Abstract
Maternal homozygosity for three independent mutant hecate alleles results in embryos with reduced expression of dorsal organizer genes and defects in the formation of dorsoanterior structures. A positional cloning approach identified all hecate mutations as stop codons affecting the same gene, revealing that hecate encodes the Glutamate receptor interacting protein 2a (Grip2a), a protein containing multiple PDZ domains known to interact with membrane-associated factors including components of the Wnt signaling pathway. We find that grip2a mRNA is localized to the vegetal pole of the oocyte and early embryo, and that during egg activation this mRNA shifts to an off-center vegetal position corresponding to the previously proposed teleost cortical rotation. hecate mutants show defects in the alignment and bundling of microtubules at the vegetal cortex, which result in defects in the asymmetric movement of wnt8a mRNA as well as anchoring of the kinesin-associated cargo adaptor Syntabulin. We also find that, although short-range shifts in vegetal signals are affected in hecate mutant embryos, these mutants exhibit normal long-range, animally directed translocation of cortically injected dorsal beads that occurs in lateral regions of the yolk cortex. Furthermore, we show that such animally-directed movement along the lateral cortex is not restricted to a single arc corresponding to the prospective dorsal region, but occur in multiple meridional arcs even in opposite regions of the embryo. Together, our results reveal a role for Grip2a function in the reorganization and bundling of microtubules at the vegetal cortex to mediate a symmetry-breaking short-range shift corresponding to the teleost cortical rotation. The slight asymmetry achieved by this directed process is subsequently amplified by a general cortical animally-directed transport mechanism that is neither dependent on hecate function nor restricted to the prospective dorsal axis.
Author Summary
One of the earliest and most crucial events in animal development is the establishment of the embryonic dorsal axis. In amphibians and fish, this event depends on the transport of so-called “dorsal determinants” from one region of the egg, at the pole opposite from the site where the oocyte nucleus lies, towards the site of axis induction. There, the dorsal determinant activates the Wnt signaling pathway, which in turn triggers dorsal gene expression. Dorsal determinant transport is mediated by the reorganization of a cellular network composed of microtubules. We determine that hecate, a zebrafish gene active during egg formation that is essential for embryonic axis induction, is required for an early step in this microtubule reorganization. We find that hecate corresponds to glutamate receptor interacting protein 2a, which participates in other animal systems in Wnt-based pathways. We also show that the microtubule reorganization dependent on hecate results in a subtle symmetry-breaking event that subsequently becomes amplified by a more general transport process independent of hecate function. Our data reveal new links between glutamate receptor interacting protein 2a, Wnt signaling and axis induction, and highlights basic mechanisms by which small changes early in development translate into global changes in the embryo.
Introduction
Dorsoventral axis formation is a fundamental process in early vertebrate embryogenesis. In many vertebrates such as amphibians and teleosts, evidence indicates that maternally-supplied dorsal determinants trigger the formation of the future dorsal organizer. Embryological manipulations have indicated that the dorsal determinants are initially localized to the vegetal pole and subsequently translocate animally to the prospective dorsal side in a microtubule-dependent process in both amphibians (reviewed in [1]) and teleosts [2]–[4]. In amphibians, translocation of the signal from the vegetal pole to the dorsal side is initiated by cortical rotation, the microtubule-dependent movement of the egg cortex with respect to its core that is triggered by fertilization and is implemented by transport along microtubule tracks (reviewed in [1]).
Although not readily apparent in the zebrafish embryo, detailed analysis has indicated the existence of processes that share similarities with the amphibian cortical rotation. Early studies showed that fluorescent polystyrene beads injected at the vegetal pole were transported animally along microtubule-based cortical tracks in a microtubule dependent manner [2], and that this movement had temporal dynamics and functional requirements similar to that of the movement of putative dorsal determinants as defined by embryological manipulations [3], [4]. Subsequent work showed that maternal factors such as Syntabulin and Wnt8a, involved in axis induction, localized to the vegetal pole of the egg and upon fertilization and egg activation shifted animally towards the presumed prospective dorsal region of the embryo [5]–[7]. Further studies of microtubule rearrangements in live embryos confirmed that the tracks of bundled microtubules that form at the zebrafish vegetal pole upon egg activation become aligned in the direction of the future dorsal side of the embryo, and showed bulk cortical particle movement analogous to a cortical rotation [8].
The movement of the dorsal determinant results in the activation of the Wnt/βcatenin signaling pathway and the activation of β-catenin-dependent targets [1]. This well-known pathway is characterized by the activation of Frizzled receptors by the Wnt ligand, and an intracellular cascade involving the activation of Dishevelled and the downregulation of a β-catenin degradation complex that includes GSK3, Axin and Adenomatous polyposis coli (APC), leading to the accumulation of β-catenin in the nucleus [9]. Nuclear β-catenin in turn interacts with transcription factors of the Tcf family to activate transcription of target genes. Wnt/βcatenin pathway components and/or nuclear accumulation of β-catenin have been shown to be involved in embryonic axis determination in diverse deuterostomes such as fish, amphibians, mammals and amphioxus, as well as in lineages as basal as echinoderms, Cnidarians and planaria (reviewed in [10]) implying that the pathway was recruited for axis determination very early in animal evolution.
Although the involvement of Wnt/βcatenin activation across species is well documented, the identity of the molecules that activate the pathway in the early embryo, often referred to as dorsal determinants, remains unknown in most cases. In Xenopus, wnt11 mRNA is first located at the vegetal pole and becomes enriched at the future dorsal side after fertilization, and depletion of wnt11 mRNA results in embryos defective in dorsal axis induction [11]. Thus, Wnt11, together with ubiquitously present Wnt5 [12], [13] has been proposed to be the dorsal determinant in this amphibian species. Studies in the zebrafish exclude a function for Wnt11 or Wnt 5 in axis induction but suggest a role for Wnt8a in this process [7]. Maternal zebrafish wnt8a mRNA is localized during oogenesis to the vegetal pole of the oocyte and, upon fertilization, wnt8a mRNA experiences a shift from its original location at the vegetal pole to an off-center region thought to correspond to the dorsal side [7]. These studies suggest that, while Wnt/β-catenin pathway activation may be highly conserved in axis induction across the animal kingdom, maternally-based mechanisms that lead to the activation of the pathway vary.
Efforts from several laboratories have used forward genetics approaches to identify maternal factors essential for various aspects of early embryonic development in the zebrafish [5], [14]–[18]. Several reports have documented maternal-effect mutations affecting zebrafish dorsal axis induction [5], [15], [17], [18]. Mutations in three maternal-effect genes, ichabod, tokkaebi and hecate, cause specific ventralized phenotypes, consistent with a role for these genes in axis induction. Overexpression of Wnt signaling pathway components in ichabod mutant embryos indicate that this mutation acts within the Wnt/β-catenin signaling pathway at a level downstream of the β-catenin degradation complex [15]. These and other results show that ichabod corresponds to a β-catenin-2 gene expressed maternally and involved in axis induction [19].
Similar overexpression analysis has shown that tokkaebi and hecate, in contrast to ichabod, act upstream of the β-catenin-degradation machinery [5], [20]. Specifically, overexpression of components that activate the pathway at multiple points can rescue the hecate and tokkaebi mutant phenotypes. Rescue by an exogenous Wnt ligand suggests that the Wnt/β-catenin pathway is intact in tokkaebi and hecate mutant embryos, and suggests that, rather than being required for an integral component of Wnt/β-catenin signaling, these genes are required to regulate an endogenous signal that activates this pathway. Consistent with such a proposed role, positional cloning reveals that tokkaebi corresponds to the syntabulin (sybu) gene, which codes for a linker of the kinesin I motor protein involved in cargo transport along microtubules [6]. Both sybu mRNA and protein are localized to the vegetal pole of the egg, and Sybu protein exhibits a slight off-center shift upon egg activation [6]. These data suggest a role for Sybu in the microtubule-dependent transport of vegetally localized dorsal determinants.
Here, we present the molecular characterization of the zebrafish maternal-effect gene hecate (hec). Maternal homozygosity for three independent mutant hec alleles results in embryos with reduced expression of dorsal organizer genes and defects in the formation of dorsoanterior structures ([20]; this report). Positional cloning reveals that hec encodes the Glutamate receptor interacting protein 2a (Grip2a). We find that grip2a mRNA, like wnt8a mRNA and Sybu protein, is localized to the vegetal pole of the oocyte and early embryo, where during egg activation it is shifted off-center corresponding to the previously proposed teleost cortical rotation [8]. The Drosophila Grip homologue has recently been shown to potentiate Wnt signaling at the neuromuscular junction by interacting with the Frizzled receptor on the cytoplasmic side of endocytosing membrane vesicles [21], [22], suggesting a potential mechanism of action for zebrafish Grip2a in axis induction at the level of Frizzled receptor regulation. Unexpectedly, however, we find that hec mutants show defects in the alignment and bundling of microtubules at the vegetal cortex, which result in corresponding defects in the asymmetric movement of wnt8a mRNA and are sufficient to explain the observed axis induction defects.
The short-range shift in vegetally localized factors such as grip2a mRNA also led us to re-examine the functional significance of the previously observed animally-directed cortical transport on axis induction. We find that, although short-range shifts in vegetal signals are affected in hec mutant embryos, these mutants do not exhibit a defect in the long-range, animally directed translocation of cortically injected dorsal beads that occurs in lateral regions of the yolk cortex. Furthermore, we show that, contrary to our expectations, such movements are not restricted to a single arc corresponding to the prospective dorsal region, but occur in multiple meridional arcs even in opposite regions of the embryo. Together, our results propose a role for hec function in the reorganization and bundling of microtubules at the vegetal cortex to mediate a symmetry-breaking event likely corresponding to the teleost cortical rotation. This asymmetry is subsequently amplified by a cortical animally-directed transport mechanism that is neither dependent on hec function nor restricted to the prospective dorsal axis.
Results
Maternal-effect mutations in hecate affect dorsoanterior development
Embryos from mothers homozygous for the mutant hec allele, for simplicity referred to here as hec mutant embryos, display a range of ventralized phenotypes [17], [20], [23] (Figure 1A–E; Table 1). In the most severe cases, 24-hour mutant embryos exhibit a severe radial ventralization and lack all dorsoanterior-derived structures (V4 class, according to criteria in [24]; Figure 1A). More moderate phenotypes can also be observed, such as embryos that lack all anterior structures as well as the notochord (a dorsally-derived structure), and display an expansion of posterior somites (V3 class; Figure 1B). More weakly ventralized embryos are also observed that lack the anterior-most head structures and the notochord and exhibit expanded posterior somites (V2 class; Figure 1C), and embryos with reduced eyes and some notochord defects (class V1; Figure 1D). In many clutches, a fraction of embryos from mutant mothers are indistinguishable from wild-type embryos (Figure 1E). Such variability in phenotypes can be observed in maternal-effect mutants (some examples can be found in [17], [25], [26]), in particular those exhibiting axis induction defects [5], [15], [27], possibly by inherent variation in the maternal composition of individual eggs coupled to gene or pathway redundancy (see also Discussion). In mutant clutches with a weak expression of the phenotype, a small and variable fraction of embryos exhibits axis duplication phenotypes (Figure 1F) instead of axis formation defects (see Discussion).
Figure 1. Axis induction defects in embryos from mothers homozygous for three different hecate mutant alleles.
A–F) Side views (except as indicated in (F)) of live embryos at 24 hpf showing a range of phenotypes from most severe (A) to normal (E) and double-axis embryos (F). A) Class V4 embryo exhibiting complete radial symmetry and lacking all anterior (e.g. head structures) or dorsalmost (e.g. notochord) structures. B) Class V3 embryo exhibiting a rudimentary axis but lacking all anterior and dorsalmost structures. C) Class V2 embryo lacking anteriormost structures such as the eyes, but showing formation of more posterior head structures such as the otic vesicles (black arrowhead, also indicated in D). D) Class V1 embryo with a relatively complete axis but reduced anterior structures, such as the eyes (white arrowhead, also labeled in E), and lacking a properly formed notochord. E) Normal embryo derived from hec mutant females indistinguishable from wild-type. Note somites in (E) are chevron-shaped, while they are blocky in (B–D) indicative of defects in notochord formation, or encircling the entire embryo in (A). F) A double-axis embryo found in hec mutant clutches exhibiting weak expressivity. Insert in the lower left shows the left axis indicated by the dashed rectangle, out of focus in the main image. Insert in the upper left panel shows a dorsal view, showing the bifurcated axis. Note the lack of defined anterior structures in both axes, as well as the lack of a notochord along the trunk, also reflected by block-shaped somites in this region. Images in A–F are side views, except for upper left insert in (F). G–I) Animal views of live wild-type embryos (G) and embryos from females homozygous for each of the three hec mutant alleles. The dorsal thickening or shield (arrow) is absent in mutant embryos. K–V) In situ hybridization analysis to detect expression of dorsally-expressed genes (gsc, chd) and ventrally-expressed genes (eve 1). The expression domains of gsc and cho is reduced in hec mutant embryos, while the expression domain of eve is expanded in these embryos. (K–V) are animal view of embryos, dorsal to the right when identifiable, at the shield stage (6 hpf). Magnification bars in (F) and (V) correspond to 100 µm for panels sets (A–F) and (G–V), respectively. Dorsal view insert in panel (F, upper left) has been reduced to 75% size.
Table 1. Phenotypic strength of hecate alleles.
| Genotype | V41 (%) | V32 (%) | V23 (%) | V14 (%) | Double axis5 (%) | Normal (%) | n |
| WT6 | 0.2 | 0.2 | 0.0 | 0.1 | 0.0 | 99.5 | 1259 |
| hecp06ucal | 76.3 | 7.7 | 6.5 | 8.7 | 0.0 | 0.9 | 1812 |
| hect2800 | 46.7 | 13.1 | 9.5 | 15.0 | 0.3 | 15.4 | 655 |
| hecp08ajug | 10.9 | 18.9 | 15.4 | 28.8 | 3.1 | 22.8 | 1516 |
Embryos from females homozygous for hec mutant alleles were scored at 24 hpf for ventralization phenotypes according to [24]. Results were pooled from clutches from 15 different females for each allele.
Severe radial ventralization and lack of all dorsoanterior structures.
Absence of anterior structures, including posterior head structures such as the otoliths, lack of notochord, and expanded posterior somites.
Absence of anterior head structures including the eyes, lack of notochord, and expanded posterior somites.
Reduced anterior head structures such as the eyes but otherwise normal.
In weak mutant clutches, a small fraction of embryos exhibit a duplicated axes, each of which often exhibits deletions of the anterior-most structures.
Clutches from wild-type female can produce rare ventralized embryos.
