The SNAREs SNAP23, Syntaxin4, and VAMP7 associate to target the delivery of MT1-MMP to sites of invadopodium formation in breast tumor cells. The interaction of these SNAREs correlates with decreased phosphorylation of Syntaxin4. The targeted delivery of MT1-MMP is required for efficient ECM degradation and cell invasion.
Abstract
Movement through the extracellular matrix (ECM) requires cells to degrade ECM components, primarily through the action of matrix metalloproteinases (MMPs). Membrane type 1–matrix metalloproteinase (MT1-MMP) has an essential role in matrix degradation and cell invasion and localizes to subcellular degradative structures termed invadopodia. Trafficking of MT1-MMP to invadopodia is required for the function of these structures, and here we examine the role of N-ethylmaleimide–sensitive factor–activating protein receptor (SNARE)–mediated membrane traffic in the transport of MT1-MMP to invadopodia. During invadopodium formation in MDA-MB-231 human breast cancer cells, increased association of SNAP23, Syntaxin4, and vesicle-associated membrane protein 7 (VAMP7) is detected by coimmunoprecipitation. Blocking the function of these SNAREs perturbs invadopodium-based ECM degradation and cell invasion. Increased level of SNAP23-Syntaxin4-VAMP7 interaction correlates with decreased Syntaxin4 phosphorylation. These results reveal an important role for SNARE-regulated trafficking of MT1-MMP to invadopodia during cellular invasion of ECM.
INTRODUCTION
In tumor cells, the acquisition of an invasive phenotype, which includes the ability to navigate through an extracellular matrix (ECM), is a prerequisite for development of metastatic disease. The ECM compartmentalizes tissues, providing a scaffold for cell adhesion, and partial degradation of this scaffold and subsequent invasion into the ECM is a hallmark of tumor progression (Hanahan and Weinberg, 2011). Cellular invasion into ECM is facilitated by a family of zinc-dependent proteolytic enzymes known as matrix metalloproteinases (MMPs). Expression of MMPs, particularly membrane type 1–MMP (MT1-MMP), has been linked to aggressive phenotypes in several cancers, including ovarian, breast, and melanoma (Sodek et al., 2007; Kondratiev et al., 2008; Perentes et al., 2011). MT1-MMP degrades components of the ECM (collagens, fibronectin, laminins; Itoh and Seiki, 2006) in a process that is regulated through posttranslational modification and intracellular trafficking. Newly synthesized MT1-MMP is activated through cleavage of its prodomain by the Golgi-associated enzyme furin (Sato et al., 2005). Subsequently, MT1-MMP can be localized to the lamellipodia of migrating cells via interaction with CD44 (Mori et al., 2002) and at invadopodia via a vesicle-associated membrane protein 7 (VAMP7)–dependent pathway (Steffen et al., 2008). Invadopodia are actin-driven membrane protrusions involved in ECM degradation and tumor cell invasion (Blouw et al., 2008; Clark et al., 2009), and the goal of the present study is to elucidate the mechanisms that control MT1-MMP localization at invadopodia.
Trafficking of cellular material between intracellular compartments and the plasma membrane is dependent upon soluble N-ethylmaleimide–sensitive factor–activating protein receptors (SNAREs), a family of membrane proteins that form complexes bridging adjacent membranes during membrane fusion. SNAREs show relatively specific pairing during membrane trafficking, which allows distinct SNARE complexes to drive specific membrane fusion events (Pfeffer, 2007). Several studies support a role for SNAREs in MT1-MMP trafficking, localization, and cell invasion (Miyata et al., 2004; Steffen et al., 2008; Kean et al., 2009; Williams and Coppolino, 2011), which complements described roles for SNARES in cell migration (Proux-Gillardeaux et al., 2005; Tayeb et al., 2005). Very little is known, however, about the interactions between SNAREs that facilitate MT1-MMP-mediated invadopodium formation or how these interactions might be regulated.
To examine the roles of SNAREs in the trafficking of MT1-MMP to invadopodia, we isolated invadopodium-enriched subcellular fractions and identified SNAREs contained therein. We report that SNAP23, Syntaxin4, and VAMP7 localize at invadopodial fractions, and inhibition of these SNAREs dramatically reduces the amount of MT1-MMP at invadopodia. We also observed increased interaction of SNAP23, Syntaxin4, and VAMP7 under conditions in which cells are forming invadopodia. Of interest, the interaction between Syntaxin4 and SNAP23 correlated with decreased phosphorylation of Syntaxin4. The findings suggest that a regulated membrane trafficking pathway mediated by SNAP23, Syntaxin4, and VAMP7 is required for invadopodium formation and efficient tumor cell invasion in vitro.
