a reduced capacity to engage AO7T. The recovery of polyubiquitylating activity in reaction mixtures containing K63 Ub indicates that one or more lysines of Ub (other than K63) negatively impacts the P79A substitution and that mutation of this lysine(s) to an arginine relieves the inhibitory effect. K48 of Ub does not appear to be the primary lysine exerting an inhibitory effect on P79A as reactions using K48R Ub showed only a slight increase in activity (Figure 2B of the Supporting Information).
Figure 6.
Select noncatalytic backside residues influence the capacity of UbcM2 to synthesize polyUb chains. (A) In vitro ubiquitylation assays conducted at pH 7.4 (left) or pH 8.5 (right) to compare the patterns of [35S]AO7T ubiquitylation between wt UbcM2 and AQVQMM, a six-residue substitution mutant of UbcM2. Reaction mixtures contained either no Ub (lanes 1 and 4) or wt Ub (lanes 2, 3, 5, and 6). (B) Recombinant ubiquitylation assays using nonradiolabeled HA-AO7T were incubated for 90 min and contained either no Ub (lane 1), UbK0 (lane 2), or wt Ub (lanes 3–7), and the indicated forms of UbcM2 (listed above blots). Reaction products were analyzed by Western blotting with the antibody listed at the left of each blot. The migrations of unmodified AO7T and ubiquitylated AO7T [AO7T-(Ub)n] are shown to the right of the blots. The migration of molecular weight markers is indicated to the left of each blot. All assays were repeated a minimum of three independent times.
Remarkably, when the residue adjacent to P79 was mutated from an asparagine to a glutamine (N80Q), the chain building capacity of UbcM2 was enhanced compared to that of the wt enzyme (Figure 6B, lane 5 vs lane 3 of the anti-Ub blot). Similarly, the R94Q/L98M double mutant enhanced UbcM2 ubiquitylating capacity (Figure 6B, lane 7 vs lane 3 of the anti-Ub blot), whereas the K86V mutant had no obvious effect (Figure 6B, lane 6 vs lane 3). N80Q UbcM2 and, to a lesser extent, the R94Q/L98M mutant displayed increased K48- and K63-linked polyUb chain building capacity compared to that of the wt enzyme (Figure 6B, lanes 5 and 7 vs lane 3 of K48 and K63 chain blots). In contrast, P79A UbcM2 showed a much reduced capacity to ubiquitylate AO7T, and the modified AO7T was below the detection level of the pan Ub antibody (Figure 6B, lane 4, anti-Ub blot), whereas fully UbK0-monoubiquitylated AO7T was readily detectable (Figure 6B, lane 2, anti-Ub blot). It should be appreciated that while the signal intensity from each linkage-specific antibody can be compared among samples on a given blot, signal intensities cannot be compared between blots probed with the different antibodies as each antibody has a distinct affinity for its cognate antigen. Thus, a more intense signal on the K48 linkage blot for N80Q UbcM2 as compared to the K63 linkage blot does not indicate that this mutant is better at building K48-linked chains than K63-linked chains. Together, the results demonstrate that residues on the backside of UbcM2 (i.e., antipodal from the catalytic pocket) have opposing effects on the ubiquitylating capacity of the enzyme and that the negative effect of the P79A mutation is dominant over the enhancing effects of the N80Q, R94Q, and L98M substitutions in the context of the AQVQMM mutant. Importantly, these findings also show that the suppressive effects of Ub lysines on UbcM2 activity are at least partially mediated by three backside residues (N80, R94, and L98) as mutation of these residues relieves this inhibition and enhances the ubiquitylating capacity of the enzyme.
