Summary
HIV-1 reverse transcription represents the predominant target for pharmacological inhibition of viral replication, but cell-intrinsic mechanisms that can block HIV-1 reverse transcription in a clinically significant way are poorly defined. We find that effective HIV-1 reverse transcription depends on the phosphorylation of viral reverse transcriptase by host cyclin-dependent kinase (CDK) 2 at a highly conserved Threonin residue. CDK2-dependent phosphorylation increased the efficacy and stability of viral reverse transcriptase and enhanced viral fitness. Interestingly, p21, a cell-intrinsic CDK inhibitor that is upregulated in CD4+ T cells from “elite controllers”, potently inhibited CDK2-dependent phosphorylation of HIV-1 reverse transcriptase and significantly reduced the efficacy of viral reverse transcription. These data suggest that p21 can indirectly block HIV-1 reverse transcription by inhibiting host co-factors supporting HIV-1 replication, and identify sites of viral vulnerability that are effectively targeted in persons with natural control of HIV-1 replication.
Introduction
CD4+ T lymphocytes represent the major target cells for infection with HIV-1, but the susceptibility of these cells to HIV-1 varies considerably among different persons (Ciuffi et al., 2004). This variation appears to be primarily related to the relative presence or absence of specific host proteins that can modulate the efficacy of HIV-1 replication by inhibiting specific steps of the viral life cycle. Over the recent years, several of such host molecules have been identified (Harris et al., 2012; Malim and Bieniasz, 2012), but the influence of these host factors on clinical rates of HIV-1 disease progression and on the ability to maintain antiretroviral drug-free control of HIV-1 replication in elite controllers remains uncertain. Interestingly, a significantly reduced susceptibility of host CD4+ T cells to HIV-1 has previously been demonstrated in two geographically distinct cohorts of elite controllers who naturally maintain undetectable levels of HIV-1 replication in the absence of antiretroviral therapy (Chen et al., 2011; Saez-Cirion et al., 2011). This raises the possibility that specific host proteins can reduce viral replication steps in these patients, and contribute to a CD4+ T cell-intrinsic mechanism of HIV-1 immune defense. Yet, classical HIV-1 restriction factors with known direct inhibitory effects on HIV-1 replication, such as APOBEC3G, TRIM5α and BST2 have reduced expression levels in CD4+ T cells from elite controllers in comparison to progressors (Abdel-Mohsen et al., 2013; Rotger et al., 2009; Vigneault et al., 2011); therefore these molecules are unlikely to contribute to HIV-1 immune defense or cell-intrinsic restriction of HIV-1 replication in a clinically significant way.
Instead of blocking HIV-1 replication steps through direct interactions with the virus, specific host proteins may indirectly restrict HIV-1 replication by inhibiting host factors that are required for HIV-1 to replicate effectively. A large number of such HIV-1 “dependency factors” have been identified (Brass et al., 2008; Konig et al., 2008; Zhou et al., 2008), and the requirement of these molecules for effective HIV-1 replication may represent a specific viral vulnerability. p21 (waf-1/cip-1) is a host protein from the cyclin-dependent kinase inhibitor (CDKI) family that is uniquely upregulated in CD4+ T cells from many elite controllers in comparison to both HIV-1 negative persons and individuals with progressive infection (Chen et al., 2011; Saez-Cirion et al., 2011), and has been implicated with restriction of HIV-1 replication in CD4+ T cells (Chen et al., 2011; Elahi et al., 2012a), hematopoietic stem cells (Zhang et al., 2007) and macrophages (Allouch et al., 2013; Bergamaschi et al., 2009), although the underlying mechanisms for inhibiting HIV-1 seem to vary in each of these cell populations. One proposed hypothesis is that p21 can inhibit HIV-1 replication by blocking cyclin-dependent kinases (CDK), a group of host molecules with an emerging function as host co-factors supporting different HIV-1 replication steps. Indeed, CDK9 (Mancebo et al., 1997) and CDK2 (Breuer et al., 2012; Deng et al., 2002) have recognized roles for increasing transcriptional elongation of HIV-1 mRNA through phosphorylation of Polymerase II, and this process can be intercepted by cell-intrinsic inhibitors of CDKs, such as p21 (Chen et al., 2011). In the present study, we show that CDK2 can support HIV-1 reverse transcription through direct phosphorylation of HIV-1 reverse transcriptase (RT) at a highly-conserved amino acid residue, that this phosphorylation is functionally relevant for maintaining RT activity, stability and viral fitness, and that CDK2-dependent RT phosphorylation can be effectively blocked by p21. Thus, these data suggest an indirect mechanism for inhibition of HIV-1 reverse transcription that seems to be active in vivo in CD4+ T cells from persons with spontaneous control of HIV-1 replication.
Results
Host CDK2 supports HIV-1 reverse transcription in CD4+ T cells
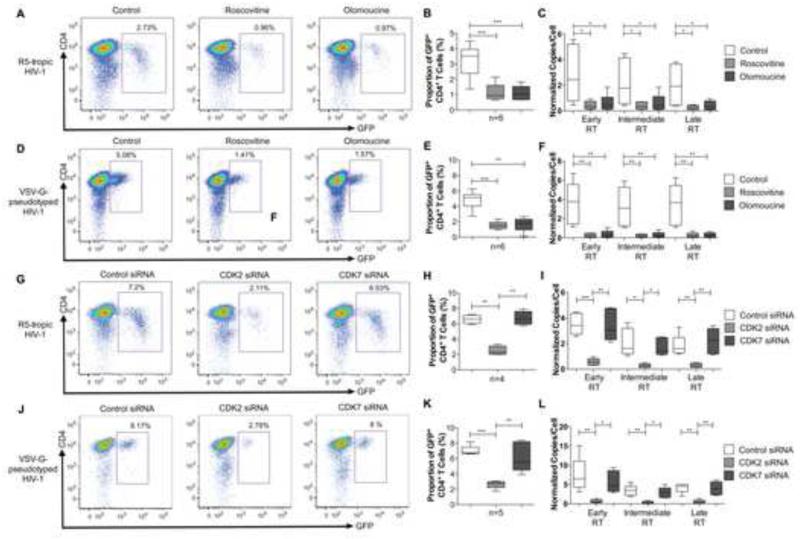
Cyclin-dependent kinases have a recognized role for supporting HIV-1 gene transcription from chromosomal DNA (Mancebo et al., 1997), but may also facilitate other steps in the HIV-1 life cycle. To investigate this, we analyzed the influence of pharmacological CDK inhibition on the HIV-1 replication cycle in CD4+ T cells. Primary CD4+ T cells from HIV-1 negative persons were ex-vivo activated with CD3/CD8 bi-specific antibodies and IL-2, followed by infection with a GFP-encoding R5-tropic HIV-1 construct in the presence or absence of Roscovitine or Olomoucine, two chemical inhibitors of CDKs that can inhibit HIV-1 mRNA transcription (Wang et al., 2001). These experiments demonstrated that pharmacological inhibition of CDKs effectively reduced the proportion of GFP-positive CD4+ T cells, and the per-cell levels of early HIV-1 reverse transcripts (minus-strand strong-stop DNA), intermediate HIV-1 reverse transcripts (minus strand DNA) and late, double-stranded HIV-1 reverse transcription products (Figure 1A-C); these data indicate blockade of HIV-1 reverse transcription by pharmacological CDK inhibitors. Similar observations were made when cells were infected with a GFP-encoding VSV-G-pseudotyped HIV-1 virus that causes single-round infections of CD4+ T cells independently of viral co-receptors (Figure 1D-F). Together, these findings imply a role of host CDKs, the target molecules for Roscovitine and Olomoucine, in supporting HIV-1 reverse transcription. To better define this, we conducted ex-vivo infection experiments of CD4+ T cells after siRNA-mediated knockdown of CDK2 and CDK7, two prominent members of the CDK family that are effectively inhibited by Roscovitine and Olomoucine and expressed in the cytoplasm of CD4+ T cells where HIV-1 reverse transcription occurs (Figure S1A). These experiments demonstrated that siRNA-mediated downregulation of CDK2, but not of CDK7, resulted in a significant reduction of the proportion of HIV-1 positive cells, and of the frequency of per-cell levels of early, intermediate and late HIV-1 reverse transcripts. This was true for cells infected with R5-tropic (Figure 1G-I) and with VSV-G-pseudotyped (Figure 1J-L) HIV-1. siRNA directed against CDK1 and CDK4 was effective in downregulating the corresponding protein expression, but had no detectable effect on the proportion of GFP-positive cells after HIV-1 infection (Figure S1B-F). Silencing of CDK9 reduced cellular susceptibility to productive HIV-1 infection, consistent with previous findings (Flores et al., 1999), but had no effect on viral reverse transcription (Figure S1B-E). Together, these experiments suggest that CDK2 is critical for supporting HIV-1 reverse transcription in CD4+ T cells.