RESULTS
SNAP23 and Syntaxin4 target VAMP7 and MT1-MMP to invadopodia
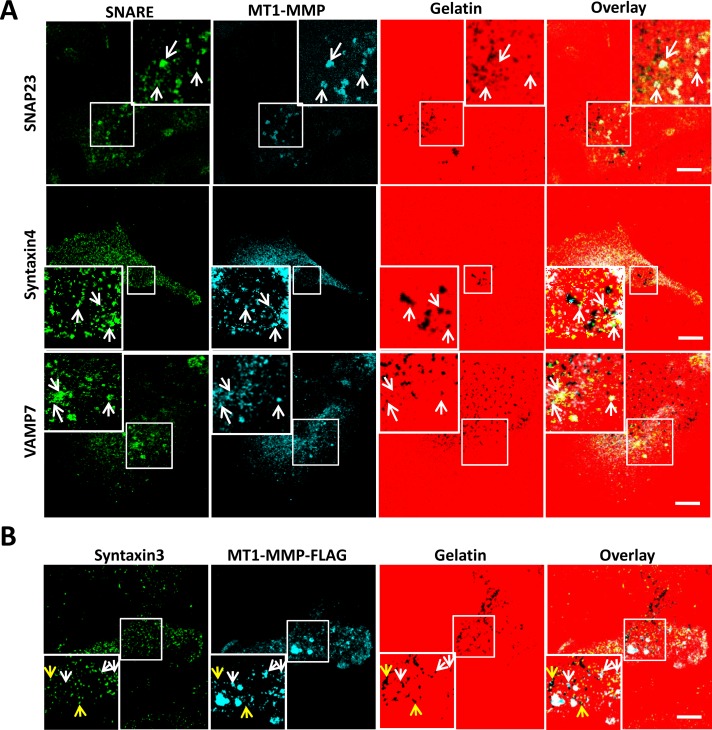
VAMP7 has been characterized as a vesicle SNARE involved in the delivery of MT1-MMP to invadopodia (Steffen et al., 2008), but the cognate SNAREs responsible for the docking of MT1-MMP–containing vesicles have yet to be described. To identify SNAREs complexed with VAMP7 during delivery of MT1-MMP to invadopodia, we fractionated MDA-MB-231 cells, derived from an invasive human breast tumor and characterized as forming stable invadopodia in vitro (Artym et al., 2006; Steffen et al., 2008), and collected invadopodial structures to determine SNARE protein content therein. Subcellular fractionation, as described previously (Mueller et al., 1992, 1999; Bowden et al., 1999, 2006), allowed isolation of invadopodium-enriched fractions (Supplemental Figures S1 and S2) containing cortactin, β1 integrin, MT1-MMP (active form only), VAMP3, VAMP7, Syntaxin3, Syntaxin4, SNAP23, and SNAP29 (Supplemental Figure S3). Roles for both Syntaxin4 and SNAP23 in the trafficking of MT1-MMP have been described (Miyata et al. 2004; Kean et al., 2009), and we found that these SNAREs were relatively abundant in invadopodial fractions compared with VAMP3, Syntaxin3, or SNAP29. Microscopic analysis revealed that SNAP23, Syntaxin4, and VAMP7 localize with MT1-MMP (Figure 1A) and F-actin structures (unpublished data) at sites of gelatin degradation. Quantification of colocalization between MT1-MMP and SNAP23, Syntaxin4, or VAMP7 yielded Pearson's r = 0.91 ± 0.08, 0.89 ± 0.05, and 0.94 ± 0.10, respectively. Analysis of Syntaxin3 localization (Figure 1B) revealed that Syntaxin3 does localize to invadopodia (yellow arrows) but does not colocalize with MT1-MMP (white arrows). Expression of SNAP29 was detected microscopically, but appreciable localization of this SNARE at invadopodia could not be demonstrated (unpublished data). We thus hypothesized that VAMP7, Syntaxin4, and SNAP23 might function to deliver MT1-MMP to invadopodia.