Structural studies with UbcH5 have demonstrated that processive polyUb chain construction is promoted by the noncovalent interaction of Ub with the backside of the E2. Relevant to our discussion here, two Ub lysine side chains (K6 and K48) contact four backside residues of UbcH5 (P17, A19, V26, and Q34; corresponding to UbcM2 residues P77, P79, K76, and R84, respectively).32 We therefore tested how these two Ub lysines impact UbcM2 activity. Wild-type UbcM2 and all of the backside mutants displayed a modest reduction in ubiquitylating activity using K6R Ub (Figure 7A, lanes 4–9). Conversely, wt UbcM2 and all of the mutants except P79A UbcM2 showed increased polyUb chain building activity with K48R Ub (Figure 7B, compare lanes 4, 5, and 7–9 to lane 3). Time course analysis of K48R Ub reactions revealed that although UbcM2 chain synthesizing activity is enhanced by mutation of K48 (Figure 7C), the pattern of chain growth is more consistent with a distributive mechanism in which chain lengths grow gradually over time, rather than the processive pattern observed for UbcH5 family members (e.g., Figure 1B). Western blot analyses of AO7T ubiquitylation by UbcM2 using K48R Ub reinforced this conclusion (Figure 7D, anti-HA blot). The increased UbcM2 activity with K48R Ub underscores the notion that in the context of wt Ub, K48 suppresses the polyubiquitylating activity of the enzyme. Importantly, the differences in polyUb chain synthesis observed with K6R Ub and K48R Ub are not a consequence of differences in UbcM2 loading with the Ub variants or in the intrinsic aminolysis activity of the UbcM2–Ub conjugates (Figure 1B,D of the Supporting Information). Thus, unlike UbcH5 family members, which interact with K6 and K48 of Ub to processively construct polyUb chains, UbcM2 activity is slightly enhanced by K6 but inhibited by K48.
Figure 7.
K6 and K48 of Ub differentially influence the ubiquitylating activity of UbcM2. (A and B) The wild type or the indicated substitution mutants of UbcM2 were incubated with K6R Ub (A) or K48R Ub (B) for 90 min at pH 7.4 (lanes 4–9). Control reaction mixtures contained either no Ub (lane 1), UbK0 (lane 2), or wt Ub (lane 3) for comparison. Ubiquitylation of [35S]AO7T was analyzed by SDS–PAGE and fluorography. (C) Kinetic assay analyzing the ubiquitylation activity of wt UbcM2 using K48R Ub at pH 7.4. Samples were incubated at 37 °C for the indicated times, solubilized, and processed for SDS–PAGE and fluorography. For panels A–C, the migrations of unmodified ([35S]AO7T) and ubiquitylated AO7T [[35S]AO7T-(Ub)n] are marked on the right and molecular weight markers on the left. (D) Similar to panel C using nonradiolabeled HA-AO7T and analyzing reaction products by Western blotting with the indicated antibodies.
Discussion
The conjugation of Ub to a target protein is accomplished by an enzyme cascade minimally consisting of an E1, an E2, and an E3. Extensive biochemical and biophysical experimentation has elaborated the general mechanism by which E1, E2, and E3 cooperate to ubiquitylate substrates (reviewed in refs (36) and (37)), yet what appears to be a relatively straightforward enzymatic cascade is in fact replete with nuance and complexities. Arguably, the greatest of these complexities is based on the recognition that substrates can be modified with Ub in a multitude of ways. These include single and multiple sites of monoUb, single and multiple sites of polyUb, and putative combinations thereof. Recent efforts to understand this panoply of Ub decorations have revealed distinct yet interconnected aspects of substrate ubiquitylation. One theme is that individual E2s can govern the topology of polyUb chains conjugated to