Microtubule orientation controls polarized trafficking in neurons. In this work, γ-tubulin is identified as a key regulator of both axonal and dendritic microtubule polarity. In addition, the idea that γ-tubulin works in dendrites by residing at Golgi outposts is tested.
Abstract
Neurons have highly polarized arrangements of microtubules, but it is incompletely understood how microtubule polarity is controlled in either axons or dendrites. To explore whether microtubule nucleation by γ-tubulin might contribute to polarity, we analyzed neuronal microtubules in Drosophila containing gain- or loss-of-function alleles of γ-tubulin. Both increased and decreased activity of γ-tubulin, the core microtubule nucleation protein, altered microtubule polarity in axons and dendrites, suggesting a close link between regulation of nucleation and polarity. To test whether nucleation might locally regulate polarity in axons and dendrites, we examined the distribution of γ-tubulin. Consistent with local nucleation, tagged and endogenous γ-tubulins were found in specific positions in dendrites and axons. Because the Golgi complex can house nucleation sites, we explored whether microtubule nucleation might occur at dendritic Golgi outposts. However, distinct Golgi outposts were not present in all dendrites that required regulated nucleation for polarity. Moreover, when we dragged the Golgi out of dendrites with an activated kinesin, γ-tubulin remained in dendrites. We conclude that regulated microtubule nucleation controls neuronal microtubule polarity but that the Golgi complex is not directly involved in housing nucleation sites.
INTRODUCTION
In all neurons in which the issue has been examined, axons and dendrites are distinguished by their arrangement of microtubules. Microtubules have a stable minus end, which is where they initiate, and dynamic plus ends that serve as sites of tubulin addition and loss. Axons always have minus-end-out microtubules, whereas at least 50% of dendritic microtubules have minus ends out (Baas and Lin, 2011; Rolls, 2011). In fact, in vivo imaging in Drosophila and Caenorhabditis elegans demonstrates that dendrites in these animals contain almost exclusively minus-end-out microtubules (Stone et al., 2008; Goodwin et al., 2012). Because differences in microtubule polarity can be used to direct different cargoes from the cell body to axons or dendrites (Kapitein et al., 2010), they are likely to be at the core of compartmental identity in neurons. It is not well understood, however, how neuronal microtubule polarity is established or maintained. One factor likely to be important to generate a comprehensive model of microtubule polarity is the source of new microtubules.
Microtubules arise primarily by γ-tubulin–mediated nucleation. γ-Tubulin serves as the core of a complex that acts as a platform for addition of α- and β-tubulin dimers (Raynaud-Messina and Merdes, 2007). In many mitotic and some interphase animal cells, γ-tubulin is localized at the centrosome and organizes microtubules into a plus-end-out array. Neurons, however, do not conform to this simple arrangement, and the site of neuronal microtubule nucleation is central to two different mechanisms that have been proposed to contribute to neuronal microtubule polarity.
The microtubule transport model posits that microtubules are nucleated in the cell body; then, small, stable microtubule seeds are released from the nucleation site and transported in a polarized way into neuronal processes (Baas and Yu, 1996; Baas and Lin, 2011). This model accounts for plus-end-out axonal microtubule polarity through the action of dynein anchored at the axon initial segment. Anchored dynein would have its motor domain free to slide plus-end-out oriented pieces of microtubules into axons and minus-end-out pieces back into the cell body. Mixed polarity in dendrites could be accounted for by the action of kinesin-6 (CHO1/MKLP1) sliding minus-end-out microtubules along plus-end-out ones (Figure 1A, model 1). The transport model was originally suggested by studies on dissociated and cultured rodent neurons (Yu et al., 1993; Sharp et al., 1995; Baas and Yu, 1996). The role of dynein in transporting plus-end-out microtubules into axons has some in vivo experimental support as well. For example, in Drosophila neurons, an increase in minus-end-out microtubules is seen in axons of dynein-subunit mutants (Zheng et al., 2008), consistent with a role for dynein in pushing these microtubules back into the cell body. However, a recent study suggested that kinesin-1 rather than dynein might drive microtubules into newly growing axons (Lu et al., 2013). There is also experimental support for transport of minus-end-out microtubules into dendrites by kinesin-6; depletion of this motor reduces numbers of dendritic minus-end-out microtubules in primary cultures of rodent neurons (Sharp et al., 1997; Lin et al., 2012). The major source of microtubules for transport in this type of model is the centrosome (Yu et al., 1993). However, in mammalian neurons, as they mature, the centrosome becomes less active for microtubule nucleation (Stiess et al., 2010), and in Drosophila neurons, nucleation does not occur at the centrosome (Nguyen et al., 2011). These studies suggest that either nucleation complexes are found at different sites in the cell body in mature neurons or the cell body becomes less important as a source for new microtubules.
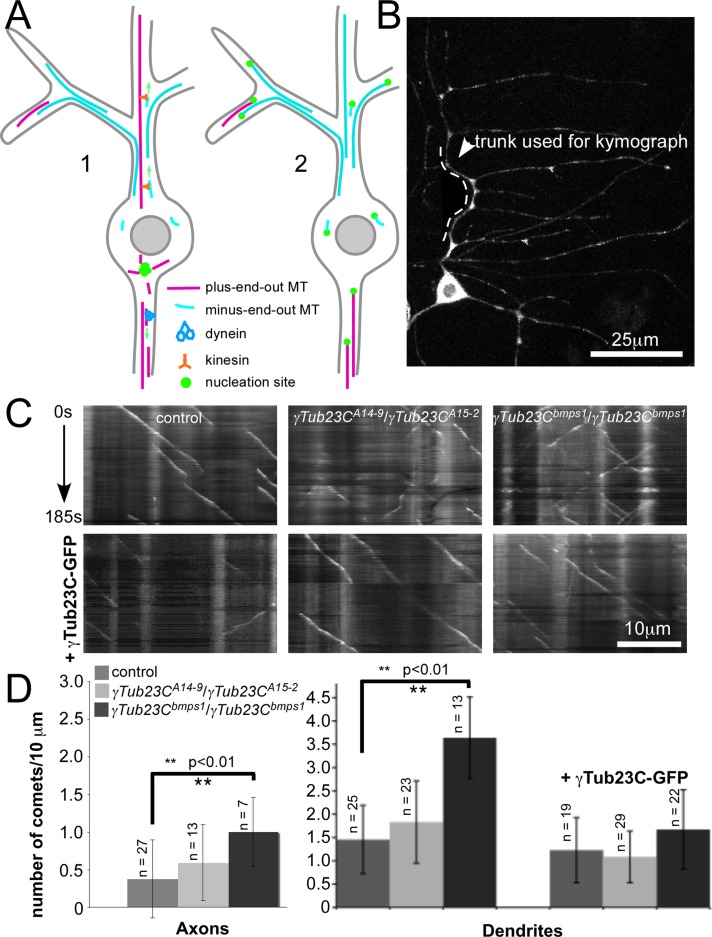
FIGURE 1:
Proposed positions of nucleation sites in neurons and changes in number of growing microtubules in different γ-tubulin mutants. (A) Left, key features of microtubule-sliding models, including nucleation of microtubules in the cell body, filtering of correctly polarized microtubule pieces at the start of the axon, and sliding minus-end-out microtubule seeds along plus-end-out ones in dendrites. Right, alternate model in which microtubules are generated locally in axons and dendrites. (B) Effect of mutations in γ-tubulin on microtubule dynamics analyzed in ddaE neurons expressing EB1-GFP under control of 221-Gal4. An example of a ddaE neuron. The dendritic analysis was performed in the main trunk (dotted line) of the dorsal comb dendrite of this cell. Axonal analysis was performed in the proximal axon. (C) Kymographs showing EB1-GFP comets over time. They were generated from time series of ddaE neurons expressing EB1-GFP. In all images the cell body is positioned at the right. (D) Quantitation of EB1-GFP comets in different genotypes. Number of EB1-GFP comets was analyzed by counting the EB1-GFP comets present in 10-μm lengths of axon in every 40th frame of movies. For dendrites, the complete number of comets was determined in the ddaE main trunk for an entire time series and then normalized to comets per 10-μm length and per 100 s. The number of animals from which the averages were quantitated is shown above the graph bars for each genotype. Error bars represent SDs, and a t test was used to determine significance.
An alternate model is that microtubules are generated locally at nucleation sites in dendrites and axons rather than in the cell body (Figure 1A, model 2). In this case, motor transport of microtubule pieces would not be required to seed microtubules, although it could still be used as a source of tubulin subunits to extend microtubules. The Golgi complex has been suggested as the local source of microtubule minus ends in Drosophila dendrites (Ori-McKenney et al., 2012), and there is precedent from other cell types for the Golgi to serve as a microtubule-organizing center (Efimov et al., 2007; Rivero et al., 2009). However, it has not been demonstrated that γ-tubulin localizes to neuronal Golgi in dendrites, and it is difficult to perform rigorous nucleation assays in neurons, as their microtubules are too stable to depolymerize. In the case of axons, the existence of local microtubule nucleation has not been intensively investigated. If microtubules are locally nucleated in axons or dendrites rather than initiated from transported seeds, then regulation of nucleation rather than action of motor proteins is likely to be critical for correct microtubule polarity.