The hect2800 allele was originally isolated in an early-pressure-based screen for recessive maternal-effect mutations [14], [17] and its effects described previously [20], [23]. Two additional alleles, hecp08ajug and hecp06ucal were identified in another maternal-effect mutant screen, based on a four-generation scheme [16], [18]. Using DNA markers linked to the hect2800 allele [20], we determined that the hecp08ajug and hecp06ucal alleles were linked to the same SSLP markers, z59658 and z24511 on chromosome 8. In addition, we carried out pair-wise crosses between individuals carrying the three mutant alleles to test for non-complementation. All crosses resulted in females that exhibited the hec mutant phenotype in their offspring in the expected proportions, i.e. Mendelian for F1 females (approximately 50% in crosses between homozygous males and heterozygous females of all allelic combinations) and maternal-dependent (near 100%) for F2 embryos (Table S1), indicating that all three mutations are part of the same complementation group.
A comparison of the phenotypes for the three hec alleles suggests that they fall within an allelic series. Embryos from mutant females carrying each of the three alleles were classified and scored at 24 hours post-fertilization (hpf; Table 1). The hecp06ucal allele shows the strongest average phenotype among the three alleles, where most embryos (76.3% from 3-month females) exhibit the strongest (V4) phenotype, while hect2800 mutants exhibit intermediate phenotype (46.7% V4 class) and hecp08ajug mutants show the weakest phenotype (10.9% V4 class). Double-axis embryos are observed only in offspring derived from females mutant for the two weaker alleles hecp08ajug and hect2800, and only in clutches with weak penetrance and expressivity (Table 1 and data not shown). In the case of ichabod and tokkaebi mutations [5], [15] axis induction defects have been reported to vary with maternal age, with younger females exhibiting stronger phenotypes. We tested embryos from 3 month old and 12 month old mutant females and find a similar trend for the effects of the three hec alleles on axis induction (Figure S1).
Previous studies have shown that the ventralized phenotype of hec mutant embryos is associated with a reduction in the embryonic shield and changes in patterns of gene expression in dorsal- and ventral-specific genes in the late blastula embryo. Expression of gene markers of the various germ layers, such as bmp2 and gata2 (ectoderm), no tail (mesendodermal precursors), and foxa2 (endoderm) was unaffected, other than regional differences due to predicted changes in dorsoventral specification [20]. As expected, embryos from females mutant for the newly isolated hec alleles, hecp06ucal and hecp08ajug, similar to the hect2800 allele [20], exhibit a reduction in the shield region corresponding to the dorsal organizer at the incipient dorsal region (Figure 1G–J), as well as a reduction in dorsal-specific gene expression (goosecoid, Figure 1K–N; chordin, Figure 1O–R) and a concomitant expansion of a ventrally-expressed gene (eve 1) (Figure 1S–V).
Together, phenotypic, linkage and complementation analysis indicate that these three mutations are alleles of the same gene.
hecate encodes the zebrafish Glutamate receptor interacting protein 2a (Grip2a)
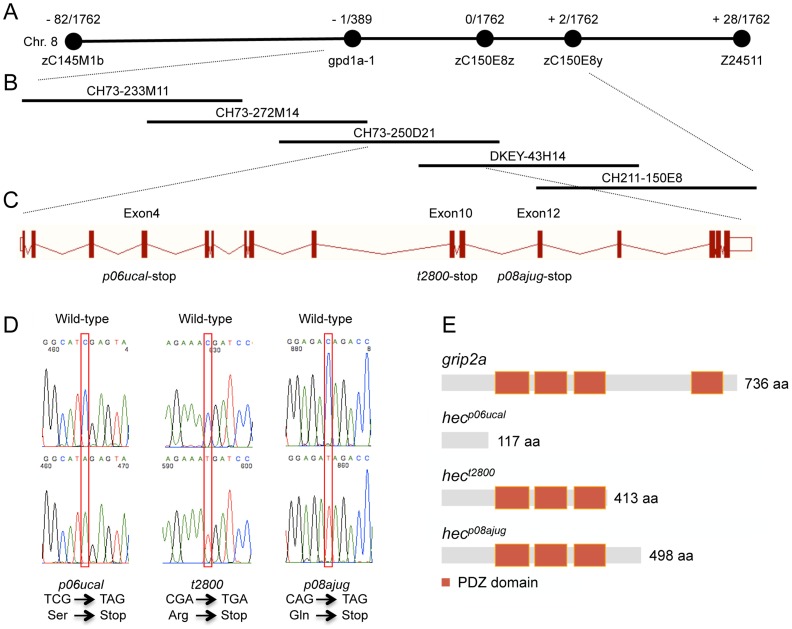
To better understand the function of the hec gene, we determined its molecular identity using a positional cloning approach. We initially identified linkage of the hec locus between SSLP markers z59658 and z24511 on chromosome 8 through mapping of the hect2800 allele [20]. Homozygous mutant males were crossed to heterozygous females to generate large numbers of fish for fine mapping. Fine mapping analysis of 1762 meioses with newly identified RFLP markers further narrowed the critical region containing hec to a genomic region between gpd1a-1 and the zC150E8y RFLP in the Ensembl database, corresponding to an interval of 383 Kb (Figure 2A). Five overlapping BAC clones were identified and aligned as a contig covering the whole critical region (Figure 2B). Within this critical region, there are 11 predicted genes according to the Ensembl database of the zebrafish genome and the GENSCAN program. Sequencing of cDNA products from wild-type and mutant alleles revealed the presence of mutations in all three hec alleles in the gene glutamate receptor interacting protein 2a (grip2a, NP_001116760.1), one of two grip2 genes in the zebrafish genome [28].
Figure 2. Molecular identification of the hecate locus.
A) Linkage map of the hec locus. The number of recombinants over the total number of analyzed meiosis is indicated. hec linkage was initially identified between SSLP markers z59658 and z24511 on chromosome 8. Fine mapping analysis with newly identified RFLP markers further narrowed the region between the gene gpd1a-1 and the RFLP zC150E8y. B) Contig of five BAC clones covering the hec critical region. CH73-233M11, CH73-272M14, CH73-250D21, DKEY-43H14 and CH211-150E8 are five sequenced and overlapping BAC clones in this interval. C) Exon-intron structure of the hec/grip2a gene, which contains 16 exons. The hecp06ucal, hect2800 and hecp08ajug alleles each cause a premature stop-codon in exon 4, exon 10 and exon 12, respectively. D) Sequence traces of the cDNA products from wild-type and the three mutant hec alleles. Nucleotide substitutions are indicated by the red box. Mutant cDNAs show a C-A transversion in codon 118 (hecp06ucal), a C-T transversion in codon 414 (hect2800), or a C-T transversion in codon 499 (hecp08ajug), all creating premature STOP codons. E) Schematic diagram showing the protein domain structures of Grip2a in the wild-type and mutant alleles. Red boxes represent conserved PDZ domains.
The zebrafish grip2a gene has 16 exons, which produces a 3,124 bp transcript and a 736 amino acid protein (Figure 2C). The three mutant alleles result in truncated forms of the Grip2a protein: hecp06ucal allele has a C-A transversion in codon 118 in exon 4, hect2800 has a C-T transversion in codon 414 in exon 10, and hecp08ajug has a C-T transversion in codon 499 in exon 12, and these three mutations generate premature nonsense (stop) codons (Figure 2D). A search in the Conserved Domain Database (CDD) in NCBI [29] indicates that Grip2a protein has four PDZ domains. The premature stop-codons for these different alleles delete the most C-terminal PDZ domain in the hect2800 and hecp08ajug alleles, and all four PDZ domains in the hecp06ucal allele (Figure 2E). The retention of PDZ domains and size of the predicted truncated proteins roughly correlates with the observed phenotypic strength in the various mutant allele backgrounds (Table 1, Figure S1), although we have not determined expression levels for the mutant proteins to confirm their relative activities. The identification of mutations in these three independently isolated alleles indicates that hec encodes Grip2a. This is further substantiated by the localization of grip2a mRNA in the region of the embryo affected by the hec mutation (see below).
Using BLAST searches on Ensembl and NCBI genome databases, homologous grip1 and grip2 genes were found for all vertebrate species, such as fish, amphibians, birds, and mammals. In invertebrate lineages, a distantly related Grip gene was identified only in Drosophila. Grip1 and Grip2 protein sequences among eight representative species were used to construct a phylogenetic tree using ClustalW (Figure S2). grip2 occurs as a single copy in amphibians, birds and mammals but is duplicated in the zebrafish and other fish species such as fugu and medaka, likely a consequence of an extra round of whole genome duplication in the ray-finned fish lineage [30], [31]. Drosophila and all vertebrate Grip1 and Grip2 proteins contain PDZ domains but zebrafish Hec/Grip2a and Fugu Grip2b contain 4 predicted PDZ domains instead of the 7 PDZ domains predicted in other members of this family (Figure S2 and not shown).
grip2a mRNA is localized to the vegetal region of the oocyte, developing an early asymmetry upon egg activation
Quantitative RT-PCR analysis of mRNA from wild-type embryos spanning early development indicates highest levels of grip2a mRNA in the 1-cell stage embryo, gradually declining to negligible levels at 50% epiboly (5.25 hpf) and thereafter (Figure S3). In adults, expression can be detected in wild-type females and isolated ovaries, but not in males or female carcasses where the ovaries have been removed (Figure S3). Thus, at our level of analysis, hec/grip2a is specifically expressed in ovaries as a maternal-specific transcript, which is consistent with the strict maternal effect observed in females homozygous for the three hec mutant alleles.
We examined the spatial expression pattern of hec/grip2a at various developmental stages during embryogenesis using whole mount in situ hybridization (Figure 3). grip2a mRNA is detected in the vegetal pole region of the yolk in early zygotes and cleavage-stage embryos (Figure 3A–E). Similar to the case of wnt8a mRNA [7] and Sybu protein [6], the grip2a mRNA localization domain is not precisely aligned with the vegetal pole in activated eggs or early embryos. Instead, grip2a mRNA is consistently located slightly off-center in the post-activation stages examined, from the early 1-cell stage embryo 10 minutes post-fertilization (mpf) until late cleavage stages (Figure 3A–D). This off-center shift is not observed in manually extruded mature, inactive eggs, where the grip2a mRNA localization domain is instead located at the vegetal pole in a radially symmetric manner (data not shown). In early embryos the extent of asymmetry of the grip2a mRNA localization domain, appears similar throughout the cleavage and blastula stages until mRNA levels become markedly reduced in the late blastula embryo (sphere stage; 4 hpf; Figure 3E). Localized grip2a mRNA can no longer be detected starting at the onset of epiboly (30% epiboly; 4.66 hpf; not shown). In contrast to its Xenopus homologue [32], zebrafish grip2a mRNA does not localize to the zebrafish germ plasm (Figure 3C and data not shown), present at the furrows corresponding to the first and second blastomeric divisions [33]–[37], nor does it become incorporated into the primordial germ cells (Figure 3D,E and data not shown), which form four cell clusters during the late cleavage stages ([33], [35], [38]; see below).
Figure 3. Whole mount in situ hybridization analysis of the expression of zebrafish grip2a mRNA in early embryos.
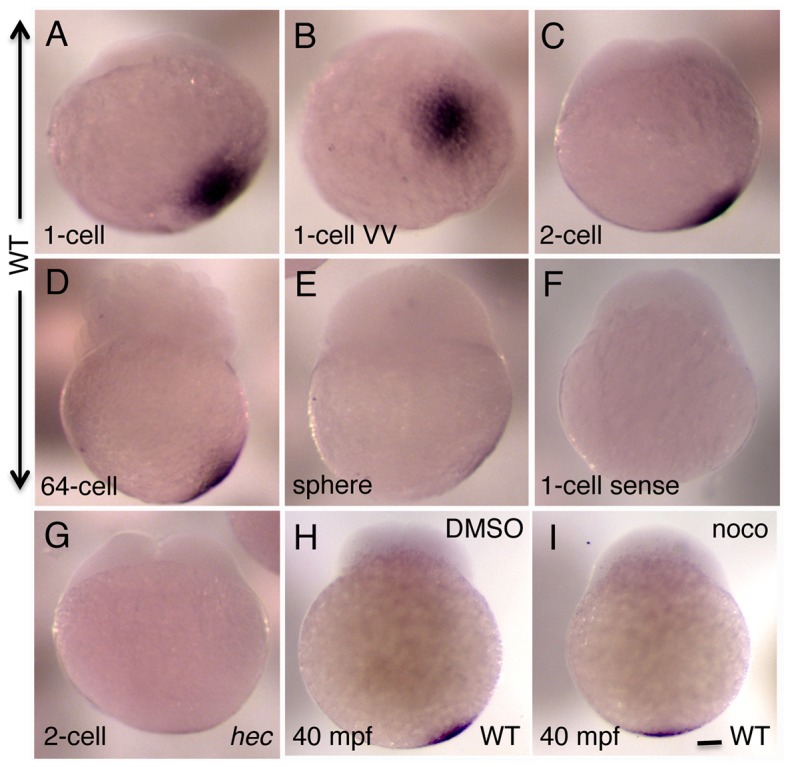
A–E) grip2a mRNA is localized in a slightly off-center position at the vegetal pole of early embryos, and remains in that position until the sphere stage when it becomes undetectable. F) Control sense probe. G) The domain of grip2a mRNA localization in hec mutants is reduced in intensity and appears to lack an off-center shift. H,I) Representative control (DMSO)- and nocodazole-treated embryos showing the lack of off-center shift in nocodazole-treated embryos (see Figure S4 for details). All panels are side views (except for (B), which is a vegetal view of the embryo in (A)) at the following stages: A,B: 1-cell (20 mpf), C,F,G: 2-cell (45 mpf), D: 64-cell (2 hpf), E: sphere (4 hpf). (H,I) are at 40 mpf, which approximately corresponds to the 2-cell stage (C) in untreated embryos. Magnification bar in (I) corresponds to 100 µm for all panels.
To further confirm the slight, off-center shift in the grip2a mRNA localization domain, and to determine whether this shift, like that of Sybu protein and wnt8a mRNA, depends on an intact microtubule network, we tested the effect of early nocodazole treatment on the grip2a mRNA localization pattern in wild-type embryos. Embryos were treated at 5 mpf and fixed at 30 and 40 mpf for in situ hybridization. For both time points, control (solvent-treated) embryos show an off-center shift in the domain of grip2a mRNA localization so that it is located within an arc at 0–20° from the true vegetal pole of the embryo, while nocodazole-treated embryos do not exhibit a discernable shift in mRNA localization (Figure 3H, 3I; Figure S4). Thus, grip2a mRNA is located at the vegetal pole of the embryo in mature oocytes, but upon egg activation (typically coupled to fertilization) this mRNA exhibits a short-range, off-center translocation within the vegetal region of the embryo. Once in an off-center position grip2a mRNA remains static until its degradation at late cleavage stages.