FIGURE 1:
Localization of SNAP23, Syntaxin4, VAMP7, and MT1-MMP at invadopodia. (A) Cells were plated on 594-labeled gelatin, fixed, permeabilized, and stained for SNAP23, Syntaxin4, or VAMP7 (green) along with MT1-MMP (blue). (B) Cells transfected with MT1-MMP-FLAG were plated on 594-labeled gelatin, fixed, permeabilized, and stained for Syntaxin3 (green) and FLAG epitope (for MT1-MMP, blue). Invadopodia are marked by the colocalization of MT1-MMP and dark spots of degraded gelatin. Scale bar, 10 μm.
To test the requirement for SNAP23, Syntaxin4, and SNAP23 function in the trafficking of MT1-MMP to invadopodia, we inhibited these SNAREs by expressing mutant SNARE constructs. These constructs exert dominant-negative effects and have been used extensively in several experimental systems to impair membrane trafficking (Hirling et al., 2000; Huang et al., 2001; Collins et al., 2002; Kean et al., 2009; Williams and Coppolino, 2011). SNAREs lacking their transmembrane domain—for example, Syntaxin4cyto (Syn4C) and VAMP7cyto (V7C)— form SNARE complexes with cognate SNAREs, blocking interactions with endogenous membrane-associated SNARE partners. SNAP23CΔ9 (S23C) lacks nine C-terminal amino acids and inhibits SNAP23-mediated traffic by forming nonfunctional complexes with its cellular binding partners.
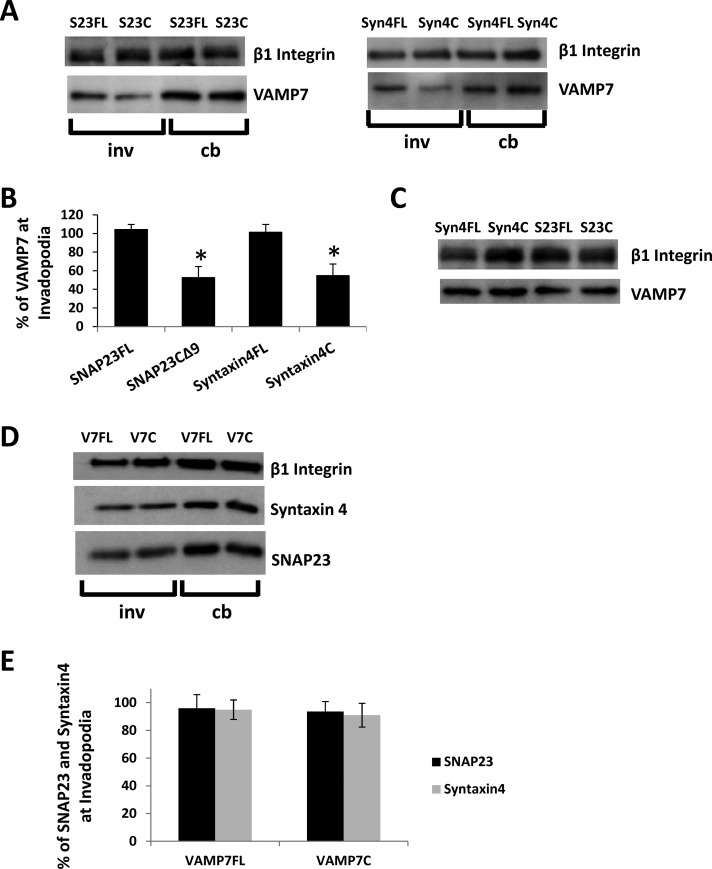
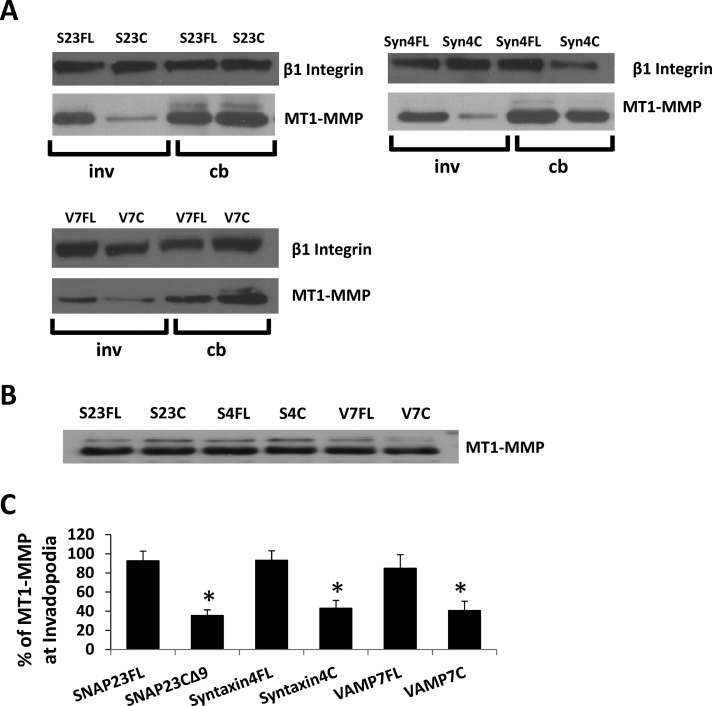
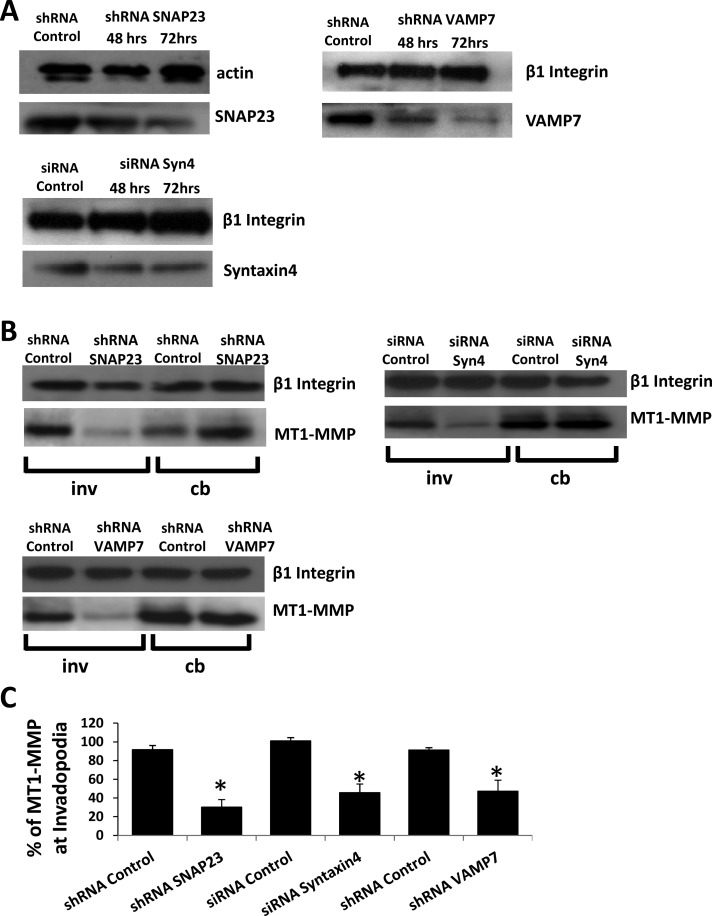