To determine whether microtubule nucleation plays a role in axonal or dendritic microtubule polarity, we analyzed neurons in animals with different types of γ-tubulin mutations. We found that both loss- and gain-of-function γ-tubulin alleles altered axonal and dendritic microtubule polarity. This result suggests a close tie between nucleation and polarity, perhaps by local nucleation sites in axons and dendrites; changes in nucleation occurring in the cell body would not be expected to alter polarity in axons and dendrites, as motors would still filter only correctly oriented microtubule seeds into neurites. To further probe the possibility of local nucleation, we assayed localization of endogenous and tagged γ-tubulin in axons and dendrites; both were present in distinctive spots at dendrite branch points and presynaptic terminals. Because the Golgi complex was previously suggested to be the site of dendritic nucleation (Ori-McKenney et al., 2012), we used several different approaches, including dragging the Golgi out of dendrites, to test the link between the Golgi complex and microtubule nucleation. Our data suggest that nucleation sites are not localized to the Golgi in dendrites. We therefore propose that tight regulation of microtubule nucleation in both axons and dendrites controls microtubule polarity without localization of nucleation sites to the Golgi complex.
RESULTS
Both loss of γ-tubulin and overactive γ-tubulin affect microtubule orientation in axons and dendrites of class I ddaE neurons
To determine whether γ-tubulin plays a role in controlling polarity of microtubules in axons and dendrites, we assayed microtubule orientation in neurons from animals with mutant versions of the γ-tubulin23C gene. In Drosophila, there are two γ-tubulin genes: γ-tubulin37C encodes a protein that is maternally expressed and involved in early embryonic development, and γ-tubulin23C encodes the major somatic γ-tubulin (Wiese, 2008). Microtubule orientation was assessed using EB1–green fluorescent protein (GFP) dynamics; EB1-GFP comets mark growing microtubules, and so the direction of comet movement indicates polarity of the microtubule (Stepanova et al., 2003). In normal Drosophila da neurons, axonal microtubules are oriented with almost 100% of their plus ends distal to the soma, whereas the opposite orientation is seen in dendrites, which have >90% of microtubule minus ends distal to the soma (Rolls et al., 2007; Stone et al., 2008). To assay the effects of changes in γ-tubulin on microtubule orientation, we looked at the class I ddaE neuron. The branches of the dorsal ddaE dendrite emerge at close to right angles from the main trunk of the dendrite, making this cell particularly sensitive to changes in regulation of microtubule polarity (Mattie et al., 2010).
We first looked at the effects of different γ-tubulin alleles on the number of growing microtubule ends marked with EB1-GFP. The number of microtubule plus ends reflects overall levels of microtubule nucleation in both cultured cells (Piehl et al., 2004) and Drosophila neurons (Chen et al., 2012). The null allele of γ-tubulin, γTub23CA15-2 (stop codon at amino acid 104; Vazquez et al., 2008), is homozygous lethal, so we used transheterozygotes of γTub23CA15-2 with the hypomorphic allele, γTub23CA14-9 (amino acid substitution R217H; Vazquez et al., 2008). In addition to the loss-of-function mutants, we tested the γTub23Cbmps1 mutant, which has dominant eye and wing defects (Vazquez et al., 2008). The original isolate of γTub23Cbmps1 contained a suppressor mutation (Vazquez et al., 2008), which we removed for this analysis. Although γTub23Cbmps1-homozygous adults do not survive, homozygotes do survive into larval life. We observed no difference in number of growing microtubule plus ends in γTub23CA15-2/γTub23CA14-9 neurons (Figure 1, C and D); however, we did notice some changes in microtubule polarity in the trunk of the dendrite used for analysis (Figure 1, B and C). We previously showed, however, that fewer growing microtubules can be induced by axon injury in γTub23CA15-2/+ animals, so we hypothesize that in uninjured neurons, a very small amount of functional γ-tubulin can support normal microtubule growth. In animals homozygous for γTub23Cbmps1, the number of growing microtubules was increased in axons and dendrites (Figure 1D). This γ-tubulin allele has not been extensively characterized, but it is dominant and contains a single amino acid substitution (M382I; Mahoney et al., 2006). In these larvae, we observed an increase in the number of EB1-GFP comets in axons and dendrites (Figure 1, C and D). This increase is consistent with the γTub23Cbmps1 allele acting in a gain-of-function manner to increase microtubule nucleation. To confirm that this phenotype was due to the mutation in γ-tubulin, we added a GFP-tagged γ-tubulin transgene that we previously demonstrated to be capable of rescuing mutant phenotypes (Chen et al., 2012). This transgene suppressed the comet increase in γTub23Cbmps1 neurons (Figure 1D), consistent with the phenotype being due to the point mutation in γ-tubulin. Because γ-tubulin acts in a multimeric complex, overexpressed γ-tubulin23C-GFP may outcompete the mutant allele for incorporation into the active nucleation complex restoring normal function. γ-Tubulin activity has also been associated with dendrite branching (Ori-McKenney et al., 2012). We therefore also assayed arbor complexity in both genetic backgrounds. In the loss-of function background, class I ddaE neurons had no change in number of dendrite branch points (Supplemental Figure S1). However, in the γTub23Cbmps1 background, branch point number was significantly increased (Supplemental Figure S1), again consistent with increased nucleation activity in this allele. We conclude that the γTub23Cbmps1 allele is likely to be an overactive mutant of γ-tubulin.
We next tested whether microtubule polarity was altered in the γTub23C loss- or gain-of-function genetic backgrounds. In control neurons, axonal microtubule polarity was ∼95% plus end out; however, in transheterozygous loss-of-function mutant axons, EB1-GFP comets traveled away from the cell body only 85% of the time (Figure 2, A and B, and Supplemental Movies S1 and S3). In control dendrites, EB1-GFP comets traveled away the cell body <10% of the time. However, loss-of-function mutant dendrites were significantly more mixed, with ∼25% of EB1-GFP comets moving toward the soma (Figure 2, A and C, and Supplemental Movies S2 and S4).
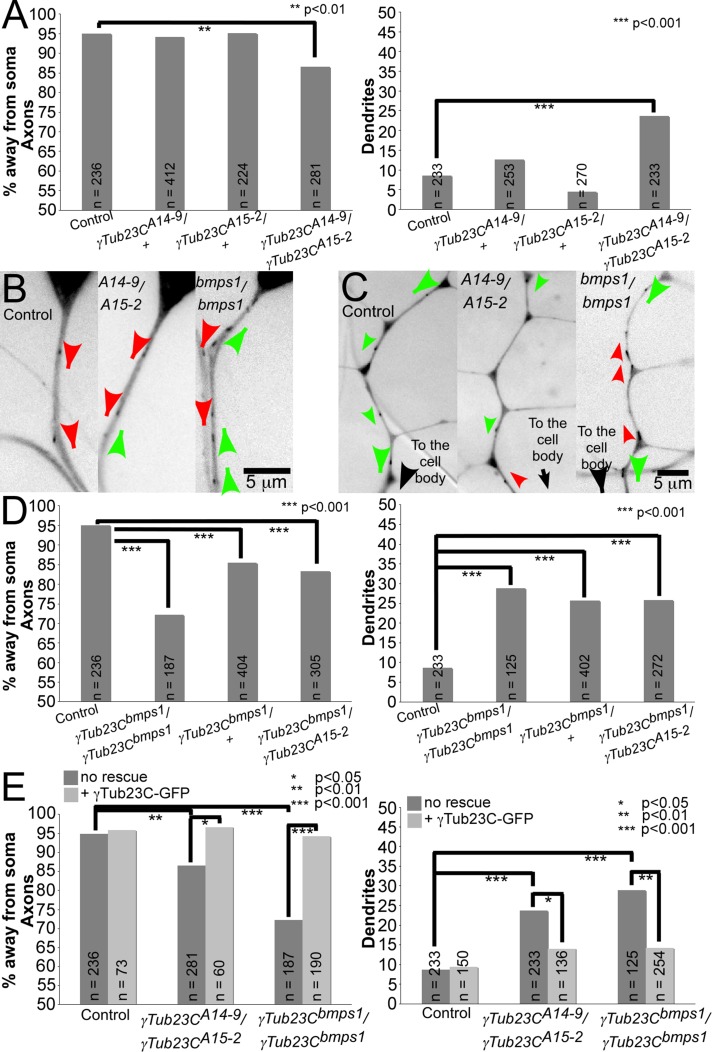
FIGURE 2:
γ-Tubulin mutants have defects in neuronal microtubule polarity. EB1-GFP was expressed in larval class I da neurons using 221-Gal4 in different mutant backgrounds. The direction of EB1-GFP comet movement was quantified in the dorsal comb dendrite of ddaE neurons. n, number of EB1-GFP comets analyzed for each genotype. (A) γ-Tubulin loss-of-function alleles analyzed as heterozygotes or transheterozygotes for axonal polarity (left) and dendritic polarity (right). (B, C) Examples of EB1-GFP comets in axons (B) and dendrites (C). Comets moving away from the cell body are indicated with red arrowheads, and comets moving toward the cell body are indicated with green arrowheads. In all images dorsal is up. Supplemental Movies S1–S6 show the EB1-GFP movement illustrated by the stills. (D) Both heterozygous and homozygous γTub23Cbmps1 mutants were analyzed for microtubule polarity in axons (left) and dendrites (right). (E) γTub23C-GFP was expressed under Gal4-UAS control in different genetic backgrounds, and microtubule polarity was analyzed in axons (left) and dendrite (right). Supplemental Movies S7–S12 illustrate the EB1-GFP movement in each genetic background. A Fisher's exact test was used to calculate p values.
