Alp7–Alp14 (TACC–TOG) is a microtubule-associated protein complex that undergoes nucleocytoplasmic shuttling. Alp7 and Alp14 have the NLS and NES responsible for the shuttling, respectively. At mitotic entry, the cyclin-dependent kinase phosphorylates Alp7, resulting in accumulation of the complex to the nucleus, which promotes spindle assembly.
Abstract
As cells transition from interphase to mitosis, the microtubule cytoskeleton is reorganized to form the mitotic spindle. In the closed mitosis of fission yeast, a microtubule-associated protein complex, Alp7–Alp14 (transforming acidic coiled-coil–tumor overexpressed gene), enters the nucleus upon mitotic entry and promotes spindle formation. However, how the complex is controlled to accumulate in the nucleus only during mitosis remains elusive. Here we demonstrate that Alp7–Alp14 is excluded from the nucleus during interphase using the nuclear export signal in Alp14 but is accumulated in the nucleus during mitosis through phosphorylation of Alp7 by the cyclin-dependent kinase (CDK). Five phosphorylation sites reside around the nuclear localization signal of Alp7, and the phosphodeficient alp7-5A mutant fails to accumulate in the nucleus during mitosis and exhibits partial spindle defects. Thus our results reveal one way that CDK regulates spindle assembly at mitotic entry: CDK phosphorylates the Alp7–Alp14 complex to localize it to the nucleus.
INTRODUCTION
Microtubules dramatically change their organization throughout the cell cycle (Kirschner and Mitchison, 1986; Hagan and Hyams, 1988; Hayles et al., 1994). Cytoplasmic arrays of microtubules are formed during interphase but are disassembled and reorganized into the spindle microtubule upon mitotic entry. A small GTPase of the Ras superfamily, Ran, plays an essential role in the spatiotemporal microtubule reorganization (Dasso, 2001; Clarke and Zhang, 2008). Ran is known to dictate the nucleocytoplasmic transport of a number of biomolecules. Guanosine diphosphate (GDP)–bound Ran predominantly exists in the cytoplasm but is converted to the GTP form (Ran-GTP) in the nucleus (Bischoff and Ponstingl, 1991; Matsumoto and Beach, 1991; Zhang and Clarke, 2001). The differential localization of the GDP- and GTP-bound forms of Ran separated by the nuclear envelope generates bidirectional transport between the cytoplasm and the nucleus.
At mitotic entry, the nuclear envelope breaks down in higher eukaryotes, but the molecular gradient of Ran-GTP remains near the chromosomes (Carazo-Salas et al., 1999, 2001; Kalab et al., 1999; Ohba et al., 1999; Wilde and Zheng, 1999). Several microtubule-associated proteins (MAPs), including TPX2, NuMA, and HURP, are called spindle assembly factors (SAFs), which are “cargo” proteins regulated by Ran-GTP (Gruss et al., 2001; Nachury et al., 2001; Wiese et al., 2001; Koffa et al., 2006; Sillje et al., 2006). They are released from the nuclear import complex in the Ran-GTP–rich area around chromosomes after nuclear envelope breakdown, which induces microtubule assembly around chromosomes (Karsenti and Vernos, 2001).
The fission yeast Schizosaccharomyces pombe undergoes a closed mitosis in which the nuclear envelope remains intact. Persistence of the nuclear envelope enables the Ran-GTP–dependent transport to remain as active during mitosis as during interphase (Matsusaka et al., 1998; Sato and Toda, 2007; Nakazawa et al., 2008). At the same time, this means that spindle microtubules must be assembled in the compartmentalized nucleus of the yeast. The spatial barrier necessitates that several MAPs be imported into the mitotic nucleus to promote spindle assembly. This suggests that the Ran-dependent system for microtubule assembly in open-mitosis organisms might have evolved from closed-mitosis organisms such as yeast. To investigate how the spindle is formed at mitotic onset, it is tempting to focus on the primitive Ran system in yeast, particularly on how SAFs overcome the nuclear envelope as a spatial barrier and accomplish their mitosis-specific enrichment to the spindle.
The orthologue of the human transforming acidic coiled-coil (TACC) protein, Alp7 (also called Mia1), undergoes nucleocytoplasmic shuttling, accumulates in the nucleus during mitosis, and localizes to spindle microtubules (Oliferenko and Balasubramanian, 2002; Sato et al., 2003), as well as to kinetochores in both mitosis and meiosis (Kakui et al., 2013; Tang et al., 2013). Alp7 has a nuclear localization signal (NLS) at the N-terminal part and is a crucial target of Ran-GTP–dependent microtubule formation (Sato and Toda, 2007, 2010). Thus Alp7 is a crucial and unique SAF that accumulates in the nucleus, and we have focused on the mechanism for Alp7 nuclear accumulation.
The significance of the nuclear function of Alp7 is demonstrated by the alp7-RARA mutant, which has mutations in the intrinsic NLS. Alp7-RARA fails to enter the nucleus and causes severe mitotic defects, such as an abnormal monopolar spindle, indicating that nuclear import of Alp7 during mitosis is indispensable for spindle integrity. The accumulation of Alp7 in the nucleus is influenced by cyclin-dependent kinase (CDK; Sato et al., 2009a), suggesting that nuclear accumulation of Alp7 is a key step linking cell-cycle control by CDK to spindle assembly.
It is also known that TACC proteins interact with their binding partner, tumor overexpressed gene (TOG; also known as XMAP215, the Xenopus orthologue), through the C-terminal TACC domain. The TACC–TOG complex is required for microtubule organization from spindle poles in various eukaryotes. TACC controls localization of TOG to the spindle microtubules and poles (Cullen and Ohkura, 2001; Lee et al., 2001; Bellanger and Gönczy, 2003; Le Bot et al., 2003; Srayko et al., 2003; Sato et al., 2004; Peset and Vernos, 2008). S. pombe has two TOG proteins, Dis1 (Nabeshima et al., 1995) and Alp14 (also called Mtc1; Garcia et al., 2001; Nakaseko et al., 2001). Both proteins have two conserved TOG domains, which consist of paddle-like HEAT repeats and are required for gripping the tubulin dimer (Neuwald and Hirano, 2000; Ohkura et al., 2001). Dis1/TOG localizes to microtubules, spindle pole bodies (SPBs; the yeast centrosome equivalent), and mitotic kinetochores (Nabeshima et al., 1995; Nakaseko et al., 2001). Dis1 is phosphorylated by CDK, which is essential for translocating Dis1 from spindle microtubules to kinetochores (Nabeshima et al., 1995; Aoki et al., 2006). TOG proteins in most eukaryotes interact with TACC proteins (Peset and Vernos, 2008), but the TACC counterpart that interacts with Dis1 has not been found.
The Alp14-deletion mutant (alp14∆) shows disorganized and fragile microtubules (Garcia et al., 2001; Nakaseko et al., 2001), indicating its role in microtubule stabilization. In line with this, TOG proteins, including Alp14, are predominantly located in dynamic plus ends of microtubules and catalyze microtubule polymerization using TOG domains (Brouhard et al., 2008; Al-Bassam and Chang, 2011; Al-Bassam et al., 2012; Widlund et al., 2011). Alp14 (TOG) interacts with Alp7 (TACC) through the C-terminal domain of Alp14 (Sato et al., 2004). The localization dependence of Alp14 on Alp7 implies that Alp7 regulates the localization, whereas Alp14 controls the dynamics of microtubules as the Alp7–Alp14 complex. It is therefore necessary to consider whether Alp14 could be responsible for nuclear accumulation of the Alp7–Alp14 complex. In this study, we construct and analyze the localization of a series of mutants of Alp7 and Alp14 throughout the cell cycle to identify the mechanism responsible for mitosis-specific nuclear localization.
RESULTS
Domain analysis of Alp14
It is necessary to learn how the Alp7–Alp14 complex is imported to the nucleus and exported to the cytoplasm in order to clarify the molecular machinery of the temporal regulation of the nuclear accumulation. We previously demonstrated that Alp7 has the NLS in its N-terminal half through truncation analyses of Alp7 (Sato and Toda, 2007). We next sought to identify the nuclear export signal (NES) of the Alp7–Alp14 complex.