Inhibition of SNAP23 or Syntaxin4 decreased the amount of VAMP7 (47.4 ± 11.9 and 46.2 ± 12.6%, respectively) in invadopodial fractions (Figure 2, A and B) without altering total cellular levels of VAMP7 (Figure 2C). Inhibition of VAMP7 did not significantly alter the amount of either SNAP23 or Syntaxin4 in invadopodial fractions (Figure 2D; quantification in Figure 2E). The amount of MT1-MMP in the invadopodial fractions was also reduced as a result of inhibiting SNAP23 (35 ± 5.6%), Syntaxin4 (42 ± 8.1%), or VAMP7 (40.7 ± 9.7%; Figure 3A; quantification in Figure 3C). Inhibition of SNAP23, Syntaxin4, or VAMP7 had no effect on total cellular MT1-MMP (Figure 3B; actin loading control not shown). Similar reductions in the amount of MT1-MMP in invadopodial fractions were also observed as a result of knocking down expression of SNAP23, Syntaxin4, or VAMP7 using RNA interference (Figure 4; quantification in Figure 4C). To confirm antibody specificity, we used immunofluorescence microscopy of cells transfected with small interfering RNA (siRNA) Syntaxin4 or short hairpin RNA (shRNA) SNAP23. Using the indicated primary antibodies to Syntaxin4 and SNAP23, we observed reduced signals in cells transfected with corresponding siRNA or shRNA constructs (Supplemental Figure S4A). We also assessed the localization of our green fluorescent protein (GFP)–tagged constructs and found that they overlapped with antibody staining of endogenous SNAP23, Syntaxin4, and VAMP7 (Supplemental Figure S4B).