Whole mount in situ hybridization of embryos from mutant female mothers homozygous for each of the three hec alleles show significantly reduced levels of localized grip2a mRNA during the early cleavage stages, ranging from reduced to nearly undetectable levels (Figure 3G and data not shown). In those embryos where grip2a mRNA localization can be discerned, the domain of localization appears centered at the vegetal pole, consistent with the absence of an off-center shift for vegetally localized products in hec mutants (see below). Quantitation of total grip2a mRNA levels in mutant embryos at the 2-cell stage indicates that, for all alleles, grip2a mRNA abundance is drastically reduced to approximately 15–25% that in wild-type embryos (Figure S3). It is possible that hec/grip2a function is required for the localization of its own mRNA, which when not localized is unstable. Alternatively, the reduction in apparent grip2a mRNA localization in hec mutant embryos might be a consequence of a decrease in grip2a mRNA abundance in these embryos, possibly by non-sense mediated mRNA decay as has been proposed for other maternal transcripts [39], [40].
Vegetal localization of grip2a mRNA is initiated during oogenesis and depends on oocyte polarity genes
To visualize how the pattern of grip2a mRNA localization is established during oogenesis, we carried out in situ hybridization analysis of wild-type oocytes at various stages using a grip2a antisense probe (Figure 4). In early stage I oocytes, grip2a transcripts appear sharply localized to a compact, spherical region at one region of the oocyte (Figure 4A, 4D). By late stage I, grip2a mRNA acquires a more spread, subcortical localization pattern still centered in one region of the oocyte (Figure 4A, 4E). This cortical localization is maintained until the end of oogenesis (Figure 4A, 4F and data not shown).
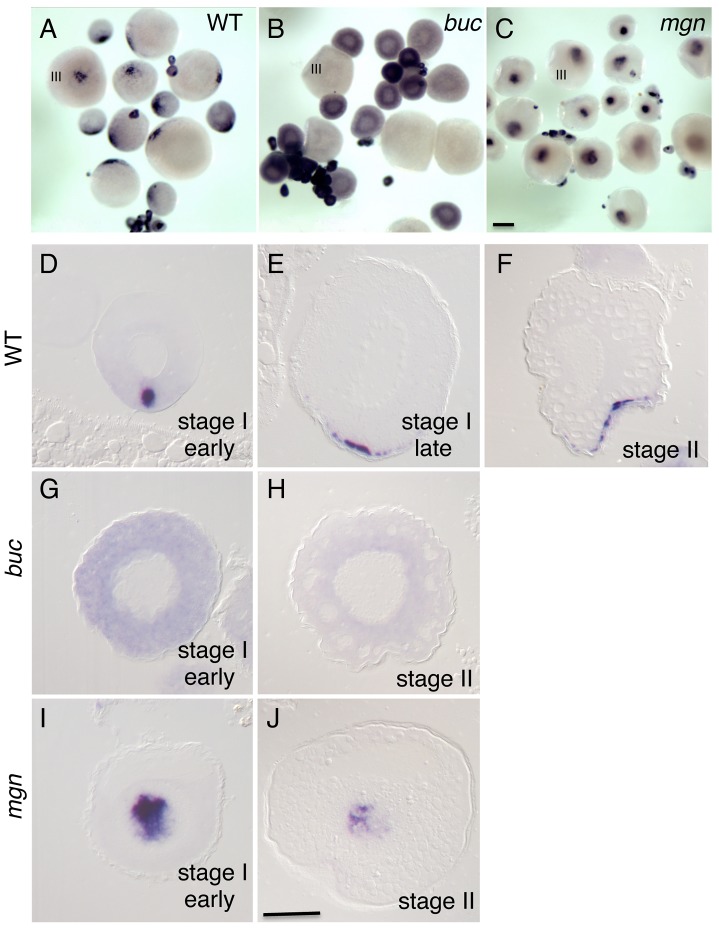
Figure 4. Localization of grip2a mRNA in wild-type and mutant oocytes.
A–C) Whole mount in situ hybridization of dissected ovaries from wild-type (A), bucky ball (buc, B) and magellan (mgn, C) mutant females. In (A–C) oocytes at stage III of development are indicated. Smaller oocytes are at stages I and II, which are difficult to differentiate in whole mounts at this magnification. The grip2a mRNA localization domain is observed in an asymmetric cortical position in wild-type oocytes (A) but is unlocalized and diffuse in buc oocytes (B) and internally-located in mgn mutants (C). D–J) Sections of wild-type and mutant oocytes at the indicated stages after labeling to detect grip2a mRNA. Stages are as indicated in the panel and were determined by size and oocyte morphology according to [101]. D–F) Wild-type oocytes showing localization to the presumptive Balbiani Body (D) and subsequent localization to a cortical domain of the oocyte corresponding to the presumptive vegetal pole (E–F). G,H) buc mutant oocytes lack the Balbiani body [39], [43] and the grip2a mRNA subcellular localization domain in stage I and II oocytes. I) mgn mutant stage I oocytes exhibit an enlarged Balbiani body [40] and displayed an enlarged grip2a mRNA localization domain. J) Stage II mgn mutant oocytes fail to localize transcripts to the vegetal pole which instead persist in an internal domain [40], as observed also for grip2a mRNA. Number of oocytes examined were as follows: wild-type: early stage I: 13, stage II: 12; buc: early stage I: 35, stage II: 26; mgn: early stage I: 23, stage II: 18. Magnification bar in (C) corresponds to 250 µm for panels (A–C), and in (J) to 50 µm for panels (D–J).
The localization pattern of grip2a mRNA in stage I oocytes is reminiscent of the zebrafish Balbiani body, a conserved aggregate of organelles present in animal oocytes shown to anchor subcellularly localized oocyte mRNAs [41]–[43]. We therefore tested whether grip2a mRNA localization is dependent on Balbiani body formation, using zebrafish mutants that affect this structure. Oocytes mutant for the gene bucky ball lack the Balbiani body [39], [43], and we found that bucky ball mutant oocytes lack grip2a mRNA subcellular localization during oogenesis (Figure 4B, 4G, 4H). Moreover, oocytes mutant for the cytoskeletal linker protein magellan (macf1), which exhibit an enlarged Balbiani body with an abnormal location [40], exhibit grip2a mRNA mislocalization (Figure 4C, 4I, 4J) similar to other Balbiani-localized transcripts. These results indicate that grip2a mRNA becomes localized to the vegetal cortex during oogenesis by a Balbiani body-dependent mechanism.
hecate/grip2a is required for microtubule rearrangements at the vegetal pole
The rescue of hec mutant embryos by overexpression of Wnt pathway components has suggested that hec likely activates signaling at an upstream step of the pathway [20]. Given that an early event in the pathway leading to Wnt signaling activation is the reorganization of microtubules at the vegetal pole required for the transport of local determinants, we tested whether this reorganization is affected in hec mutant embryos (Fig. 5; Figure S5). Consistent with previous studies [2], [8], [27], we find that microtubules at the vegetal cortex in wild-type appear as parallel tracks of bundled microtubules at 20 mpf (Figure 5A). In hec mutant embryos, such parallel tracks of bundled microtubules are not observed (Figure 5B). In these mutants, unbundled microtubules typically appear to radially emanate from one or more aster-like structures at the vegetal pole region (Figure 5C–F). Exposure to the microtubule-stabilizing drug taxol [44] during the first cell cycle (5 to 35 mpf) does not influence the degree of residual axis induction in hec mutants (Figure S6), suggesting that the observed defects may not be simple consequence of altered microtubule dynamics. Labeling of the F-actin cortical network in the vegetal cortex region shows a similar appearance in wild-type and mutant embryos (Figure S7), including the presence of F-actin rich protrusions as previously described [45].
Figure 5. Microtubule reorganization at the vegetal cortex is affected in hecate mutants.
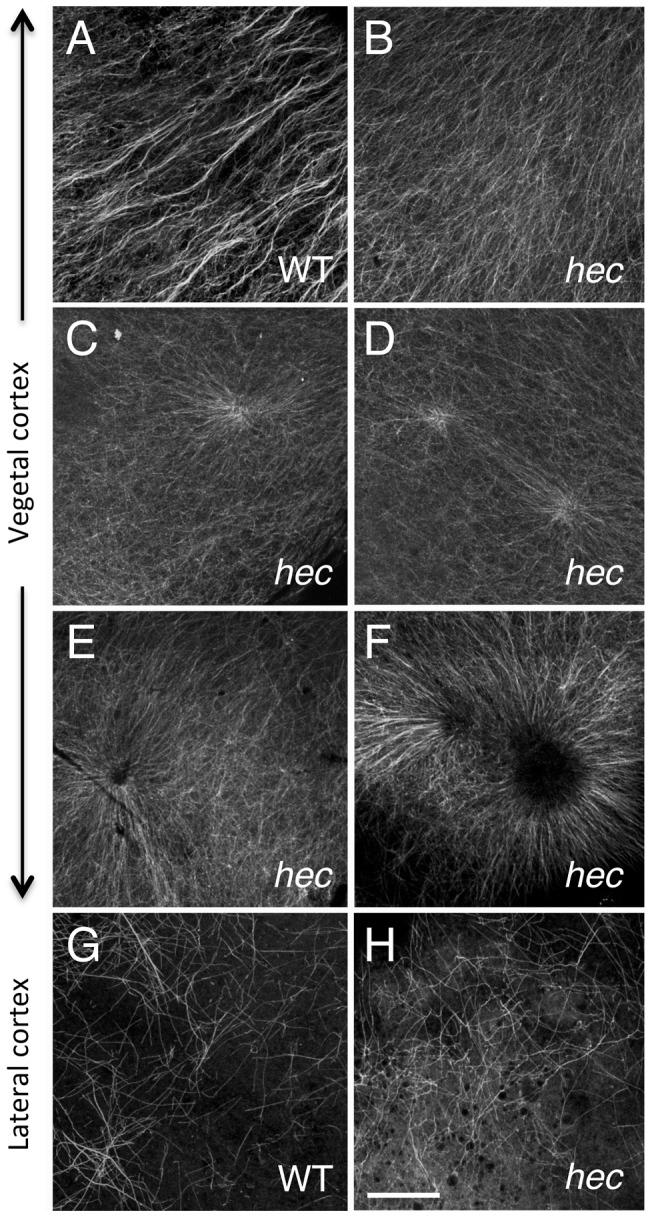
A–F) Cortical microtubule network at the vegetal pole in wild-type (A) and hec mutant (B–F) embryos at 20 mpf. Microtubules appear oriented in the same direction and bundled in wild-type embryos (A). The extent of co-orientation and bundling is greatly reduced in hec mutant embryos (B), where microtubules form multiple aster-like structures which can have a well focused-center (C,D) or can exhibit a central microtubule-free zone (E,F) and often overlap (F) or interdigitate (D). The relatively unbundled microtubule arrangement shown in (B) also corresponds to a sector of a large aster-like structure emanating from a not shown central core. Up to 6 aster-like structures were observed in the vegetal cortex of a single embryo. G,H) Cortex in mediolateral regions shows a loose and apparently random network of microtubules which appears similar in both wild-type (G) and hec mutant (H) embryos (n = 8 for wild-type and mutants). All images are z-axis projections of confocal image stages. The phenotype was fully (100%) penetrant according to the two main categories (wild-type, aligned and bundled microtubules; mutant, radialy oriented and unbundled microtubules, with 10 wt and 25 hec mutant embryos imaged. Magnification bar in (H) corresponds to 40 µm for all panels.
Short-range symmetry breaking and anchoring of vegetally localized factors are affected in hec mutant embryos
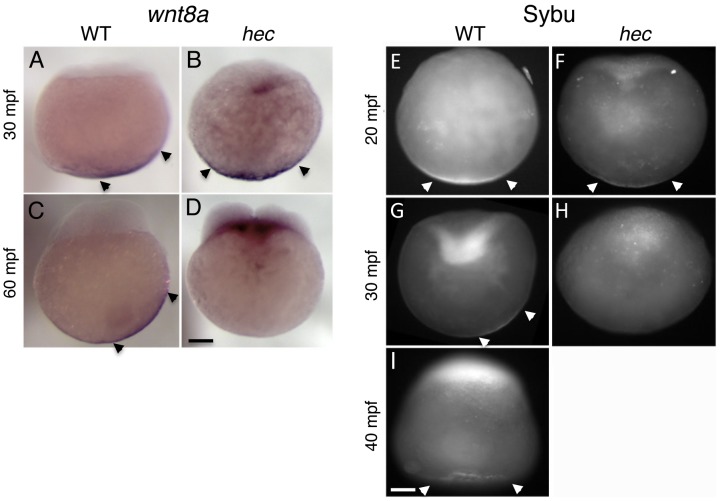
Since vegetal cortical microtubules have been proposed to mediate the off-center shift of factors involved in axis induction that are initially localized to the vegetal pole, such as grip2a mRNA (this report), wnt8a mRNA ([7]; Figure 6A, 6C) and Sybu protein ([6]; Figure 6E, 6G), we tested whether these shifts were affected in hec mutant embryos. As noted above the grip2a mRNA localization domain in hec mutants, when still detectable, fails to undergo an off-center shift (Figure 3G). In situ hybridization analysis to detect wnt8a mRNA shows this mRNA also fails to undergo a noticeable off-center shift in one-cell (30 mpf) hec mutant embryos (Figure 6B). At the 4-cell stage (60 mpf), wnt8a mRNA localization at the vegetal pole is significantly reduced or undetectable (Figure 6D), although this defect is associated with an overall decrease in the relative expression of wnt8a mRNA (Figure S8). In the case of Sybu protein, wild-type embryos show localization centered at the vegetal pole until 20 mpf (Figure 6E) and an off-center shift of protein localization by 30 mpf (Figure 6G), as previously reported [6]. In hec mutants, the Sybu protein localization domain can be initially detected centered at the vegetal pole of hec mutants (Figure 6F). However, Sybu protein is no longer detectable by 30 mpf (Figure 6H), precluding testing an effect on Sybu protein off-center movement. These data indicate that hec/grip2a is essential for the short-range, symmetry-breaking transport of vegetally-localized factors, and are consistent with hec function being essential for microtubule reorganization in this region. The reduction in vegetally localized wnt8a mRNA and Sybu protein in hec mutants contrasts with the perduring vegetal localization of these factors in embryos with a perturbed microtubule network ([6], [7]; Figure 3I, 6I).
Figure 6. Defects in the vegetal localization of wnt8a mRNA and Sybu protein.