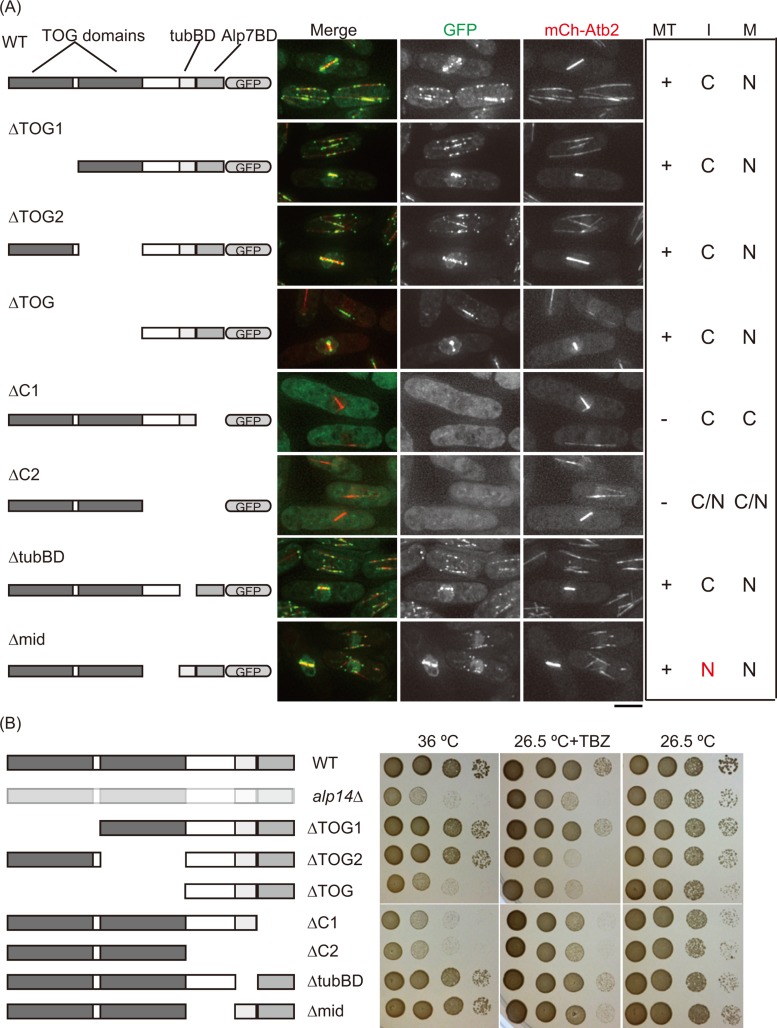
Regarding the NES, we previously found that the mutant protein Alp7-L461A accumulates in the nucleus even during interphase (Ling et al., 2009; Sato et al., 2009a). Although residues around L461 could be good candidates for the NES, the Alp7-L461A mutant protein failed to associate with Alp14 (Sato et al., 2009a). We therefore could not exclude the possibility that L461 is a binding site for Alp14, and the binding partner Alp14 might have a real NES sequence. To test this possibility, we constructed seven truncation mutants that contained a short version of alp14 genes with the green fluorescent protein (GFP) gene fused to the C-terminus and expressed them from the endogenous promoter. The localization was observed together with mCherry-Atb2, a marker for microtubules. As seen before, Alp14-GFP localized to cytoplasmic microtubules during the interphase of wild-type (WT) cells, and in mitosis, it accumulated in the nucleus and localized to spindle microtubules (Garcia et al., 2001; Nakaseko et al., 2001; Figure 1A).
FIGURE 1:
Domain analysis of Alp14 reveals NES activity in the middle region. (A) Seven alp14 truncation mutant genes were fused with the GFP gene and expressed from the native alp14+ promoter: TOG1 (∆1–244), TOG2 (∆245–500), ∆TOG (∆1–500), ∆C1 (∆696–809), ∆C2 (∆501–809), ∆tubBD (∆642–695), and ∆mid (∆501–641). Wild type and alp14 mutants were grown at 25°C and visualized together with mCherry-Atb2, a microtubule marker. Alp7BD, Alp7-binding domain; tubBD, tubulin-binding domain. Right, localization of Alp14 (wild-type and mutant proteins) to microtubules (MT), in interphase (I), and in mitosis (M). +, localized; –, not localized; C, cytoplasm; N, nucleus. Scale bar, 5 μm. (B) Three truncated mutants (∆TOG, ∆C1, and ∆C2) exhibited temperature and TBZ sensitivities. The 1/10 serial dilutions of wild- type cells (WT; top), the alp14-deletion mutant (second from top), and alp14 truncation mutants were spotted on rich medium (YE5S) incubated at 26.5 or 36°C and YE5S plate containing TBZ (10 μg/ml) incubated at 26.5°C.
Three truncation mutant proteins that lack the N-terminal region (∆TOG, ∆TOG1, and ∆TOG2) also localized to cytoplasmic and spindle microtubules. ∆TOG-GFP lacking the entire TOG domains was able to localize to cytoplasmic microtubules during interphase and to spindle microtubules during mitosis (Figure 1A), although its localization pattern appeared distinct from that of WT, possibly because the overall microtubule array was disorganized in this mutant. The ∆TOG mutant showed severe sensitivity to both high temperature and thiabendazole (TBZ), meaning that this mutant lost its function (Figure 1B). Neither ∆TOG1 (lacking the first TOG domain) nor ∆TOG2 (lacking the second TOG domain) showed temperature sensitivity. ∆TOG2 showed TBZ sensitivity, whereas ∆TOG1 did not. The sensitivity in ∆TOG2 may be due to structural inflexibility caused by an artificial connection between the TOG1 and the C-terminal domains. Alternatively, as reported previously (Al-Bassam et al., 2012), TOG2 is more important than TOG1 for Alp14 function. Thus one TOG domain is dispensable for microtubule formation and maintenance.
The behavior of C-terminal truncation mutants confirmed the importance of the C-terminal region for localization to microtubules (Sato et al., 2004). The C-terminal end (amino acids 696–809) interacts with Alp7 (Sato et al., 2004). ∆C1 and ∆C2, which lack the binding region, could not localize to microtubules. The microtubule-binding domain (tubBD; 642–695) is believed to combine directly with tubulin/microtubule, as previously shown by an in vitro assay (Nakaseko et al., 2001). However, ∆tubBD-GFP still localized to microtubules, although the fluorescence signal was weaker than seen in the wild-type Alp14-GFP. The ∆tubBD mutant showed little sensitivity to high temperature or TBZ (Figure 1B), implying that the ∆tubBD protein might be loaded into microtubules in vivo via interaction with Alp7.
Of interest, the last mutant, Alp14-∆mid (∆501-641)-GFP, accumulated in the nucleus not only during mitosis but also during interphase (Figure 1A). The ∆mid mutant has no temperature or TBZ sensitivity (Figure 1B).
Taken together, the results show that TOG domains are dispensable for the association of Alp14 with microtubules; the Alp7 interaction domain, but not the microtubule binding domain, is crucial for this association; and the domain 501–641 has NES activity.
L615 is critical for the NES activity of Alp14
Because the constitutive nuclear localization seen in the alp14-∆mid mutant indicates a lack of NES, we next tried to narrow down the region. Seven truncation mutants that lack narrow regions in 501–641 were created to pinpoint the crucial region for the NES activity. Each mutant lacks ∼20 residues sequentially in 501–641 and was fused with GFP to observe its localization with mCherry-Atb2. As shown in Figure 2A, six mutant proteins localized to cytoplasmic microtubules during interphase in the same way as the wild-type Alp7. The ∆601-620 mutant, however, showed nuclear accumulation during interphase, suggesting the existence of NES activity within 601–620. To confirm that NES activity resided in this region, we fused these 20 residues with the reporter protein glutathione S-transferase (GST)–GFP and expressed and the resulting fusion protein GST-GFP-14NES in WT cells. As shown in Figure 2B, GST-GFP exists in both the nucleus and the cytoplasm. Quantification of the average GFP fluorescence intensity in the nucleus and the cytoplasm verified that the nuclear GST-GFP intensity was 1.24-fold greater than the cytoplasmic intensity (Figure 2C). In contrast, the GST-GFP-14NES fusion protein predominantly localized to the cytoplasm (Figure 2B), and the nuclear/cytoplasmic signal ratio of GST-GFP-14NES dropped to approximately one-third that of GST-GFP (Figure 2C). This demonstrates that 601–620 of Alp14 retains the NES activity.
FIGURE 2:
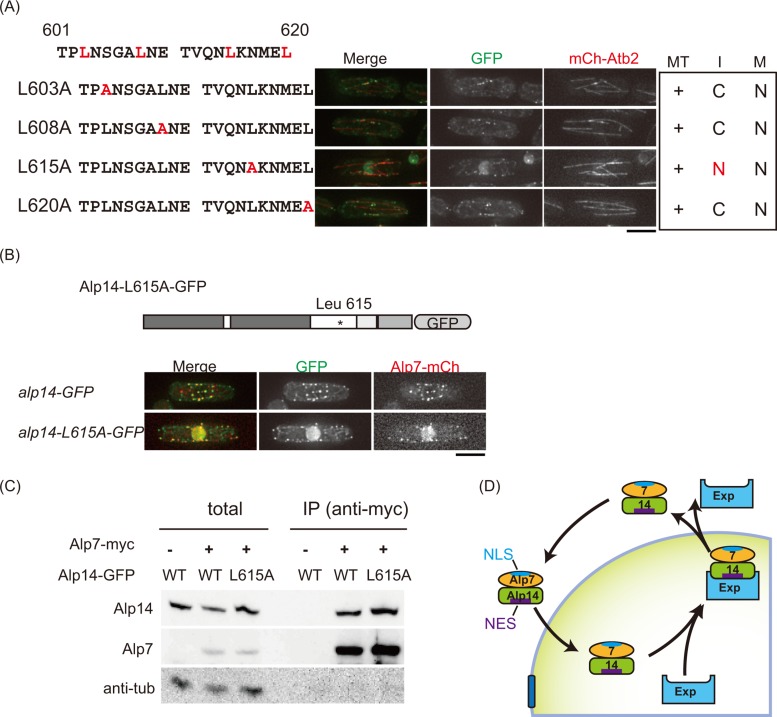
The region 601–620 of Alp14 has NES activity. (A) Localization of Alp14-GFP (WT) and truncation mutant proteins listed on the left. Each mutant lacks ∼20 residues sequentially in 501–641. Strains were cultured at 25°C and observed with mCherry-Atb2. Right, their localization to microtubules (MT), in interphase (I), and in mitosis (M). +, localized; C, cytoplasm; N, nucleus. (B) A sequence of residues 601–620 of Alp14 was fused with reporter protein (GST-GFP) and expressed in a Cut11-3mRFP strain. Top, actual sequence of 601–620. Bottom, schematic illustration of the reporter constructs. (C) Ratio of GFP fluorescence intensity (nucleus/cytoplasm) quantified for GST-GFP and GST-GFP-14NES. n = 15 cells, *p = 2.20 × 10−16 < 0.0001 (Student's t test).