FIGURE 2:
Inhibition of Syntaxin4 or SNAP23 alters localization of VAMP7 in invadopodia fractions. (A, D) Representative Western blots of cell body and invadopodial fractions. Cells were transfected with constructs of SNAP23-full length (S23FL), SNAP23-C∆9 (S23C), Syntaxin4-full length (S4FL), Syntaxin4-cytoplasmic domain (S4C), VAMP7-full length (V7FL), or VAMP7-cytoplasmic domain (V7C). Cells were fractionated 2.5 h after plating on gelatin and probed for VAMP7 (A) or SNAP23 and Syntaxin4 (D) in cell body (cb) or invadopodial (inv) fractions. We used β1 integrin as a loading control. Quantification of triplicate Western blots showing amount of VAMP7 (B) or SNAP23 and Syntaxin4 (E) in indicated invadopodial fractions. Asterisks denote values significantly different from control; p < 0.05. (C) Whole-cell lysates from transiently transfected cell samples were probed for VAMP7.
FIGURE 3:
Inhibition of Syntaxin4 or SNAP23 alters localization of MT1-MMP in invadopodial fractions. (A) Representative Western blots of cell body and invadopodial fractions. Cells were transfected with constructs of SNAP23-full length (S23FL), SNAP23-C∆9 (S23C), Syntaxin4-full length (S4FL), Syntaxin4-cytoplasmic domain (S4C), VAMP7-full length (V7FL), or VAMP7-cytoplasmic domain (V7C). Cells were fractionated 2.5 h after plating on gelatin and probed for MT1-MMP in cell body (cb) or invadopodial (inv) fractions. β1 integrin was used as a loading control. (C) Quantification of triplicate Western blots showing amount of MT1-MMP in indicated invadopodial fractions. Asterisks denote values significantly different from control; p < 0.05. (B) Whole-cell lysates from transiently transfected cell samples were probed for MT1-MMP.
FIGURE 4:
RNA interference (RNAi)–mediated knockdown of SNAP23, Syntaxin4, or VAMP7 reduces MT1-MMP localization in invadopodial fractions. (A) Western blot analysis of SNAP23, VAMP7, and Syntaxin4 expression after RNAi-mediated knockdown. Cells were transfected with shRNA control, shRNA SNAP23, or shRNA VAMP7 constructs or with either siRNA control or siRNA Syntaxin4. At 48 and 72 h posttransfection, cells were lysed and analyzed for total SNAP23, VAMP7, or Syntaxin4 by Western blot. Actin or integrin was used as a loading control. (B) Distribution of MT1-MMP in cell fractions after knockdown of SNARE expression. Cells were transfected (as in A) for 72 h, followed by cell fractionation 2.5 h after plating on gelatin. Cell body (cb) and invadopodial (inv) fractions were then prepared and probed for MT1-MMP protein by Western blot. β1 integrin was used as a loading control. (C) Quantification of triplicate Western blots showing amount of MT1-MMP in indicated invadopodial fractions. Asterisks denote values significantly different from control; p < 0.05.