Figure 1. Host CDK2 supports HIV-1 reverse transcription.
Activated CD4+ T cells were ex-vivo infected with GFP-encoding, R5-tropic HIV-1 or GFP-encoding VSV-G-pseudotyped HIV-1 in the presence or absence of the pharmaceutical CDK inhibitors Roscovitine and Olomoucine, or after transfection with siRNA directed against CDK2 or CDK7. (A/D/G/J): Representative flow cytometry plots showing infected GFP+ CD4+ cells. (B/E/H/K): Proportions of GFP+ CD4+ T cells after exposure to indicated CDK inhibitors or CDK-directed siRNAs. (C/F/I/L): Quantitative analysis of early, intermediate and late HIV-1 RT transcripts in CD4+ T cells from the indicated study groups. All statistical comparisons were performed using Student’s t tests. *, p<0.05. **, p<0.01. ***,p<0.001. See also Figure S1.
HIV-1 RT is a substrate for CDK2-dependent phosphorylation
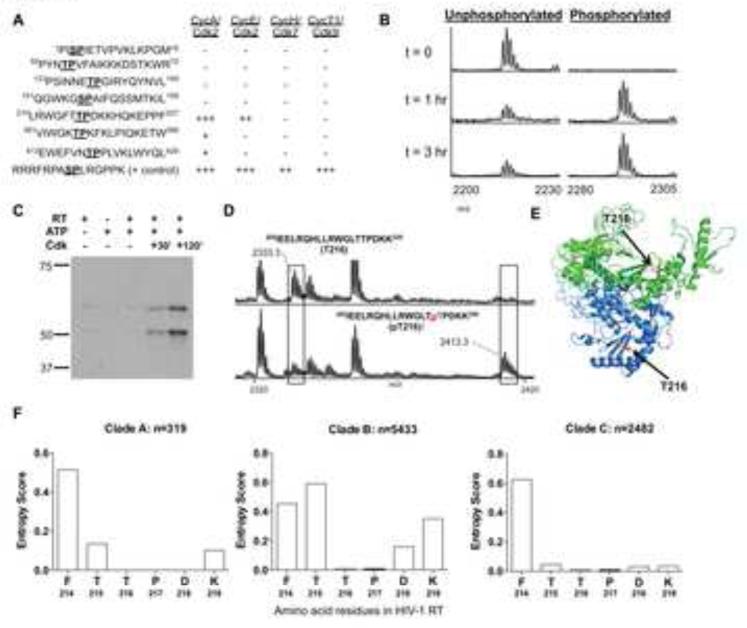
HIV-1 gene products can represent direct targets for phosphorylation by host kinases (Francis et al., 2011). We hypothesized that CDK2 can support HIV-1 reverse transcription through phosphorylation of HIV-1 reverse transcriptase (RT). CDKs are requisite proline-directed serine/threonine kinases with preference for basic amino acids located c-terminal to the phosphate acceptor (Nigg, 1993); therefore the canonical CDK substrate motif is Ser/Thr-Pro-X-Arg/Lys, where X can be any amino acid(s). The HIV-1 RT protein contains seven such CDK motifs (Figure S2A). Synthetic peptides were generated with sequences derived from these motifs and their surrounding amino acids for an initial rapid screen of potential CDK phosphorylation sites. Each of these peptides was assayed as a CDK2 substrate by incubating with either CDK2/cyclin A or CDK2/cyclin E and ATP (Figure 2A), followed by mass spectrometric analysis. Robust phosphorylation of the synthetic peptide containing the motif at Thr216 (corresponding to HIV-1 RT amino acid residues [210-227]) was observed using both CDK2/cyclin A and CDK2/cyclin E, and trace phosphorylation was observed at two other peptides with CDK2/cyclin A (Figure 2A). Phosphorylation was not observed with any other HIV-1 RT peptides harboring CDK phosphorylation motifs (data not shown). When the [210-227] peptide was incubated with CDK2/cyclin A for 0, 1 and 3 hours, a time-dependent increase in mass spectral signal at +80 mass units (corresponding to covalent addition of HPO3) was observed (Figure 2B). When similar assays were performed using CDK7 or CDK9, the positive control peptide, but no HIV-1 RT peptides, were phosphorylated (Figure 2A and Figure S2B), which indicates a selective susceptibility to phosphorylation by CDK2 and corresponds to our prior observation that HIV-1 reverse transcription in CD4+ T cells is most dominantly influenced by CDK2 (Figure 1G-L).
Figure 2. HIV-1 RT is a substrate for CDK2-dependent phosphorylation.
(A): Level of phosphorylation observed by semi-quantitative MALDI mass spectrometry on synthetic peptides derived from HIV-1 RT after incubation with indicated CDKs and ATP for 3 hours. “+” denotes phosphopeptide levels < 1-25%; “++” indicates phosphopeptide levels of 25%-50%; “+++” reflects phosphopeptide levels >50% of total peptide signal intensity. (B): Time course for in vitro phosphorylation reaction of synthetic peptide [210-227] incubated with ATP and cyclin A/CDK2, assayed by MALDI-selected ion monitoring mass spectrometry. (C): Western blot probed by anti-[phospho-210-227] primary antibody showing time-dependence of phosphorylation on recombinant HIV-1 RT protein incubated with cyclin A/CDK2 and ATP. (D): MALDI mass spectra of endoproteinase Lys-C proteolzyed unphosphorylated (top panel) and in vitro cyclin A/CDK2 phosphorylated (bottom panel) HIV-1 RT protein. The signal with m/z corresponding to the Thr216 phosphorylated proteolytic peptide is observed in the phosphorylated but not the unphosphorylated digest. (E): Solvent-exposed position of Thr216 in crystal structures of HIV-1 RT. (F): Entropy of amino acid residues at position 214-219 in indicated clade A, B and C HIV-1 RT sequences obtained from the Los Alamos HIV-1 sequence database. See also Figure S2.
To further investigate the potential phosphorylation site at Thr216, the CDK2-phosphorylated [210-227] peptide was subjected to tandem mass spectrometry (MS2). Specific fragmentation of the phosphorylated peptide showed a strong signal at −98 mass units from the parent peptide peak, corresponding to the neutral loss of phosphoric acid, a diagnostic characteristic of Ser/Thr phosphorylated peptide ions (Figure S2C). Furthermore, using backbone fragmentation, we found that the y”11 fragment ion, which runs from Pro217 to Phe227 and does not contain Thr216, is observed at a mass indicating no phosphate. In contrast, the y”12 fragment ion, which runs from Thr216 to Phe227, is observed at a mass indicating the presence of a phosphate adduct (Figure S4). Because the two fragments both arise from the singly phosphorylated [210-227] peptide, and are identical in amino acid sequence except for the Thr216 residue that is selectively contained in the y”12 fragment ion, these data indicate that Thr216 is indeed the CDK2 phosphorylation site (Figure S2C).