Having confirmed that the 601–620 region is sufficient for the NES activity of Alp14, we further pursued the critical residue for the NES activity in this region. There are four hydrophobic residues in 601–620 (leucines L603, L608, L615, and L620; Figure 3A), each of which was converted to alanine by mutagenesis. Three mutants (L603A, L608A, and L620A) behaved normally, but Alp14-L615A-GFP showed nuclear accumulation during interphase. Thus the L615 residue is critical for the NES activity of Alp14.
FIGURE 3:
Leucine 615 is critical for the NES activity of Alp14. (A) Four point mutants of Alp14-GFP were observed at 25°C. Each mutant (L603A, L608A, L615A, and L620A) has single–amino acid conversion (leucine to alanine) in 601–620. (B) Strains expressing Alp7-mCherry with Alp14-GFP (WT) or Alp14-L615A-GFP were observed at 25°C. (C) The expression level of Alp14-L615A-GFP and its interaction with Alp7-myc were examined using coimmunoprecipitation assay followed by Western blotting. Alp7-myc was immunoprecipitated, and Alp14-GFP and Alp14-L615A-GFP were detected with the anti-GFP antibody. –, untagged strain; +, myc-tagged strains. (D) A model describing how the Alp7–Alp14 complex shuttles between the nucleus and the cytoplasm in interphase. Alp7 and Alp14 shuttles as a complex: NLS in Alp7 is responsible for the nuclear import of the complex, and NES in Alp14 is in charge of exporting the complex to the cytoplasm. Exp, exportin.
We next observed localization of the TACC protein Alp7 when the mutant TOG protein Alp14-L615A-GFP accumulated in the nucleus during interphase. Alp7 was then tagged with mCherry and endogenously expressed in the alp14-L615A-GFP mutant. Like Alp14-L615A-GFP, Alp7-mCherry also accumulated in the nucleus throughout the cell cycle (Figure 3B). Cytoplasmic microtubule organization appeared almost normal in the mutants lacking NES activity (alp14-∆mid, ∆601-620, and L615A), even though most of the Alp7–Alp14 complex accumulated in the nucleus (Figures 1A, 2A, and 3, A and B), indicating that some fraction of the proteins still localized to the cytoplasm.
The protein expression levels of Alp14-L615A-GFP and its affinity to Alp7 were comparable to those of Alp14-WT-GFP (Figure 3C). It is notable that Alp14 failed to enter the nucleus in the NLS-deficient alp7-RARA mutant (Sato et al., 2009a), indicating that the nuclear entry of the Alp7–Alp14 complex relies on the NLS in Alp7. Taken together, these results suggest that Alp14 and Alp7 shuttle between the nucleus and the cytoplasm as a complex during interphase and that Alp7 contributes to importing Alp14 in the nucleus, whereas the NES in Alp14 is responsible for exporting the complex during interphase (Figure 3D).
Alp7-∆61-116-GFP shows reduced nuclear accumulation
Having clarified how the nuclear import and export of the Alp7–Alp14 complex is achieved, we next sought to illuminate the molecular mechanism of how the complex accumulates, specifically during mitosis. We then searched for mutants of Alp7 and Alp14 that do not exhibit nuclear accumulation during mitosis. First, we suspected the possibility that the NES activity of Alp14 is inhibited during mitosis in order to achieve the nuclear accumulation of Alp7–Alp14.
To this end, we created a number of truncation mutants of Alp14, but none of them lost mitosis-specific nuclear accumulation except the C-terminally-deleted mutants that could not bind to Alp7 (Sato et al., 2004; Figure 1A and unpublished data). Next we tested the possibility that Alp14 is phosphorylated by CDK, possibly leading to inhibition of NES activity. We then performed an in vitro phosphorylation assay, but we did not find any hint of Alp14 phosphorylation by CDK (Supplemental Figure S1A). Because Alp14 possesses seven consensus sequences of CDK phosphorylation, we made the alp14 mutant by substituting the seven serine and threonine residues with alanine (alp14-7A-GFP; Supplemental Figure S1B). The localization of Alp14-7A-GFP, however, was normal (Supplemental Figure S1, B and C), reducing the possibility of phosphorylation-dependent regulation of localization.
Because we did not find any evidence for phosphorylation-based localization control of Alp14, we next focused on analyses of the binding partner, Alp7. We constructed truncation mutants of Alp7 tagged with GFP and observed with mCherry-Atb2 as a microtubule marker (Figure 4A). It was already shown that the C-terminal coiled-coil domain (TACC domain) of Alp7 is crucial for interaction with Alp14 and that the N-terminal region contains the NLS (Sato et al., 2004; Sato and Toda, 2007). The truncated protein lacking the entire TACC domain was located in both the nucleus and the cytoplasm (Sato et al., 2004; Sato and Toda, 2007), possibly because of passive diffusion. To block effects of diffusion, we constructed the mutant alp7-∆210-418-GFP, which lacks the middle C-terminal domain, with the far end (419–474) corresponding to the Alp14-binding domain left intact (Figure 4A and unpublished data). The mutant protein accumulated in the mitotic nucleus, although it did not localize to microtubules. This indicates that the middle region (210–418) of Alp7 does not contribute to mitosis-specific nuclear accumulation.
FIGURE 4:
Alp7-∆61-116 fails to accumulate in the nucleus. (A) Truncated proteins of Alp7 (∆210–418, ∆1–60, ∆61–116, ∆127–144, ∆145–164, ∆165–187, and ∆188–209) were expressed from alp7 native promoter and observed with mCherry-Atb2. I, interphase cells; M, mitotic cells. All strains are cultured at 25°C. Right, their localization to microtubules (MT), in interphase, and in mitosis. +, localized; C, cytoplasm; N, nucleus. (B) alp7-GFP nda3-KM311 cells were arrested at metaphase at 17°C for 6.5 h, and Alp7-GFP was immunoprecipitated with anti-GFP antibody and treated with λ-phosphatase (PPase) in the presence or absence of the phosphatase inhibitor. The upper band (black arrowhead) corresponds to phosphorylated Alp7-GFP, whereas the lower band (white arrowhead) is nonphosphorylated Alp7-GFP.
We then tested whether the critical element for mitosis-specific nuclear accumulation resides in the N-terminus. Of interest, we found that Alp7-∆61-116-GFP was reduced in nuclear accumulation during mitosis and during localization to the spindle, whereas other mutants behaved normally, in the same way as wild-type cells (Figure 4A). This indicates that the N-terminal region 61–116 right before the NLS (117–126) is responsible for mitotic nuclear accumulation of Alp7.
Our previous results demonstrated that mitotic nuclear accumulation of Alp7 is controlled by CDK (Sato et al., 2009a). Taking these results together, we hypothesized that CDK phosphorylates Alp7, which causes mitotic accumulation of Alp7–Alp14. To test whether Alp7 is phosphorylated in vivo, we prepared a mitotic extract from the cold-sensitive mutant of β-tubulin, nda3-KM311 (Toda et al., 1983; Umesono et al., 1983; Hiraoka et al., 1984), and purified Alp7-GFP expressed therein via immunoprecipitation followed by SDS–PAGE gel mobility shift assay. As shown in Figure 4B, Alp7-GFP was detected as a double band. The higher band with low mobility in the gel shifted to the lower band when treated with λ protein phosphatase. Addition of a phosphatase inhibitor canceled the mobility shift (Figure 4B), suggesting that the double band reflects phosphorylation of some fraction of Alp7 in mitotic cells.
Five residues in the N-terminus of Alp7 are phosphorylated by CDK during mitosis
To test whether Alp7 is directly phosphorylated by CDK, we performed an in vitro kinase assay using Alp7 peptides arrayed on a sheet (Figure 5A). Twenty-residue peptides placed every two residues from the N-terminus to the C-terminus of Alp7 were designed to cover the entire length of the Alp7 protein sequence with overlaps, and each kind of peptide was placed on a membrane. When the membrane with arrayed peptides was incubated with Cdc2-GST purified from mitotic cells and radioactive ATP, phosphorylation occurred mainly in two clusters of the Alp7 N-terminus array (clusters 1 and 2, Figure 5A).
FIGURE 5:
Alp7 is phosphorylated by CDK. (A) Kinase assay was performed with the membrane on which Alp7 peptides are sequentially spotted. Each spot contains peptides of 20-residue length. CDK was pulled down from mitotic S. pombe extract and incubated with radioactive ATP and the membrane. Two clusters showed intense phosphorylation (clusters 1 and 2, dotted boxes); the regions correspond to the bold letters. Candidate residues (serine and threonine) for CDK phosphorylation are highlighted (boxes). (B) Schematic illustration of Alp7-5A. Asterisks indicate position of five serine/threonine residues (S50, S51, T100, T116, and T131) mutagenized to alanine. (C) In vitro kinase assay using recombinant proteins GST-Alp7-WT and GST-Alp7-5A. Proteins were incubated with radioactive ATP and Cdc2-GST purified from mitotic S. pombe extract. GST-Alp7-WT was efficiently phosphorylated by CDK, whereas GST-Alp7-5A was not. (D) alp7-GFP nda3-KM311 and alp7-5A-GFP nda3-KM311 cells were arrested in mitosis (17°C) or incubated as asynchronous cultures (30°C). Cells were lysed, and protein extracts were resolved on the SDS–PAGE gel, followed by Western blotting using the anti-GFP antibody. The upper bands (filled arrowheads) correspond to phosphorylated Alp7-GFP, whereas the lower band (open arrowhead) is nonphosphorylated Alp7-GFP.