substrates. The underlying premise is that each E2 has an intrinsic preference(s) for synthesizing a specific Ub–Ub linkage(s) and this preference is conferred by particular residues within the enzyme, often with a contribution from residues of Ub16 and/or the partner E3.10,38 A second theme is that a subset of E2s have been consigned to conjugate monoUb(s) to substrates and other E2s may then be recruited to extend polyUb chains from these primed sites. While recent studies have defined some of the unique mechanisms governing the polyubiquitylating properties of particular E2s (e.g., refs (22) and (32)), understanding how other E2s are restricted to attaching monoUb is limited.31,39,40
Our biochemical characterization of UbcM2, a highly conserved E2 associated with numerous cellular processes, reveals that although UbcM2 is largely restricted to modifying a proxy substrate with monoUb, it possesses an intrinsic capacity to construct polyUb chains. In particular, UbcM2 can synthesize K63-linked and, to a lesser extent, K6- and K48-linked chains. This latent capacity was uncovered in reaction mixtures incubated for extended times and was further highlighted when single-lysine Ub variants were used in place of wt Ub (Figure 2). Notably, these data indicate that one or more Ub lysines actively suppress the polyUb capacity of UbcM2. Mutation of the suppressive lysine(s) relieves the suppression, and polyUb chains are synthesized, albeit nonprocessively. Lys48 of Ub is the major suppressive lysine as UbcM2 shows enhanced polyUb synthesizing activity in reaction mixtures containing K48R Ub (e.g., Figure 7B–D). In contrast, reaction mixtures containing K6R Ub showed a mild suppression of UbcM2 activity (Figure 7A), indicating that K6 of Ub promotes ubiquitylation by the enzyme.
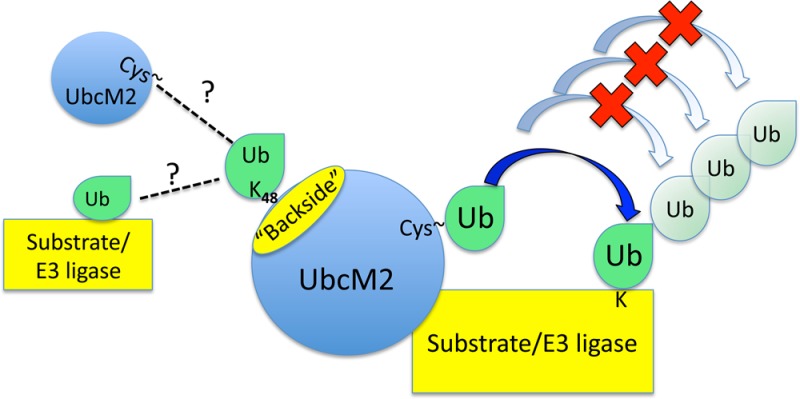
Our findings highlight key distinctions between UbcM2 and the closely related UbcH5 family of E2s. Although the Ubc domain of UbcM2 is 66% identical and 78% similar to the UbcH5 Ubc domain, UbcM2 is a monoubiquitylating enzyme, and under conditions that permit polyUb construction, chain formation occurs in a stepwise fashion indicative of a distributive mechanism (e.g., Figure 7C). In contrast, UbcH5 family members processively synthesize polyUb chains once the priming mono-Ub transfers have occurred (e.g., Figure 1B). This processivity requires a patch of backside residues that cooperate to bind Ub and promote multimerization of the Ub-charged enzyme.32 Interestingly, the counterparts of several of these residues in UbcM2 differentially impact activity (Figure 6B). In UbcM2, P79 promotes activity whereas N80, R94, and L98 restrict it. The underlying mechanism for these opposing effects is not clear, although one possibility is that the substitution mutants increase the affinity of a noncovalent interaction between UbcM2 and Ub. NMR experiments show that wt UbcM2 displays very weak backside binding to wt Ub (R. Klevit, unpublished data), and via the incorporation of substitutions that make UbcM2 more “UbcH5-like”, the backside binding of Ub to UbcM2 becomes stronger, albeit very modestly. However, this increase does not translate into an increase in UbcM2 activity in our assays. In fact, when all six substitutions are incorporated into a single molecule, as in the AQVQMM mutant of UbcM2, the ubiquitylating capacity of UbcM2 is suppressed and this suppression is specifically attributable to the P79A substitution (Figure 6). These data imply that depending on the particular E2, noncovalent backside binding to Ub can stimulate polyUb chain synthesis, as in the case of UbcH5, or relegate an enzyme to monoubiquitylation, as in the case of UbcM2.