When we assayed microtubule orientation in homozygous γTub23Cbmps1 larvae, we found that, as in the loss-of-function mutants, microtubules became more mixed in polarity in axons and dendrites (Figure 2, B–D, and Supplemental Movies S5 and S6). In addition, animals with only one copy of the γTub23Cbmps1 allele exhibited mixed microtubule polarity, consistent with a gain of function of this allele (Figure 2D). In either type of mutant background, microtubule polarity was rescued by overexpression of γ-tubulin23C-GFP (Figure 2Eand Supplemental Movies S7–S12). Because either decreased or increased activity of γ-tubulin disrupted microtubule polarity, we conclude that regulated microtubule nucleation is required for axonal and dendritic microtubule polarity.
Localization of γ-tubulin in axons and dendrites
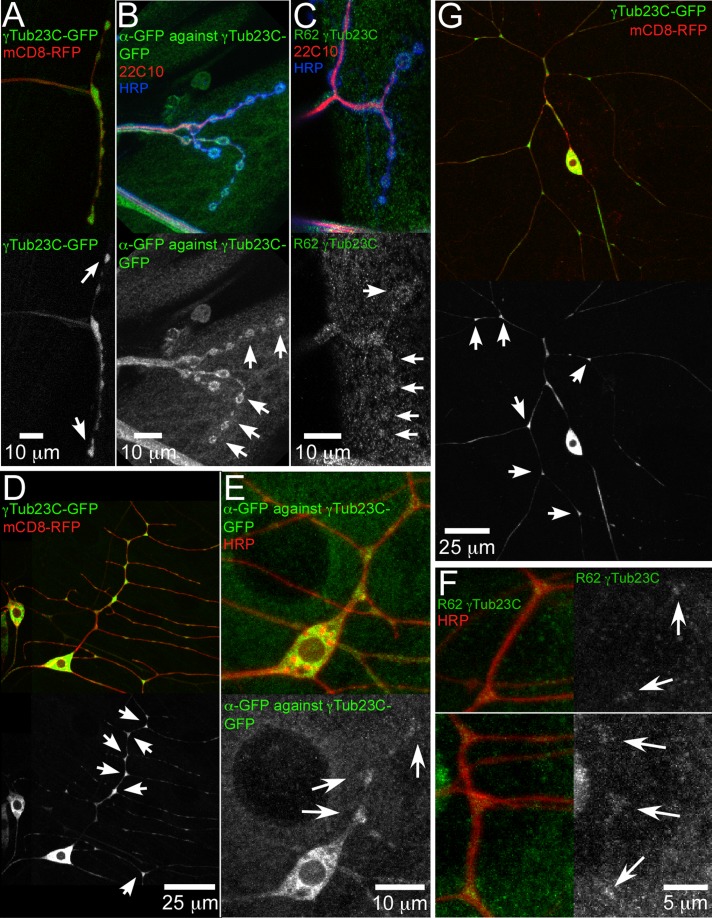
Disruption of microtubule polarity in axons and dendrites by different types of γ-tubulin alleles suggested a close relationship between microtubule polarity and microtubule nucleation and was consistent with local nucleation mediated by γ-tubulin in these compartments. Although γ-tubulin localization is easy to detect when it is concentrated on the centrosome, it can be much more elusive when distributed to other sites in the cell. Nevertheless, we tried to determine the localization of both endogenous and GFP-tagged γ-tubulin. In the cell body, γ-tubulin23C-GFP has a diffuse pattern (Nguyen et al., 2011). Diffuse tagged and endogenous γ-tubulin was seen throughout axons of da neurons. In addition, motor neuron axons are present in the body wall under the da neurons, and we observed concentrations of γ-tubulin in their large presynaptic boutons (Figure 3, A–C). In dendrites, endogenous and tagged γ-tubulin was concentrated at dendrite branch points (Figure 3, D–G). For comparison, we examined a soluble fluorescent protein in the same neurons and calculated the ratio of the fluorescence intensity at the branch points and between branch points (Supplemental Figure S2). This ratio was close to 1 for the soluble protein and >2 for γ-tubulin23C-GFP, indicating that this marker is significantly concentrated at the branch point. Although staining for endogenous γ-tubulin was quite faint and somewhat variable, more signal was present within branch points than between branch points in dendrites (Figure 3F and Supplemental Figure S3). Relative branch point fluorescence was lower in the γTub23CA15-2/γTub23CA14-9 neurons, consistent with reduced but not absent γ-tubulin in this background, whereas branch point concentration was similar to control neurons in γTub23Cbmps1 animals (Supplemental Figure S3). Overall the localization of tagged and endogenous γ-tubulin to axons and dendrites is consistent with it controlling nucleation outside the cell body. In particular, the concentration at dendrite branch points suggested that nucleation activity might be concentrated at these junctions.
FIGURE 3:
γ-Tubulin localization in motor axon terminals and da neuron dendrites. (A–C) Localization of γ-tubulin in motor axons. (A) γ-Tubulin23C-GFP and the membrane marker mCD8-RFP were expressed with 477-Gal4, which labels a few motor neurons in addition to class IV da neurons. Motor axon terminals have large synaptic boutons that are very useful for localization studies. Arrows indicate tips of motor axons. (B) Larvae expressing γ-tubulin23C-GFP were fixed and stained with antibodies to GFP, to the microtubule binding protein Futsch (22C10 antibody), and with anti–horseradish peroxidase (HRP), which stains neuronal membranes in Drosophila. (C) Larvae without tagged γ-tubulin were fixed and stained with 22C10, anti-HRP, and anti–γ-tubulin23C antibodies. In B and C, arrows indicate synaptic boutons. (D–G) Localization of γ-tubulin in dendrites of da neurons. (D, E) γ-Tubulin23C-GFP and mCD8-RFP were expressed in class I da neurons with 221-Gal4, and cells were either visualized in whole live animals (D) or fixed and stained (E). (F) Animals without overexpressed γ-tubulin were fixed and stained with antibodies to γ-tubulin23C and HRP. A portion of the comb dendrite of ddaE. (G) γ-Tubulin23C-GFP in class IV and mCD8-RFP expressed in class IV da neurons with 477-Gal4 and imaged in living animals. In D–G, arrows indicate dendrite branch points.
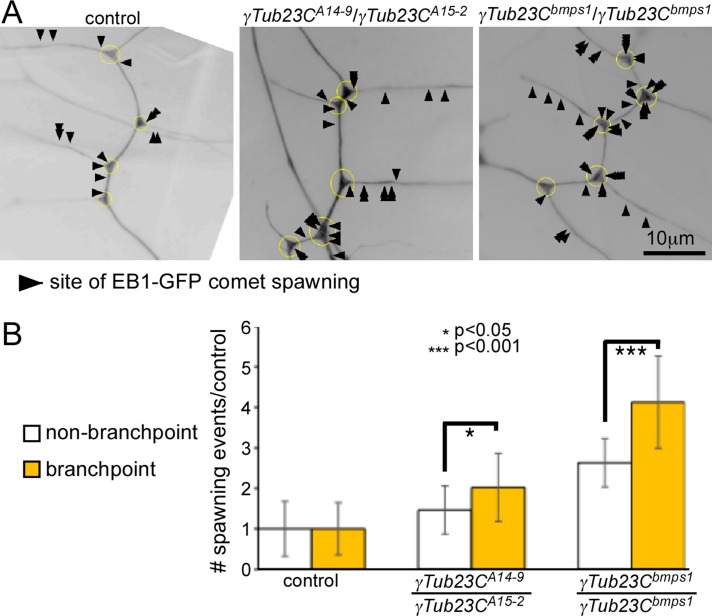
Localization of nucleation activity in neurons is very challenging. Nucleation assays typically rely on depolymerizing microtubules completely and then assaying the site where microtubules first start regrowing. Neuronal microtubules are incredibly stable, and we have not been able to depolymerize them in neurons in vivo in Drosophila or in tissue explants and thus have not been able to perform traditional nucleation assays. Although the number of EB1-GFP comets is a rough readout for overall level of microtubule nucleation activity, in neurons with long, stable microtubules, individual plus ends may be far away from the nucleation site. Similarly, although appearance of new EB1-GFP comets was previously counted as a readout of microtubule nucleation (Ori-McKenney et al., 2012), most new EB1-GFP comets in neurons are likely to represent catastrophe rescue rather than nucleation events. However, we reasoned that the sites where appearance of comets was preferentially increased in the overactive γTub23Cbmps1 allele would be at least loosely indicative of nucleation sites. We therefore assayed appearance of new EB1-GFP comets (or comet spawning) in neurons with different γTub23C alleles (Figure 4 and Supplemental Movies S13–S15). We found that comet spawning was increased more at branch points than between branch points when nucleation was up-regulated in the γTub23Cbmps1 background (Figure 4 and Supplemental Movie S15). Surprisingly, branch point spawning was also preferentially increased in the loss-of-function background, perhaps because remaining nucleation sites were activated as much as possible to make up for the reduction in γ-tubulin (Figure 4 and Supplemental Movie S14). The preferential increase in spawning at branch points when γ-tubulin activity was altered suggests a link between dendrite branch points and microtubule nucleation activity, which is consistent with the concentration of endogenous and tagged γ-tubulin at branch points.