The first cluster corresponds to amino acids 43–74, and the second cluster corresponds to 97–126, which includes the NLS (117–126). These clusters contain a number of serine/threonine residues, four sites of which (S50, S51, T100, T116) appear to be highly phosphorylated, judging by the assay. The second cluster includes two consensus phosphorylation sites (TP; T100 and T116) near the NLS. The N-terminal half of Alp7 (1–209) has one more consensus site (T131), which is slightly separated from these two clusters but right after NLS. We suspected possible phosphorylation to these five sites, and to test this, we purified the phosphodeficient recombinant proteins GST-Alp7-5A and GST-Alp7 (WT) from Escherichia coli (Figure 5B). The in vitro phosphorylation assay of these proteins, using Cdc2-GST purified from yeast and radioactive ATP, revealed that CDK phosphorylates GST-Alp7 but not GST-Alp7-5A (Figure 5C). Thus Alp7 is phosphorylated in vitro by CDK in the N-terminus, particularly at five serine or threonine residues (S50, S51, T100, T116, and T131).
To investigate whether these sites are phosphorylated in vivo, we prepared lysates from cells expressing Alp7-GFP or Alp7-5A-GFP from both asynchronous and mitotically arrested nda3-KM311 cells. We then resolved the cell extracts on the SDS–PAGE gel, followed by mobility shift assays with the anti-GFP antibody. We found mobility shifts of Alp7-GFP in the M-phase–arrested extract (17°C; Figure 5D), whereas we recognized no band shifts for Alp7-5A-GFP in either the asynchronous or the mitotic extracts (Figure 5D). This strongly indicates that five residues in the N-terminus of Alp7 are phosphorylated in vivo and that phosphorylation only occurs during mitosis. Taking all of the evidence together, we conclude that CDK phosphorylates five serine/threonine residues in the N-terminus of Alp7 during mitosis.
Alp7-5A-GFP retained nuclear import activity but caused reduction of mitotic accumulation
Next we examined whether Alp7-5A-GFP accumulates normally in the nucleus during mitosis. We replaced the chromosomal alp7+ gene with the mutant gene alp7-5A-GFP and observed its localization. Strikingly, nuclear accumulation during mitosis decreased more in Alp7-5A-GFP than with Alp7-GFP (WT; Figure 6A). Quantitative assays for the nuclear/cytoplasmic ratio of Alp7-GFP verified that the ratio of Alp7-5A-GFP remarkably decreased compared with that of Alp7-WT-GFP (Figure 6B). The defects of Alp7-5A were not due to protein instability or loss of interaction with Alp14, judging from Western blotting and coimmunoprecipitation assays (Figure 6C). The cytoplasmic retention of Alp7-5A-GFP may reflect loss of ability for Alp7 in mitosis-specific nuclear accumulation, as expected. It is possible, however, that this could have been due to complete loss of NLS activity caused by the point mutations created near NLS (Figure 6A). To examine the NLS activity of the 5A mutant, we added leptomycin B (LMB), an inhibitor of the nuclear export factor Crm1/exportin (Kudo et al., 1998), to living cells expressing Alp7-GFP and Alp7-5A-GFP and monitored the behavior (Figure 6, D and E). Alp7-GFP (WT) started to accumulate in the nucleus 30 min after LMB addition and finished accumulation in 90% of the interphase nuclei within the next 10 min (Figure 6, D and E). Alp7-5A-GFP started to accumulate in the nucleus 40 min after LMB addition, and it accumulated in >90% of interphase nuclei within the next 10 min. The timing of initial nuclear accumulation was delayed, but the kinetics of accumulation was comparable in those two strains (Figure 6E). These results suggest that Alp7-5A retains the ability for nuclear import, although its efficiency is slightly decreased.
FIGURE 6:
The phosphodeficient alp7-5A mutant displays spindle defects. (A) Localization of Alp7-GFP (WT) and Alp7-5A-GFP observed at 25°C with Cut11-3mRFP as a nuclear envelope maker. I, interphase; M, mitosis. Nuclear accumulation in mitotic cells was reduced in the alp7-5A-GFP mutant (white arrowhead). (B) The ratio of the nuclear GFP intensity to the cytoplasmic intensity was quantified in both mitotic alp7-GFP and alp7-5A-GFP cells. n = 15 cells, *p = 5.35 × 10−35 < 0.0001 (Student's t test). (C) Alp7-5A-GFP interacts with Alp14-HA. Cell extracts were prepared from cells expressing Alp14-HA with either Alp7-WT-GFP or Alpl7-5A-GFP. Alp7-GFP and Alp7-5A-GFP were immunoprecipitated with anti-GFP antibody, and Alp14-HA was detected with anti-hemagglutinin antibody. –, untagged Alp7 was expressed; +, Alp14-HA was expressed. (D) alp7-GFP (WT) and alp7-5A-GFP strains were observed with 10-min intervals after addition of leptomycin B (200 ng/ml), a Crm1/exportin inhibitor. (E) Time-course kinetics of population of cells in which GFP fluorescence is accumulated in the nucleus or in which GFP fluorescence was recognized on cytoplasmic microtubules. In alp7-5A-GFP, the timing of nuclear accumulation of GFP signal was delayed ∼10 min compared with WT. (F) Temperature sensitivity of the indicated strains was tested by spotting cells in 1/10 serial dilution. (G) Localization of Alp7-GFP and Alp7-5A-GFP in metaphase cells observed with mCherry-Atb2 at 36°C. Abnormally extended cytoplasmic astral microtubules were observed in alp7-5A-GFP cells (arrowheads). (H) A typical example of alp7-5A-GFP cells with mitotic defects at 36°C. Monopolar spindles and abnormally extended cytoplasmic microtubules are observed.
We also found that the alp7-5A mutant exhibits temperature sensitivity at 36°C, which was suppressed by an addition of a canonical classical NLS derived from a T antigen of SV40 (Figure 6F). This suggests that defects in alp7-5A are due to loss of nuclear accumulation but not to other types of functional retardation. To investigate the effect of Alp7-5A on microtubule organization, we observed Alp7-5A-GFP at 36°C with mCherry-Atb2. As shown in Figure 6G, mitotic alp7-5A-GFP cells displayed cytoplasmic astral microtubules abnormally extended from SPBs. Note that Alp7-5A-GFP localized to the cytoplasmic astral microtubules but not to the spindle formed in the nucleus. It is possible that cytoplasmic microtubules are hyperstabilized because of a higher population of cytoplasmic Alp7 proteins in alp7-5A-GFP than that in wild-type cells. This also indicates that Alp7-5A is fully functional concerning microtubule stabilization at the restrictive temperature. As a result, live imaging revealed a prolonged mitotic prophase caused by the monopolar spindle in alp7-5A-GFP (Figure 6H). Thus the nonphosphorylated form of Alp7 causes reduction of Alp7 nuclear accumulation in mitosis, which results in defects of spindle assembly during early mitosis. We conclude that the nuclear accumulation of Alp7 is critical for spindle integrity during mitosis.
Alp7 binds to importin α/Cut15
It is known that the nuclear import of proteins containing a classical NLS depends on importin α. S. pombe has two importin α proteins, Cut15 and Imp1 (Matsusaka et al., 1998; Umeda et al., 2005). We next investigated the interaction between Alp7 and Cut15 using the yeast two-hybrid system. We found that the N-terminal half of Alp7 containing the NLS (Alp7-N-WT) interacted with Cut15-80-522, which is believed to lack the importin-β–binding domain at the N-terminus, as well as the C-terminal unstructured region (Figure 7, A and B; Conti et al., 1998; Umeda et al., 2005). We also detected an interaction of Cut15-80-522 with Alp7-N-5A, although the interaction was slightly decreased, whereas the phosphomimetic protein Alp7-N-5E interacted with Cut15-80-522 as strongly as WT did. This suggests that N-terminal phosphorylation sites of Alp7 affected the interaction between Alp7 and Cut15 at least in budding yeast cells.
DISCUSSION
How Alp7–Alp14 is transported during the cell cycle
We propose a model for the mechanism of Alp7–Alp14 nucleocytoplasmic transport in Figure 7C. Alp7–Alp14 shuttles between the nucleus and the cytoplasm throughout the cell cycle, and the balance of NLS in Alp7 and NES in Alp14 determines the localization. During interphase, the NES of Alp14 dominates the localization of Alp7–Alp14. On mitotic entry, CDK phosphorylates serine/threonine residues close to the NLS of Alp7, in which phosphorylation enhances the interaction between Alp7 and Cut15. Then NLS activity prevails over NES activity, and, as a result, the Alp7–Alp14 complex accumulates in the nucleus and localizes to spindle microtubules to promote spindle assembly.
FIGURE 7:
Association of Alp7 to Cut15/importin α is weakened by the phosphodeficient 5A mutation. (A, B) Two-hybrid assay showing interaction between Cut15/importin-α and Alp7-N terminus (amino acids 1–209). Alp7-N-WT, Alp7-N-5A, and Alp7-N-5E physically interacted with Cut15-80-522 in the SC–His+3AT medium (low-stringency conditions) (A), whereas Alp7-N-5A was reduced in affinity to Cut15-80-522 in the SC–His–Ade medium (high-stringency conditions) (B). T and 53 represent T-antigen and p53 used as control, respectively. (C) A schematic model for transport of the Alp7–Alp14 complex in interphase and mitosis. Alp7 plays a role in import of the complex, whereas Alp14 plays a role in export. The import efficiency of the complex may be upregulated by enhancing the interaction of Alp7 and Cut15 through phosphorylation of Alp7 by CDK.