A–D) Off-center shift of wnt8a mRNA is affected in hec mutants. Whole mount in situ hybridization of wild-type embryos (A,C) and hec mutant embryos (B,D) at the 1- (A,B, 30 mpf) and 4- (C,D, 60 mpf) cell stages. Images show representative embryos. A majority of wild-type embryos showed a clear off-center shift (85%, n = 27 at 30 mpf and 74%, n = 47 at 60 mpf). A majority of hec mutant embryos showed vegetal localization without a shift at 30 mpf (79%, n = 33, remaining embryos show no localization) and absence of localization at 60 mpf (89%, n = 38, remaining embryos show reduced vegetal localization without a shift). The apparent label at the base of the blastodisc is observed in a majority of mutant embryos (71%, n = 38) but not in wild-type (C) or control embryos labeled with other probes (not shown) and may reflect remaining wnt8a mRNA that has lost anchoring at the vegetal pole and has moved animally through the action of axial streamers [83]. E–I) Localization of Sybu protein is affected in hec mutants. Whole mount immunofluorescence to detect Sybu protein of untreated wild-type (E,G) and hec mutant (F,H,) embryos and nocodazole-treated wild-type embryos (I) at the indicated stages. In wild-type embryos, an off-center shift in Sybu protein can be observed starting at 30 mpf (G). In hec mutants, Sybu protein becomes undetectable levels by this same time point (H). Patterns of localization of Sybu protein at 10 mpf and 20 mpf time points (combined n: 32 WT, 19 mutant for 10–20 mpf), and 30 mpf and 40 mpf time points were similar and have been combined. 59% (n = 32) of wild-type and 63% (n = 19) of hec mutant embryos showed centered vegetal localization during 10–20 mpf. At 30–40 mpf, the percent of embryos that showed vegetal localization, now with an off-center shift, was reduced to 25% (n = 28) in wild-type, and 0% (n = 25) of hec mutants showed any localization at these time points. Treatment of wild-type embryos with nocodazole inhibits the shift but does not result in delocalization from the vegetal cortex (I, embryo at 40 mpf), as previously shown [6]. Magnification bars in (D) and (I) correspond to 100 µm for panels sets (A–D) and (E–I), respectively.
Long-range animally-directed transport is independent of hecate/grip2a function and not restricted to the prospective dorsal region
The short-range off-center shift observed in the case of Sybu protein and grip2a mRNA, which occurs within the confines of the vegetal region, contrasts with the long-range transport thought to be involved in transporting a putative dorsal signal to blastomeres at the animal region [2]. wnt8a mRNA has been observed to reach the base of the blastomeres by the 16-cell stage (1.5 hpf; [7]), although in our experiments this RNA exhibits a relatively static off-center shift throughout the first 60 mpf (Figure 6A, 6C), similar to the short-range movement of Sybu protein and grip2a mRNA. The animally-directed translocation of a putative dorsal signal is thought to be reflected in the microtubule-dependent, animally-oriented movement of small (0.2 µm) polystyrene fluorescent beads during the first several cell cycles. When injected into the vegetal region, these beads reach the base of the blastomeres at the animal region by traveling through cortical paths [2].
Using the transport of microinjected fluorescent beads as an assay for this long-range transport mechanism, we tested whether long-range vegetal-to-animal movement along the cortex might be affected in hec mutant embryos. As previously reported [2], in wild-type embryos bead movement from the vegetal region is observed along a meridional arc along the cortex reaching the base of the blastomeres at the animal pole (41% (n = 87); Figure 7A,A′). In hec mutant embryos, beads appear to be transported to a similar extent as in wild-type, also reaching the base of the blastodisc (Figure 7B,B′) and at a similar observed frequency (39% (n = 51)). Thus, hec function does not appear to be required for long-range animally-directed transport along the lateral cortex. The apparently normal movement of animally-directed beads in hec mutants appears to conflict with the role of this gene in early microtubule reorganization but is consistent with the observed presence of multiple microtubule populations [2], [8]: aligned bundles of short microtubules at the vegetal region, which we find to be dependent on hec function, and a more dispersed and randomly oriented network in more medial regions, which appears unaffected in hec mutants (Figure 5G, 5H and data not shown).
Figure 7. Long-range animally-directed transport is not affected in hecate mutants.
A–C) Paths of injected 0.2 um fluorescent beads after injection into the vegetal pole with a single (A,B) or double (C) injection in wild-type (A,C) or hec mutant (B) embryos. (A′–C′) show merged imaged including the fluorescent channel (shown in A–C) and corresponding DIC optics at low intensity. The extent and frequency of bead transport appeared similar in wild-type and mutant embryos (A,B, see text). Injections into two opposite sides of the vegetal pole results in multiple animally-directed paths, indicating that the entirety of the mediolateral cortex is competent for bead movement. Arrowheads and arrows in (A–C) indicate site of injection in the vegetal region and animally-directed paths along mediolateral regions, respectively. Magnification bar in (C′) corresponds to 100 µm for all panels.
The direction of aligned short microtubule bundles at the vegetal pole has been shown to correlate with the dorsal axis but only occurs in a limited area within the vegetal half of the early embryo [8]. Given that directionality of a long-range movement might depend on an earlier oriented symmetry-breaking event, we wondered whether the long-range animally directed movement was specific to the putative dorsal region, or whether the entirety of the cortex was competent to support such transport. To test whether long-range animally-directed transport was specific to the prospective dorsal region of the embryo or was a general property of the lateral cortex, we carried out two slightly off-center fluorescent bead injections on opposite sides of wild-type embryos. Such doubly-injected embryos show animally-directed bead transport along meridional arches in opposite regions of the embryo (Figure 7C,C′), an observation inconsistent with only the prospective dorsal region mediating long range vegetal-to-animal cortical transport. Instead, our data suggest that the entirety of the cortex can mediate long-range animally-directed movement.
The apparently normal long-range transport of beads in hec mutant embryos, in the presumed absence of an early short-range symmetry-breaking process, may reflect asymmetries in the location of the beads during injection, followed by the action of this hec-independent long-range transport mechanism. Our data suggest that the transport of dorsal determinants to the prospective site of dorsal induction depends on two sequential processes: (i) an initial short-range transport dependent on hec-mediated formation of short aligned microtubule bundles that results in determinant asymmetry at the vegetal region of the embryo, and (ii) a subsequent long-range transport through lateral cortical regions, which is independent of hec function and not specific to the prospective dorsal region.
Divergence of grip function in zebrafish and Xenopus
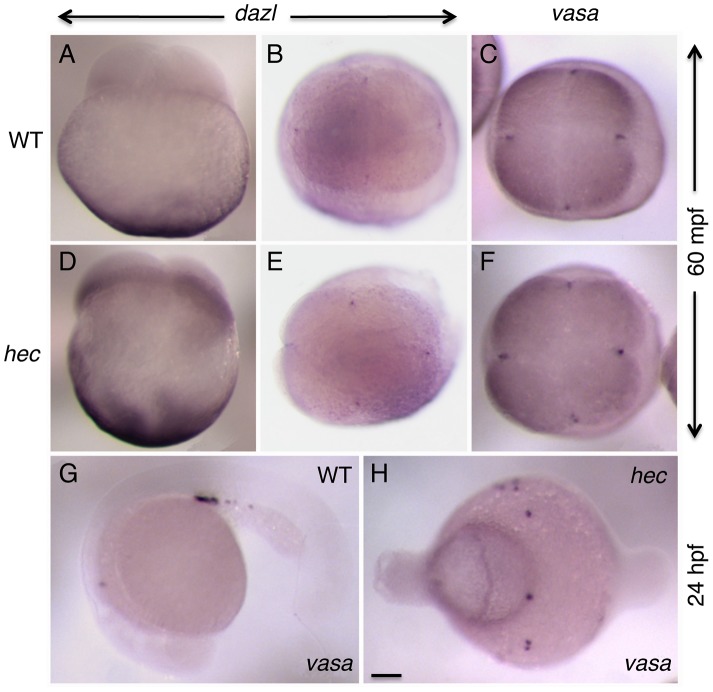
In Xenopus, the homologous gene grip2 (previously referred to as grip2.1), like zebrafish hec/grip2a, is expressed maternally and its mRNA is localized to the vegetal pole of the egg and early embryo [32]. Following this vegetal localization, Xenopus grip2 mRNA becomes incorporated into primordial germ cells (PGCs) where it plays a role in their migration and survival [32]. In contrast, although localized to the vegetal pole, zebrafish grip2a mRNA does not localize to the zebrafish germ plasm or PGCs (Figure 3 and data not shown). We used whole mount in situ hybridization to test whether germ plasm localization or PGC development may be affected in hec mutant embryos (Figure 8). The localization patterns of dazl mRNA, a germ plasm component initially localized to the vegetal pole of the egg ([36], [46], [47]; Figure 8A), is not affected in these mutants (Figure 8D). During the first two cell cycles, dazl mRNA localizes normally to the furrows in hec mutants (Figure 8B, 8E), as does vasa mRNA (Figure 8C, 8F), an animal germ plasm component already present in the animal cortical region during egg activation [33], [36], [38], [48]. During embryonic development, although PGC migration is abnormal in strong hec mutants due to their radially symmetric, ventroposteriorized morphology, the average number of induced PGCs (as determined by cells expressing vasa in the 10.5 hpf embryo [33], [38], [48]) is similar to that in wild-type embryos (Figure 8G, 8H; Figure S9). Thus, as opposed to the case of Xenopus grip2, our observations do not support a role for zebrafish hec/grip2a in PGC development.
Figure 8. Germ plasm recruitment and PCG determination appears unaffected in hecate mutants.
A–F) Whole mount in situ hybridization of 4-cell stage (60 mpf) embryos to label the germ line-specific genes dazl1 (A,B,D,E), and vasa (C,F) in wild-type (A–C) and hec mutant (D–F) embryos. Early localization of dazl mRNA to the vegetal pole (A,D, side views), and dazl mRNA and vasa mRNA to the germ plasm as it becomes recruited to the furrows of the first and second cleavage cycles (B,C,E,F, animal views). Localization of dazl domains of recruitment at the furrow is not detected in the side views as the levels of mRNA in these domains are relatively low and the focal plane is not optimal for their visualization. G,H) PGC determination, as determined by vasa expression in 24-hour embryos (G), appears unaffected in hec mutants (H), although the PGCs in mutant embryos do not become clustered as in wild-type due to the aberrant cell specification in these mutants. Quantification of the number of vasa-expressing cells is presented in Figure S9. Magnification bar in (H) corresponds to 100 µm for all panels.
Two other zebrafish grip-related genes, grip1 and grip2b, exhibit maternal expression as assayed by RT-PCR of RNA from 1–4 cell embryos but do not show localization to the vegetal pole or germ plasm at these early stages, or in PGCs in the 24-hour embryo ([28]; our own data). Together, these data indicate that, in spite of similar mRNA localization patterns at the vegetal pole of the egg, grip homologues in Xenopus and zebrafish have divergent roles in PGC development and axis induction, respectively.
Discussion
A gynogenesis-based forward genetic screen in the zebrafish led to the isolation of a mutation in hec, which results in axis induction defects [17], [20], [23]. Here, we report the isolation of two additional alleles of hec, identified through an F3 inbreeding screen, and the molecular identification of this gene as encoding Glutamate receptor interacting protein 2a. This molecular assignment, as well as the localization pattern of its products, suggests that vegetally localized grip2a mRNA in zebrafish acts at the vegetal pole in the early events of dorsal axis induction. Accordingly, we find that hec functions to reorganize and align microtubule bundles that are involved in the symmetry-breaking transport of factors localized to the vegetal cortex, which is essential for axis induction.
Role of hecate/grip2a in cortical microtubule rearrangements and axis induction
In both teleosts and amphibians, dorsal determinants are initially localized at the vegetal pole and then translocate to the prospective dorsal side in a microtubule-dependent manner to initiate the dorsal cell fate program (reviewed in [1], [49], [50]). Genetic and molecular searches have identified factors localized to the vegetal cortex with a role in axis induction in the zebrafish, such as the kinesin-1 linker Syntabulin [5], [6] and the wnt8a mRNA [7]. We show that the mRNA product for zebrafish hec/grip2a is another zebrafish maternal factor involved in axis induction, whose product is also localized to the vegetal pole of the oocyte and early embryo.
Zebrafish hec/grip2a is expressed solely during oogenesis as a maternal transcript, which is consistent with the identification of multiple maternal-effect mutant alleles of this gene, all of which lack associated zygotic defects. In particular, fish homozygous for the hecp06ucal allele, predicted to lack all four PDZ domains present in the wild-type protein and therefore likely a null, exhibit a highly penetrant maternal-effect phenotype yet are themselves viable. These observations indicate that hec/grip2a has a dedicated function in early axis determination. While we cannot rule out that hec/grip2a is expressed in older embryos or adults at levels below the sensitivity of our assays, other related genes such as grip1 or grip2b are expressed at later stages of development ([28]; our own data) when they may provide essential zygotic functions.
Mechanisms inducing microtubule bundling and alignment, essential for the establishment of the primary body axis, remain incompletely understood. In Xenopus, cortical rotation and microtubule reorganization are dependent on both kinesin and dynein motor activity [51], [52] and other factors such as Trim36, a ubiquitin ligase whose mRNA is localized to the vegetal egg cortex [53], Dead end, an RNA binding factor needed for trim36 mRNA vegetal cortex localization [54] and the lipid droplet component Perilipin 2, whose mRNA is also localized to the Xenopus vegetal pole [55], [56]. We find that zebrafish hec/grip2a function is required for the reorganization of vegetal cortex microtubules into bundles normally directed towards the prospective dorsal axis [2], [8]. The lack of microtubule network alignment in hec mutants and associated defects in the transport of putative dorsal determinants such as wnt8a mRNA likely result in the axis induction defects observed in these mutants.
Grip was originally identified as a factor interacting with AMPA-type glutamate receptors [57] and its multiple PDZ domains are thought to facilitate protein-protein interactions within large macromolecular complexes, including the surface presentation and trafficking of transmembrane proteins [58]–[60]. grip genes have been implicated in epithelial development in both mouse and zebrafish embryos ([28], [61], reviewed in [62]). Other studies have implicated Drosophila Grip as a mediator of Wnt ligand activity in the postsynaptic terminal of the neuromuscular junction [21], [22].
Our findings suggest parallels between subcellular transport at the vegetal pole of the zebrafish zygote and transport of neurotransmitter receptors in neurons. In the zebrafish zygote, transport of wnt8a mRNA depends on microtubules and occurs concomitantly with the movement of the kinesin adaptor Syntabulin [6], [7]. Similarly, in dendrites Glutamate receptors associated with mammalian GRIP1 are driven by kinesin along microtubules [63], and Syntabulin has been shown to be required for axonal transport [64]–[66]. In neurons, glutamate receptors and GRIP associate with membrane vesicles [67], [68]. Although membrane vesicles have not been reported to be associated with dorsal determinants in zebrafish, studies in Xenopus implicate membrane vesicles in the transport of dorsal determinants [69]–[72]. Further studies will be required to determine mechanisms driving the reorganization of vegetal cortex microtubules, the precise role of Grip2a in this process and whether Grip factors have an analogous cytoskeletal restructuring function in other systems.