FIGURE 4:
EB1-GFP comet spawning is preferentially upregulated at dendrite branch points in γ-tubulin mutant animals. (A) Sites of EB1-GFP comet appearance are marked with arrowheads in time-lapse movies of the dendrite arbor of ddaE neurons in different genetic backgrounds. All comet initiation events occurring within a 175-s segment from time-lapse movies are shown. Yellow circles indicate branch points. Comets that initiated within these circles were scored as branch point spawning. (B) Number of spawning events within and outside branch points normalized to control neurons. In both γ-tubulin mutant backgrounds, the overall level of spawning increased, but it increased more within the branch points than between them. Error bars show SDs. Number of animals analyzed for each genotype was as follows: control, 25; γTub23CA14-9/γTub23CA15-2 mutants, 21; γTub23Cbmps1 mutants, 12.
Golgi outposts are positioned at proximal dendrite branch points in neurons with large arbors
Golgi outposts have been suggested to be sites of acentrosomal microtubule nucleation at dendrite branch points of Drosophila da neurons (Ori-McKenney et al., 2012). The Golgi is thus a good candidate for housing dendritic γ-tubulin. However, because the Golgi is found only in dendrites and not axons (Craig and Banker, 1994; Horton and Ehlers, 2003; Ye et al., 2007), it would be unlikely to house any γ-tubulin involved in controlling axonal microtubule organization.
To determine whether γ-tubulin might coincide with Golgi outposts, we analyzed the localization of Golgi markers in class I (ddaE) and class IV (ddaC) da neurons. Previous descriptions of Golgi outposts in Drosophila neurons relied exclusively on tagged markers expressed in the ddaC neuron, so we focused our analysis on these characterized fusion proteins. We expressed GalT-YFP, a trans-Golgi marker, and ManII-GFP, a medial/trans-Golgi marker, in da neurons (Ye et al., 2007; Zheng et al., 2008; Ori-McKenney et al., 2012). Both Golgi markers labeled the larger Golgi outposts seen in proximal dendrite branch points of large class IV neurons (Supplemental Figure S4, A–C). Smaller, dimmer spots of fluorescence were seen in both axons and dendrites, but because axons do not contain Golgi, we considered that these spots were most likely due to leakage of overexpressed markers into some other compartment. When counting Golgi outposts in dendrites, we included only punctae that were larger and brighter than these fluorescent spots seen throughout the neuron. For most analyses we used GalT-YFP because it had less background expression, including complete exclusion from the nucleus.
Surprisingly, we noticed a difference in dendritic Golgi outpost localization between class I and class IV da neurons. Compared to class IV da neurons, class I da neurons have simple dendrite arbors. Although prominent Golgi complexes were seen in the cell body in class I neurons, most had no dendritic Golgi outposts (Supplemental Figure S4, D and E). This lack of Golgi at the branch points of class I neurons was in stark contrast to γ-tubulin localization, which was seen at most branch points in class I neurons (Figure 3, D–F). Thus, although γ-tubulin seems to be localized to dendrites, its position does not match well with that of Golgi outposts.
To test whether the Golgi might colocalize with γ-tubulin when present in the same large, proximal branch points of ddaC neurons, we used two-color live imaging of the Golgi and γ-tubulin. To mark the Golgi, we used γCOP–red fluorescent protein (RFP), as γCOP is part of the vesicle coat complex that mediates retrograde transport from the Golgi to the endoplasmic reticulum and concentrates on the Golgi. Spots of γCOP-RFP did not colocalize well with γ-tubulin23C-GFP even in the same branch point (Supplemental Figure S5).
The Golgi complex can be disrupted in neurons in vivo by expressing kinesin motor domains fused with Golgi-binding domains
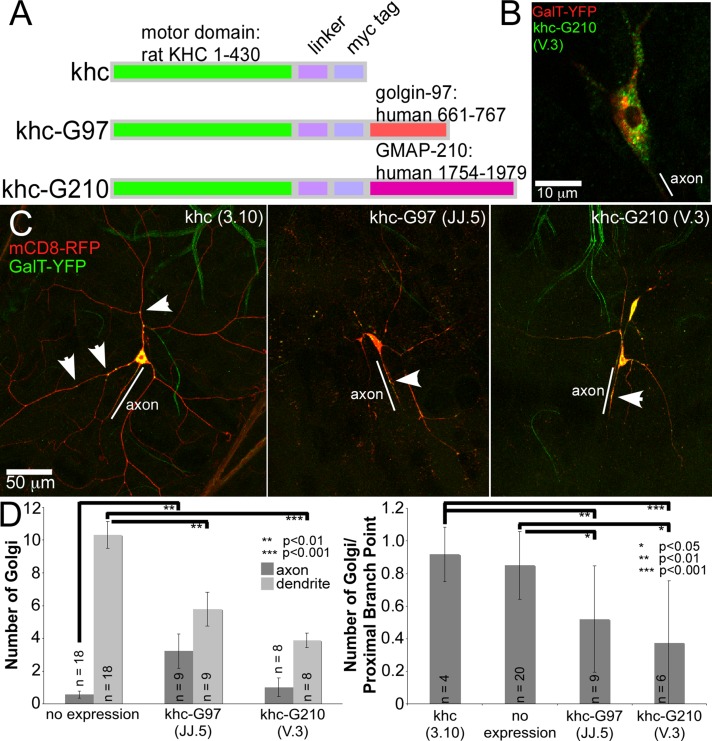
Even though the localization of tagged Golgi proteins suggested that the Golgi was not a strong candidate for housing nucleation sites, we considered that we could have missed very small Golgi outposts in small dendrite arbors. To probe more directly the function of the Golgi in neuronal microtubule organization, we developed a strategy to drag the Golgi out of dendrites. A number of large coiled-coil proteins localize to the Golgi complex, and the Golgi-binding region of several of these has been identified (Gillingham and Munro, 2003). GMAP-210 localizes to the cis-Golgi using a region near its C-terminus (Chen et al., 1999), and golgin-97 localizes to the trans-Golgi through its GRIP domain (Barr, 1999; Kjer-Nielsen et al., 1999; Munro and Nichols, 1999). We attached the Golgi-binding domains from these two proteins to truncated kinesin heavy chain because truncated forms are constitutively activated (Verhey et al., 1998). To test these fusion proteins, we expressed them in cultured mammalian cells that have the Golgi complex positioned adjacent to the centrosomal microtubule-organizing center. In these cells, microtubule minus ends are found in the middle of the cell, and plus ends are near the cell periphery. In control cells without the kinesin-Golgi fusions, a Golgi marker formed a tight clump in the middle of the cell, whereas in cells that expressed kinesin-Golgi fusions, the Golgi was fragmented and moved to the cell periphery, presumably by being dragged out toward microtubule plus ends (Supplemental Figure S6). The fusion to the GMAP-210 domain colocalized with a cis-Golgi marker as expected but was able to relocalize both cis- and trans-Golgi compartments (Supplemental Figure S6). Overexpression of a mutant version of the golgin-97–binding domain that does not target the Golgi had no effect on the Golgi complex, and the effect of the kinesin-G97 fusion was specific to the Golgi complex, as lysosomes and endoplasmic reticulum were unaffected (Supplemental Figure S6). We next generated transgenic flies expressing these constructs (Figure 5A), since the Golgi-binding domains are well conserved. To determine whether these transgenes were being expressed, we stained larvae with anti-myc antibodies; myc staining was seen in a punctate pattern in neurons (Figure 5B).
FIGURE 5:
Expression of kinesin-Golgi fusions affects Golgi localization, Golgi morphology, and neuronal morphology. (A) Diagram of kinesin fusion constructs expressed in Drosophila neurons. (B) Larvae expressing GalT-YFP and myc-tagged kinesin-Golgi fusions in ddaC neurons with the 477-Gal4 driver were stained with anti-myc antibody. (C) mCD8-RFP and GalT-YFP were expressed with the kinesin-Golgi fusions in larvae using 477-Gal4. Specific transgene insertions are indicated with designations JJ.5 and V.3. Arrows indicate Golgi punctae. Dendrites had severely reduced arbors in neurons expressing either kinesin-Golgi fusion. (D) Left, Golgi outposts counted in ddaC dendrites and proximal axons. Average numbers of Golgi in each compartment. Right, numbers of Golgi normalized to the number of proximal branch points. p values were calculated with a t test. Error bars show SD; n, number of cells analyzed in each genotype.
To test for activity of kinesin-Golgi fusions in Drosophila neurons, we analyzed cell shape, as well as localization of the Golgi complex. Defects in Golgi-based trafficking within neurons have negative effects on dendrite growth, branching, and maintenance (Ye et al., 2007), so we tested whether the kinesin-Golgi fusions altered dendrite morphology. The dendrite arbor was significantly reduced in both class I ddaE and class IV ddaC neurons that expressed kinesin-Golgi fusions (Figure 5C and Supplemental Figures S7 and S8). This reduction in dendrite arbor is similar to that observed in mutants of dar3, the Drosophila homologue of the endoplasmic reticulum-to-Golgi transport regulator Sar1, and in mutants of a dynein subunit in which Golgi outposts are mislocalized (Ye et al., 2007; Zheng et al., 2008) and suggests that the Golgi is mislocalized or disrupted by expression of kinesin-Golgi fusions.