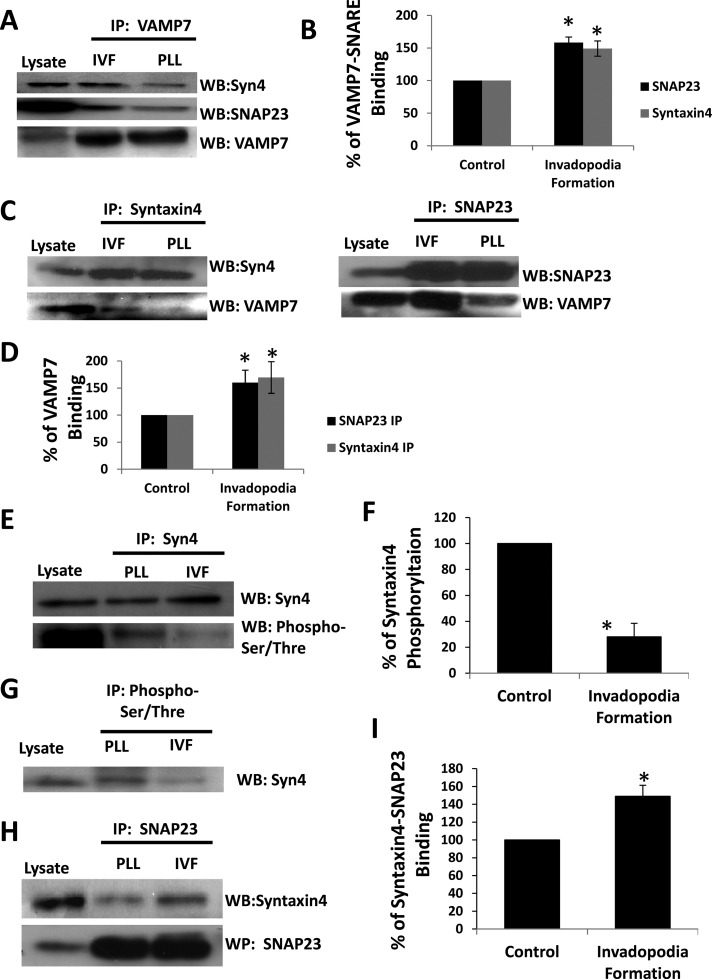
SNAP23-Syntaxin4-VAMP7 interaction is increased during invadopodium formation
SNARE complex formation has been shown to change in response to glucose (Kalwat et al., 2012), protein kinase C (Aquino et al., 2012) and protein kinase A (Foster et al., 1998) activity, mast cell degranulation (Sander et al., 2008), and adherence to ECM (Skalski et al., 2010). We therefore tested whether SNARE interactions are altered during invadopodium formation. Cells were plated on 0.2% gelatin to stimulate invadopodium formation or on poly-l-lysine (PLL) as control, and SNARE interactions were biochemically assessed. VAMP7 immunoprecipitates were found to contain increased amounts of Syntaxin4 (149 ± 11%) and SNAP23 (153 ± 9%) when cells were forming invadopodia compared with PLL controls (Figure 5A; quantification in Figure 5B). Similarly, syntaxin4 and SNAP23 immunoprecipitates were found to contain increased amounts of VAMP7 when cells were forming invadopodia (169 ± 29 and 160 ± 23%, respectively; Figure 5C; quantification in Figure 5D).
FIGURE 5:
Alterations to SNARE complex formation during invadopodium formation. Cells were plated on PLL (control) or gelatin (invadopodium formation [IVF]) for 2.5 h, collected, lysed, and analyzed by immunoprecipitation/SDS–PAGE/Western blot. (A) VAMP7 immunoprecipitates were probed for SNAP23, Syntaxin4, and VAMP7. (B) Quantification of the amount of Syntaxin4 or SNAP23 that was immunoprecipitated with VAMP7 in A. (C) Syntaxin4 and SNAP23 immunoprecipitates were probed for VAMP7 and Syntaxin4 or SNAP23. (D) Quantification of the amount of VAMP7 that was immunoprecipitated with SNAP23 or Syntaxin4 in C. (E) Syntaxin4 immunoprecipitates were probed for phospho-Ser/Thr, stripped, and reprobed for Syntaxin4. (F) Quantification of the proportion of immunoprecipitated Syntaxin4 that was phosphorylated in E. (G) Immunoprecipitations using phospho-Ser/Thr antibody were probed for Syntaxin4. (H) SNAP23 immunoprecipitates were probed for Syntaxin4. (I) Quantification of the amount of Syntaxin4 that was immunoprecipitated with SNAP23 in H. In B, D, F, and I, triplicate experiments were analyzed; asterisks denote values significantly different from control cells (p < 0.05).