The C-terminal part of Alp14 controls its localization
Our truncation analyses show that the C-terminal part of Alp14 is critical for localization of the MAP to the microtubule lattice (Figure 1A; Sato et al., 2004). Al-Bassam et al. (2012) performed plasmid-based domain analyses in which they expressed several Alp14 mutant fragments by plasmids in the alp14∆ mutant. From their analyses, it remained unclear whether the C-terminal part of Alp14 (∆TOG) localizes to microtubule plus tips. The present study's observation is based on integrated genes with GFP expressed at the endogenous level and succeeded in visualizing the localization of Alp14-∆TOG. We found that Alp14-∆TOG-GFP is able to localize to microtubules, but the localization pattern is distinct from that of wild-type Alp14-GFP. Namely, Alp14-∆TOG-GFP seems to localize to the microtubule lattice rather than the plus tip, where wild-type Alp14-GFP preferentially localizes (Figure 1A). Al-Bassam et al. (2012) also suggested that Alp14 harboring mutations in the second TOG domain (TOG2) failed to rescue the loss of microtubule bundles in the alp14∆ mutant. Our ∆TOG2 did not show growth defects at 36°C but did exhibit hypersensitivity to TBZ like alp14∆. This suggests that TOG1 of Alp14, even without TOG2, retains the function to some extent, although it is compromised under some conditions.
Alp14 is responsible for nuclear export of the Alp7–Alp14 complex
Previous studies suggested that NES activity exists in Alp7 (Ling et al., 2009; Sato et al., 2009a), and the present study shows that the activity resides in the Alp14-binding domain. Indeed, the Alp7-L461A mutant protein displayed two properties: constant nuclear accumulation and loss of Alp14 binding (Sato et al., 2009a). It had been unclear whether Alp14 and Exportin/Crm1 shared L461 of Alp7 as their docking site or Alp14 bound to this site has the intrinsic NES. This study demonstrates that the latter is the case: the intrinsic NES resides in Alp14 (L615), which is responsible for the nuclear export of the Alp7–Alp14 complex. Experimental and computational analyses show that the consensus sequence of NES is relatively loose: ϕ-X2-3-ϕ-X2-3-ϕ-X-ϕ, where ϕ is L, V, I, F, or M, and X is any amino acid (Bogerd et al., 1996; Fukuda et al., 1996; Henderson and Eleftheriou, 2000; Engelsma et al., 2004; Fu et al., 2013). The sequence in Alp14, VQNLKNMEL (612–620), that was deemed necessary for the nuclear export of the complex (Figures 2B and 3A) matches the aforementioned consensus sequence. Indeed, a single mutation in the second hydrophobic residue therein, L615A, caused abnormal nuclear accumulation of Alp14 together with Alp7. Therefore L615 of Alp14, rather than L461 of Alp7, plays a critical role for the NES of the MAP complex. The interaction between 601 and 620 of Alp14 and Crm1 would be the next challenge, although NES activity might not be regulated in a temporal manner during the cell cycle. In summary, we conclude that Alp7 and Alp14 behave as a complex and shuttle between the nucleus and the cytoplasm throughout the cell cycle and that the complex predominantly localizes to the cytoplasm during interphase in order to balance NLS in Alp7 and NES in Alp14.
Alp7 is phosphorylated by CDK
Our previous study demonstrated that localization of Alp7 is under control by CDK, but it remained unclear how the localization was regulated. Global phosphoproteomic assays to search for phosphorylation sites have been conducted and so far identify only serine 17 as a phosphorylation site (Wilson-Grady et al., 2008; Beltrao et al., 2009). In this study, a peptide-array–based in vitro kinase assay revealed that the N-terminus of Alp7 is phosphorylated by CDK. A further kinase assay successfully pinpointed the phosphorylation sites: five serine or threonine residues around the NLS. Mobility shift assays using S. pombe protein extracts from asynchronous and mitotically arrested cells confirmed that these five residues are phosphorylated in vivo and only during mitosis (Figure 5D). These results significantly point out the missing link of the cell cycle and Ran-GTP–dependent microtubule formation.
Localization of other MAPs is also controlled by CDK. The other TOG in fission yeast, Dis1, is phosphorylated by CDK (Aoki et al., 2006). Phosphorylation of Dis1 by CDK is believed to enhance Dis1’s association with kinetochores and/or its removal from the microtubule lattice during metaphase (Aoki et al., 2006). The phosphodeficient mutant protein Dis1-6A, in which six consensus sites of CDK phosphorylation were substituted with alanine, localized to the spindle in the nucleus (Aoki et al., 2006), suggesting that phosphorylation by CDK did not affect the nuclear localization of Dis1. It is also of note that Dis1 has no canonical classic NLS, and a TACC counterpart may not exist for Dis1. We therefore speculate that distinct molecular systems operate for the nucleocytoplasmic transport of Dis1.
Ase1 (human PRC1) is a microtubule-bundling factor (Pellman et al., 1995; Loïodice et al., 2005; Yamashita et al., 2005), and its nuclear entry is promoted by CDK (Fu et al., 2009), although it remains uncharacterized how the nuclear entry is enhanced. Ase1 interacts with Klp9, the kinesin-6 family protein, to recruit Klp9 to the spindle midzone for spindle elongation in anaphase, and the interaction is inhibited by CDK's phosphorylation of Klp9 until anaphase onset (Fu et al., 2009). Association of nuclear and spindle–associated protein (NuSAP) with the spindle in metaphase is also inhibited by CDK's phosphorylation of NuSAP (Chou et al., 2011). In those cases, phosphorylation by CDK exerts inhibitory effects on the interaction between the MAPs and spindle microtubules. This study provides a molecular mechanism by which phosphorylation by CDK serves to enhance nuclear accumulation of a MAP complex and its association with spindle microtubules, thereby promoting spindle assembly in the initial stage of mitosis.
CDK controls mitosis-specific nuclear accumulation of the Alp7–Alp14 complex
The alp7-5A mutant exhibited defects in bipolar microtubule formation at the restrictive temperature. Addition of an external NLS rescued the temperature-sensitive alp7-5A mutant, indicating that the defect in the alp7-5A mutant lies in an ability to localize to the nucleus. Our microscopy indeed demonstrated that Alp7-5A-GFP failed to accumulate to the nucleus during mitosis. Alp7-5A-GFP, however, accumulated to the nucleus when LMB was added to inhibit nuclear exclusion. This is in sharp contrast to the NLS-deficient mutant protein Alp7-RARA-GFP, which did not accumulate to the nucleus in the presence of LMB (Sato and Toda, 2007; Sato et al., 2009a). We therefore conclude that Alp7-5A-GFP retains the ability to enter the nucleus but fails to accumulate efficiently, and phosphorylation at these sites is required to shift the balance of Alp7–Alp14 shuttling from the cytoplasm to the nucleus.
It is possible that phosphorylation by CDK enhances the NLS activity of Alp7 to accomplish nuclear accumulation in mitosis. Two-hybrid analysis in budding yeast cells implied that the interaction between Cut15/importin α and Alp7 was weakened in the presence of 5A mutations. Thus we speculate that the NLS activity of Alp7-5A is kept at the minimal level irrespective of cell cycle stage.
Klp5 and Klp6, fission yeast kinesin-8 family proteins that form a heterodimer, are known to undergo nucleocytoplasmic shuttling (Unsworth et al., 2008). Of interest, Klp5 and Klp6 each have an NLS, and they migrate to the nucleus independently, despite their dependence on localization to spindle microtubules. In addition, both Klp5 and Klp6 have CDK phosphorylation consensus sequences close to their NLS sequences. It would be valuable to analyze whether those NLS are also enhanced by phosphorylation, as in the Alp7–Alp14 complex. Temporal enhancement of NLS might be a universal mechanism for nucleocytoplasmic shuttling proteins that change their localization in a temporal manner.
MATERIALS AND METHODS
Yeast genetics and strains
Strains used this study are listed in Supplemental Table S1. We used standard methods for yeast genetics and gene tagging (Moreno et al., 1991; Bähler et al., 1998; Sato et al., 2005). For visualization of mCherry-tubulin and cyan fluorescent protein (CFP)–tubulin, we used Z2-mCherry-atb2(α2-tubulin)-hph and Z2-CFP-atb2-nat strains, respectively. As previously described for Patb2-GFP-atb2-kan (Kakui et al., 2013), those strains were made as follows: Patb2-mCherry-atb2-hph and Patb2-CFP-atb2-nat DNA fragment was prepared and integrated into gene-free region adjacent to zfs1+ on the chromosome II as an extra copy of endogenous atb2+. Truncation mutants were made as follows: construction of truncated alp7 and alp14 genes was done on plasmids harboring alp7+ and alp14+ genes, respectively. Those truncated DNA fragments were amplified with PCR and transformed into S. pombe cells to integrate them into the original locus of the gene. The truncation mutants therefore do not possess the wild-type allele of the gene and express the mutant genes from the native promoter. For reporter protein assay (Figure 2B), genes for fusion proteins GST-GFP and GST-GFP-14NES (residues 601–620 of Alp14 were added to the C-terminus of GST-GFP) were placed under the adh1 promoter (Padh1) on the plasmid pFA6a containing the hph marker gene. The resulting plasmids containing Padh1-GST-GFP-hph and Padh1-GST-GFP-14NES-hph were linearized and integrated into the arg1 gene locus of the strain expressing Cut11-3mRFP as a nuclear envelope marker.