We favor the idea that strengthened backside binding of Ub negatively impacts UbcM2 activity by enhancing the suppressive effect of Ub Lys48 and possibly other Ub lysines. Support for this comes from the observation that the activities of AQVQMM and the individual UbcM2 mutants were increased in assays containing K48R Ub (Figure 7B). Although the enhancement for P79A UbcM2 was rather modest (Figure 7B, lane 6), it was confirmed in time course assays (Figure 2B of the Supporting Information). Lys48 likely does not act alone; an additional lysine(s) of Ub contributes to the suppression of UbcM2 because both the AQVQMM mutant and P79A UbcM2 exhibited robust polyUb chain building in reaction mixtures containing K63 Ub (Figure 2A of the Supporting Information), a variant in which all lysines of Ub except K63 have been mutated to arginine.
An additional feature revealed by these studies is that the ability of UbcM2 to synthesize polyUb chains appears to correlate with how tightly the enzyme binds to the E3/substrate/proxy substrate. For example, UbcM2 readily builds K63-linked polyUb chains on AO7T using K63 Ub but still only monoubiquitylates BRCA1-BARD1 with this Ub variant (Figure 4A). Accordingly, GST-UbcM2 coprecipitates AO7T but not BRCA1-BARD1 (Figure 4B). One explanation for these findings is that the tighter binding of AO7T to UbcM2 better promotes the “closed” conformation of the UbcM2–Ub conjugate and thereby enhances the reactivity of the thioester bond for nucleophilic attack by Ub lysines. This in turn promotes polyUb chain synthesis. In contrast, the closed conformation is not promoted by relatively weak binding E3 ligases such as BRCA1-BARD1, and as a consequence, UbcM2 transfers only monoUb. A second but not mutually exclusive possibility is that tight binding of AO7T to UbcM2 may limit the inhibitory Ub interaction that restricts UbcM2 activity whereas weak BRCA1-BARD1 binding is more permissive for Ub Lys48 inhibition. A third possibility is that relatively weak binding of UbcM2 to particular E3 ligases corresponds to a short residency time of docking of the E2–Ub conjugate to the E3 and that this is conducive to a single Ub transfer followed by E3 dissociation. Statistically, an unmodified E3 is more likely to bind to the reloaded E2–Ub conjugate, again producing a mono-Ub product prior to dissociation. In pairings in which UbcM2 binds more tightly to its partner E3, even weak self-association of the UbcM2–Ubs conjugate with itself would increase the local concentration of the charged enzyme, perhaps allowing for chain formation.
An additional finding from this work is that the N-terminal extension of UbcM2 is not responsible for the inhibition of polyUb chain extension in reactions with AO7T (Figure 5), in contrast to results for several other RING E3 ligases, including CIAP2, Mdm2, and CARD2.31 As our reaction conditions differed from those of the published study, we repeated the experiments with CIAP2 and confirmed that the N-terminal extension of UbcM2 inhibits polyubiquitylation of the CIAP2 RING under both sets of reaction conditions (data not shown). We also compared the binding of UbcM2 to AO7 versus CIAP2 and found that the enzyme forms a stable complex with AO7 but not with CIAP2 (Figure 3 of the Supporting Information). These observations support a model in which the N-terminal extension represses the polyubiquitylating capacity of UbcM2 when the enzyme partners with E3 ligases to which it binds relatively weakly. In contrast, when UbcM2 engages an E3 ligase in a more stable complex, the effect of the N-terminal extension is neutralized. Tight binding may restrict access or flexibility of the N-terminal extension such that it cannot exert its inhibitory effect on catalysis. These conclusions further support the emerging model in which the ubiquitylating capacity of E2s is dictated both by specific residues within the enzyme and by interactions with specific E3 ligase partners (e.g., refs (10) and (38)).