Since our data suggested that Thr216 represents the only site in HIV-1 RT susceptible to CDK-dependent phosphorylation, we performed phosphorylation analyses on the intact HIV-1 RT protein. Recombinant HIV-1 RT protein was phosphorylated in vitro and subjected to analysis using western blots. For this purpose, we generated rabbit-derived antibodies that specifically recognized the HIV-1 RT [210-227] peptide sequence phosphorylated at position Thr216 (Figure S2D). In western blot experiments, two RT bands (p51 and p66 subunits) were detected with anti-[phospho-210-227] antibodies after incubation with ATP and CDK2/Cyclin A for 30 min (Figure 2C). An increase in the intensity of the phospho-RT bands was observed after incubation for 120 min. To conclusively determine the phosphorylation site in the RT protein, phosphorylated HIV-1 RT was digested by endoproteinase Lys-C and the digests were analyzed by mass spectrometry. A mass spectral peak at m/z 2413 was observed in the spectrum arising from the CDK2-phosphorylated but not the unphosphorylated HIV-1 RT digest. This peak corresponds to the phosphorylated Lys-C peptide [202-220], containing the Thr216 CDK phosphorylation site (Figure 2D, bottom panel). Phosphorylation at Thr216 was confirmed by MS2 and MS3 fragmentation (Figure S2E). In contrast, the spectrum of the unphosphorylated HIV-RT digest contained a significant peak at m/z 2334 corresponding to unphosphorylated peptide [202-220] (Figure 2D, top panel); that peak appears substantially weaker in the spectrum from the CDK2-phopsphorylated RT digest. No other peaks corresponding to phosphorylation at CDK consensus motifs were observed in either digest (data not shown). Together, these data strongly indicate CDK2-dependent phosphorylation specifically at Thr216 in HIV-1 RT.
We subsequently analyzed the structural position of the targeted Thr216 residue, using prior crystal structures of HIV-1 RT (Jager et al., 1994; Kohlstaedt et al., 1992). We observed that Thr216 lies in β strand 11 for both the p51 and p66 subunits, situated near the interface between the fingers and palm of the structure (Figure 2E). This solvent-exposed position of Thr216 at the molecular surface is consistent with its role as a target for cytoplasmic host kinases. In addition, we found that the targeted CDK phosphorylation motif (Thr216 and adjacent Pro217) was highly conserved across more than 8000 viral clade A, B or C sequences derived from the Los Alamos HIV-1 sequence database (www.hiv.lanl.gov), while adjacent amino acid residues were more variable (Figure 2F). Together, these observations support the notion that the Thr216 residue in HIV-1 RT is susceptible to CDK phosphorylation, and functionally and/or structurally important for HIV-1 replication.
Substitution of Thr216 by Alanine reduces HIV-1 reverse transcription
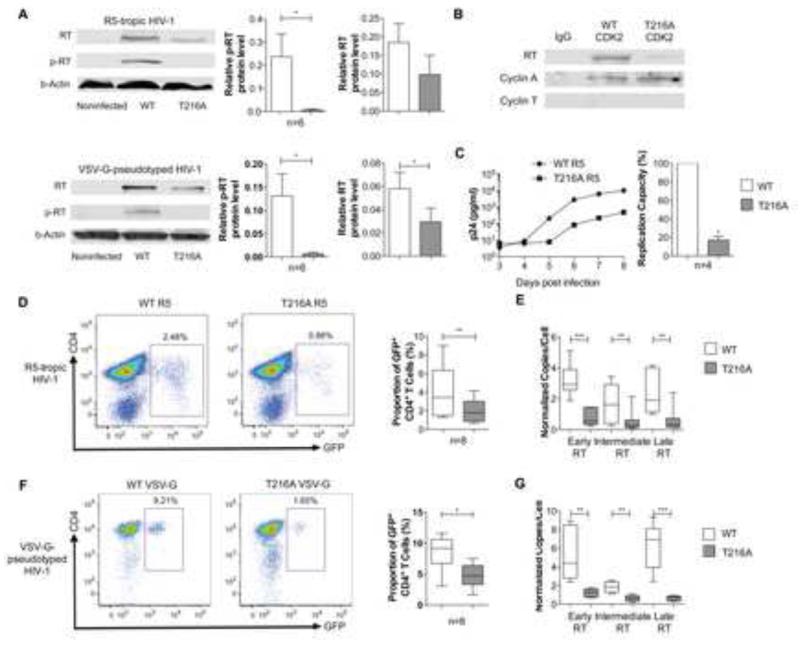
To investigate the functional relevance of CDK2-dependent phosphorylation of HIV-1 RT, we used site-directed mutagenesis to construct a GFP-encoding R5-tropic HIV-1 variant in which the targeted Threonine residue at position 216 is replaced by Alanine (T216A); this leads to removal of a hydroxyl group and abrogates susceptibility to CDK2-dependent phosphorylation at this position. Using western blot experiments with antibodies selectively recognizing the RT peptide (amino acids 210-227) encompassing phosphorylated Thr216 (Figure S6), we observed a clear signal corresponding to the detection of phosphorylated RT in CD4+ T cells infected with the wild-type virus; however, no evidence for phosphorylation of HIV-1 RT was noted following infection with the variant HIV-1 construct, despite detection of total HIV-1 RT, although at slightly weaker intensity compared to cells infected with wild-type virus (Figure 3A). Similar findings were made when CD4+ T cells were infected with a VSV-G-pseudotyped wild-type (wt) virus and the corresponding variant harboring the T216A mutation. Using co-immunoprecipitation experiments, we also observed that CDK2 extracted from CD4+ T cells infected with the wt virus, but not with the T216A variant, closely associated with HIV-1 RT (Figure 3B). These data indicate that phosphorylation of HIV-1 RT at position Thr216 can occur in ex vivo-infected CD4+ T cells.
Figure 3. HIV-1 RT T216A mutation decreases HIV-1 reverse transcription.
Activated CD4+ T cells were ex-vivo infected with GFP-encoding R5-tropic or VSV-G-pseudotyped HIV-1 and with corresponding variants with replacement of Threonine by Alanine at position 216 (T216A). (A): Detection of HIV-1 RT using western blots with antibodies directed against unphosphorylated HIV-1 RT (upper row), antibodies directed against the [phospho-210-227] RT peptide (middle row), or β-actin-specific antibodies (lower row). Left Panel shows one experimental example, right panel summarizes relative signal intensities of indicated bands from n=6 different experiments. (B): Whole protein lysates from CD4+ T cells infected with R5-tropic wild-type HIV-1 or the T216A variant were precipitated with CDK2 or IgG antibody, followed by interrogation with antibodies directed against HIV-1 RT, or against cyclin A or T as controls. One representative experiment out of 3 is shown. (C): HIV-1 p24 antigen production in the supernatant of activated CD4+ T cells infected with similar titers of R5-tropic wild-type HIV-1 or the corresponding T216A variant. Data were analyzed by fitting a linear model to the log-transformed p24 concentration using maximum likelihood models. Left panel demonstrates one representative experiment, right panel summarizes the relative replication capacity of wild-type and variant viruses, calculated based on the slope for the p24 production curves in n=4 experiments. (D/F): Proportions of GFP+ CD4+ T cells after infection with GFP-encoding R5-tropic (D) or VSV-G-pseudotyped (F) HIV-1 wt and corresponding T216A variants. (E/G): Quantitative analysis of early, intermediate and late HIV-1 RT transcripts in activated CD4+ cells infected with either wild-type R5-tropic (E) or VSV-G-pseudotyped (G) HIV-1 or the corresponding T216A variants.
We subsequently assessed the replicative activity of the wild-type and the variant virus in ex-vivo activated CD4+ T cells. A direct comparison of the replicative activity of the wild-type and the variant virus over an 8-day period demonstrated a significantly reduced ability of the variant virus to produce HIV-1 p24 antigen; this is consistent with reduced viral fitness (Figure 3C). We also observed that infection of CD4+ T cells with the T216A variant lead to reduced proportions of GFP-positive CD4+ T cells in comparison to CD4+ T cells infected with the wild-type virus under identical conditions (Figure 3D). The lower replicative activity of the variant virus was associated with reduced levels of early, intermediate and late HIV-1 reverse transcripts, suggesting that ineffective reverse transcription is the major replicative defect in this viral variant (Figure 3E). Lower proportions of viral reverse transcripts were also observed after infection of CD4+ T cells with a VSV-G-pseudotyped virus harboring the T216A variant, confirming a lower replicative competence of the T216A variant, and demonstrating that the T216A mutation was associated with defective HIV-1 reverse transcription independently of viral entry mechanisms (Figure 3F-G). Together, these findings suggest that phosphorylation of HIV-1 RT critically influences viral fitness through modulation of viral reverse transcription.