Microscopy and LMB treatment
Cells were grown in yeast extract medium supplemented with adenine, uracil, leucine, histidine, and lysine (YE5S) at 25°C and observed in minimal medium + N with supplements at 25°C (Figures 1A, 2, A and B, 3, A and B, 4A, and 6, A and D) or at 36°C (3 h; Figure 6, G and H). For experiments in Figure 6D, LMB was added at the final concentration of 200 ng/ml immediately after acquisition of images at the first time point, followed by time-lapse imaging. To acquire images, the DeltaVision SoftWoRx system (GE Healthcare, Little Chalfont, UK) was used as described previously (Sato et al., 2009b). Images were acquired as serial sections along the z-axis and stacked using the Quick projection algorithm in SoftWoRx. The captured images were processed with Photoshop CS5, version 15.0 (Adobe, San Jose, CA).
Quantification of signal intensity
Fluorescence intensity was measured in Figures 2C and 6B and Supplemental Figure S1C. Average fluorescence intensities of GFP in 6 × 6 pixels in the nucleus, the cytoplasm, and the background (extracellular region) were quantified using the Data Inspector command of the SoftWoRx. Background intensity was subtracted from nuclear intensity and cytoplasmic intensity, and then the ratio of the nuclear to the cytoplasmic intensity was calculated.
Protein extraction, immunoprecipitation, and λ-phosphatase treatment
For Figures 3C and 6C, cells were collected and lysed with glass beads in HB buffer (25 mM 3-(N-morpholino)propanesulfonic acid, pH 7.2, 15 mM MgCl2, 5 mM ethylene glycol tetraacetic acid [EGTA], 60 mM β-glycerophosphate, 0.1 mM Na orthovanadate, 15 mM p-nitrophenyl phosphate, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 0.2% NP-40, complete protease inhibitor [Roche, Penzberg, Germany]), and the supernatant after centrifuge was collected as cell extract. Cell extract and protein G–coupled Dynabeads (Life Technologies, Waltham, MA) were mixed with the anti-Myc antibody (9E10; Sigma-Aldrich, St. Louis, MO; Figure 3C) or the anti-GFP antibody (A11122; Life Technologies; Figure 6C) and then incubated for 1.5 h at 4°C for immunoprecipitation. After that, beads were washed and resuspended with HB buffer containing the SDS sample buffer and boiled. Proteins were resolved on SDS–PAGE gels (10% acrylamide gel), followed by Western blotting with monoclonal anti-GFP (1:400; 7.1 and 13.1; Roche), anti-hemagglutinin (16B12; Life Technologies), and anti–α-tubulin (B5-1-2; Sigma-Aldrich) antibodies.
For Figure 4B, the alp7-GFP nda3-KM311 (the cold-sensitive β-tubulin mutant) strain was cultured at 30°C for 18 h and then shifted to 17°C for 6.5 h in the YE5S liquid medium. Cells were then collected and lysed with glass beads in HB buffer, and the supernatant after centrifuge was collected as cell extract. Cell extract was incubated with the anti-GFP antibody (A11122; Life Technologies) and protein G–coupled Dynabeads for 1.5 h at 4°C to immunoprecipitate Alp7-GFP. After the incubation, beads were washed with HB buffer and resuspended in λ-phosphatase buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.1 mM EGTA, pH 8.0, 2 mM dithiothreitol, 0.01% Brij-35, 2 mM MnCl2). Then λ-protein phosphatase (New England Biolabs, Ipswich, MA) with or without phosphatase inhibitor (10 mM EGTA, 10 mM Na orthovanadate, 20 mM β-glycerophosphate, 15 mM p-nitrophenylphosphate) was added, and samples were incubated for 20 min at 30°C. After that, phosphatase buffer was removed, and the pellet was resuspended with HB buffer followed by SDS sample buffer and boiled. Proteins were resolved on the SDS–PAGE gel (10% acrylamide gel; mono:bis = 29.8:0.2), followed by Western blotting with the anti-GFP monoclonal antibody (1:400; 7.1 and 13.1; Roche).
For Figure 5D, strains alp7-GFP nda3-KM311 and alp7-5A-GFP nda3-KM311 were cultured at 30°C for 18 h and then shifted to 17°C or kept at 30°C for 6 h in the YE5S liquid medium. Harvested cells were lysed with glass beads in HB buffer. Cell extracts were separated through SDS–PAGE (8% acrylamide gel supplemented with the final concentration of 50 μM Phos-Tag [Wako, Osaka, Japan]) and followed by Western blotting with anti-GFP monoclonal antibody (1:400; 7.1 and 13.1; Roche).
Preparation of CDK and GST-fused proteins
For Figure 5A, the nda3-KM311 strain was cultured at 30°C for 18 h and then shifted to 17°C for 13 h in YE5S to induce mitotic arrest. Cells were collected and lysed with glass beads in HB buffer. Cell extracts were incubated with agarose-conjugated p13/suc1 (Millipore, Billerica, MA) for 1.5 h at 4°C. After washing with HB buffer, the pellet was resuspended with HB buffer and used as the CDK solution.
For the assay in Figure 5C, The Cdc2-GST fusion protein was purified from S. pombe cells using a GST pull-down method. First, the cdc2+ gene was tagged with the GST gene using the standard method (Bähler et al., 1998) in the nda3-KM311 strain. The resultant strain, nda3-KM311 cdc2-GST, was cultured at 30°C for 18 h and then shifted to 17°C for 13 h. Cell extraction was done as described. Crude extracts were then incubated with glutathione Sepharose 4B (GE Healthcare) for 1.5 h at 4°C. After washing with HB buffer, the pellet was resuspended with HB buffer and used as the CDK solution. Activity of the purified Cdc2-GST was confirmed by an in vitro kinase assay using histone H1 (Calbiochem, San Diego, CA) as a substrate.
To express recombinant GST–Alp7-WT and GST–Alp7-5A in E. coli BL21 (DE3) cells, coding sequences for alp7+ and alp7-5A were amplified with PCR and cloned into the GST-containing vector pGEX-4T (GE Healthcare).
In vitro kinase assay using a peptide array or recombinant substrates
For the kinase assay using a peptide array (Figure 5A), peptides were synthesized and spotted onto a cellulose membrane. This membrane was activated by treatment with 50% ethanol and 10% acetic acid for 1 h at room temperature and then washed with phosphate-buffered saline (PBS). The membrane was incubated for 2 h in the phosphorylation buffer (10 mM Tris-HCl, pH 8.0, 50 μM ATP, 100 μM MgSO4, 100 μg/ml bovine serum albumin [BSA]; Ohta et al., 2012). For the kinase reaction, we added 0.84 μM (10 μCi) [γ-32P]ATP and CDK solution (Cdc2 purified from S. pombe cells) and incubated for 3 h. After the incubation, the membrane was washed with PBS and dried, and radioactivity was detected with autoradiography.
For the in vitro kinase assay using recombinant substrates (Figure 5C), CDK solution (Cdc2-GST purified from S. pombe cells) and substrates (GST-Alp7-WT and GST-Alp7-5A) were incubated in HB buffer containing 20 μg/ml BSA and 200 μM ATP at 30°C for 30 min in the presence of 50 μM (1.5 μCi) [γ-32P]ATP. Reactions were terminated by addition of SDS sample buffer. Proteins were separated with SDS–PAGE, and the gel was stained with Biosafe-Coomassie (Bio-Rad, Hercules, CA), followed by autoradiography.
Yeast two-hybrid assay
Plasmids used in two-hybrid assays are listed in Supplemental Table S2. Genes encoding Alp7-N(1-209), Alp7-N(1-209)-5A, and Alp7-N(1-209)-5E were amplified with PCR and cloned into the vector pGBKT7 (Clontech, Mountain View, CA) and were used as bait. The gene for Cut15-80-522 was similarly cloned into the vector pGADT7 (Clontech) and used as prey. Plasmids pGBKT7-T (cloned T-antigen) and pGADT7-53 (cloned p53) provided by Clontech were used as control. The Saccharomyces cerevisiae strain AH109 was cultured in the liquid medium YPDA (rich in yeast extract) and transformed with the plasmids. Transformants were grown on the synthetic medium SC lacking both leucine and tryptophan (SC–L–W). As selective media, SC plates lacking histidine and containing 5 mM of 3-amino-1,2,4-triazole (–His+3AT) and SC plates lacking both histidine and adenine (–His–Ade) were used.
Supplementary Material
Acknowledgments
We thank Y. Iino for support, M. Yoshida for LMB, Y. Kakui for plasmids, and R. Arai for comments. We are grateful to the Peptide Synthesis Unit of Cancer Research UK for preparing the peptide-arrayed sheet. N.O. is a research fellow of the Japan Society for the Promotion of Science. This study was supported by the Japan Society for the Promotion of Science KAKENHI Grants-in-Aid for Young Scientists (A) (21687015) and for Scientific Research (B) (25291041) to M.S. and for Scientific Research (S) (21227007) to M.Y.; the Sumitomo Foundation, the Naito Foundation, the Kato Memorial Bioscience Foundation, and a Kishimoto Grant from the Senri Life Science Foundation to M.S.; and Cancer Research UK to T.T.