In neurons expressing the kinesin-Golgi fusions, we expected that the kinesin motors would pull the Golgi to microtubule plus ends, meaning that the Golgi would be pulled from the dendrites into the soma and possibly out of the soma and into the axons. To test whether the fusions decreased the amount of Golgi outposts seen in dendrites, we expressed UAS-GalT-YFP in conjunction with the membrane marker UAS-mCD8-RFP and assayed the number of Golgi present in the axons and dendrites of class IV da neurons (Figure 5, C and D). Neurons expressing kinesin-Golgi fusions had fewer Golgi outposts in dendrites than neurons that did not express the fusions (Figure 5D). The reduced number of Golgi outposts was likely not simply a result of the morphological changes, as most Golgi outposts in control neurons were found in proximal dendrites, which remained in the simplified dendrite arbor (Figure 5, C and D). To make sure that the decrease in Golgi in the arbors was not secondary to decreased number of branch points, we quantitated the number of Golgi and normalized it to the number of proximal branch points where the Golgi resides. This calculation also showed reduced dendritic Golgi when kinesin-Golgi fusions were expressed (Figure 5C, right). In addition, the kinesin-G97 fusion was able to pull the GalT-YFP trans-Golgi marker into axons, perhaps because it targets the trans-Golgi, whereas the kinesin-G210 targets the cis-Golgi (Figure 5, C and D).
Along with changes in Golgi localization, we noted changes in Golgi morphology. Expression of either khc-G97 or khc-G210 resulted in less distinct GalT-YFP punctae in the cell body and in dendrites and more small spots (Figure 5C). This was similar to the Golgi fragmentation observed in cultured cells (Supplemental Figure S6). These results indicate that the kinesin-Golgi fusions can reduce dendritic Golgi and impair dendrite growth, consistent with overall disruption and mislocalization of the Golgi complex.
Golgi disruption results in small changes in microtubule polarity in dendrites but not axons of class I and class IV da neurons
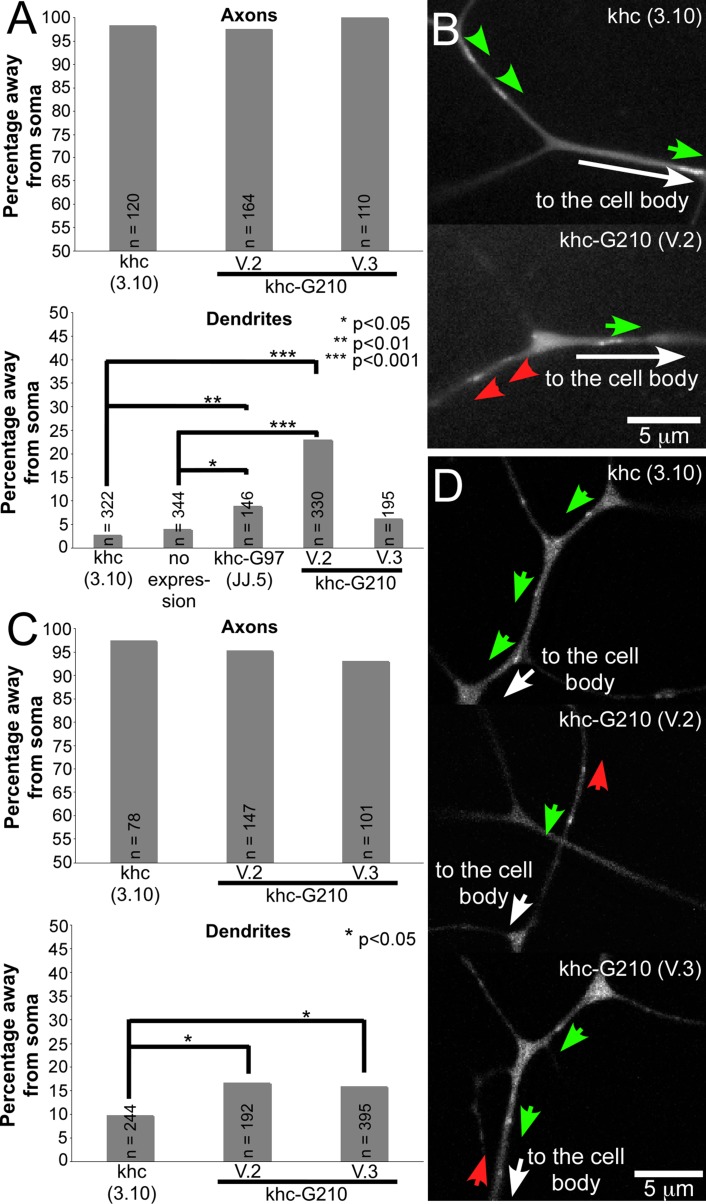
If the Golgi houses microtubule nucleation sites, then we might expect to see similar phenotypes from Golgi disruption and γ-tubulin23C mutants. To examine microtubule polarity in neurons expressing kinesin-Golgi fusions, we analyzed EB1-GFP comet movement. In control axons of class IV da neurons expressing only the kinesin motor domain, microtubules were primarily plus end out, as expected (Figure 6A). This plus-end-out orientation did not significantly change with expression of the kinesin-Golgi fusions. In dendrites, we found a significant increase in the percentage of plus-end-out microtubules with two of the three kinesin-Golgi fusion insertions tested (Figure 6, A and B). This effect was much more variable between lines than the effect on dendrite morphology, perhaps indicating that microtubule polarity is less sensitive to Golgi disruption than other dendritic features.
FIGURE 6:
Microtubule polarity in neurons expressing kinesin-Golgi fusions. (A) EB1-GFP and the indicated kinesin constructs were expressed using 477-Gal4, and microtubule polarity was assayed in the ddaC neuron. n, number of EB1-GFP comets analyzed for each condition. p values were calculated with a Fisher's exact test. (B) Stills from movies showing examples of EB1-GFP movement in ddaC dendrites. Green arrows indicate EB1-GFP comets moving to the cell body; red arrows indicate EB1-GFP comets moving away from the cell body. (C) Microtubule polarity was assayed by quantitating direction of EB1-GFP comet movement in the main branch of the comb dendrite of the ddaE neuron. n and p are as in A. (D) Stills from movies showing examples of EB1-GFP movement in ddaE comb dendrites. Green arrows indicate EB1-GFP comets moving to the cell body; red arrows indicate EB1-GFP comets moving away from the cell body.
To probe the relationship between the Golgi and polarity further, we assayed microtubule polarity in class I ddaE neurons that do not have obvious Golgi outposts. We saw a subtle yet significant mixing of microtubule orientation in these dendrites (Figure 6, C and D). On the basis of these results, we think it most likely that disruption of the Golgi affects microtubule polarity in dendrites indirectly, perhaps by altering trafficking of a microtubule regulator. Unlike mutations in γ-tubulin23C, disruption of the Golgi never altered axonal microtubule orientation.
Golgi disruption alters localization of Apc2-GFP in Drosophila da neurons
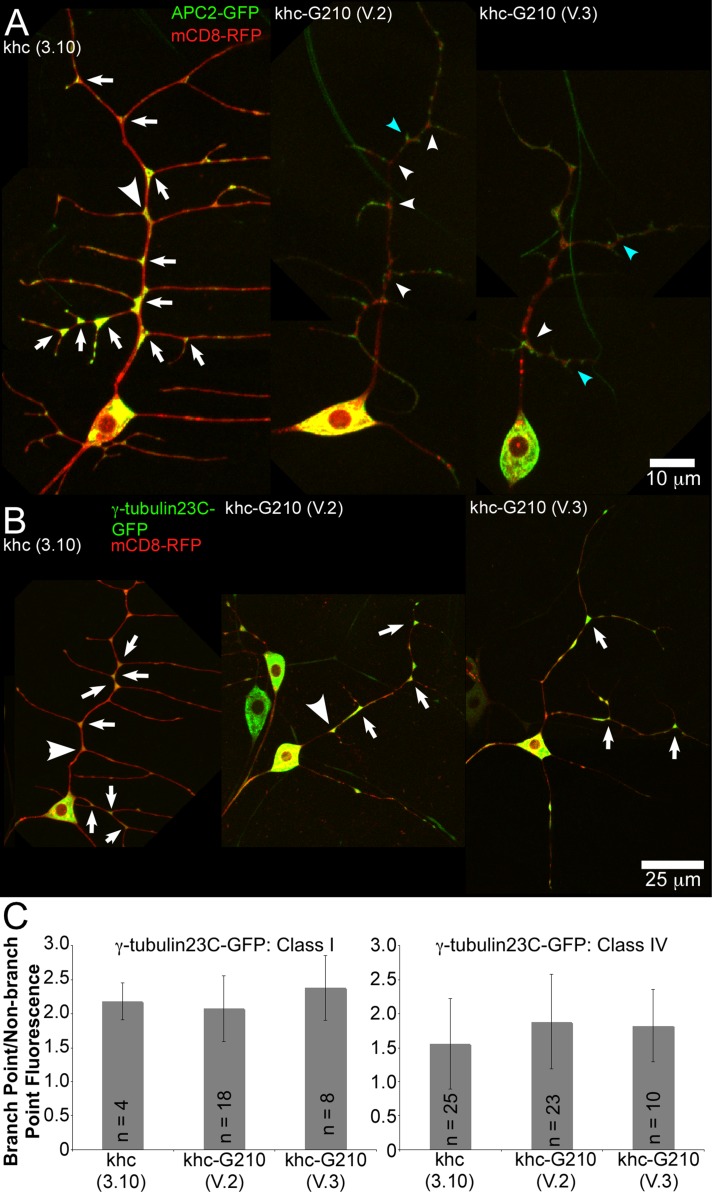
To probe whether Golgi disruption might alter identified regulators of dendritic microtubule polarity, we examined the distribution of Apc2-GFP in the presence of the kinesin-Golgi fusions. Maintenance of dendrite microtubule orientation in Drosophila da neurons has been found to require kinesin-2, EB1, Apc, and Apc2, which are proposed to work together to direct growing microtubules through branch points toward the cell body (Mattie et al., 2010). Apc2-GFP localizes very specifically to dendrite branch points, and knockdown of Apc2 alters microtubule orientation in da neurons (Mattie et al., 2010). To test whether the changes in dendrite microtubule orientation seen in the kinesin-Golgi fusions might be a result of mislocalization of Apc2, we looked for changes in Apc2-GFP distribution in the presence of the fusions. Control neurons that expressed only the kinesin motor domain had Apc2-GFP at dendrite branch points (Figure 7A and Supplemental Figure S9A). In neurons expressing kinesin-Golgi fusions, Apc2-GFP was much less concentrated at the branch point in ddaE and ddaC neurons (arrowheads, Figure 7A and Supplemental Figure S9A). Mistargeting of Apc2-GFP could account for the subtle change in microtubule polarity induced by expression of kinesin-Golgi fusions.