Because binding of Syntaxin4 to cognate SNAREs is altered by its phosphorylation (Foster et al., 1998), we investigated changes to Syntaxin4 phosphorylation in our system. Syntaxin4 was immunoprecipitated from cells plated on PLL or 0.2% gelatin and examined for phosphorylation (Figure 5, E and F). A 71.9 ± 10.4% reduction in the amount of phosphorylated Sytnaxin4 was observed in cells during invadopodium formation compared with PLL control (Figure 5, E and F). Consistent with this, a decrease in the amount of Syntaxin4 pulled down in anti–phospho-Ser/Thr immunoprecipitations was observed (Figure 5G). Furthermore, we found a 49 ± 12% increase in the amount of Syntaxin4 in SNAP23 immunoprecipitates from cells forming invadopodia compared with cells on PLL (Figure 5, H and I). Together the data point to an association between Syntaxin4 and SNAP23, possibly in a VAMP7-containing complex, during invadopodium formation. This association correlates with decreased phosphorylation of Syntaxin4.
Expression and function of syntaxin4, SNAP23, and VAMP7 are required for invadopodium formation and cell invasion
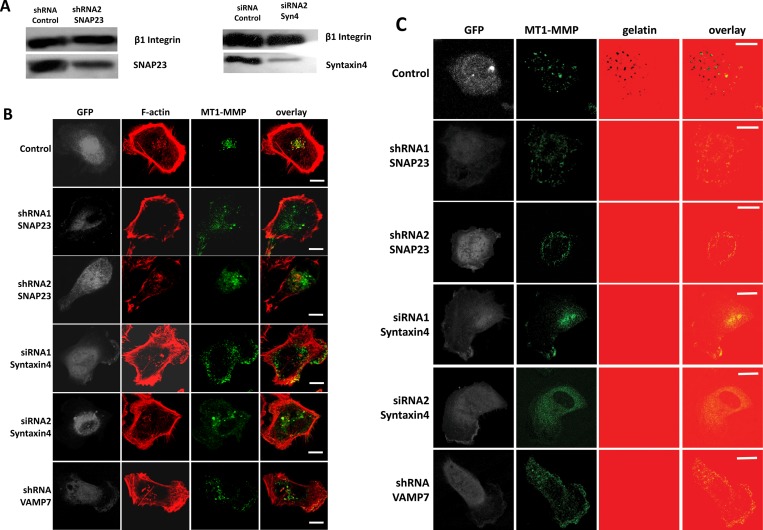
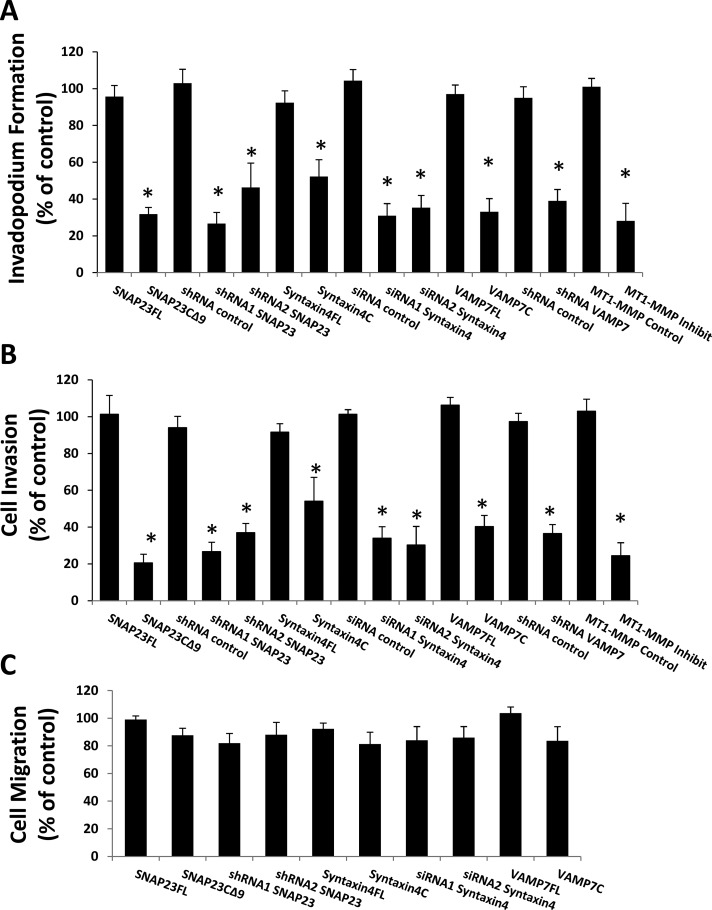
We next examined the effect of reducing expression of Syntaxin4, SNAP23, or VAMP7, using RNA interference, on invadopodium-based gelatin degradation and cell invasion. Knockdown of SNAP23, Syntaxin4, or VAMP7 altered MT1-MMP localization at F-actin punctae and perturbed invadopodium-based degradation of gelatin (Figure 6, B and C). MT1-MMP was found to localize at F-actin punctae in control cells (Figure 6, B and C, top), whereas knockdown of SNAP23, VAMP7, or Syntaxin4 expression reduced the amount of MT1-MMP in F-actin punctae (Figure 6B). Degradation of gelatin was also reduced under these conditions (Figure 6C). Similar results were obtained when knockdown of SNAP23 or Syntaxin4 was repeated with different siRNA and shRNA constructs (Figure 6, A–C). Inhibiting the function of Syntaxin4, SNAP23, or VAMP7, using the foregoing inhibitory constructs also significantly reduced the percentage of cells forming active invadopodia by 48.8 ± 9.2, 68.2 ± 3.8, and 66.9 ± 7.7%, respectively (Figure 7A). Quantitatively, knockdown of Syntaxin4, SNAP23, or VAMP7 reduced the percentage of cells forming active invadopodia by >50% (Figure 7A). Consistent with this, inhibiting the function of these SNAREs or knocking down their expression reduced cell invasion in Matrigel invasion