Is Grip2a required for downstream events in Wnt signaling involved in axis induction?
Our previous studies have shown that manipulations to activate Wnt signaling, including the overexpression of wnt8 mRNA, can rescue the hec mutant phenotype, which suggests that hec acts in an upstream event required for Wnt signaling activation during axis induction [20]. Our identification of a role for Grip2a on cytoskeletal events needed for the relocation of dorsal determinants is consistent with such an upstream role. However, our studies do not rule out a more direct role for Grip2a as a regulator of Wnt pathway components. We note that the off-center shift of vegetally localized grip2a mRNA upon egg activation could provide an asymmetric source of Grip2a protein to influence Wnt signaling activity at the prospective dorsal region. Drosophila Grip is known to interact with the Wnt receptor Frizzled-2, promoting the trafficking of a Frizzled-2 C-terminal fragment to the nucleus to activate target genes [21], [22], [73], [74], and it is possible that some of these interactions are conserved in the zebrafish embryo. It is also possible that Grip2a regulates non-canonical Wnt signaling, such as Wnt/calcium signaling, which in turn influences axis induction. hec mutants exhibit an increased frequency of intracellular calcium transients in blastula stage embryos (2.00–3.33 hpf; [20]), and the resulting intracellular calcium increase has been proposed to attenuate Wnt/β-catenin signaling pathway activity [75]–[79]. Further studies will be necessary to determine whether hec/grip2a, in addition to functioning in cytoskeletal organization in the early zebrafish embryo, has a direct role in the regulation of Wnt/β-catenin signaling and axis induction.
A multiple-step mechanism for the transport of dorsal determinants in zebrafish
Multiple studies in Xenopus have indicated the formation of long tracks of cortical microtubules associated with the cortical rotation [80]–[82]. While in this organism the cortical rotation involves the concerted movement of the cortex along a distance corresponding to a 30° arc, microtubule tracks and so-called fast transport of subcellular components, such as membrane organelles and specific factors, encompass a longer distance corresponding to a 60–90° arc (reviewed in [1]). Thus, in Xenopus both the cortical rotation and fast transport may participate in the relocation of dorsal determinants. In zebrafish embryos, a cortical rotation-like process results in the displacement of granules along a 20° arc from the vegetal pole of the embryo [8], with mediolateral regions of the cortex exhibiting a loose meshwork of microtubules independent of dorsoventral position [2], [8]. The restriction of microtubule bundling and alignment to the vegetal region of the zebrafish embryo raises the question of how long-range transport of dorsal determinants to the animal pole is achieved.
We found that injected beads reach the base of blastomeres in hec mutant embryos, which lack a cortical rotation-like movement, with a frequency similar to that observed in wild-type embryos. This suggests that transport of beads along the mediolateral cortex is independent of hec/grip2a function and aligned vegetal microtubules. Our finding that beads are able to move animally along opposite sides in multiply injected embryos, further suggests that the entirety of the mediolateral cortex, not just the prospective dorsal region, is competent for long-range vegetal-to-animal transport. Previous studies have identified animally-directed transport movement of cytoplasmic particles along cortical “meridional” streamers, hypothesized to mediate transport of vegetally-injected fluorescent beads [83]. This meridional transport along the mediolateral cortex may depend on various possible structures or processes, such as perpendicular bundles aligned along the animal-vegetal axis in deep regions of the cortex [8], incipient yolk cytoplasmic layer (YCL) microtubules emanating from marginal blastomeres into the yolk [84], an emerging property of the loose network of cortical microtubules found in this region of the embryo [2], [8], or other insofar unidentified cytoskeletal networks. Further analysis of the dynamic aspects of cytoskeletal networks in this region will be required to understand the mechanistic basis of this meridional transport system and its role in axis induction. In addition, our results do not exclude the possibility that diffusion of a translated protein such as Wnt8a may also contribute to long-range transport, as previously suggested [8].
Together, these data indicate that the transport of the putative dorsal determinant in the early zebrafish embryo involves at least two separate mechanisms: a short-range transport dependent on hec/grip2a function and aligned microtubule bundles, and a subsequent long range transport relying on the mediolateral cytoskeletal network (Figure 9A). We hypothesize that the former generates an off-center, symmetry-breaking shift in initially symmetrically localized putative dorsal determinants, while the latter acts as a more general conduit that amplifies the early asymmetry. In hec mutants, the initial symmetry-breaking event is affected, so that even with a functional long-range animal transport mechanism, in most embryos an insufficient amount of dorsal determinants reaches the blastomeres at the animal pole (Figure 9B). A dual mechanism of dorsal determinant transport may also explain how a fraction of embryos from females homozygous for the presumptive null allele, hecp06ucal, which lacks all 4 conserved PDZ domains, can develop a normal dorsal axis. In such embryos small fluctuations may occur in the position of the vegetally localized dorsal determinant, which could be amplified by the mediolateral transport system that is unaffected in these mutants. The presence of an embryo-wide pathway directing long-range transport towards the animal pole may also explain the appearance of double-axis embryos observed only in the weakest hec mutant clutches. In these cases, an aberrantly organized vegetal microtubule network may result in the off-center vegetal shift of dorsal determinants in more than one direction, leading to their animally-directed transport along multiple paths and resulting in supernumerary or expanded regions of axis induction. A wider distribution of dorsal determinants in weak hec mutants would result in their reduced concentration in animal regions and reduced Wnt pathway activation, consistent with the observed lack of anterior-most structures in the resulting double-axis embryos. This dual transport model suggests mechanisms by which small, directed changes, like the specific early short range symmetry-breaking event, can be amplified during early development by an embryo-wide mechanism to result in large differences in cell fate specification.
Figure 9. Amplification of hecate/grip2a-dependent symmetry breaking event by a general animal-directed long-range transport system.
A) Cortical shifts of various vegetally localized components, including wnt8a mRNA, Sybu protein and grip2a mRNA ([6], [7], [8]; this report) are short-range and dependent on microtubule bundling and alignment, itself dependent on hec function. In wild-type embryos, such a short-range shift generates a symmetry breaking event that is subsequently amplified by long-range, animally-directed transport mechanism independent of hec function and not restricted to the prospective dorsal axis. B) In hec mutant embryos, neither reorganization of vegetal microtubules into aligned bundles nor a short-range shift occur, so that, even though long-range transport remains intact, vegetal determinant transport to the animal pole is affected. The mechanistic basis for the long-range transport, occurring in the region of a loosely organized mediolateral microtubule cortical network remains to be determined (see Discussion).
hecate may represent a gene duplication adapted for axis induction
Using BLAST searches on Ensembl and NCBI genome databases, homologous grip1 and grip2 genes were found for all vertebrate species. In Drosophila, there is an ancient single Grip gene. All other species containing recognizable grip genes are vertebrates. In amphibians, birds and mammals, there is one grip1 gene and one grip2 gene. On the other hand, zebrafish have one grip1 gene and two grip2 genes, likely due to an ancestral genome duplication in the teleost lineage [30], [31]. Following published nomenclature [28], we refer to the grip2 gene corresponding to hec, located in chromosome 8, as grip2a, and the copy located in chromosome 22, as grip2b. Phylogenetic analysis indicates that zebrafish grip2a is only present in fish species (zebrafish, fugu and medaka). Drosophila Grip and all vertebrate Grip1 and Grip2 proteins (including zebrafish Grip2b) contain seven conserved PDZ domains, while zebrafish Grip2a contains only 4 PDZ domains.
The amphibian grip2 homolog was identified as a novel vegetally-localized mRNA in Xenopus oocytes that is present in the mitochondrial cloud (Balbiani body) and subsequently in the germ plasm throughout oogenesis and early embryogenesis [32], [85], [86]. In Xenopus grip2 morphants, PGC numbers are significantly reduced, and PGCs are also present at ectopic locations along the anteroposterior axis in tailbud stage embryos [32], [86]. However, unlike the case of Xenopus grip2, zebrafish grip2a mRNA does not localize to the germ plasm or PGCs, and the number of PGCs is unaffected in strongly ventralized mutants (i.e. radially symmetric ventralized embryos). These observations suggest that in zebrafish PGCs are determined independently of hec/grip2a function. Therefore, it appears that Xenopus grip2 and zebrafish grip2a mRNA, in spite of a similar localization at the vegetal pole of the oocyte, have distinct functions in early embryogenesis: Xenopus grip2 is involved in PGC development, while zebrafish grip2a appears to be devoted to dorsal axis formation.
Despite this divergent function, the fact that zebrafish hec/grip2a and Xenopus grip2 are maternally expressed and localized to the vegetal pole during oogenesis, the site of localization of both germ plasm components and dorsal determinants in these two lineages, suggests that an ancestral grip gene may have functioned in both axis induction and PGC development. There is precedent for a relationship between these two processes, notably in Drosophila [87] but also in systems as basal as planaria [88], [89] and annelids [90]. This relationship is also supported by the recent finding that the germ cell-specific factor Dead end is required for microtubule rearrangements and cortical rotation in Xenopus [54]. The presence of a single localization system involved in both the induction of the primary embryonic axis and the separation between germ cell and soma may constitute a simple mechanism for the species patterning and propagation. Studies of the role of grip genes in other organisms may shed light on the relationship between axis induction and PGC determination during evolution.
Materials and Methods
Ethics statement
All animals were handled in strict accordance with good animal practice as defined by the relevant national and/or local animal welfare bodies, and all animal work was approved by the appropriate committee (University of Wisconsin – Madison assurance number A3368-01).
Fish maintenance and genetic lines
Fish stocks were raised and maintained under standard conditions at 28.5° [91]. The hect2800 allele was originally isolated in an early-pressure-based screen for recessive maternal-effect mutations [14], [17], while the hecp06ucal and hecp08ajug alleles were found in a four-generation scheme based on natural crosses ([16], [18], see also [92]). The hect2800 allele was induced in an AB/Tübingen hybrid background [14], [17], which was further hybridized with the WIK line during linkage mapping. The hecp08ajug and hecp06ucal alleles were induced in a Tübingen background, which was hybridized to an AB line in an F4 genetic screen coordinated with linkage mapping [16], [18]. Homozygous mutant hec fish were identified by genotyping the flanking SSLP markers z59658 and z24511, which are 1.2 cM apart on linkage group 8 and both of which were polymorphic for all three alleles. Mutant embryos were obtained by crossing homozygous hec females to AB males. Embryos from females homozygous mutant for the hecp06ucal allele were used unless otherwise specified. Wild-type control embryos were derived from either the AB line or heterozygous sibling females. Clutches were synchronized through 5-minute collections during natural spawning. Oocytes were collected from wild-type, bucky ballp106re [39], [43] and magellanp6cv [40] mutants. Embryos were collected and developed in E3 embryonic medium [14] and were staged according to the age and morphological standards described in [93].
For complementation tests of hect2800, hecp08ajug and hecp06ucal mutants, homozygous mutant males of one allele were crossed with heterozygous females of another allele to produce offspring, which were raised to adulthood. Female adult fish were crossed with wild-type males and phenotyped as wild-type or ventralized mutant by examining the resulting clutches at 24 hpf. Only those clutches producing more than 50 embryos were scored and non-complementation was indicated by the presence of ventralization phenotypes similar in expressivity and penetrance to those in clutches from mutant females from the original three mutant alleles on their own.
Isolation and genotyping of genomic DNA
Fish were anesthetized with MESAB (0.014%) and the tail fin was clipped using a razor blade and placed into 100 ul DNA lysis buffer (10 mM Tris, pH 8.0; 10 mM EDTA, pH 8.0; 200 mM NaCl; 1% Triton X-100) containing 5 µg 10 mg/ml Proteinase K. Tissue lysates were incubated overnight at 55°C, and were incubated at 94°C for 10 minutes to inactivate Proteinase K. Lysates were diluted 1∶6 with water, and 2.5 µl of this genomic DNA diluted lysate was used per 10 µl PCR reaction. For a 10 µl PCR reaction, 2 µl of GoTaq green Buffer, 0.2 µl of dNTPs, 0.05 µl of GoTaq DNA polymerase (Promega), 2.5 ul genomic DNA diluted lysate, and 1 µl each of 10 µM forward and reverse primers were used. For SSLP markers, PCR products were analyzed on a 2% high-resolution agarose gel right after the PCR reaction. For RFLP markers, PCR products were used for FastDigest Restriction Enzyme (Fermentas) digestion for 30 min, and then analyzed on a 1.5% regular agarose gel.
Positional cloning and sequence analysis
Initial linkage was identified by bulk segregant analysis with SSLP markers. Once initial linkage of the mutation was obtained, genotypically identified homozygous mutant males were crossed to heterozygous females to generate large numbers of fish for fine mapping [92].
Chromosome walking was conducted by screening the CHORI-211 BAC library using marker z67047 and zC150E8z (0 recombination/1762 genomes). PCR-based screening of the primary pool and secondary pools identified 2 positive BAC clones. Individual BAC clones were ordered from BACPAC resources center (http://bacpac.chori.org).
To find the mutation in hec/grip2a, 5 fragments of hec/grip2a cDNA from mutant and wild-type 1-cell embryo cDNA were amplified by RT-PCR with 5 primer pairs, which cover the entire hec/grip2a coding region:
1. 5′-ATGTCCTGCATCTTGCTTCCAGAG-3′ and 5′-CCTCAGTGGGAATCCCATTAATGG-3′
2. 5′-TGGAGTGTTACAAGTTGGCGACAG-3′ and 5′-TGAATGGCTTCGCTCAGAGGTTTG-3′
3. 5′-TTCATATCGGTGACCGAGTTTTGG-3′ and 5′-GACATTATTGTAGCCTCAAGCTCG-3′
4. 5′-GAGACCTGCGGTCAGTCAGAAATC-3′ and 5′-GTGCTCTGTGTTTCTCATTTGTGG-3′
5. 5′-AGGACACTTCCCAACAGTCTGCAC-3′ and 5′-ACCTGATCACTTCTAACCCAACAG-3′
All PCR products were cloned into pGEM-T easy vector and sequenced.
For Phylogenetic analysis, homologous grip1 and grip2 genes were found using BLAST searches on Ensembl and NCBI genome databases. A phylogenetic tree was constructed and drawn using ClustalW in the MegAlign program from Lasergen. PDZ domains were identified using CD-search in the Conserved Domain Database (CDD) in NCBI [29]. Schematic diagram of the protein domain structures for each gene were drawn using DomainDraw [94].