FIGURE 7:
Localization of Apc2-GFP and γ-tubulin23C-GFP in neurons with disrupted Golgi. (A) mCD8-RFP was expressed with either Apc2-GFP or γ-tubulin23C-GFP in class I neurons with 221-Gal4. Kinesin constructs were expressed as indicated. Neurons expressing Apc2-GFP. Dendrite branch points containing Apc2-GFP fluorescence are indicated with arrows. Dendrite branch points that lack clear Apc2-GFP punctae are indicated with white arrowheads, and ectopic projections that contain Apc2-GFP are indicated with blue arrowheads. (B) Localization of γ-tubulin23C-GFP in ddaE neurons. Branch points containing concentrations of γ-tubulin23C-GFP are indicated with arrows. (C) Average fluorescence intensity of γ-tubulin23C-GFP within branch points and outside branch points was measured. The ratio of these measurements is shown for class I (ddaE) and class IV (ddaC) neurons. n, number of cells in class I neurons, with one branch point examined per cell. In class IV neurons, which have more variability in γ-tubulin23C-GFP concentration, n is the number of branch points, with at least five cells examined in each condition. Error bars show SD.
Localization of γ-tubulin23C-GFP to dendrite branch points is not affected by Golgi disruption
To test whether disruption of the Golgi affected localization of γ-tubulin23C-GFP in a similar manner to Apc2-GFP, we expressed both γ-tubulin23C-GFP and mCD8-mRFP in the kinesin-Golgi fusion background. We found that, unlike Apc2-GFP, localization of γ-tubulin23C-GFP to dendrite branch points was not altered by the kinesin-Golgi fusions (Figure 7, B and C). This was seen in both class I ddaE neurons, which do not contain obvious dendritic Golgi outposts, and class IV ddaC neurons, which contain dendritic Golgi outposts (Supplemental Figure S9). We conclude that targeting of γ-tubulin23C-GFP to dendrite branch points is independent of the Golgi complex and the role of microtubule nucleation in controlling microtubule polarity does not require association with the Golgi.
DISCUSSION
To investigate the relationship between microtubule nucleation and polarized organization of microtubules in neurons, we examined the effects of different γ-tubulin mutants on axonal and dendritic microtubule polarity. Surprisingly, loss- and gain-of-function γ-tubulin alleles altered both axonal and dendritic microtubule polarity. Because this suggested a close link between microtubule nucleation and polarity control, we examined localization of endogenous and tagged γ-tubulin. Whether in fixed or living animals, γ-tubulin was observed in axons and dendrites and was particularly concentrated at dendrite branch points and axon termini.
The localization of γ-tubulin to axons and dendrites, as well as the close tie between microtubule polarity and γ-tubulin activity, argues that many microtubules in mature neurons are likely to be generated locally rather than transported from the cell body. Although transport of microtubule pieces from the cell body is important for initial axon outgrowth (Yu et al., 1993; Lu et al., 2013) and early dendritic development (Sharp et al., 1995, 1997; Yu et al., 2000; Lin et al., 2012), its role in more-mature neurons is not as clear. Indeed, kinesin-6, the motor that is involved in microtubule transport into dendrites, is down-regulated as neurons mature (Lin et al., 2012), and kinesin-1–mediated microtubule sliding is important only in the earliest stages of axon initiation in Drosophila (Lu et al., 2013).
Local nucleation of microtubules in dendrites was recently proposed to play a role in complex branching of class IV dendritic arborization neurons and was suggested to occur at Golgi outposts (Ori-McKenney et al., 2012). Although our results support an important role for microtubule nucleation in dendrites, we were unable to find any evidence that nucleation sites were housed on the Golgi complex. First, changes in microtubule nucleation also affected axonal polarity even though Golgi outposts are not found in axons. Second, using well-established Golgi markers, including the one used in the previous dendritic nucleation study (Ori-McKenney et al., 2012), we found that Golgi outposts were only present in large dendrite arbors, although minus-end-out microtubules and a link between nucleation and polarity were found in small arbors (ddaE) as well as large arbors (ddaC). Third, whereas dragging the Golgi out of dendrites with kinesin-Golgi fusions disrupted branch point localization of Apc2-GFP, it did not alter γ-tubulin23C-GFP concentration at dendrite branch points. Thus, although we favor a model in which regulated nucleation locally controls microtubule polarity in axons and dendrites, we do not find support for the idea that this regulated nucleation occurs at the Golgi complex. The Golgi is only one of several cellular membranes known to house nucleation sites; in muscle cells, γ-tubulin nucleates microtubules at the nuclear envelope (Bugnard et al., 2005), and in the C. elegans gonad, γ-tubulin–mediated nucleation occurs at the plasma membrane (Zhou et al., 2009). It is therefore possible that γ-tubulin in dendrite branch points is associated with either the plasma membrane or some other internal membrane structure such as the endoplasmic reticulum.
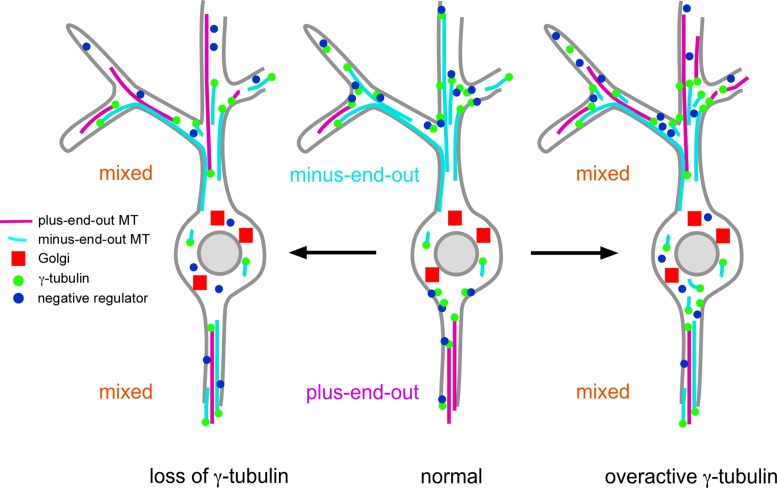
How γ-tubulin might control microtubule polarity in axons and dendrites will be extremely interesting to determine. On the basis of our γ-tubulin mutant results and the previous finding that microtubule dynamics in axons and dendrites was massively upregulated by axon injury or stress through nucleation (Stone et al., 2010; Chen et al., 2012), we propose a model in which γ-tubulin in axons and dendrites is tightly negatively regulated (Figure 8). The proposed negative regulator not only would control number of active nucleation sites, but it also would need to link nucleation to polarity in some way to account for the polarity changes in loss- and gain-of-function γ-tubulin mutant backgrounds. Loss of interaction with the negative regulator could explain both the increase in dynamics and polarity disruption seen in the γTub23Cbmps1 overactive mutants. Involvement of a negative regulator to control polarized use of nucleation sites could also explain why reduction of γ-tubulin affects microtubule orientation. When γ-tubulin levels are decreased, the negative regulator may be inactivated in order for the remaining γ-tubulin to become active (Figure 8). As a trade-off, the newly activated γ-tubulin would be dysregulated. Of course, the existence of such a regulator is speculative at this point. However, it is clear that understanding how microtubule nucleation is regulated in neurons will be critical for generating complete models of neuronal microtubule polarity.
FIGURE 8:
Model for the regulation of γ-tubulin in Drosophila neurons. On the basis of localization and mutant phenotypes, we propose that γ-tubulin localizes throughout axons and dendrites, with concentrations at specific places, including dendrite branch points. Under normal circumstances, a negative regulator of nucleation could ensure that new microtubules are generated in the correct orientation in axons and dendrites. When levels of γ-tubulin are reduced (left), the negative regulator is inactivated so that microtubules can still be generated, but polarity control is disrupted. The overactive γ-tubulin mutant may have increased activity because it fails to interact with the negative regulator (right). In many cells in which γ-tubulin activity is linked to microtubule polarity in axons and dendrites, the Golgi complex is found only in the cell body.