In summary, this work advances our understanding of the highly conserved E2, UbcM2, by identifying multiple noncatalytic residues of the Ubc domain, antipodally situated from the active site, that impact the ubiquitylating activity of the enzyme. We have also uncovered a novel role for Lys48 of Ub as a modulator of E2 activity. Collectively, these findings provide additional mechanisms by which an E2 can be primarily relegated to monoubiquitylating its cognate targets despite possessing the inherent capacity to synthesize polyUb chains. Such a limitation provides a guarantee that the monoubiquitylating E2 is kept from generating even small amounts of the polyUb chain on its AO7-targeted substrates.
Acknowledgments
We thank Emily Duncan for preparation of proteins for NMR binding experiments and members of the Plafker laboratory for helpful discussions and encouragement.
Glossary
Abbreviations
- K
lysine
- Ub
ubiquitin
- E1
ubiquitin-activating enzyme
- E2
ubiquitin-conjugating enzyme
- E3
ubiquitin protein ligase
- polyUb
polymers of ubiquitin
- RING
really interesting new gene
- HECT
homologous to E6-AP carboxyl-terminal domain
- Ubc
catalytic core domain
- RFP
red fluorescent protein
- wt
wild type
- IPTG
isopropyl β-d-1-thiogalactopyranoside
- BD/BC
BARD/BRCA1
- SDS–PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- GST
glutathione S-transferase
- GSH
glutathione
- CBB
Coomassie Brilliant Blue
- HA
hemaglutinin
- H6T-UbcM2
His6-T7-tagged UbcM2
- UbK0
lysine-less ubiquitin.
Supporting Information Available
Lysine reactivity assays demonstrating that wt UbcM2 is similarly charged with the indicated variants of Ub and that each variant is comparably discharged from the E2 (Figure 1A,B,D), evidence that wt and P79A UbcM2 behave comparably with respect to Ub loading and discharge (Figure 1C), evidence that the two backside mutants, AQVQMM and P79A UbcM2, both retain the capacity to synthesize K63-linked chains using K63 Ub despite having reduced ubiquitylating activity in reaction mixtures containing wt Ub (Figure 2A), evidence that P79A UbcM2 has a modest increase in ubiquitylating activity when using K48R Ub as compared to wt Ub (Figure 2B), and evidence that UbcM2 can form a stable complex with AO7 but not with the RING finger-containing protein CIAP2 (Figure 3). This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
This work was supported by National Institutes of Health Grant R01 GM092900 (to S.M.P.) and Grant HR13-182 from The Oklahoma Center for the Advancement of Science and Technology (to S.M.P.).
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Olsen S. K.; Lima C. D. (2013) Structure of a ubiquitin E1-E2 complex: Insights to E1-E2 thioester transfer. Mol. Cell 49, 884–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y.; Rape M. (2009) Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 10, 755–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau V.; Tobias J. W.; Bachmair A.; Marriott D.; Ecker D. J.; Gonda D. K.; Varshavsky A. (1989) A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243, 1576–1583. [DOI] [PubMed] [Google Scholar]
- Finley D.; Sadis S.; Monia B. P.; Boucher P.; Ecker D. J.; Crooke S. T.; Chau V. (1994) Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Mol. Cell. Biol. 14, 5501–5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrower J. S.; Hoffman L.; Rechsteiner M.; Pickart C. M. (2000) Recognition of the polyubiquitin proteolytic signal. EMBO J. 19, 94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. T.; Kim K. P.; Lledias F.; Kisselev A. F.; Scaglione K. M.; Skowyra D.; Gygi S. P.; Goldberg A. L. (2007) Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J. Biol. Chem. 282, 17375–17386. [DOI] [PubMed] [Google Scholar]