RT-PCR and quantitative RT-PCR
Total RNA was isolated from whole embryos using TRIzol reagent (Invitrogen). cDNA was synthesized using random primers (Invitrogen) and AMV Reverse Transcriptase (Promega). RT-PCR reactions were performed with primer pairs derived from hec/grip2a and ef1α, using 30 cycles at an annealing temperature of 58°C (in the semi-quantitative range). Absence of genomic contamination was verified by a negative control RT reaction without the Reverse Transcriptase. The following primers were used for the amplifications:
hec/grip2a, 5′-GAGACCTGCGGTCAGTCAGAAATC-3′ and 5′-TATGAAGCTCTAGAGGCACTGACG-3′,
wnt8a, 5′-CGGAAAAATGGGTGGTCGTG-3′ and 5′-AGTCGACCAGCTTCGTTGTT-3′,
ef1α, 5′-ACCGGCCATCTGATCTACAA-3′ and 5′-CAATGGTGATACCACGCTCA-3′.
Quantitative (q) RT-PCR was performed on an iCycler machine (Bio-Rad) using iQ SYBR Green Supermix (Bio-Rad). The thermal profile used for amplification is: 95°C for 3 min, 40 cycles of 94°C for 30 s, 58°C for 30 s and 72°C for 30 s. The relative mRNA level was quantified and normalized to ef1α.
In situ hybridizations and antibody labeling
In situ hybridizations of embryos were carried out as described previously [95]. Probes for in situ included goosecoid [96], chordin [97], even skipped 1 [98], vasa [33], dazl [46] and wnt8a [7], [99]. For the grip2a in situ probe, a fragment of grip2a cDNA was cloned into pGEM-T easy vector as described in the positional cloning section. Five different probes were tested against 5 different cDNA sequences, all of which showed the same expression pattern. Subsequently, all the expression data was acquired using one of the probes. Antisense digoxygenin probe was generated by linearizing and transcribing with SacII and SP6 RNA polymerase, while sense probe control was generated by linearization with SpeI and transcription with T7 RNA polymerase. Images were acquired with a Leica-FLIII microscope and a color camera (Diagnostic Instruments Spot Insight).
Ovaries were dissected from euthanized females and fixed overnight at 4°C in 4% paraformaldehyde. Fixed ovaries were then dehydrated in MeOH.
Whole mount in situ hybridization of oocytes was performed as previously described [100]. Following staining, oocytes were embedded in JB-4 Plus Plastic resin and 7 micron sections were cut using a microtome. Stained sections were coated with Permount (Fisher) prior to addition of a coverslip.
Antibody labeling of microtubules was as previously described [36]. Prior to the labeling procedure, embryos were dissected using fine dissecting forceps to generate two halves. To image cortical microtubules, bisections were carried out along an equatorial plane, the vegetal halves were labeled and mounted in 50% glycerol with DABCO reagent to prevent bleaching, with the vegetal cortex facing the coverslip. For imaging of mediolateral microtubules, bisections were carried out along a meridional plane and each mediolateral halves were labeled and mounted as above with the mediolateral cortex facing the coverslip. Images were acquired using an upright Zeiss LSM510 confocal microscope using an oil immersion 63× objective and collected as single 1.5 µm optical sections with a pinhole diameter of about 1 Airy unit, a low scan speed (preset 6) and noise filtering through a 4-pass line mode average. The resulting images were analyzed with Fiji software.
Antibody labeling of embryos to detect Sybu protein was carried in whole mount embryos using whole embryos as described previously [6], and images were acquired using a Zeiss Axioplan2 fluorescent microscope and OpenLab software.
Nocodazole treatment was carried out through exposure by 10 mpf of dechorionated embryos to a final concentration of 4 µg/ml nocodazole in E3 (diluted from a 5 mg/ml solution in DMSO), followed by fixation at the indicated periods.
Phalloidin labeling was carried out as in [36], with the exception that dechorionated embryos were labeled whole and equatorially bisected prior to mounting with the vegetal pole facing the coverslip.
Fluorescent beads injection
Fluorescent beads injection experiments were carried as previously described [2]. A suspension of 0.2 mm fluorescent polystyrene beads (1 ul; Polysciences) was diluted in 23 ml water and colored with trace amounts of phenol red (0.05%). The injection solution is microinjected into the embryos near the vegetal pole using a Phemtojet microinjector (Eppendorf). Embryos had been injected at the 2-cell stage and imaged at 2 hours later. Embryos were mounted in methyl cellulose and imaged with an upright fluorescence microscope (Zeiss, Axioplan II) and a black and white digital camera (Zeiss, Axiocam).
Supporting Information
Age-dependency of the hec mutant phenotype. The hecate mutant phenotype is stronger in younger females. Also note that at both ages the strength of the alleles is hecp06ucal>hect2800>hecp08ajug. Homozygous mutant females at 3 and 12 month after birth were crossed against wild-type males to produce clutches of mutant embryos. Phenotypes were classified at 24 hpf as in [24] and Table 1. Results were pooled from clutches from 15 different females for each allele. Number of embryos is as follows: 3-month females: WT, 1440; hecp06ucal, 1178; hect2800, 575; hecp08ajug, 1399; 12-month females: WT, 1259; hecp06ucal, 1812; hect2800, 655; hecp08ajug, 1516. Pearson's Chi-squared test shows statistical significant differences between 3-month and 12-month females for all alleles (p-values: hecp06ucal, 0.002477; hect2800, <2.2e-16; hecp08tajug, <2.2e-16) but not for wild-type females (p-value 0.3421). Pair-wise comparisons between all three alleles are also significantly different using the same analysis (p-values: hecp06ucal vs. hect2800 <2.2e-16; hecp06ucal vs. hecp08ajug <2.2e-16; hect2800 vs. hecp08ajug <2.2e-16).
(TIFF)
Phylogenetic tree of Grip1 and Grip2 proteins among Drosophila and vertebrate species and number of PDZ domains in the predicted protein. Left: phylogenetic tree using ClustalW. Gene-ID from NCBI or Ensembl-genome databases: GRIP2 rat: NP_612544.2; GRIP2 mouse: NP_001152979.1; GRIP2 human: NP_001073892.1; Grip2 Xenopus: NP_001091382.1; GRIP2 chicken: ENSGALP00000010397; Grip2b zebrafish: XP_001922281.1; Grip2b medaka: ENSORLP00000012064; Grip2b fugu: ENSTRUP00000040982; Grip2a fugu: ENSTRUP00000024040; Grip2a medaka: ENSORLP00000012637; Grip2a zebrafish: NP_001116760.1; GRIP1 mouse: NP_083012.1; GRIP1 rat: ENSRNOP00000061369; GRIP1 human: ENSP00000352780; GRIP1 chicken: ENSGALP00000016069; Grip1 Xenopus: ENSXETP00000015955; Grip1 fugu: ENSTRUP00000012549; Grip1 medaka: ENSORLP00000021825; Grip1 zebrafish: NP_001038316.1; Grip fruitfly: NP_572285.2. Right: Diagrams of the overall structure of the respective proteins (gray) highlighting the number of PDZ domains (orange), based on Ensembl annotation.
(TIFF)
Expression of zebrafish grip2a mRNA in wild-type and hecate mutant embryos. A) RT-PCR analysis of grip2a mRNA and ef1α control expression in wild-type and hec mutant embryos, as well as wild-type ovaries and wild-type adults (male, female, female with removed ovaries). B) Quantitative RT-PCR analysis shows grip2a mRNA expression levels, relative to ef1α expression at the same stages. Maternal grip2a mRNA levels are reduced in embryos mutant for all hec alleles.
(TIFF)
Quantification of the off-center shift of grip2a mRNA localization domain in control (DMSO-treated) and nocodazole-treated embryos fixed at 40 mpf. A) Diagram indicating angles of the grip2a mRNA localization domain landmarks with respect to the vegetal pole of the embryo. LB: left boundary; RB: right boundary; S: total span of domain; M: midpoint of domain. B) Quantification parameters, with brackets indicating standard deviation. In control embryos, but not in nocodazole-treated embryos, left and right boundaries and midpoint of the grip2a mRNA localization domain experience a similar off-center shift, while the span of the domain appears unchanged. Blind analysis of 38 and 41 DMSO- and nocodazole-treated embryos, respectively. Similar changes were observed in embryos fixed at 30 mpf, although differences at this time point were less pronounced than at 40 mpf (data not shown).
(TIF)
Distribution of microtubule organization phenotypes at the vegetal cortex of wild-type and hecate mutant embryos. A–G) Examples of various vegetal cortex microtubule arrangements at 20 mpf: normally aligned (A), partially aligned (B, C), unbundled and lacking organization (D, E) and exhibiting aster-like structures (F, G). (A) is from a wild-type embryo and (B–G) from hec mutants. Magnification bar in (G) corresponds to 40 µm for panels (A–G). H) Distribution of phenotypes. Wild-type embryos exhibit highly aligned microtubule network while hec mutants show disorganization, lack of bundling and aster-like structures. The two distributions are significantly different (Fisher's Exact Test, p-value = 5.335e-09).
(TIF)
Treatment with taxol does not affect the hec mutant phenotype. Phenotypic distribution of axial defects (classification as in [24]) of taxol- and solvent (DMSO)-treated wild-type and hec mutant embryos. Data is derived from two to three different experiments after a 30 minute exposure to 10 µM taxol starting at 10 mpf. There are no statistically significant phenotypic differences between taxol-treated and control embryos (student t-test, 2-tailed, unpaired).
(TIFF)
F-actin cortex at the vegetal pole is similar in wild-type and hec mutant embryos. At 20 mpf, both wild-type (A) and mutant (B) eggs show F-actin rich folds and villi-like structures, which may correspond to previously described microplicae [45]. Number of embryos tested: 18 wt (from a pool of four females) and 24 mutants (from two different mutant females). A fraction (21%, n = 24) of hec mutant embryos show radial F-actin enrichments (C), correlating with aster-like microtubule structures in these embryos (Figure S5). Magnification bar in (C) corresponds to 40 µm in all panels.
(TIFF)
Expression of wnt8a mRNA in hec mutant embryos. Quantitative RT-PCR analysis of wnt8a mRNA levels relative to ef1α mRNA. wnt8a relative expression in hec mutants is close to wild-type at 30 mpf but becomes reduced at 60 mpf.
(TIFF)
PGC determination is not adversely affected in hecate mutant embryos. The number of PGCs (as identified by vasa expression through whole mount in situ hybridization) was determined in wild-type and hec mutant embryos at the 3- through 5-somite stages (ca. 10.5 hpf). At these stages, PGCs have not yet reached the prospective gonad location and appear relatively scattered, which facilitated quantification analysis. The average number of PGCs was 17.9 +/− 3.5 in hec mutant embryos (n embryos = 158), compared to 13.6 +/− 3.6 in wild-type embryos (n embryos = 214). The slightly higher number of PGCs in mutants was statistically significant (Student's t-test, p-value = 0.02). It is possible, however, that this difference reflects a bias in our ability to count individual PGCs in hec mutants, where PGCs are dispersed throughout the span of the embryos, compared to wild-type embryos, where PGCs are concentrated along the dorsal axis. Regardless of this uncertainty, the data indicate that hec function is not essential for PGCs determination.
(TIFF)
Three identified hec mutant alleles do not complement each other in complementation crosses. Top: Homozygous mutant males of one allele were crossed with heterozygous females of another allele to produce offsprings, which are raised to adulthood. Female adult F1 progeny were tested for hec-associated maternal effects by crossing them against wild-type males and scoring the resulting F2 clutches as wild-type or mutant. Only test crosses with more than 50 embryos were scored, and were scored as mutant when the fraction of V1–V4 categories was ≥50%. F2 clutches scored as wild-type exhibited ≤2% of embryos with V1–V4 phenotypes. If mutations are allelic, crosses are expected to yield F1 females exhibiting wild-type (from heterozygote females) or mutant (from transheterozygote females) F2 phenotypes in a 1∶1 ratio. Bottom: Summary of analysis showing non-complementation of three hec alleles.
(DOC)
Acknowledgments
We thank all members of the Pelegri Lab for critical comments during various stages of this work. We also thank Drs. Arne Lekven (Texas A & M University) and Masahiko Hibi (Nagoya University) and other members of the zebrafish research community for generously providing reagents.