CDK2-dependent phosphorylation increases function and stability of HIV-1 RT
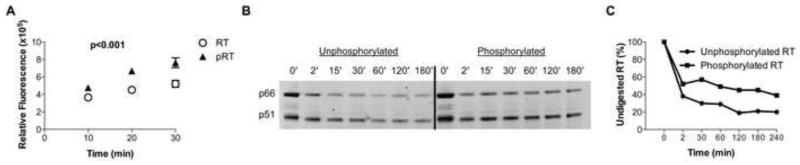
We subsequently determined whether CDK phosphorylation of HIV-1 RT directly modulates the enzymatic activity of RT. For this purpose, HIV-1 RT was exposed to CDK2/cyclin A, which resulted in 92% and 62% phosphorylation of the p51 and p66 RT subunits, respectively (Figure S3A-C); unphosphorylated HIV-1 RT was used as a control. Either HIV-1 RT was incubated with a poly (A) template, oligo dT primer and dTTP, followed by monitoring of RT activity via the formation of RNA-DNA heteroduplexes. These experiments demonstrated an increased enzymatic activity of phosphorylated HIV-1 RT (Figure 4A), suggesting that CDK2 phosphorylation supports the functional activity of the enzyme.
Figure 4. CDK2-dependent phosphorylation enhances activity and stability of HIV-1 RT.
(A) Assessment of the functional activity of phosphorylated and unphosphorylated HIV-1 RT in a cell-free system. One representative experiment out of 4 is shown. (B) SDS-PAGE gel of limited proteolysis of unphosphorylated (left) and cyclin A/CDK2 phosphorylated (right) HIV-1 RT time course. Degradation of the p66 and p51 subunits of HIV-1 RT (major upper and lower bands, respectively) is shown as a function of time and phosphorylation state. (C) Quantitative comparison of the rates of proteolysis for unphosphorylated and phosphorylated HIV-1 RT as measured by SDS-PAGE gel densitometry of Sypro Ruby-stained HIV-1 RT. See also Figures S3.
Since phosphorylation often influences the stability of proteins (Nishi et al., 2011), we considered whether CDK2 might serve to modulate the resistance of HIV-1 RT to proteolytic degradation. Limited proteolysis was employed as an in vitro tool for probing HIV-1 RT stability. CDK2/cyclin A-phosphorylated and unphosphorylated HIV-1 RT was incubated with subtilisin protease - known to be sensitive to the three-dimensional conformation of its substrate - at different reaction times ranging from 2 min to 4 hr (Figure 4 B/C). These experiments demonstrated a markedly more rapid decrease in band intensity for the unphosphorylated HIV-1 RT. Indeed, after 180 minutes of exposure to subtilisin, gel densitometry indicated that 45% of the phosphorylated RT, versus only 20% of the unphosphorylated RT, remained undigested, demonstrating that CDK2 phosphorylation protects HIV-1 RT from protease cleavage. Moreover, CD4+ T cell infection with the T216A variant in the presence of proteasome inhibitors (Schwartz et al., 1998) enhanced viral replication and reverse transcription to levels observed in cells infected with wild-type virus, suggesting that proteasomal degradation of unphosphorylated HIV-1 RT contributes to replicative defects of the T216A variant (Figure S3D-E). Together, these data indicate that CDK2 phosphorylation of Thr216 protects HIV-1 RT from protease activity, and suggests that its mechanism of action may be to stabilize HIV-1 RT against intracellular proteases, perhaps via conformational modulation.
p21 (waf-1/cip-1) suppresses CDK2-dependent HIV-1 RT phosphorylation
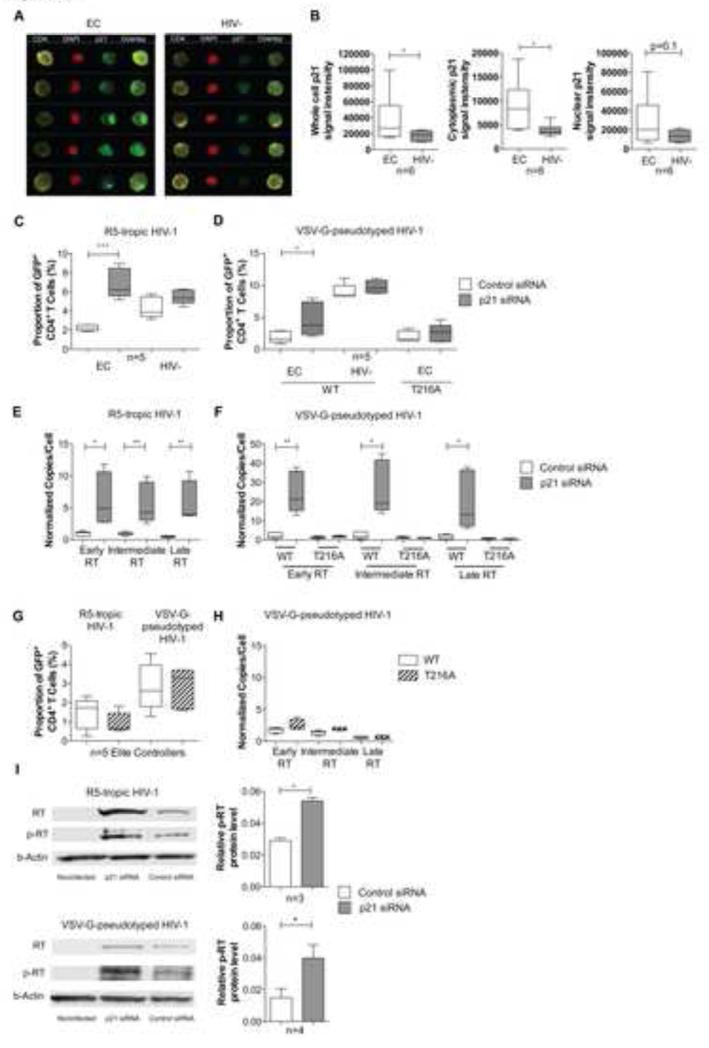
The cell-intrinsic CDK inhibitor p21 (waf-1/cip1) is upregulated in CD4+ T cells from many persons with natural control of HIV-1 replication (“elite controllers”)(Chen et al., 2011; Saez-Cirion et al., 2011), raising the possibility that p21 has a functional role for inhibiting CDK2-dependent phosphorylation of RT and reducing the efficacy of viral reverse transcription in CD4+ T cells from these patients. To investigate this, we first analyzed the subcellular localization of p21 in CD4+ T cells from elite controllers, using high-throughput imaging flow cytometry, an approach that combines standard flow cytometry with high-resolution fluorescence microscopy. For this purpose, CD4+ T cells from elite controllers and HIV-1 negative persons were intracellularly stained with antibodies directed against p21, and with the nuclear dye DAPI. This analysis demonstrated that p21 was located both in the nuclear and in the cytoplasmic subcellular compartments (Figure 5A/B), and did not change substantially after in vitro activation (Figure S4A). Importantly, we observed that total, nuclear and cytoplasmic cell-associated p21 was expressed at significantly higher levels in CD4+ T cells from elite controllers compared to HIV-1 negative persons, but differences were more obvious for cytoplasmic p21 than for nuclear p21 (Figure 5A-B). This suggests that p21 may be functionally relevant for inhibiting CDK2-dependent phosphorylation of HIV-1 RT in the cytoplasm of CD4+ T cells from elite controllers. To further investigate this, we analyzed HIV-1 reverse transcription in CD4+ T cells from elite controllers after electroporation with p21-specific siRNA, which resulted in effective reduction of p21 protein expression (Figure S4B). We found that following infection of CD4+ T cells from elite controllers with either R5-tropic or VSV-G-pseudotyped HIV-1, siRNA-mediated reduction of p21 expression resulted in enhanced levels of HIV-1 positive cells, and in increased levels of viral reverse transcripts. Interestingly, such effects were not visible in CD4+ T cells from HIV-1 negative persons, likely due to lower baseline expression of p21 that minimizes biological effects of p21 silencing (Figure 5C-F). Moreover, p21 knockdown did not affect HIV-1 replication in CD4+ T cells from elite controllers infected with the T216A variant, supporting the notion that p21 inhibition of HIV-1 reverse transcription depends on phosphorylation of Thr216 (Figure 5C-F). In addition, we observed that in CD4+ T cells from elite controllers, the wild-type virus and the T216A variant showed a similar replicative activity (Figure 5G/H), which represented a sharp contrast to the replicative advantage of the wild-type virus in CD4+ T cells from HIV-1 negative individuals (Figure 3D-G); this indicates that the increased viral fitness associated with RT phosphorylation at position Thr216 is not visible in cells with high-level p21 expression. Finally, we noted that p21 knockdown led to increased levels of phosphorylated HIV-1 RT in CD4+ T cells from EC infected with R5-tropic or VSV-G pseudotyped HIV-1, providing formal evidence for an inhibitory efect of p21 on CDK2-dependent RT phosphorylation (Figure 5I). Notably, p21 silencing did not substantially affect levels of total HIV-1 RT during single-round infection with VSV-G-pseudotyped virus, but appeared to increase levels of total RT in cells infected with R5-tropic HIV-1 in which the release of p21-dependent viral inhibition is amplified during multiple rounds of infection. Taken together, these data suggest that high-level p21 expression can indirectly inhibit viral reverse transcription through blockade of CDK2-dependent phosphorylation of viral RT.