- Kim H. T.; Kim K. P.; Uchiki T.; Gygi S. P.; Goldberg A. L. (2009) S5a promotes protein degradation by blocking synthesis of nondegradable forked ubiquitin chains. EMBO J. 28, 1867–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J.; Schwartz D.; Elias J. E.; Thoreen C. C.; Cheng D.; Marsischky G.; Roelofs J.; Finley D.; Gygi S. P. (2003) A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21, 921–926. [DOI] [PubMed] [Google Scholar]
- Christensen D. E.; Brzovic P. S.; Klevit R. E. (2007) E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat. Struct. Mol. Biol. 14, 941–948. [DOI] [PubMed] [Google Scholar]
- David Y.; Ternette N.; Edelmann M. J.; Ziv T.; Gayer B.; Sertchook R.; Dadon Y.; Kessler B. M.; Navon A. (2011) E3 ligases determine ubiquitination site and conjugate type by enforcing specificity on E2 enzymes. J. Biol. Chem. 286, 44104–44115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiger G.; Hao B.; Mohl D. A.; Deshaies R. J. (2009) The acidic tail of the Cdc34 ubiquitin-conjugating enzyme functions in both binding to and catalysis with ubiquitin ligase SCFCdc4. J. Biol. Chem. 284, 36012–36023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiger G.; Saha A.; Lewis S.; Kuhlman B.; Deshaies R. J. (2009) Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell 139, 957–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markin C. J.; Saltibus L. F.; Kean M. J.; McKay R. T.; Xiao W.; Spyracopoulos L. (2010) Catalytic proficiency of ubiquitin conjugation enzymes: Balancing pKa suppression, entropy, and electrostatics. J. Am. Chem. Soc. 132, 17775–17786. [DOI] [PubMed] [Google Scholar]
- Pastushok L.; Spyracopoulos L.; Xiao W. (2007) Two Mms2 residues cooperatively interact with ubiquitin and are critical for Lys63 polyubiquitination in vitro and in vivo. FEBS Lett. 581, 5343–5348. [DOI] [PubMed] [Google Scholar]
- Pierce N. W.; Kleiger G.; Shan S. O.; Deshaies R. J. (2009) Detection of sequential polyubiquitylation on a millisecond timescale. Nature 462, 615–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo-Brenni M. C.; Foster S. A.; Morgan D. O. (2010) Catalysis of lysine 48-specific ubiquitin chain assembly by residues in E2 and ubiquitin. Mol. Cell 39, 548–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A.; Lewis S.; Kleiger G.; Kuhlman B.; Deshaies R. J. (2011) Essential role for ubiquitin-ubiquitin-conjugating enzyme interaction in ubiquitin discharge from Cdc34 to substrate. Mol. Cell 42, 75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N.; Wang P.; Jeffrey P. D.; Pavletich N. P. (2000) Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102, 533–539. [DOI] [PubMed] [Google Scholar]
- Dou H.; Buetow L.; Sibbet G. J.; Cameron K.; Huang D. T. (2012) BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat. Struct. Mol. Biol. 19, 876–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plechanovova A.; Jaffray E. G.; Tatham M. H.; Naismith J. H.; Hay R. T. (2012) Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature 489, 115–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda J. N.; Littlefield P. J.; Soss S. E.; Nordquist K. A.; Chazin W. J.; Brzovic P. S.; Klevit R. E. (2012) Structure of an E3:E2 approximately Ub Complex Reveals an Allosteric Mechanism Shared among RING/U-box ligases. Mol. Cell 47, 933–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger M. B.; Liang Y. H.; Das R.; Mariano J.; Li S.; Li J.; Kostova Z.; Byrd R. A.; Ji X.; Weissman A. M. (2013) A Structurally Unique E2-Binding Domain Activates Ubiquitination by the ERAD E2, Ubc7p, through Multiple Mechanisms. Mol. Cell 50, 516–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagola K.; von Delbruck M.; Dittmar G.; Scheffner M.; Ziv I.; Glickman M. H.; Ciechanover A.; Sommer T. (2013) Ubiquitin Binding by a CUE Domain Regulates Ubiquitin Chain Formation by ERAD E3 Ligases. Mol. Cell 50, 528–539. [DOI] [PubMed] [Google Scholar]