Funding Statement
This work was supported by NIH grants RO1GM065303 to FP and R01GM56326 and R01HD050901 to MCM, an American Heart Association predoctoral fellowship 0710181Z to XG, Advanced Opportunity Fellowship through SciMed Graduate Research Scholars at the University of Wisconsin – Madison and NIH Training Grant T32-GM007133 to EW, NIH Training Grant T32-HD007516 and NRSA postdoctoral fellowship 5F32GM77835 to EWA, an American Cancer Society postdoctoral fellowship #PF-05-041-01-DDC to TG and Damon Runyan postdoctoral fellowship DRG1826-04 to FLM. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Houston DW (2012) Cortical rotation and messenger RNA localization in Xenopus axis formation. WIREs Dev Biol 1: 371–388. [DOI] [PubMed] [Google Scholar]
- 2. Jesuthasan S, Strähle U (1997) Dynamic microtubules and specification of the zebrafish embryonic axis. Curr Biol 7: 31–42. [DOI] [PubMed] [Google Scholar]
- 3. Mizuno T, Yamaha E, Kuroiwa A, Takeda H (1999) Removal of vegetal yolk causes dorsal deficiencies and impairs dorsal-inducing ability of the yolk cell in zebrafish. Mech Dev 81: 51–63. [DOI] [PubMed] [Google Scholar]
- 4. Ober EA, Schulte-Merker S (1999) Signals from the yolk cell induce mesoderm, neuroectoderm, the trunk organizer, and the notochord in zebrafish. Dev Biol 215: 167–181. [DOI] [PubMed] [Google Scholar]
- 5. Nojima H, Shimizu T, Kim C-H, Yabe T, Bae Y-K, et al. (2004) Genetic evidence for involvement of maternally derived Wnt canonical signaling in dorsal determination in zebrafish. Mech Dev 121: 371–386. [DOI] [PubMed] [Google Scholar]
- 6. Nojima H, Rothhämel S, Shimizu T, Kim C-H, Yonemura S, et al. (2010) Syntabulin, a motor protein linker, controls dorsal determination. Development 137: 923–933. [DOI] [PubMed] [Google Scholar]
- 7. Lu F-I, Thisse C, Thisse B (2011) Identification and mechanism of regulation of the zebrafish dorsal determinant. Proc Natl Acad Sci USA 108: 15876–15880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tran LD, Hino H, Quach H, Lim S, Shindo A, et al. (2012) Dynamic microtubules at the vegetal cortex predict the embryonic axis in zebrafish. Development 139: 3644–3652. [DOI] [PubMed] [Google Scholar]
- 9. Gordon MD, Nusse R (2006) Wnt signaling: multiple pathways, multiple receptors, and multiple transcription factors. J Biol Chem 281: 22429–22433. [DOI] [PubMed] [Google Scholar]
- 10. Petersen CP, Reddien PW (2009) Wnt signaling and the polarity of the primary body axis. Cell 139: 1056–1058. [DOI] [PubMed] [Google Scholar]
- 11. Tao Q, Yokota C, Puck H, Kofron M, Birsoy B, et al. (2005) Maternal Wnt11 activates the canonical Wnt signaling pathway required for axis formation in Xenopus embryos. Cell 120: 857–871. [DOI] [PubMed] [Google Scholar]
- 12. Cha S-W, Tadjuidje E, Tao Q, Wylie C, Heasman J (2008) Wnt5a and Wnt11 interact in a maternal Dkk1-regulated fashion to activate both canonical and non-canonical signaling in Xenopus axis formation. Development 135: 3719–3729. [DOI] [PubMed] [Google Scholar]
- 13. Cha S-W, Tadjuidje E, White JG, Wells J, Mayhew C, et al. (2009) Wnt11/5a complex formation caused by tyrosine sulfation increases canonical signaling activity. Curr Biol 19: 1573–1580. [DOI] [PubMed] [Google Scholar]
- 14. Pelegri F, Schulte-Merker S (1999) A gynogenesis-based screen for maternal-effect genes in the zebrafish,. Meth Cell Biol 60: 1–20. [DOI] [PubMed] [Google Scholar]
- 15. Kelly C, Chin AJ, Leatherman JL, Kozlowski DJ, Weinberg ES (2000) Maternally controlled β-catenin-mediated signaling is required for organizer formation in the zebrafish. Development 127: 3899–3911. [DOI] [PubMed] [Google Scholar]
- 16. Dosch R, Wagner DS, Mintzer KA, Runke G, Wiemelt AP, et al. (2004) Maternal control of vertebrate development before the midblastula transition: mutants from the zebrafish I. Dev Cell 6: 771–780. [DOI] [PubMed] [Google Scholar]
- 17. Pelegri F, Dekens MPS, Schulte-Merker S, Maischein H-M, Weiler C, et al. (2004) Identification of recessive maternal-effect mutations in the zebrafish using a gynogenesis-based method. Dev Dyn 231: 325–336. [DOI] [PubMed] [Google Scholar]
- 18. Wagner DS, Dosch R, Mintzer KA, Wiemelt AP, Mullins MC (2004) Maternal control of development at the midblastula transition and beyond: mutants from the zebrafish II. Dev Cell 6: 781–790. [DOI] [PubMed] [Google Scholar]
- 19. Bellipanni G, Varga M, Maegawa S, Imai Y, Kelly C, et al. (2006) Essential and opposing roles of zebrafish β-catenins in the formation of dorsal axial structures and neuroectoderm. Development 133: 1299–1309. [DOI] [PubMed] [Google Scholar]
- 20. Lyman-Gingerich J, Westfall TA, Slusarski DC, Pelegri F (2005) hecate, a zebrafish maternal effect gene, affects dorsal organizer induction and intracellular calcium transient frequency. Dev Biol 286: 427–439. [DOI] [PubMed] [Google Scholar]
- 21. Ataman B, Ashley J, Gorczyca D, Gorczyca M, Mathew D, et al. (2006) Nuclear trafficking of Drosophila Frizzled-2 during synapse development requires the PDZ protein dGRIP. Proc Natl Acad Sci USA 103: 7841–7846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Korkut C, Ataman B, Ramachandran P, Ashley J, Barria R, et al. (2009) Trans-synaptic transmission of vesicular Wnt signals through Evi/Wntless. Cell 139: 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lyman-Gingerich J, Lindeman R, Putiri E, Stolzmann K, Pelegri F (2006) The analysis of axis induction mutant embryos reveals morphogenetic events associated with zebrafish yolk extension formation. Dev Dyn 235: 2749–2760. [DOI] [PubMed] [Google Scholar]
- 24. Kishimoto Y, Lee K-H, Zon L, Hammerschmidt M, Schulte-Merker S (1997) The molecular nature of zebrafish swirl: BMP2 function is essential during early dorsoventral patterning. Development 124: 4457–4466. [DOI] [PubMed] [Google Scholar]
- 25. Mohler J, Wieschaus EF (1986) Dominant maternal effect mutations of Drosophila melanogaster causing the production of double-abdomen embryos. Genetics 112: 808–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lehmann R, Nüsslein-Volhard C (1991) The maternal gene nanos has a central role in posterior pattern formation of the Drosophila embryo. Development 112: 679–691. [DOI] [PubMed] [Google Scholar]
- 27. Mei W, Lee KW, Marlow FL, Miler AL, Mullins MC (2009) hnRNP I is required to generate the Ca2+ signal that causes egg activation in zebrafish. Development 136: 3007–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Carney TJ, Feitosa NM, Sonntag C, Slanchev K, Kluger J, et al. (2010) Genetic analysis of fin development in zebrafish identifies Furin and Hemicentin1 as potential novel Fraser Syndrome disease genes. PLoS Genet 6: e1000907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Marchler-Bauer A, Bryant SH (2004) CD-Search: protein domain annotations on the fly. Nuc Acids Res 32: W327–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Christoffels A, Koh EG, Chia JM, Brenner S, Aparicio S, et al. (2004) Fugu genome analysis provides evidence for a whole-genome duplication early during the evolution of ray-finned fishes. Mol Biol Evol 21: 1146–1151. [DOI] [PubMed] [Google Scholar]
- 31. Sémon M, Wolfe KH (2007) Reciprocal gene loss between Tetraodon and zebrafish after whole genome duplication in their ancestor. Trends Genet 23: 108–112. [DOI] [PubMed] [Google Scholar]
- 32. Tarbashevich K, Koebernick K, Pieler T (2007) XGRIP2.1 is encoded by a vegetally localizing, maternal mRNA and functions in germ cell development and anteroposterior PGC positioning in Xenopus laevis . Dev Biol 311: 554–565. [DOI] [PubMed] [Google Scholar]
- 33. Yoon C, Kawakami K, Hopkins N (1997) Zebrafish vasa homologue RNA is localized to the cleavage planes of 2- and 4-cell-stage embryos and is expressed in the primordial germ cells. Development 124: 3157–3165. [DOI] [PubMed] [Google Scholar]
- 34. Pelegri F, Knaut H, Maischein H-M, Schulte-Merker S, Nüsslein-Volhard C (1999) A mutation in the zebrafish maternal-effect gene nebel affects furrow formation and vasa RNA localization. Curr Biol 9: 1431–1440. [DOI] [PubMed] [Google Scholar]
- 35. Knaut H, Pelegri F, Bohmann K, Schwarz H, Nüsslein-Volhard C (2000) Zebrafish vasa RNA but not its protein is a component of the germ plasm and segregates asymmetrically prior to germ line specification. J Cell Biol 149: 875–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Theusch EV, Brown KJ, Pelegri F (2006) Separate pathways of RNA recruitment lead to the compartmentalization of the zebrafish germ plasm. Dev Biol 292: 129–141. [DOI] [PubMed] [Google Scholar]
- 37. Eno C, Pelegri F (2013) Gradual recruitment and selective clearing generate germ plasm aggregates in the zebrafish embryo. Bioarchitecture 3: 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Braat AK, Zandbergen T, van de Water S, Goos HJT, Zivkovic D (1999) Characterization of zebrafish primordial germ cells: morphology and early distribution of vasa RNA. Dev Dyn 216: 153–167. [DOI] [PubMed] [Google Scholar]
- 39. Bontems F, Stein A, Marlow F, Lyautey J, Gupta T, et al. (2009) Bucky ball organizes germ plasm assembly in zebrafish. Curr Biol 19: 414–422. [DOI] [PubMed] [Google Scholar]
- 40. Gupta T, Marlow FL, Ferriola D, Mackiewicz K, Dapprich J, et al. (2010) Microtubule actin crosslinking factor 1 regulates the balbiani body and animal-vegetal polarity of the zebrafish oocyte. PLoS Genet 6: e1001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cox RT, Spradling AC (2003) A Balbiani body and the fusome mediate mitochondrial inheritance during Drosophila oogenesis. Development 130: 1579–1590. [DOI] [PubMed] [Google Scholar]
- 42. Pepling ME, Wilhelm JE, O'Hara AL, Gephart GW, Spradling AC (2007) Mouse oocytes within germ cell cysts and primordial follicles contain a Balbiani body. Proc Natl Acad Sci USA 104: 187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Marlow FL, Mullins MC (2008) Bucky ball functions in Balbiani body assembly and animal-vegetal polarity in the oocyte and follicle cell layer in zebrafish. Dev Biol 321: 40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. De Brabander M, Geuens G, Nuydens R, Willebrords R, De Mey J (1961) Taxol induces the assembly of free microtubules in living cells and blocks the organizing capacity of the centrosomes and kinetochores. Proc Natl Acad Sci USA 78: 5608–5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Donovan MJ, Hart NH (1986) Cortical granule exocytosis is coupled with membrane retrieval in the egg of Brachydanio . J Exp Zool 237: 391–405. [DOI] [PubMed] [Google Scholar]
- 46. Maegawa S, Yasuda K, Inoue K (1999) Maternal mRNA localization of zebrafish DAZ-like gene. Mech Dev 81: 223–226. [DOI] [PubMed] [Google Scholar]
- 47. Hashimoto Y, Maegawa S, Nagai T, Yamaha E, Suzuki H, et al. (2004) Localized maternal factors are required for zebrafish germ cell formation. Dev Biol 268: 152–161. [DOI] [PubMed] [Google Scholar]
- 48. Olsen LC, Aasland R, Fjose A (1997) A vasa-like gene in zebrafish identifies putative primordial germ cells. Mech Dev 66: 95–105. [DOI] [PubMed] [Google Scholar]
- 49. De Robertis EM, Kuroda H (2004) Dorsal-ventral patterning and neural induction in Xenopus embryos. Annu Rev Cell Dev Biol 20: 285–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pelegri F (2003) Maternal factors in zebrafish development. Dev Dyn 228: 535–554. [DOI] [PubMed] [Google Scholar]
- 51. Marrari Y, Terasaki M, Arrowsmith V, Houliston E (2000) Local inhibition of cortical rotation in Xenopus eggs by an anti-KRP antibody. Dev Biol 224: 250–262. [DOI] [PubMed] [Google Scholar]
- 52. Marrari Y, Rouviere C, Houliston E (2004) Complementary roles for dynein and kinesins in the Xenopus egg cortical rotation. Dev Biol 271: 38–48. [DOI] [PubMed] [Google Scholar]
- 53. Cuykendall TN, Houston DW (2009) Vegetally localized Xenopus trim36 regulates cortical rotation and dorsal axis formation. Development 136: 3057–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mei W, Jin Z, Lai F, Schwend T, Houston DW, et al. (2013) Maternal Dead-End1 is required for vegetal cortical microtubule assembly during Xenopus axis specification. Development 140: 2334–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chan AP, Kloc M, Etkin LD (1999) fatvg encodes a new localized RNA that uses a 25-nucleotide element (FVLE1) to localize to the vegetal cortex of Xenopus oocytes. Development 126: 4943–4953. [DOI] [PubMed] [Google Scholar]
- 56. Chan AP, Kloc M, Larabell CA, LeGross M, Etkin LD (2007) The maternally localized RNA fatvg is required for cortical rotation and germ cell formation. Mech Dev 124: 350–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dong H, Zhang P, Song I, Petralia RS, Liao D, et al. (1999) Characterization of the Glutamate receptor-interacting proteins GRIP1 and GRIP2. J Neurosci 19: 6930–6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lee HJ, Zheng JJ (2010) PDZ domains and their binding partners: structure, specificity, and modification. Cell Commun Signal 8: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bard L, Groc L (2011) Glutamate receptor dynamics and protein interaction: lessons from the NMDA receptor. Mol Cell Neurosci 48: 298–307. [DOI] [PubMed] [Google Scholar]
- 60. Ivarsson Y (2012) Plasticity of PDZ domains in ligand recognition and signaling. FEBS Lett 586: 2638–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Takamiya K, Kostourou V, Adams S, Jadeja S, Chalepakis G, et al. (2004) A direct functional link betwen the multi-PDZ domain protein GRIP1 and the Fraser sydrome protein Fras1. Nat Genet 36: 172–177. [DOI] [PubMed] [Google Scholar]
- 62. Sugiura T, Shimizu T, Kijima A, Minakata S, Kato Y (2011) PDZ adaptors: their regulation of epithelial transporters and involvement in human diseases. J Pharm Sci 100: 3620–3635. [DOI] [PubMed] [Google Scholar]
- 63. Setou M, Seog DH, Tanaka Y, Kanai Y, Takei Y, et al. (2002) Glutamate-receptor-interacting protein GRIP1 directly steers kinesin to dendrites. Nature 417: 83–87. [DOI] [PubMed] [Google Scholar]
- 64. Su Q, Cai Q, Gerwin C, Smith CL, Sheng ZH (2004) Syntabulin is a microtubule-associated protein implicated in syntaxin tranport in neurons. Nat Cell Biol 6: 941–953. [DOI] [PubMed] [Google Scholar]
- 65. Cai Q, Gerwin C, Sheng ZH (2005) Syntabulin-mediated anterograde transport of mitochondria along neuronal processes. J Cell Biol 170: 959–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cai Q, Pen PY, Sheng ZH (2007) Syntabulin-kinesin-1 family member 5B-mediated axonal tranport contributes to activity-dependent presynaptic assembly. J Neurosci 27: 7284–7296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Fagni L, Ango F, Peroy J, Bockaert J (2004) Identification and functional roles of metabotropic glutamate receptor-interacting proteins. Sem Cell Dev Biol 15: 289–298. [DOI] [PubMed] [Google Scholar]