Abbreviations used:
- CDK
cyclin-dependent kinase
- CFP
cyan fluorescent protein
- GDP
guanosine diphosphate
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- GTP
guanosine triphosphate
- LMB
leptomycin B
- MAP
microtubule-associated protein
- NES
nuclear export signal
- NLS
nuclear localization signal
- SAF
spindle assembly factor
- SPB
spindle pole body
- TACC
transforming acidic coiled-coil
- TBZ
thiabendazole
- TOG
tumor overexpressed gene
- tubBD
tubulin-binding domain
- YE5S
yeast extract medium supplemented with adenine, uracil, leucine, histidine, and lysine
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-11-0679) on April 30, 2014.
The authors declare no competing financial interests.
REFERENCES
- Al-Bassam J, Chang F. Regulation of microtubule dynamics by TOG-domain proteins XMAP215/Dis1 and CLASP. Trends Cell Biol. 2011;21:604–614. doi: 10.1016/j.tcb.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bassam J, Kim H, Flor-Parra I, Lal N, Velji H, Chang F. Fission yeast Alp14 is a dose-dependent plus end-tracking microtubule polymerase. Mol Biol Cell. 2012;23:2878–2890. doi: 10.1091/mbc.E12-03-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K, Nakaseko Y, Kinoshita K, Goshima G, Yanagida M. Cdc2 phosphorylation of the fission yeast Dis1 ensures accurate chromosome segregation. Curr Biol. 2006;16:1627–1635. doi: 10.1016/j.cub.2006.06.065. [DOI] [PubMed] [Google Scholar]
- Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Bellanger JM, Gönczy P. TAC-1 and ZYG-9 form a complex that promotes microtubule assembly in C. elegans embryos. Curr Biol. 2003;13:1488–1498. doi: 10.1016/s0960-9822(03)00582-7. [DOI] [PubMed] [Google Scholar]
- Beltrao P, Trinidad JC, Fiedler D, Roguev A, Lim WA, Shokat KM, Burlingame AL, Krogan NJ. Evolution of phosphoregulation: comparison of phosphorylation patterns across yeast species. PLoS Biol. 2009;7:e1000134. doi: 10.1371/journal.pbio.1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff FR, Ponstingl H. Mitotic regulator protein RCC1 is complexed with a nuclear ras-related polypeptide. Proc Natl Acad Sci USA. 1991;88:10830–10834. doi: 10.1073/pnas.88.23.10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd HP, Fridell RA, Benson RE, Hua J, Cullen BR. Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol Cell Biol. 1996;16:4207–4214. doi: 10.1128/mcb.16.8.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouhard GJ, Stear JH, Noetzel TL, Al-Bassam J, Kinoshita K, Harrison SC, Howard J, Hyman AA. XMAP215 is a processive microtubule polymerase. Cell. 2008;132:79–88. doi: 10.1016/j.cell.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carazo-Salas RE, Gruss OJ, Mattaj IW, Karsenti E. Ran-GTP coordinates regulation of microtubule nucleation and dynamics during mitotic-spindle assembly. Nat Cell Biol. 2001;3:228–234. doi: 10.1038/35060009. [DOI] [PubMed] [Google Scholar]
- Carazo-Salas RE, Guarguaglini G, Gruss OJ, Segref A, Karsenti E, Mattaj IW. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature. 1999;400:178–181. doi: 10.1038/22133. [DOI] [PubMed] [Google Scholar]
- Chou HY, Wang TH, Lee SC, Hsu PH, Tsai MD, Chang CL, Jeng YM. Phosphorylation of NuSAP by Cdk1 regulates its interaction with microtubules in mitosis. Cell Cycle. 2011;10:4083–4089. doi: 10.4161/cc.10.23.18200. [DOI] [PubMed] [Google Scholar]
- Clarke PR, Zhang C. Spatial and temporal coordination of mitosis by Ran GTPase. Nat Rev Mol Cell Biol. 2008;9:464–477. doi: 10.1038/nrm2410. [DOI] [PubMed] [Google Scholar]
- Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- Cullen CF, Ohkura H. Msps protein is localized to acentrosomal poles to ensure bipolarity of Drosophila meiotic spindles. Nat Cell Biol. 2001;3:637–642. doi: 10.1038/35083025. [DOI] [PubMed] [Google Scholar]
- Dasso M. Running on Ran: nuclear transport and the mitotic spindle. Cell. 2001;104:321–324. doi: 10.1016/s0092-8674(01)00218-5. [DOI] [PubMed] [Google Scholar]
- Engelsma D, Bernad R, Calafat J, Fornerod M. Supraphysiological nuclear export signals bind CRM1 independently of RanGTP and arrest at Nup358. EMBO J. 2004;23:3643–3652. doi: 10.1038/sj.emboj.7600370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C, Ward JJ, Loïodice I, Velve-Casquillas G, Nedelec FJ, Tran PT. Phospho-regulated interaction between kinesin-6 Klp9p and microtubule bundler Ase1p promotes spindle elongation. Dev Cell. 2009;17:257–267. doi: 10.1016/j.devcel.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu SC, Huang HC, Horton P, Juan HF. ValidNESs: a database of validated leucine-rich nuclear export signals. Nucleic Acids Res. 2013;41:D338–D343. doi: 10.1093/nar/gks936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M, Gotoh I, Gotoh Y, Nishida E. Cytoplasmic localization of mitogen-activated protein kinase kinase directed by its NH2-terminal, leucine-rich short amino acid sequence, which acts as a nuclear export signal. J Biol Chem. 1996;271:20024–20028. doi: 10.1074/jbc.271.33.20024. [DOI] [PubMed] [Google Scholar]
- Garcia MA, Vardy L, Koonrugsa N, Toda T. Fission yeast ch-TOG/XMAP215 homologue Alp14 connects mitotic spindles with the kinetochore and is a component of the Mad2-dependent spindle checkpoint. EMBO J. 2001;20:3389–3401. doi: 10.1093/emboj/20.13.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss OJ, Carazo-Salas RE, Schatz CA, Guarguaglini G, Kast J, Wilm M, Le Bot N, Vernos I, Karsenti E, Mattaj IW. Ran induces spindle assembly by reversing the inhibitory effect of importin α on TPX2 activity. Cell. 2001;104:83–93. doi: 10.1016/s0092-8674(01)00193-3. [DOI] [PubMed] [Google Scholar]
- Hagan IM, Hyams JS. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1988;89:343–357. doi: 10.1242/jcs.89.3.343. [DOI] [PubMed] [Google Scholar]
- Hayles J, Fisher D, Woollard A, Nurse P. Temporal order of S phase and mitosis in fission yeast is determined by the state of the p34cdc2-mitotic B cyclin complex. Cell. 1994;78:813–822. doi: 10.1016/s0092-8674(94)90542-8. [DOI] [PubMed] [Google Scholar]
- Henderson BR, Eleftheriou A. A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp Cell Res. 2000;256:213–224. doi: 10.1006/excr.2000.4825. [DOI] [PubMed] [Google Scholar]
- Hiraoka Y, Toda T, Yanagida M. The NDA3 gene of fission yeast encodes beta-tubulin: a cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell. 1984;39:349–358. doi: 10.1016/0092-8674(84)90013-8. [DOI] [PubMed] [Google Scholar]
- Kakui Y, Sato M, Okada N, Toda T, Yamamoto M. Microtubules and Alp7-Alp14 (TACC-TOG) reposition chromosomes before meiotic segregation. Nat Cell Biol. 2013;15:786–796. doi: 10.1038/ncb2782. [DOI] [PubMed] [Google Scholar]
- Kalab P, Pu RT, Dasso M. The ran GTPase regulates mitotic spindle assembly. Curr Biol. 1999;9:481–484. doi: 10.1016/s0960-9822(99)80213-9. [DOI] [PubMed] [Google Scholar]
- Karsenti E, Vernos I. The mitotic spindle: a self-made machine. Science. 2001;294:543–547. doi: 10.1126/science.1063488. [DOI] [PubMed] [Google Scholar]
- Kirschner M, Mitchison T. Beyond self-assembly: from microtubules to morphogenesis. Cell. 1986;45:329–342. doi: 10.1016/0092-8674(86)90318-1. [DOI] [PubMed] [Google Scholar]
- Koffa MD, Casanova CM, Santarella R, Kocher T, Wilm M, Mattaj IW. HURP is part of a Ran-dependent complex involved in spindle formation. Curr Biol. 2006;16:743–754. doi: 10.1016/j.cub.2006.03.056. [DOI] [PubMed] [Google Scholar]
- Kudo N, Wolff B, Sekimoto T, Schreiner EP, Yoneda Y, Yanagida M, Horinouchi S, Yoshida M. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp Cell Res. 1998;242:540–547. doi: 10.1006/excr.1998.4136. [DOI] [PubMed] [Google Scholar]
- Le Bot N, Tsai MC, Andrews RK, Ahringer J. TAC-1, a regulator of microtubule length in the C. elegans embryo. Curr Biol. 2003;13:1499–1505. doi: 10.1016/s0960-9822(03)00577-3. [DOI] [PubMed] [Google Scholar]
- Lee MJ, Gergely F, Jeffers K, Peak-Chew SY, Raff JW. Msps/XMAP215 interacts with the centrosomal protein D-TACC to regulate microtubule behaviour. Nat Cell Biol. 2001;3:643–649. doi: 10.1038/35083033. [DOI] [PubMed] [Google Scholar]