- Kim H. C.; Huibregtse J. M. (2009) Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol. Cell. Biol. 29, 3307–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matuschewski K.; Hauser H. P.; Treier M.; Jentsch S. (1996) Identification of a novel family of ubiquitin-conjugating enzymes with distinct amino-terminal extensions. J. Biol. Chem. 271, 2789–2794. [DOI] [PubMed] [Google Scholar]
- Pestov D. G.; Grzeszkiewicz T. M.; Lau L. F. (1998) Isolation of growth suppressors from a cDNA expression library. Oncogene 17, 3187–3197. [DOI] [PubMed] [Google Scholar]
- Plafker K. S.; Farjo K. M.; Wiechmann A. F.; Plafker S. M. (2008) The human ubiquitin conjugating enzyme, UBE2E3, is required for proliferation of retinal pigment epithelial cells. Invest. Ophthalmol. Visual Sci. 49, 5611–5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanjiang X.; Hongjuan H.; Tiantian G.; Yan Z.; Zhijun H.; Qiong W. (2012) Expression patterns of ubiquitin conjugating enzyme UbcM2 during mouse embryonic development. Gene Expression 15, 163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plafker K. S.; Nguyen L.; Barneche M.; Mirza S.; Crawford D.; Plafker S. M. (2010) The ubiquitin-conjugating enzyme UbcM2 can regulate the stability and activity of the antioxidant transcription factor Nrf2. J. Biol. Chem. 285, 23064–23074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David Y.; Ziv T.; Admon A.; Navon A. (2010) The E2 ubiquitin-conjugating enzymes direct polyubiquitination to preferred lysines. J. Biol. Chem. 285, 8595–8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher F. R.; Wilson G.; Day C. L. (2013) The N-terminal extension of UBE2E ubiquitin-conjugating enzymes limits chain assembly. J. Mol. Biol. 425, 4099–4111. [DOI] [PubMed] [Google Scholar]
- Brzovic P. S.; Lissounov A.; Christensen D. E.; Hoyt D. W.; Klevit R. E. (2006) A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol. Cell 21, 873–880. [DOI] [PubMed] [Google Scholar]
- Lorick K. L.; Jensen J. P.; Fang S.; Ong A. M.; Hatakeyama S.; Weissman A. M. (1999) RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. U.S.A. 96, 11364–11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C. M.; Rose I. A. (1985) Functional heterogeneity of ubiquitin carrier proteins. J. Biol. Chem. 260, 1573–1581. [PubMed] [Google Scholar]
- Wenzel D. M.; Lissounov A.; Brzovic P. S.; Klevit R. E. (2011) UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature 474, 105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger M. B.; Pruneda J. N.; Klevit R. E.; Weissman A. M. (2013) RING-type E3 ligases: Master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta 1, 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel D. M.; Stoll K. E.; Klevit R. E. (2011) E2s: Structurally economical and functionally replete. Biochem. J. 433, 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodson C.; Purkiss A.; Miles J. A.; Walden H. (2014) Structure of the Human FANCL RING-Ube2T Complex Reveals Determinants of Cognate E3-E2 Selection. Structure 22, 337–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoll K. E.; Brzovic P. S.; Davis T. N.; Klevit R. E. (2011) The essential Ubc4/Ubc5 function in yeast is HECT E3-dependent, and RING E3-dependent pathways require only monoubiquitin transfer by Ubc4. J. Biol. Chem. 286, 15165–15170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Zhang J.; Bauer A.; Zhang L.; Selinger D. W.; Lu C. X.; Ten Dijke P. (2013) Fine-tuning BMP7 signalling in adipogenesis by UBE2O/E2-230K-mediated monoubiquitination of SMAD6. EMBO J. 32, 996–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.