Figure 5. Inhibition of p21 (waf-1/cip-1) increases HIV-1 reverse transcription in CD4+ T cells from elite controllers.
(A): Analysis of the subcellular localization of p21 in CD4+ T cells. PBMCs from Elite controllers or HIV-1 negative patients were stained with antibodies against CD4 (yellow pseudocolor), p21 (green pseudocolor) and DAPI (red pseudocolor) and subjected to ImageStream analysis. Selected images of single-cell analysis plots highlighting differences between an HIV-1 elite controller and an HIV-1 negative patient are shown. (B): Cumulative average p21 signal intensity in CD4+ T cells from Elite controllers (n=6) and HIV-1 negative persons (n=6) in total, cytoplasmic and nuclear compartments of CD4+ T cells. (C/D): Proportion of HIV-1 positive CD4+ T cells from EC and HIV-1 negative persons after ex-vivo infection with R5-tropic or VSV-G-pseudotyped HIV-1 in the presence of siRNA-mediated knock-down of p21. The T216A variant virus was used where indicated. (E/F): Corresponding analysis of early, intermediate and late RT transcripts in CD4 T cells from elite controllers (n=5). (G-H): Comparison of HIV-1 replication patterns of wild-type HIV-1 and the T216A variant in CD4+ T cells from EC. (G) demonstrates proportion of infected CD4+ T cells, (H) reflects levels of early, intermediate and late reverse transcripts. (I): HIV-1 RT phosphorylation in CD4+ T cells after experimental p21 knock-down. CD4+ T cells were transfected with siRNA directed against p21 or control siRNA, infected with R5-tropic or VSV-G pseudotyped HIV-1 and hybridized with antibody against HIV-1 RT, anti-[phospho-210-227] RT or β-actin. See also Figure S4.
Discussion
CDKs represent host molecules with a wide range of functions for regulating cell proliferation, survival and gene expression, but also have recognized roles for modulating replication of viruses in human cells. This seems to be particularly true for human Herpesviruses (Durand and Roizman, 2008; Zydek et al., 2010), some of which encode for viral CDK orthologs to avoid dependence on host CDKs for supporting critical viral replication steps (Kuny et al., 2010). In HIV-1 infection, CDKs have mostly been recognized for their role in promoting transcriptional elongation of nascent HIV-1 mRNA through phosphorylation of host Polymerase II (Flores et al., 1999; Mancebo et al., 1997) and, possibly, direct phosphorylation of HIV-1 Tat (Ammosova et al., 2006); however, a possible impact of CDKs on viral pre-integration steps remained unknown. Using a combination of proteomic, biochemical and reverse phosphorylation approaches, our data provide evidence that the process of HIV-1 reverse transcription is under tight regulatory control of CDK2. These data support the emerging recognition of host kinases as modulators of HIV-1 replication in human CD4+ T cells, and suggest that inter-individual differences in the CD4+ T cell susceptibility to HIV-1 may, at least to some extent, be related to a fine-tuned interplay between host kinases and virally-encoded substrate proteins. In line with this, prior studies demonstrated that host kinases from the ERK/MAPK family can enhance chromosomal HIV-1 integration through the induction of phosphorylation-induced conformational changes in HIV-1 integrase (Manganaro et al., 2010). Conformational changes that may result from CDK2-mediated phosphorylation of RT remain unknown at present but might involve structural alterations that improve the nucleotide binding capacity of HIV-1 RT. In this regard, it is interesting that the targeted Thr216 residue is in immediate proximity to the RT amino acid residues T215 and K219 with known roles for influencing the incorporation of nucleotides and pharmaceutical nucleotide analogues into evolving HIV-1 reverse transcripts (Chamberlain et al., 2002; Tu et al., 2010). Moreover, recent work has shown that CDK1, an alternative member of the cyclin-dependent kinase family, can influence HIV-1 replication through phosphorylation-dependent inactivation of SAMHD1, a host restriction factor that can otherwise inhibit HIV-1 replication in myeloid and lymphoid cells (Cribier et al., 2013; White et al., 2013). Although CDK1 failed to exert marked effects on HIV-1 replication in ex-vivo activated primary CD4+ T cells in our experiments, these data concur in demonstrating a significant role of CDKs for modulating HIV-1 replication in human cells.
The role of p21 for cell-intrinsic restriction of HIV-1 has become increasingly evident over the recent years, and is further supported by findings described in this manuscript. So far, p21 has been implicated in inhibition of HIV-1 replication steps in macrophages (Allouch et al., 2013; Bergamaschi et al., 2009), hematopoietic stem cells (Zhang et al., 2007) and CD4+ T cells (Chen et al., 2011; Elahi et al., 2012a), but the proposed underlying mechanisms and the specific viral replication steps that are targeted seemed to differ in each cell population. In macrophages, p21 was recently suggested to block HIV-1 reverse transcription through a blockade of host molecules responsible for maintaining adequate dNTP supply, which in resting cells is critical for supporting viral replicative activity (Allouch et al., 2013). The mechanism of p21-mediated HIV-1 restriction in CD4+ T cells seems to be of particular interest due to the selective upregulation of this protein in elite controllers and its association with reduced cellular susceptibility of CD4+ T cells to HIV-1 infection (Chen et al., 2011; Elahi et al., 2012b). Our data suggest that p21 can effectively inhibit HIV-1 reverse transcription in CD4+ T cells, but the underlying mechanism is conceptually different from pharmacological inhibition of RT and relies on blockade of CDK2 as a host factor or “HIV-1 dependency factor” that is required for effective viral reverse transcription in CD4+ T cells. A similar indirect mechanism of viral inhibition appears to occur at the level of HIV-1 mRNA transcription, where p21 can block CDK9-mediated phosphorylation of polymerase II and reduce transcriptional elongation of evolving viral mRNA transcripts (Chen et al., 2011). As such, p21 seems to be able to restrict at least two critical HIV-1 replication steps through a combined blockade of host CDKs supporting HIV-1 replication in CD4+ T cells. This indirect mechanism of viral inhibition appears less effective than the pharmacodynamic activities of traditional antiretroviral agents used for clinical purposes, but avoids a direct interaction with HIV-1 gene products and in this way may be less susceptible to viral mutational escape that can very effectively abrogate effects of pharmacological RT inhibitors. In line with this, prior observations demonstrated no selection of viral escape variants in ex-vivo cultures of HIV-1 infected CD4+ T cells exposed to suboptimal concentrations of pharmacological CDK inhibitors (Wang et al., 2001). Notably, by indirectly blocking CDK-dependent HIV-1 replication steps, p21 seems to differ from a variety of alternative cell-intrinsic host restriction factors that target HIV-1 genes or gene products directly and for which a role in natural immune defense in HIV-1 elite controllers is less evident. Whether p21 can also inhibit CDK1-dependent phosphorylation of SAMHD1 (White et al., 2013) and block HIV-1 replication by reducing CDK1-mediated SAMHD1 inactivation will be an important aspect of future studies.