- Ling YC, Vjestica A, Oliferenko S. Nucleocytoplasmic shuttling of the TACC protein Mia1p/Alp7p is required for remodeling of microtubule arrays during the cell cycle. PLoS One. 2009;4:e6255. doi: 10.1371/journal.pone.0006255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loïodice I, Staub J, Setty TG, Nguyen NP, Paoletti A, Tran PT. Ase1p organizes antiparallel microtubule arrays during interphase and mitosis in fission yeast. Mol Biol Cell. 2005;16:1756–1768. doi: 10.1091/mbc.E04-10-0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T, Beach D. Premature initiation of mitosis in yeast lacking RCC1 or an interacting GTPase. Cell. 1991;66:347–360. doi: 10.1016/0092-8674(91)90624-8. [DOI] [PubMed] [Google Scholar]
- Matsusaka T, Imamoto N, Yoneda Y, Yanagida M. Mutations in fission yeast Cut15, an importin alpha homolog, lead to mitotic progression without chromosome condensation. Curr Biol. 1998;8:1031–1034. doi: 10.1016/s0960-9822(07)00425-3. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Nabeshima K, Kurooka H, Takeuchi M, Kinoshita K, Nakaseko Y, Yanagida M. p93dis1, which is required for sister chromatid separation, is a novel microtubule and spindle pole body-associating protein phosphorylated at the Cdc2 target sites. Genes Dev. 1995;9:1572–1585. doi: 10.1101/gad.9.13.1572. [DOI] [PubMed] [Google Scholar]
- Nachury MV, Maresca TJ, Salmon WC, Waterman-Storer CM, Heald R, Weis K. Importin β is a mitotic target of the small GTPase Ran in spindle assembly. Cell. 2001;104:95–106. doi: 10.1016/s0092-8674(01)00194-5. [DOI] [PubMed] [Google Scholar]
- Nakaseko Y, Goshima G, Morishita J, Yanagida M. M phase-specific kinetochore proteins in fission yeast: microtubule-associating Dis1 and Mtc1 display rapid separation and segregation during anaphase. Curr Biol. 2001;11:537–549. doi: 10.1016/s0960-9822(01)00155-5. [DOI] [PubMed] [Google Scholar]
- Nakazawa N, Nakamura T, Kokubu A, Ebe M, Nagao K, Yanagida M. Dissection of the essential steps for condensin accumulation at kinetochores and rDNAs during fission yeast mitosis. J Cell Biol. 2008;180:1115–1131. doi: 10.1083/jcb.200708170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF, Hirano T. HEAT repeats associated with condensins, cohesins, and other complexes involved in chromosome-related functions. Genome Res. 2000;10:1445–1452. doi: 10.1101/gr.147400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba T, Nakamura M, Nishitani H, Nishimoto T. Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science. 1999;284:1356–1358. doi: 10.1126/science.284.5418.1356. [DOI] [PubMed] [Google Scholar]
- Ohkura H, Garcia MA, Toda T. Dis1/TOG universal microtubule adaptors—one MAP for all. J Cell Sci. 2001;114:3805–3812. doi: 10.1242/jcs.114.21.3805. [DOI] [PubMed] [Google Scholar]
- Ohta M, Sato M, Yamamoto M. Spindle pole body components are reorganized during fission yeast meiosis. Mol Biol Cell. 2012;23:1799–1811. doi: 10.1091/mbc.E11-11-0951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliferenko S, Balasubramanian MK. Astral microtubules monitor metaphase spindle alignment in fission yeast. Nat Cell Biol. 2002;4:816–820. doi: 10.1038/ncb861. [DOI] [PubMed] [Google Scholar]
- Pellman D, Bagget M, Tu YH, Fink GR, Tu H. Two microtubule-associated proteins required for anaphase spindle movement in Saccharomyces cerevisiae. J Cell Biol. 1995;130:1373–1385. doi: 10.1083/jcb.130.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peset I, Vernos I. The TACC proteins: TACC-ling microtubule dynamics and centrosome function. Trends Cell Biol. 2008;18:379–388. doi: 10.1016/j.tcb.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Sato M, Dhut S, Toda T. New drug-resistant cassettes for gene disruption and epitope tagging in Schizosaccharomyces pombe. Yeast. 2005;22:583–591. doi: 10.1002/yea.1233. [DOI] [PubMed] [Google Scholar]
- Sato M, Koonrugsa N, Vardy L, Tournier S, Millar JB, Toda T. Deletion of Mia1/Alp7 activates Mad2-dependent spindle assembly checkpoint in fission yeast. Nat Cell Biol. 2003;5:764–766. doi: 10.1038/ncb0903-764. [DOI] [PubMed] [Google Scholar]
- Sato M, Okada N, Kakui Y, Yamamoto M, Yoshida M, Toda T. Nucleocytoplasmic transport of Alp7/TACC organizes spatiotemporal microtubule formation in fission yeast. EMBO Rep. 2009a;10:1161–1167. doi: 10.1038/embor.2009.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Toda T. Alp7/TACC is a crucial target in Ran-GTPase-dependent spindle formation in fission yeast. Nature. 2007;447:334–337. doi: 10.1038/nature05773. [DOI] [PubMed] [Google Scholar]
- Sato M, Toda T. Space shuttling in the cell: nucleocytoplasmic transport and microtubule organization during the cell cycle. Nucleus. 2010;1:231–236. doi: 10.4161/nucl.1.3.11443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Toya M, Toda T. Visualization of fluorescence-tagged proteins in fission yeast: the analysis of mitotic spindle dynamics using GFP-tubulin under the native promoter. Methods Mol Biol. 2009b;545:185–203. doi: 10.1007/978-1-60327-993-2_11. [DOI] [PubMed] [Google Scholar]
- Sato M, Vardy L, Garcia MA, Koonrugsa N, Toda T. Interdependency of fission yeast Alp14/TOG and coiled coil protein Alp7 in microtubule localization and bipolar spindle formation. Mol Biol Cell. 2004;15:1609–1622. doi: 10.1091/mbc.E03-11-0837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillje HH, Nagel S, Korner R, Nigg EA. HURP is a Ran-importin β-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr Biol. 2006;16:731–742. doi: 10.1016/j.cub.2006.02.070. [DOI] [PubMed] [Google Scholar]
- Srayko M, Quintin S, Schwager A, Hyman AA. Caenorhabditis elegans TAC-1 and ZYG-9 form a complex that is essential for long astral and spindle microtubules. Curr Biol. 2003;13:1506–1511. doi: 10.1016/s0960-9822(03)00597-9. [DOI] [PubMed] [Google Scholar]
- Tang NH, Takada H, Hsu KS, Toda T. The internal loop of fission yeast Ndc80 binds Alp7/TACC-Alp14/TOG and ensures proper chromosome attachment. Mol Biol Cell. 2013;24:1122–1133. doi: 10.1091/mbc.E12-11-0817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Umesono K, Hirata A, Yanagida M. Cold-sensitive nuclear division arrest mutants of the fission yeast Schizosaccharomyces pombe. J Mol Biol. 1983;168:251–270. doi: 10.1016/s0022-2836(83)80017-5. [DOI] [PubMed] [Google Scholar]
- Umeda M, Izaddoost S, Cushman I, Moore MS, Sazer S. The fission yeast Schizosaccharomyces pombe has two importin-α proteins, Imp1p and Cut15p, which have common and unique functions in nucleocytoplasmic transport and cell cycle progression. Genetics. 2005;171:7–21. doi: 10.1534/genetics.105.042598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umesono K, Toda T, Hayashi S, Yanagida M. Cell division cycle genes NDA2 and NDA3 of the fission yeast Schizosaccharomyces pombe control microtubular organization and sensitivity to anti-mitotic benzimidazole compounds. J Mol Biol. 1983;168:271–284. doi: 10.1016/s0022-2836(83)80018-7. [DOI] [PubMed] [Google Scholar]
- Unsworth A, Masuda H, Dhut S, Toda T. Fission yeast kinesin-8 Klp5 and Klp6 are interdependent for mitotic nuclear retention and required for proper microtubule dynamics. Mol Biol Cell. 2008;19:5104–5115. doi: 10.1091/mbc.E08-02-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widlund PO, Stear JH, Pozniakovsky A, Zanic M, Reber S, Brouhard GJ, Hyman AA, Howard J. XMAP215 polymerase activity is built by combining multiple tubulin-binding TOG domains and a basic lattice-binding region. Proc Natl Acad Sci USA. 2011;108:2741–2746. doi: 10.1073/pnas.1016498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C, Wilde A, Moore MS, Adam SA, Merdes A, Zheng Y. Role of importin-β in coupling Ran to downstream targets in microtubule assembly. Science. 2001;291:653–656. doi: 10.1126/science.1057661. [DOI] [PubMed] [Google Scholar]
- Wilde A, Zheng Y. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 1999;284:1359–1362. doi: 10.1126/science.284.5418.1359. [DOI] [PubMed] [Google Scholar]
- Wilson-Grady JT, Villen J, Gygi SP. Phosphoproteome analysis of fission yeast. J Proteome Res. 2008;7:1088–1097. doi: 10.1021/pr7006335. [DOI] [PubMed] [Google Scholar]
- Yamashita A, Sato M, Fujita A, Yamamoto M, Toda T. The roles of fission yeast Ase1 in mitotic cell division, meiotic nuclear oscillation, and cytokinesis checkpoint signaling. Mol Biol Cell. 2005;16:1378–1395. doi: 10.1091/mbc.E04-10-0859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Clarke PR. Roles of Ran-GTP and Ran-GDP in precursor vesicle recruitment and fusion during nuclear envelope assembly in a human cell-free system. Curr Biol. 2001;11:208–212. doi: 10.1016/s0960-9822(01)00053-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.