- 68. Furukawa J (2012) Structure and function of glutamate receptor amino terminal domains. J Physiol 590: 63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Larabell CA, Torres M, Rowning BA, Yost C, Miller JR, et al. (1997) Establishment of the dorsoventral axis in Xenopus embryos is presaged by early asymmetries in β-catenin that are modulated by the Wnt signalling pathway. J Cell Biol 136: 1123–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rowning BA, Wells J, Wu M, Gerhart JC, Moon RT, et al. (1997) Microtubule-mediated transport of organelles and localization of β–catenin to the future dorsal side of Xenopus eggs. Proc Natl Acad Sci USA 94: 1224–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Miller JR, Rowning BA, Larabell CA, Yang-Snyder JA, Bates R, et al. (1999) Establishment of the dorsal-ventral axis in Xenopus embryos coincides with the dorsal enrichment of dishevelled that is dependent on cortical rotation. J Cell Biol 146: 427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Weaver C, Far GH III, Pan W, Rowning BA, Wang J, et al. (2003) GBP binds kinesin light chain and translocates during cortical rotation in Xenopus eggs. Development 130: 5425–5436. [DOI] [PubMed] [Google Scholar]
- 73. Swan LE, Wichmann C, Prange U, Schmid A, Schmidt M, et al. (2004) A glutamate receptor-interacting protein homolog organizes muscle guidance in Drosophila. Genes Dev 18: 223–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Swan LE, Schmidt M, Schwarz T, Ponimaskin E, Prange U, et al. (2006) Complex interaction of Drosophila GRIP PDZ domains and Echinoid during muscle morphogenesis. EMBO J 25: 3640–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kume S, Inoue T, Mikoshiba K (2000) Gαs family G proteins activate IP3-Ca2+ signaling via Gβγ and transduce ventralizing signals in Xenopus . Dev Biol 226: 88–103. [DOI] [PubMed] [Google Scholar]
- 76. Saneyoshi T, Kume S, Amasaki Y, Mikoshiba K (2002) The Wnt/calcium pathway activates NF-AT and promotes ventral cell fate in Xenopus embryos. Nature 417: 295–299. [DOI] [PubMed] [Google Scholar]
- 77. Westfall TA, Brimeyer R, Twedt J, Gladon J, Olberding A, et al. (2003) Wnt5/pipetail functions in vertebrate axis formation as a negative regulator of Wnt/β-catenin activity. J Cell Biol 162: 889–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Westfall TA, Hjertos B, Slusarski DC (2003) Requirement for intracellular calcium modulation in zebrafish dorsal-ventral patterning. Dev Biol 259: 380–391. [DOI] [PubMed] [Google Scholar]
- 79. Wu S-Y, Shin J, Sepich DS, Solnica-Krezel L (2012) Chemokine GPCR signaling inhibits β-catenin during zebrafish axis formation. PLoS Biol 10: e1001403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Elinson RP, Rowning B (1988) Transient array of parallel microtubules in frog eggs: potential tracks for a cytoplasmic rotation that specifies the dorso-ventral axis. Dev Biol 128: 185–197. [DOI] [PubMed] [Google Scholar]
- 81. Houliston E, Elinson RP (1991) Patterns of microtubule polymerization relating to cortical rotation in Xenopus laevis eggs. Development 112: 107–117. [DOI] [PubMed] [Google Scholar]
- 82. Schroeder MM, Gard DL (1992) Organization and regulation of cortical microtubules during the first cell cycle of Xenopus eggs. Development 114: 699–709. [DOI] [PubMed] [Google Scholar]
- 83. Fuentes R, Fernández J (2010) Ooplasmic segregation in the zebrafish zygote and early embryo: pattern of ooplasmic movements and transport pathways. Dev Dyn 239: 2172–2189. [DOI] [PubMed] [Google Scholar]
- 84. Solnica-Krezel L, Driever W (1994) Microtubule arrays of the zebrafish yolk cell: organization and function during epiboly. Development 120: 2443–2455. [DOI] [PubMed] [Google Scholar]
- 85. Kaneshiro K, Miyauchi M, Tanigawa Y, Ikenishi K, Komiya T (2007) The mRNA coding for Xenopus glutamate receptor interacting protein 2 (XGRIP2) is maternally transcribed, transported through the late pathway and localized to the germ plasm. Biochem Biophys Res Commun 355: 902–906. [DOI] [PubMed] [Google Scholar]
- 86. Kirilenko P, Weierud FK, Zorn AM, Woodland HR (2008) The efficiency of Xenopus primordial germ cell migration depends on the germplasm mRNA encoding the PDZ domain protein Grip2. Differentiation 76: 392–403. [DOI] [PubMed] [Google Scholar]
- 87. St. Johnston D, Nüsslein-Volhard C (1992) The origin of pattern and polarity in the Drosophila embryo. Cell 68: 201–219. [DOI] [PubMed] [Google Scholar]
- 88. Rebscher N, Zelada-González F, Banisch TU, Raible F, Arendt D (2007) Vasa unveils a common origin of germ cells and of somatic stem cells from the posterior growth zone in the polychaete Platynereis dumerilii . Dev Biol 306: 599–611. [DOI] [PubMed] [Google Scholar]
- 89. Wang Y, Zayas RM, Guo T, Newmark PA (2007) nanos function is essential for development and regeneration of planarian germ cells. Proc Natl Acad Sci USA 104: 5901–5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Gazave E, Béhague J, Laplane L, Guillou A, Préau L, et al. (2013) Posterior elongation in the annelid Platynereis dumerilii involves stem cells molecularely related to primordial germ cells. Dev Biol 382: 246–267. [DOI] [PubMed] [Google Scholar]
- 91.Brand M, Granato M, Nüsslein-Volhard C (2002) Keeping and raising zebrafish. In: Nüsslein-Volhard C, Dahm R, editors. Zebrafish - A Practical Approach. Oxford: Oxford University Press. 7–37 p. [Google Scholar]
- 92. Pelegri F, Mullins M (2011) Genetic screens for mutations affecting adult traits and parental-effect genes. Meth Cell Biol 104: 83–120. [DOI] [PubMed] [Google Scholar]
- 93. Kimmel C, Ballard WW, Kimmel SR, Ullmann B, Schilling TF (1995) Stages of embryonic development in the zebrafish. Dev Dyn 203: 253–310. [DOI] [PubMed] [Google Scholar]
- 94. Fink JL, Hamilton N (2007) DomainDraw: a macromolecular feature drawing program. In Silico Biol 7: 145–150. [PubMed] [Google Scholar]
- 95. Pelegri F, Maischein H-M (1998) Function of zebrafish β-catenin and TCF-3 in dorsoventral patterning. Mech Dev 77: 63–74. [DOI] [PubMed] [Google Scholar]
- 96. Schulte-Merker S, Hammerschmidt M, Beuchle D, Cho K, DeRobertis EM, et al. (1994) Expression of the zebrafish goosecoid and no tail gene products in wild-type and mutant ntl embryos. Development 120: 843–852. [DOI] [PubMed] [Google Scholar]
- 97. Schulte-Merker S, Lee KJ, McMahon AP, Hammerschmidt M (1997) The zebrafish organizer requires chordino . Nature 387: 862–863. [DOI] [PubMed] [Google Scholar]
- 98. Joly J-S, Joly C, Schulte-Merker S, Boulkebache H, Condamine H (1993) The ventral and posterior expression of the homeobox gene eve1 is perturbed in dorsalized and mutant embryos. Development 119: 1261–1275. [DOI] [PubMed] [Google Scholar]
- 99. Lekven AC, Thorpe CJ, Waxman JS, Moon RT (2001) Zebrafish wnt8 encodes two wnt8 proteins on a bicistronic transcript and is required for mesoderm and neuroectoderm pattterning. Dev Cell 1: 103–114. [DOI] [PubMed] [Google Scholar]
- 100. Thisse C, Thisse B, Schilling TF, Postlethwait JH (1993) Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development 119: 1203–1215. [DOI] [PubMed] [Google Scholar]
- 101. Selman K, Wallace RA, Sarka A, Qi X (1993) Stages of oocyte development in the zebrafish, Brachydanio rerio . J Morphol 218: 203–224. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Age-dependency of the hec mutant phenotype. The hecate mutant phenotype is stronger in younger females. Also note that at both ages the strength of the alleles is hecp06ucal>hect2800>hecp08ajug. Homozygous mutant females at 3 and 12 month after birth were crossed against wild-type males to produce clutches of mutant embryos. Phenotypes were classified at 24 hpf as in [24] and Table 1. Results were pooled from clutches from 15 different females for each allele. Number of embryos is as follows: 3-month females: WT, 1440; hecp06ucal, 1178; hect2800, 575; hecp08ajug, 1399; 12-month females: WT, 1259; hecp06ucal, 1812; hect2800, 655; hecp08ajug, 1516. Pearson's Chi-squared test shows statistical significant differences between 3-month and 12-month females for all alleles (p-values: hecp06ucal, 0.002477; hect2800, <2.2e-16; hecp08tajug, <2.2e-16) but not for wild-type females (p-value 0.3421). Pair-wise comparisons between all three alleles are also significantly different using the same analysis (p-values: hecp06ucal vs. hect2800 <2.2e-16; hecp06ucal vs. hecp08ajug <2.2e-16; hect2800 vs. hecp08ajug <2.2e-16).
(TIFF)
Phylogenetic tree of Grip1 and Grip2 proteins among Drosophila and vertebrate species and number of PDZ domains in the predicted protein. Left: phylogenetic tree using ClustalW. Gene-ID from NCBI or Ensembl-genome databases: GRIP2 rat: NP_612544.2; GRIP2 mouse: NP_001152979.1; GRIP2 human: NP_001073892.1; Grip2 Xenopus: NP_001091382.1; GRIP2 chicken: ENSGALP00000010397; Grip2b zebrafish: XP_001922281.1; Grip2b medaka: ENSORLP00000012064; Grip2b fugu: ENSTRUP00000040982; Grip2a fugu: ENSTRUP00000024040; Grip2a medaka: ENSORLP00000012637; Grip2a zebrafish: NP_001116760.1; GRIP1 mouse: NP_083012.1; GRIP1 rat: ENSRNOP00000061369; GRIP1 human: ENSP00000352780; GRIP1 chicken: ENSGALP00000016069; Grip1 Xenopus: ENSXETP00000015955; Grip1 fugu: ENSTRUP00000012549; Grip1 medaka: ENSORLP00000021825; Grip1 zebrafish: NP_001038316.1; Grip fruitfly: NP_572285.2. Right: Diagrams of the overall structure of the respective proteins (gray) highlighting the number of PDZ domains (orange), based on Ensembl annotation.
(TIFF)
Expression of zebrafish grip2a mRNA in wild-type and hecate mutant embryos. A) RT-PCR analysis of grip2a mRNA and ef1α control expression in wild-type and hec mutant embryos, as well as wild-type ovaries and wild-type adults (male, female, female with removed ovaries). B) Quantitative RT-PCR analysis shows grip2a mRNA expression levels, relative to ef1α expression at the same stages. Maternal grip2a mRNA levels are reduced in embryos mutant for all hec alleles.
(TIFF)
Quantification of the off-center shift of grip2a mRNA localization domain in control (DMSO-treated) and nocodazole-treated embryos fixed at 40 mpf. A) Diagram indicating angles of the grip2a mRNA localization domain landmarks with respect to the vegetal pole of the embryo. LB: left boundary; RB: right boundary; S: total span of domain; M: midpoint of domain. B) Quantification parameters, with brackets indicating standard deviation. In control embryos, but not in nocodazole-treated embryos, left and right boundaries and midpoint of the grip2a mRNA localization domain experience a similar off-center shift, while the span of the domain appears unchanged. Blind analysis of 38 and 41 DMSO- and nocodazole-treated embryos, respectively. Similar changes were observed in embryos fixed at 30 mpf, although differences at this time point were less pronounced than at 40 mpf (data not shown).
(TIF)
Distribution of microtubule organization phenotypes at the vegetal cortex of wild-type and hecate mutant embryos. A–G) Examples of various vegetal cortex microtubule arrangements at 20 mpf: normally aligned (A), partially aligned (B, C), unbundled and lacking organization (D, E) and exhibiting aster-like structures (F, G). (A) is from a wild-type embryo and (B–G) from hec mutants. Magnification bar in (G) corresponds to 40 µm for panels (A–G). H) Distribution of phenotypes. Wild-type embryos exhibit highly aligned microtubule network while hec mutants show disorganization, lack of bundling and aster-like structures. The two distributions are significantly different (Fisher's Exact Test, p-value = 5.335e-09).
(TIF)
Treatment with taxol does not affect the hec mutant phenotype. Phenotypic distribution of axial defects (classification as in [24]) of taxol- and solvent (DMSO)-treated wild-type and hec mutant embryos. Data is derived from two to three different experiments after a 30 minute exposure to 10 µM taxol starting at 10 mpf. There are no statistically significant phenotypic differences between taxol-treated and control embryos (student t-test, 2-tailed, unpaired).
(TIFF)
F-actin cortex at the vegetal pole is similar in wild-type and hec mutant embryos. At 20 mpf, both wild-type (A) and mutant (B) eggs show F-actin rich folds and villi-like structures, which may correspond to previously described microplicae [45]. Number of embryos tested: 18 wt (from a pool of four females) and 24 mutants (from two different mutant females). A fraction (21%, n = 24) of hec mutant embryos show radial F-actin enrichments (C), correlating with aster-like microtubule structures in these embryos (Figure S5). Magnification bar in (C) corresponds to 40 µm in all panels.
(TIFF)
Expression of wnt8a mRNA in hec mutant embryos. Quantitative RT-PCR analysis of wnt8a mRNA levels relative to ef1α mRNA. wnt8a relative expression in hec mutants is close to wild-type at 30 mpf but becomes reduced at 60 mpf.
(TIFF)
PGC determination is not adversely affected in hecate mutant embryos. The number of PGCs (as identified by vasa expression through whole mount in situ hybridization) was determined in wild-type and hec mutant embryos at the 3- through 5-somite stages (ca. 10.5 hpf). At these stages, PGCs have not yet reached the prospective gonad location and appear relatively scattered, which facilitated quantification analysis. The average number of PGCs was 17.9 +/− 3.5 in hec mutant embryos (n embryos = 158), compared to 13.6 +/− 3.6 in wild-type embryos (n embryos = 214). The slightly higher number of PGCs in mutants was statistically significant (Student's t-test, p-value = 0.02). It is possible, however, that this difference reflects a bias in our ability to count individual PGCs in hec mutants, where PGCs are dispersed throughout the span of the embryos, compared to wild-type embryos, where PGCs are concentrated along the dorsal axis. Regardless of this uncertainty, the data indicate that hec function is not essential for PGCs determination.
(TIFF)
Three identified hec mutant alleles do not complement each other in complementation crosses. Top: Homozygous mutant males of one allele were crossed with heterozygous females of another allele to produce offsprings, which are raised to adulthood. Female adult F1 progeny were tested for hec-associated maternal effects by crossing them against wild-type males and scoring the resulting F2 clutches as wild-type or mutant. Only test crosses with more than 50 embryos were scored, and were scored as mutant when the fraction of V1–V4 categories was ≥50%. F2 clutches scored as wild-type exhibited ≤2% of embryos with V1–V4 phenotypes. If mutations are allelic, crosses are expected to yield F1 females exhibiting wild-type (from heterozygote females) or mutant (from transheterozygote females) F2 phenotypes in a 1∶1 ratio. Bottom: Summary of analysis showing non-complementation of three hec alleles.
(DOC)