Elite controllers represent a heterogeneous group of individuals, and underlying immune defense mechanisms against HIV-1 in these patients seem to vary (Saez-Cirion and Pancino, 2013; Walker and Yu, 2013). In many of these patients, highly-functional HIV-1-specific CD8+ T cell responses appear to represent the major correlate of immune protection, specifically when restricted by protective HLA class I alleles (Pereyra et al., 2010). Yet, a subgroup of elite controllers is able to maintain undetectable levels of HIV-1 replication in the absence of strong HIV-1-specific CD8+ T cells (Emuet al., 2008). Weak HIV-1-specific CD8+ T cells are also observed in a group of patients who maintain undetectable levels of viral replication after being exposed to a course of antiretroviral therapy in primary HIV-1 infection (Saez-Cirion et al., 2013). A better understanding of synergisms between cell-intrinsic and CD8 T cell mediated adaptive immunity in elite controllers may help to induce a drug-free remission or functional cure of HIV-1 infection in broader patient populations.
Experimental Procedures
Patients
PBMCs from HIV-1-infected individuals and HIV-1-negative individuals were used for this study according to protocols approved by the Institutional Review Board of the Massachusetts General Hospital. All study subjects gave written consent to participate.
HIV-1 viruses and constructs
The GFP-encoding R5-tropic and VSV-G-pseudotyped HIV-1 plasmids were kindly provided by Dr. Dan R. Littman (New York University, New York, New York, USA). Viral particles were produced by transfecting 293T cells (NIH AIDS Reagent Program) with the respective HIV-1 plasmids and, if applicable, with pCG-VSV-G, using TransIT-293 (Mirus) in OptiMEM. R5-tropic or VSV-G-pseudotyped viral variants encoding for a T216A mutation were generated by site-directed mutagenesis by a commercial supplier (Genewiz); the correct sequence of the variant constructs was confirmed by repeated viral sequencing. Supernatants containing infectious retroviruses were harvested 48 hours after transfection, centrifuged and filtered. Viral titers were determined by measuring p24 levels in the supernatant of infected 293 T cells, and confirmed by measuring TCID50 levels in TZM-bl cells (NIH AIDS Reagent Program).
In vitro infection assays
PBMCs were stimulated in RPMI medium containing 10% FCS, recombinant IL-2 (50 U/ml), and an anti-CD3/CD8 bispecific antibody (0.5 μg/ml). A homogenous population of activated CD4+ T cells were infected on day 5 with the indicated HIV-1 viruses for 4h (R5) or 2h (VSV-G) at 37°C. After 2 washes, cells were resuspended and plated at 1×106 cells/well in a 24-well plate. The CD4+ T cells were subjected to flow cytometric analysis of GFP+ CD4+ T cells at 96h after infection with GFP-encoding R5-tropic HIV-1 or 48h after infection with VSV-G-pseudotyped HIV-1 virus. When indicated, proteasome inhibitors (MG-132, Millipore) were added at a concentration of 1μg/ml. For replication capacity assays, CD4+ T cells were infected with either wild-type or T216A-variant R5-tropic HIV-1 viruses. At indicated time intervals, supernatants were analyzed by p24 ELISA kit (Perkin Elmers). Replication capacities were calculated by fitting a linear model to the log-transformed p24 concentration using maximum likelihood models.
siRNA-mediated gene knockdown
CD4+ T cells were suspended in 100 μl transfection solution (Lonza), and target-specific or control siRNA (Dharmacon) was added at a concentration of 4 nmol/ml. Samples were then transferred into nucleofection cuvettes and transfected using program T-023 in a Nucleofector device (Lonza). Afterwards, cells were resuspended in medium supplemented with 20% FCS; IL-2 (50 U/ml) was added 2h after transfection. Cells were infected with indicated HIV-1 strains at 24h after transfection.
Flow cytometry
The proportion of CD3+ CD4+ GFP-positive cells was analyzed on an LSRII flow cytometer (BD Biosciences); data analysis was performed using FlowJo software (TreeStar). For imaging flow cytometry experiments, PBMCs were stained with antibodies directed against CD3 and CD4, fixed and permeabilized. Subsequently, intracellular staining was performed with p21-specific antibodies (clone F-5, Santa Cruz Biotechnology). The samples were run on an ImageStream Cytometer (Amnis); data visualization and co-localization analysis were performed using IDEAS Software (Amnis).
Detection of HIV-1 reverse transcripts
Cells were harvested 18h after infection of activated cells with VSV-G-pseudotyped HIV-1 and 48h after infection of activated cells with R5-tropic HIV-1. RT products were amplified from cell lysates with primers and probes hRU5-F2, hRU5-R, hRU5-P (early RT transcripts), FST-F1, SS-R4, P-HUS-SS1 (intermediated RT transcripts) and GagF1, GagR1, P-HUS-103 (late RT transcripts) (Mbisa et al., 2009). CCR5 was amplified as a housekeeping gene and used for the quantification of input cell numbers (Chen et al., 2011). Serial dilutions of DNA from cell lysates of the HIV-1–infected cell line 293T (provided by Dr. Frederic Bushman, University of Pennsylvania) were used for reference purposes.
In vitro kinase assays
5μM HIV-1 RT peptides or RT proteins were incubated with 2 mM ATP and 550nM of indicated recombinant CDK (Life Technologies) in NE Buffer. The kinase reaction was stopped by addition of SDS-PAGE loading buffer. Subsequently, proteins or peptides were analyzed by western blot or mass spectrometry.
Peptide analysis
Mass spectrometric analysis of ex-vivo kinase assay products was performed by a Thermo LTQ XL linear ion trap mass spectrometer (Thermo Scientific) equipped with a vacuum MALDI source. Synthetic HIV-1 RT peptide samples were mixed with saturated α-Cyano-4-hydroxycinnamic acid, CHCA (Sigma-Aldrich) in 0.1% trifluoroacetic acid: acetonitrile (70:30, v/v). A 1.0 μL aliquot of sample/matrix mixture was dispensed onto the MALDI plate, and the solvent evaporated at room temperature.
In gel digestion
RT samples were separated by NuPAGE and the protein bands were cut out and digested by lysyl endopeptidase (Lys-C, Promega) at 37 °C for 4h. The enzyme Lys-C was added to a final substrate-to-enzyme ratio of 40:1 with 50 mM ammonium bicarbonate, pH 8.5. After digestion, POROS 20 R2 resin (Applied Biosystems) was added into the digested RT samples for extraction at 4°C for 4h. Prior to MALDI mass spectrometric analysis, the digests were desalted using ZipTip (Millipore) and eluted with CHCA MALDI matrix solution from resins directly onto a MALDI plate, and analyzed in single-stage, MS2 and MS3 modes. Protein bands were identified using Mascot (Matrix Science). Identification of phosphopeptide peaks and interpretation of MS2 and MS3 spectra was performed manually, with the assistance of PAWS sequence analysis software (Genomic Solutions) and ProteinInfo (Rockefeller University, NY; http://prowl.rockefeller.edu) to calculate theoretical molecular weights.