MATERIALS AND METHODS
Drosophila stocks
The Bloomington Drosophila Stock Center (Bloomington, IN) provided all Gal4 driver lines, all γ-tubulin mutant lines, and all fluorescently tagged marker lines except those noted further. For γ-tubulin mutant studies, we took out the l(1)dd4su(Pl) suppressor from the fly stock to obtain γTub23Cbmps1/CyO flies. The Golgi markers UAS-GalT-YFP and UAS-ManII-EGFP were obtained from Bing Ye (University of Michigan, Ann Arbor, MI) and Kassandra Ori-McKenney (University of California, San Francisco, San Francisco, CA). UAS-EB1-GFP lines were previously described (Rolls et al., 2007), as was generation of UAS-γ-tubulin23C-GFP (Nguyen et al., 2011).
Generation of kinesin-Golgi fusions and transgenic fly lines
Mammalian vectors containing the motor domain of rat Kif5b (Khc) connected to either the C-terminal region of the cis-Golgi protein GMAP210 or the C-terminus of the trans-Golgi protein golgin-97 via a linker and myc tag were generated. A mutant version of the golgin-97 targeting domain that cannot localize to the Golgi was also used (G97-Y697A) for validation (Munro and Nichols, 1999). These fusions were tested in mammalian COS cells first. Kinesin fusions were transiently expressed in COS cells from plasmids GRIP22 (khc-G210), GRIP26 (khc-G97), and GRIP27 (khc-G97-Y697A). Cells were transfected, fixed, and processed for immunofluorescence as described previously (Levine and Munro, 1998). Once tested, the fusions were placed into the fly vector pUAST::3YCT. Transgenic flies were obtained using standard genetic procedures. Five transgenic fly lines were generated that were used in this study: 1) khc, which is a control fusion without the Golgi-binding domain; 2) khc-G97 (JJ.2), which we also use as a control, as lack of c-myc staining indicates that the fusion is not being expressed; 3) khc-G97 (JJ.5); 4) khc-G210 (V.2); and 5) khc-G210 (V.3).
Larval immunostaining
Immunostaining of third-instar larvae was performed as previously described (Nguyen et al., 2011). Images of larval fillets were acquired with a Zeiss LSM 510 confocal microscope (Carl Zeiss, Jena, Germany), and the appropriate stacks were combined to make composite images. The following primary antibodies were used: mouse anti-c-myc (Developmental Studies Hybridoma Bank), 22C10 (Developmental Studies Hybridoma Bank), Alexa Fluor–conjugated goat anti-HRP (Jackson ImmunoResearch Laboratories, West Grove, PA), rabbit anti-GFP (Abcam, Cambridge, MA), and R62 against γ-tubulin23C (a gift from Brigitte Raynaud-Messina, Institute of Pharmacology and Structural Biology, Toulouse, France; Raynaud-Messina et al., 2001). The appropriate secondary fluorescent antibodies were obtained from Jackson ImmunoResearch.
Localization experiments
In all in vivo localization and morphology experiments, a time series was taken for each cell, and the stage and focus were moved incrementally in order to capture all of the dendrites. The resulting time-series images were then broken into stacks for each area focused on and then pasted together to make a composite image of the cell.
For comparison of endogenous and GFP-tagged γ-tubulin concentrations at dendrite branch points versus non–branch points, an ImageJ (National Institutes of Health, Bethesda, MD) plug-in was created to measure fluorescence intensity. The plug-in averaged all of the pixels in a specified quantification area, except for any pixel lower than a specified background value. Only one frame per movie, where the branch point and non–branch point were most in focus, was analyzed, and the same quantification areas were used between the control and the endogenous and GFP-tagged γ-tubulin analyses.
For quantitation of Golgi outposts, larval neurons expressing UAS-GalT-YFP were imaged live using an Olympus FluoView FV1000 confocal microscope (Olympus, Tokyo, Japan). The Golgi outposts were manually counted in the axons and dendrites of each cell.
Analysis of dendrite morphology
The class I da neurons of larvae expressing EB1-GFP were imaged in vivo using a Zeiss LSM 510 confocal microscope. Morphology of the proximal axon, cell body, and the entire dendrite arbor was imaged in the same manner as the localization experiments. To measure dendrite complexity, the total number of dendrite branch points in each class I neuron was determined, and the average number of branch points per class I neuron was calculated.
The class IV da neurons of larvae expressing EB1-GFP were imaged in vivo using a Zeiss AxioImager M2 microscope with Colibri LED light source (Carl Zeiss). To measure dendrite radius, the distance from the center of the soma to the most-distal dendrite tip was quantitated. To measure dendrite complexity, a circle with a 100 μm radius was drawn, with the center of the circle as the center of the soma. For each neuron analyzed, the dendrite branches that crossed through the border of each of these circles were counted.
Analysis of microtubule orientation and dynamics
EB1-GFP directionality experiments were performed as previously described (Nguyen et al., 2011). For analysis of EB1-GFP dynamics, larvae were imaged live using an Olympus FluoView FV1000. For axons, the visible comets were counted every 40 frames (2 s/frame) over a 10-μm length and the weighted mean calculated. For dendrites, the visible comets within the dendritic trunk were counted and normalized to comets per 10-μm length per 100 s. All images were analyzed using ImageJ.
Analysis of EB1-GFP spawning at dendrite branch points
For EB1-GFP comet-spawning analysis, image stacks were manually processed to include only those regions of the dendrite that remained in focus for the duration. Branch points were circled on an overlay, and the total branch point number was recorded, as well as the non–branch point dendrite length. EB1-GFP comets were then counted as they appeared over the course of the video. Any comet whose origination could not be seen was excluded from the analysis. The appearance of each new comet was classified as either a branch point comet or a non–branch point comet. Values were normalized by branch point number for branch point comets and by dendrite length for non–branch point comets and pooled for each subset.
Supplementary Material
Acknowledgments
We are very thankful to everyone who sent us fly stocks and antibodies, including Bing Ye, Kassandra Ori-McKenney, Annette Parks and Tim Megraw, Fumiko Kawasaki and Richard Ordway, and Brigitte Raynaud-Messina. We also thank Greg Kothe for help with cloning. We are also grateful to the Bloomington Drosophila Stock Center and the Vienna Drosophila RNAi Center for serving as important resources to scientists studying Drosophila and to the Developmental Studies Hybridoma Bank for serving as an important resource to the scientific community. Finally, we appreciate the other members of the Rolls lab for their encouragement and input throughout the experimental process. Funding for this work was provided by National Institutes of Health Grant R01 GM085115.
Abbreviation used:
- RFP
red fluorescent protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-09-0515) on May 7, 2014.
M.M.N. performed all γ-tubulin–related experiments, except those noted in what follows, as well as microtubule orientation assays in class I da neurons with kinesin-Golgi fusions, Apc2-GFP localization assays, and Golgi marker comparison studies. C.J.M. cloned the kinesin-Golgi fusions into fly vectors, generated transgenic fly lines from these vectors, tested for expression of the fusions in larvae, and performed the Golgi localization experiments. E.S.M. performed microtubule orientation assays in the class IV da neurons with the kinesin-Golgi fusions and analyzed dendrite morphology in class IV neurons with the fusions. D.J.G. generated kymographs for γ-tubulin mutant studies and with A.T.W. performed EB1-GFP comet number and spawning analyses in the dendrites of γ-tubulin mutants. M.K.L. examined coexpression of γ-tubulin23C-GFP and γCOP-RFP in da neurons and contributed to microtubule orientation assays in the axons of class IV da neurons. N.L.M. created the ImageJ plug-in used to analyze fluorescence intensity. S.M. generated the mammalian vectors with the kinesin-Golgi fusions. M.M.R. supervised the planning of experiments and interpretation of results. M.M.N. prepared the figures and wrote the article with M.M.R.
The authors declare that they have no competing interests.
Present addresses: *Biomedical Sciences Graduate Program, Ohio State University, Columbus, OH 43210
†Program in Neuroscience, Harvard Medical School, Boston, MA 02115.