Western blots
Protein lysates were prepared from cells using radio-immunoprecipitation assay buffer (Sigma) supplemented with the Halt protease and phosphatase inhibitor cocktail (Pierce). Following denaturation, samples were resolved on Novex 4-12% Tris-Glycine mini gels (Life Technologies), electroblotted to PVDF membranes (Life Technologies) and blocked with bovine serum albumin (Sigma). Blots were hybridized with monoclonal antibodies against indicated proteins (p21: clone C-19 (Santa Cruz); CDK1: clone 5A6 (Thermo Scientific), CDK4: clone PA5-14445 (Thermo Scientific), CDK9: clone: PA5-19674 (Thermo Scientific), CDK2: clone 78B2 (Cell Signaling), CDK7: clone MO1 (Cell Signaling), Lamin B1: clone D4Q4Z (Cell Signaling), β-actin: AB8224 (Abcam)). For detection of phosphorylated HIV-1 RT, blots were hybridized with antibodies against phosphorylated HIV-1 RT, generated by vaccination of rabbits with the phosphorylated target peptide (LRWGFTpTPDKKHQKEPPF) according to standard procedures by a commercial manufacturer (ThermoFisher Scientific). Detection of non-phosphorylated HIV-1 RT was performed using a commercially available antibody (AB63911 (Abcam)). After washing, membranes were probed with Cy5-labeled secondary antibody (ECL Plex, GE Healthcare) for 1h for chemifluorescence detection by Typhoon 9410 (Amersham Biosciences), using excitation/emission wavelengths of 649/670. Alternatively, signals were visualized using secondary goat-anti-rabbit antibodies labeled with IRDye 800CW, followed by quantification of signals using Odyssey Image System (Li-Cor) and Image J software (NIH).
Co-immunoprecipitation assays
Protein lysates from CD4+ T cells were incubated in spin columns coated with anti-CDK2 antibody (clone 78B2 (Cell Signaling) for 12h at 4°C. After washing, protein complexes were eluted, boiled and subjected to SDS-PAGE followed by Western blotting using antibodies against HIV-1 RT (AB63911 (Abcam)), cyclin A (clone BF683 (Cell Signaling)) and cyclin T (clone 8744 (Cell Signaling)).
Determination of HIV-1 RT stability
Recombinant HIV-1 RT was phosphorylated by a CDK kinase assay, followed by limited proteolysis with subtilisin (Sigma). The 0.16 mg/ml RT samples were incubated with subtilisin protease (1 μg/ml) in 100 mM Tris pH 8.5 buffer at 30°C for 2, 30, 60, 120, 180 or 240 min (Serrano et al., 1984). For analysis, the collected samples were heated with SDS sample buffer at 90°C, alkylated with iodoacetamide, and separated by SDS gel electrophoresis. The gels were then fixed in 40% methanol and 10% glacial acetic acid for 1h, and stained with SYPRO® Ruby Protein Gel Stain (Life Technologies) under dark conditions overnight. After washed twice in 10% methanol and 7% glacial acetic acid for 1h, the stained gels were scanned on a Typhoon 9410 Imager, using a green 532 nm excitation laser and 610BP30 emission filter. Densitometry quantitation was performed using ImageQuant software (Molecular Dynamics).
Functional assessment of HIV-1 RT
After in vitro kinase assays, 0.25 units of RT and phospho-RT were analyzed with the EnzCheck RT assay (Life Technologies). The HIV-1 RTs were incubated with 0.125 μg/ml Oligo d(T)16 primer, 2.5 μg/ml poly(A) ribonucleotide template, and 5 μM dTTP at room temperature for 10, 20 and 30 minutes. The reaction was quenched by adding EDTA; the RT activity was measured using a fluorometric assay.
Statistical analysis
Data are expressed as mean and standard deviation/standard error, or as box and whisker plots indicating the median, the 25% and 75% percentile and the minimum and maximum of all data. Differences between different cohorts or different experimental conditions were tested for statistical significance using paired t test or unpaired t test, as appropriate. A p-value of 0.05 was considered significant.
Supplementary Material
Highlights.
Effective HIV-1 reverse transcription depends on host cyclin-dependent kinase (CDK)2
HIV-1 reverse transcriptase (RT) is a substrate for CDK2-dependent phosphorylation
CDK2-dependent phosphorylation increases RT stability and viral fitness
Upregulation of the CDK inhibitor p21 in controllers indirectly blocks HIV-1 RT
Acknowledgement
M. Lichterfeld is supported by the U.S. National Institutes of Health (AI093203, AI098487), by the Clinical Scientist Development Award from the Doris Duke Charitable Foundation (grant number 2009034), and by the American Foundation for AIDS Research (grant 108302-51-RGRL). X. G. Yu is supported by the U. S. National Institute of Health (AI089339, AI078799, AI098484). E. J. Chang is supported by the U. S. National Institutes of Health (SC2GM086343). The authors would like to acknowledge Pratikkumar Rathod and Abbas Nazir for technical assistance. Sample collection from elite controllers was supported by the Bill and Melinda Gates Foundation, the Mark and Lisa Schwartz Foundation, the Ragon Institute of MGH, MIT and Harvard and the International HIV Controller Consortium.
Footnotes
The authors declare that financial conflicts of interest do not exist.
Author contributions
Performance of experiments: JL (tissue culture, ex-vivo infection, PCR, western blots with cell lysates), HPH (mass spectrometry, protein biochemistry, in vitro kinase assays, RT stability assays) Data analysis and interpretation: JL, HPH, MJB, EJC, XGY, ML Writing of manuscript and preparation of Figures: JL, HPH, EJC, ML Contribution of PBMC samples: FP, BDW Study supervision: XGY, EJC, ML Research idea and study concept: EJC, ML
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdel-Mohsen M, Raposo RA, Deng X, Li M, Liegler T, Sinclair E, Salama MS, Ghanem HE, Hoh R, Wong JK, et al. Expression profile of host restriction factors in HIV-1 elite controllers. Retrovirology. 2013;10:106. doi: 10.1186/1742-4690-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allouch A, David A, Amie SM, Lahouassa H, Chartier L, Margottin-Goguet F, Barre-Sinoussi F, Kim B, Saez-Cirion A, Pancino G. p21-mediated RNR2 repression restricts HIV-1 replication in macrophages by inhibiting dNTP biosynthesis pathway. Proc Natl Acad Sci U S A. 2013;110:E3997–4006. doi: 10.1073/pnas.1306719110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammosova T, Berro R, Jerebtsova M, Jackson A, Charles S, Klase Z, Southerland W, Gordeuk VR, Kashanchi F, Nekhai S. Phosphorylation of HIV-1 at by CDK2 in HIV-1 transcription. Retrovirology. 2006;3:78. doi: 10.1186/1742-4690-3-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergamaschi A, David A, Le Rouzic E, Nisole S, Barre-Sinoussi F, Pancino G. The CDK inhibitor p21Cip1/WAF1 is induced by FcgammaR activation and restricts the replication of human immunodeficiency virus type 1 and related primate lentiviruses in human macrophages. J Virol. 2009;83:12253–12265. doi: 10.1128/JVI.01395-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- Breuer D, Kotelkin A, Ammosova T, Kumari N, Ivanov A, Ilatovskiy AV, Beullens M, Roane PR, Bollen M, Petukhov MG, et al. CDK2 regulates HIV-1 transcription by phosphorylation of CDK9 on serine 90. Retrovirology. 2012;9:94. doi: 10.1186/1742-4690-9-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain PP, Ren J, Nichols CE, Douglas L, Lennerstrand J, Larder BA, Stuart DI, Stammers DK. Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215. J Virol. 2002;76:10015–10019. doi: 10.1128/JVI.76.19.10015-10019.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Li C, Huang J, Cung T, Seiss K, Beamon J, Carrington MF, Porter LC, Burke PS, Yang Y, et al. CD4+ T cells from elite controllers resist HIV-1 infection by selective upregulation of p21. J Clin Invest. 2011;121:1549–1560. doi: 10.1172/JCI44539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciuffi A, Bleiber G, Munoz M, Martinez R, Loeuillet C, Rehr M, Fischer M, Gunthard HF, Oxenius A, Meylan P, et al. Entry and transcription as key determinants of differences in CD4 T-cell permissiveness to human immunodeficiency virus type 1 infection. J Virol. 2004;78:10747–10754. doi: 10.1128/JVI.78.19.10747-10754.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribier A, Descours B, Valadao AL, Laguette N, Benkirane M. Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep. 2013;3:1036–1043. doi: 10.1016/j.celrep.2013.03.017. [DOI] [PubMed] [Google Scholar]
- Deng L, Ammosova T, Pumfery A, Kashanchi F, Nekhai S. HIV-1 Tat interaction with RNA polymerase II C-terminal domain (CTD) and a dynamic association with CDK2 induce CTD phosphorylation and transcription from HIV-1 promoter. J Biol Chem. 2002;277:33922–33929. doi: 10.1074/jbc.M111349200. [DOI] [PubMed] [Google Scholar]
- Durand LO, Roizman B. Role of cdk9 in the optimization of expression of the genes regulated by ICP22 of herpes simplex virus 1. J Virol. 2008;82:10591–10599. doi: 10.1128/JVI.01242-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elahi S, Niki T, Hirashima M, Horton H. Galectin-9 binding to Tim-3 renders activated human CD4+ T cells less susceptible to HIV-1 infection. Blood. 2012a;119:4192–4204. doi: 10.1182/blood-2011-11-389585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elahi S, Niki T, Hirashima M, Horton H. Galectin-9 binding to Tim-3 renders activated human CD4+ T cells less susceptible to HIV-1 infection. Blood. 2012b doi: 10.1182/blood-2011-11-389585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emu B, Sinclair E, Hatano H, Ferre A, Shacklett B, Martin JN, McCune JM, Deeks SG. HLA class I-restricted T-cell responses may contribute to the control of human immunodeficiency virus infection, but such responses are not always necessary for long-term virus control. J Virol. 2008;82:5398–5407. doi: 10.1128/JVI.02176-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores O, Lee G, Kessler J, Miller M, Schlief W, Tomassini J, Hazuda D. Host-cell positive transcription elongation factor b kinase activity is essential and limiting for HIV type 1 replication. Proc Natl Acad Sci U S A. 1999;96:7208–7213. doi: 10.1073/pnas.96.13.7208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis AC, Di Primio C, Allouch A, Cereseto A. Role of phosphorylation in the nuclear biology of HIV-1. Curr Med Chem. 2011;18:2904–2912. doi: 10.2174/092986711796150478. [DOI] [PubMed] [Google Scholar]
- Harris RS, Hultquist JF, Evans DT. The restriction factors of human immunodeficiency virus. J Biol Chem. 2012;287:40875–40883. doi: 10.1074/jbc.R112.416925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager J, Smerdon SJ, Wang J, Boisvert DC, Steitz TA. Comparison of three different crystal forms shows HIV-1 reverse transcriptase displays an internal swivel motion. Structure. 1994;2:869–876. doi: 10.1016/s0969-2126(94)00087-5. [DOI] [PubMed] [Google Scholar]
- Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- Konig R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, Chiang CY, Tu BP, De Jesus PD, Lilley CE, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuny CV, Chinchilla K, Culbertson MR, Kalejta RF. Cyclin-dependent kinase-like function is shared by the beta- and gamma-subset of the conserved herpesvirus protein kinases. PLoS Pathog. 2010;6:e1001092. doi: 10.1371/journal.ppat.1001092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim MH, Bieniasz PD. HIV Restriction Factors and Mechanisms of Evasion. Cold Spring Harb Perspect Med. 2012;2:a006940. doi: 10.1101/cshperspect.a006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancebo HS, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Peng J, Blau C, Hazuda D, Price D, et al. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 1997;11:2633–2644. doi: 10.1101/gad.11.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manganaro L, Lusic M, Gutierrez MI, Cereseto A, Del Sal G, Giacca M. Concerted action of cellular JNK and Pin1 restricts HIV-1 genome integration to activated CD4+ T lymphocytes. Nat Med. 2010;16:329–333. doi: 10.1038/nm.2102. [DOI] [PubMed] [Google Scholar]
- Mbisa JL, Delviks-Frankenberry KA, Thomas JA, Gorelick RJ, Pathak VK. Real-time PCR analysis of HIV-1 replication post-entry events. Methods Mol Biol. 2009;485:55–72. doi: 10.1007/978-1-59745-170-3_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA. Cellular substrates of p34(cdc2) and its companion cyclin-dependent kinases. Trends Cell Biol. 1993;3:296–301. doi: 10.1016/0962-8924(93)90011-o. [DOI] [PubMed] [Google Scholar]
- Nishi H, Hashimoto K, Panchenko AR. Phosphorylation in protein-protein binding: effect on stability and function. Structure. 2011;19:1807–1815. doi: 10.1016/j.str.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, Walker BD, Ripke S, Brumme CJ, Pulit SL, Carrington M.w,, et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotger M, Bayard C, Taffe P, Martinez R, Cavassini M, Bernasconi E, Battegay M, Hirschel B, Furrer H, Witteck A, et al. Contribution of genome-wide significant single-nucleotide polymorphisms and antiretroviral therapy to dyslipidemia in HIV-infected individuals: a longitudinal study. Circ Cardiovasc Genet. 2009;2:621–628. doi: 10.1161/CIRCGENETICS.109.874412. [DOI] [PubMed] [Google Scholar]
- Saez-Cirion A, Bacchus C, Hocqueloux L, Avettand-Fenoel V, Girault I, Lecuroux C, Potard V, Versmisse P, Melard A, Prazuck T, et al. Post-treatment HIV-1 controllers with a long-term virological remission after the interruption of early initiated antiretroviral therapy ANRS VISCONTI Study. PLoS Pathog. 2013;9:e1003211. doi: 10.1371/journal.ppat.1003211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Cirion A, Hamimi C, Bergamaschi A, David A, Versmisse P, Melard A, Boufassa F, Barre-Sinoussi F, Lambotte O, Rouzioux C, et al. Restriction of HIV-1 replication in macrophages and CD4+ T cells from HIV controllers. Blood. 2011;118:955–964. doi: 10.1182/blood-2010-12-327106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Cirion A, Pancino G. HIV controllers: a genetically determined or inducible phenotype? Immunol Rev. 2013;254:281–294. doi: 10.1111/imr.12076. [DOI] [PubMed] [Google Scholar]
- Schwartz O, Marechal V, Friguet B, Arenzana-Seisdedos F, Heard JM. Antiviral activity of the proteasome on incoming human immunodeficiency virus type 1. J Virol. 1998;72:3845–3850. doi: 10.1128/jvi.72.5.3845-3850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano L, Avila J, Maccioni RB. Controlled proteolysis of tubulin by subtilisin: localization of the site for MAP2 interaction. Biochemistry. 1984;23:4675–4681. doi: 10.1021/bi00315a024. [DOI] [PubMed] [Google Scholar]
- Tu X, Das K, Han Q, Bauman JD, Clark AD, Jr., Hou X, Frenkel YV, Gaffney BL, Jones RA, Boyer PL, et al. Structural basis of HIV-1 resistance to AZT by excision. Nat Struct Mol Biol. 2010;17:1202–1209. doi: 10.1038/nsmb.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneault F, Woods M, Buzon MJ, Li C, Pereyra F, Crosby SD, Rychert J, Church G, Martinez-Picado J, Rosenberg ES, et al. Transcriptional profiling of CD4 T cells identifies distinct subgroups of HIV-1 elite controllers. J Virol. 2011;85:3015–3019. doi: 10.1128/JVI.01846-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BD, Yu XG. Unravelling the mechanisms of durable control of HIV-1. Nat Rev Immunol. 2013;13:487–498. doi: 10.1038/nri3478. [DOI] [PubMed] [Google Scholar]
- Wang D, de la Fuente C, Deng L, Wang L, Zilberman I, Eadie C, Healey M, Stein D, Denny T, Harrison LE, et al. Inhibition of human immunodeficiency virus type 1 transcription by chemical cyclin-dependent kinase inhibitors. J Virol. 2001;75:7266–7279. doi: 10.1128/JVI.75.16.7266-7279.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TE, Brandariz-Nunez A, Valle-Casuso JC, Amie S, Nguyen LA, Kim B, Tuzova M, Diaz-Griffero F. The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe. 2013;13:441–451. doi: 10.1016/j.chom.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Scadden DT, Crumpacker CS. Primitive hematopoietic cells resist HIV-1 infection via p21. J Clin Invest. 2007;117:473–481. doi: 10.1172/JCI28971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC, Stec E, Ferrer M, Strulovici B, Hazuda DJ, et al. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 2008;4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Zydek M, Hagemeier C, Wiebusch L. Cyclin-dependent kinase activity controls the onset of the HCMV lytic cycle. PLoS Pathog. 2010;6:e1001096. doi: 10.1371/journal.ppat.1001096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.