REFERENCES
- Baas PW, Lin S. Hooks and comets: the story of microtubule polarity orientation in the neuron. Dev Neurobiol. 2011;71:403–418. doi: 10.1002/dneu.20818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas PW, Yu W. A composite model for establishing the microtubule arrays of the neuron. Mol Neurobiol. 1996;12:145–161. doi: 10.1007/BF02740651. [DOI] [PubMed] [Google Scholar]
- Barr FA. A novel Rab6-interacting domain defines a family of Golgi-targeted coiled-coil proteins. Curr Biol. 1999;9:381–384. doi: 10.1016/s0960-9822(99)80167-5. [DOI] [PubMed] [Google Scholar]
- Bugnard E, Zaal KJ, Ralston E. Reorganization of microtubule nucleation during muscle differentiation. Cell Motil Cytoskeleton. 2005;60:1–13. doi: 10.1002/cm.20042. [DOI] [PubMed] [Google Scholar]
- Chen Y, Chen PL, Chen CF, Sharp ZD, Lee WH. Thyroid hormone, T3-dependent phosphorylation and translocation of Trip230 from the Golgi complex to the nucleus. Proc Natl Acad Sci USA. 1999;96:4443–4448. doi: 10.1073/pnas.96.8.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Stone MC, Tao J, Rolls MM. Axon injury and stress trigger a microtubule-based neuroprotective pathway. Proc Natl Acad Sci USA 109. 2012:11842–11847. doi: 10.1073/pnas.1121180109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig AM, Banker G. Neuronal polarity. Annu Rev Neurosci. 1994;17:267–310. doi: 10.1146/annurev.ne.17.030194.001411. [DOI] [PubMed] [Google Scholar]
- Efimov A, et al. Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev Cell. 2007;12:917–930. doi: 10.1016/j.devcel.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham AK, Munro S. Long coiled-coil proteins and membrane traffic. Biochim Biophys Acta. 2003;1641:71–85. doi: 10.1016/s0167-4889(03)00088-0. [DOI] [PubMed] [Google Scholar]
- Goodwin PR, Sasaki JM, Juo P. Cyclin-dependent kinase 5 regulates the polarized trafficking of neuropeptide-containing dense-core vesicles in Caenorhabditis elegans motor neurons. J Neurosci. 2012;32:8158–8172. doi: 10.1523/JNEUROSCI.0251-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton AC, Ehlers MD. Dual modes of endoplasmic reticulum-to-Golgi transport in dendrites revealed by live-cell imaging. J Neurosci. 2003;23:6188–6199. doi: 10.1523/JNEUROSCI.23-15-06188.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein LC, Schlager MA, Kuijpers M, Wulf PS, van Spronsen M, MacKintosh FC, Hoogenraad CC. Mixed microtubules steer dynein-driven cargo transport into dendrites. Curr Biol. 2010;20:290–299. doi: 10.1016/j.cub.2009.12.052. [DOI] [PubMed] [Google Scholar]
- Kjer-Nielsen L, Teasdale RD, van Vliet C, Gleeson PA. A novel Golgi-localisation domain shared by a class of coiled-coil peripheral membrane proteins. Curr Biol. 1999;9:385–388. doi: 10.1016/s0960-9822(99)80168-7. [DOI] [PubMed] [Google Scholar]
- Levine TP, Munro S. The pleckstrin homology domain of oxysterol-binding protein recognises a determinant specific to Golgi membranes. Curr Biol. 1998;8:729–739. doi: 10.1016/s0960-9822(98)70296-9. [DOI] [PubMed] [Google Scholar]
- Lin S, Liu M, Mozgova OI, Yu W, Baas PW. Mitotic motors coregulate microtubule patterns in axons and dendrites. J Neurosci. 2012;32:14033–14049. doi: 10.1523/JNEUROSCI.3070-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Fox P, Lakonishok M, Davidson MW, Gelfand VI. Initial neurite outgrowth in Drosophila neurons is driven by kinesin-powered microtubule sliding. Curr Biol. 2013;23:1018–1023. doi: 10.1016/j.cub.2013.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney MB, et al. Presenilin-based genetic screens in Drosophila melanogaster identify novel notch pathway modifiers. Genetics. 2006;172:2309–2324. doi: 10.1534/genetics.104.035170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattie FJ, Stackpole MM, Stone MC, Clippard JR, Rudnick DA, Qiu Y, Tao J, Allender DL, Parmar M, Rolls MM. Directed microtubule growth, +TIPs, kinesin-2 are required for uniform microtubule polarity in dendrites. Curr Biol. 2010;20:2169–2177. doi: 10.1016/j.cub.2010.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S, Nichols BJ. The GRIP domain—a novel Golgi-targeting domain found in several coiled-coil proteins. Curr Biol. 1999;9:377–380. doi: 10.1016/s0960-9822(99)80166-3. [DOI] [PubMed] [Google Scholar]
- Nguyen MM, Stone MC, Rolls MM. Microtubules are organized independently of the centrosome in Drosophila neurons. Neural Dev. 2011;6:38. doi: 10.1186/1749-8104-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori-McKenney KM, Jan LY, Jan YN. Golgi outposts shape dendrite morphology by functioning as sites of acentrosomal microtubule nucleation in neurons. Neuron. 2012;76:921–930. doi: 10.1016/j.neuron.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piehl M, Tulu US, Wadsworth P, Cassimeris L. Centrosome maturation: measurement of microtubule nucleation throughout the cell cycle by using GFP-tagged EB1. Proc Natl Acad Sci USA. 2004;101:1584–1588. doi: 10.1073/pnas.0308205100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud-Messina B, Debec A, Tollon Y, Gares M, Wright M. Differential properties of the two Drosophila gamma-tubulin isotypes. Eur J Cell Biol. 2001;80:643–649. doi: 10.1078/0171-9335-00195. [DOI] [PubMed] [Google Scholar]
- Raynaud-Messina B, Merdes A. Gamma-tubulin complexes and microtubule organization. Curr Opin Cell Biol. 2007;19:24–30. doi: 10.1016/j.ceb.2006.12.008. [DOI] [PubMed] [Google Scholar]
- Rivero S, Cardenas J, Bornens M, Rios RM. Microtubule nucleation at the cis-side of the Golgi apparatus requires AKAP450 and GM130. EMBO J. 2009;28:1016–1028. doi: 10.1038/emboj.2009.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls MM. Neuronal polarity in Drosophila: sorting out axons and dendrites. Dev Neurobiol. 2011;71:419–429. doi: 10.1002/dneu.20836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls MM, Satoh D, Clyne PJ, Henner AL, Uemura T, Doe CQ. Polarity and compartmentalization of Drosophila neurons. Neural Dev. 2007;2:7. doi: 10.1186/1749-8104-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DJ, Yu W, Baas PW. Transport of dendritic microtubules establishes their nonuniform polarity orientation. J Cell Biol. 1995;130:93–103. doi: 10.1083/jcb.130.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DJ, Yu W, Ferhat L, Kuriyama R, Rueger DC, Baas PW. Identification of a microtubule-associated motor protein essential for dendritic differentiation. J Cell Biol. 1997;138:833–843. doi: 10.1083/jcb.138.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova T, Slemmer J, Hoogenraad CC, Lansbergen G, Dortland B, De Zeeuw CI, Grosveld F, van Cappellen G, Akhmanova A, Galjart N. Visualization of microtubule growth in cultured neurons via the use of EB3-GFP (end-binding protein 3-green fluorescent protein) J Neurosci. 2003;23:2655–2664. doi: 10.1523/JNEUROSCI.23-07-02655.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiess M, Maghelli N, Kapitein LC, Gomis-Ruth S, Wilsch-Brauninger M, Hoogenraad CC, Tolic-Norrelykke IM, Bradke F. Axon extension occurs independently of centrosomal microtubule nucleation. Science. 2010;327:704–707. doi: 10.1126/science.1182179. [DOI] [PubMed] [Google Scholar]
- Stone MC, Nguyen MM, Tao J, Allender DL, Rolls MM. Global up-regulation of microtubule dynamics and polarity reversal during regeneration of an axon from a dendrite. Mol Biol Cell. 2010;21:767–777. doi: 10.1091/mbc.E09-11-0967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone MC, Roegiers F, Rolls MM. Microtubules have opposite orientation in axons and dendrites of Drosophila neurons. Mol Biol Cell. 2008;19:4122–4129. doi: 10.1091/mbc.E07-10-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez M, Cooper MT, Zurita M, Kennison JA. gammaTub23C interacts genetically with brahma chromatin-remodeling complexes in Drosophila melanogaster. Genetics. 2008;180:835–843. doi: 10.1534/genetics.108.093492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey KJ, Lizotte DL, Abramson T, Barenboim L, Schnapp BJ, Rapoport TA. Light chain-dependent regulation of kinesin's interaction with microtubules. J Cell Biol. 1998;143:1053–1066. doi: 10.1083/jcb.143.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C. Distinct Dgrip84 isoforms correlate with distinct gamma-tubulins in Drosophila. Mol Biol Cell. 2008;19:368–377. doi: 10.1091/mbc.E07-08-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye B, Zhang Y, Song W, Younger SH, Jan LY, Jan YN. Growing dendrites and axons differ in their reliance on the secretory pathway. Cell. 2007;130:717–729. doi: 10.1016/j.cell.2007.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Centonze VE, Ahmad FJ, Baas PW. Microtubule nucleation and release from the neuronal centrosome. J Cell Biol. 1993;122:349–359. doi: 10.1083/jcb.122.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Cook C, Sauter C, Kuriyama R, Kaplan PL, Baas PW. Depletion of a microtubule-associated motor protein induces the loss of dendritic identity. J Neurosci. 2000;20:5782–5791. doi: 10.1523/JNEUROSCI.20-15-05782.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Wildonger J, Ye B, Zhang Y, Kita A, Younger SH, Zimmerman S, Jan LY, Jan YN. Dynein is required for polarized dendritic transport and uniform microtubule orientation in axons. Nat Cell Biol. 2008;10:1172–1180. doi: 10.1038/ncb1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Rolls MM, Hall DH, Malone CJ, Hanna-Rose W. A ZYG-12-dynein interaction at the nuclear envelope defines cytoskeletal architecture in the C. elegans gonad. J Cell Biol. 2009;186:229–241. doi: 10.1083/jcb.200902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.