assays (Figure 7B). The inhibition of gelatin degradation and cell invasion caused by inhibiting Syntaxin4, SNAP23, or VAMP7 was similar to that caused by inhibition of MT1-MMP with 900 nM SB-3CT pMS (Figure 7, A and B). The effects of SNAP23, Syntaxin4, and VAMP7 on cell migration were also assessed, and no significant effect was observed (Figure 7C). Our results are consistent with an established role for VAMP7 in delivering MT1-MMP to invadopodia (Steffen et al., 2008) and now define the cognate SNAREs involved in this process as Syntaxin4 and SNAP23. Furthermore, this is the first demonstration of SNARE interactions that are regulated during invadopodium-based degradation of ECM.
FIGURE 6:
RNAi-mediated knockdown of SNAP23, Syntaxin4, or VAMP7 impairs MT1-MMP localization at invadopodia. Cells were cotransfected with GFP along with shRNA control, shRNA SNAP23, shRNA VAMP7, siRNA control, or siRNA Syntaxin4. A second set of constructs targeting SNAP23 (shRNA2) and Syntaxin4 (siRNA2) was used, and knockdown of the SNAREs was confirmed by Western blot (A). Cells were plated on unlabeled (B) or 594-labeled (C) gelatin, fixed, permeabilized, and stained for MT1-MMP and/or F-actin. (B) Localization of MT1-MMP (green) and F-actin (red) in control cells or cells targeted for knockdown of SNAP23, VAMP7, or Syntaxin4. (C) Invadopodium-based degradation of gelatin in control cells or cells transfected with RNAi constructs targeting SNAP23, Syntaxin4, or VAMP7. Invadopodial degradation is represented by MT1-MMP (green) overlying dark areas on the gelatin (red). Scale bar, 10 μm.
FIGURE 7:
Inhibition of SNARE function or knockdown of SNARE expression perturbs invadopodium formation and cell invasion. Cells were cotransfected with GFP along with shRNA control, shRNA1 or shRNA2 to SNAP23, shRNA to VAMP7, siRNA control, or siRNA1 or siRNA2 to Syntaxin4. Alternatively, cells were transfected with GFP-tagged constructs of SNAPFL, SNAP23C∆9, Syntaxin4FL, or Syntaxin4cyto or subjected to inhibition of MT1-MMP by treatment with SB-3CT pMS (900 nM) at the time of plating. (A) Invadopodium formation. Cells were plated on Alexa 594–labeled gelatin for 3 h and microscopically analyzed. Cells with F-actin punctae overlying dark areas of matrix degradation were counted as cells forming invadopodia. Percentages of cells forming invadopodia, normalized to control (GFP-transfected cells), are presented. Means ± SEM from three independent experiments in which 100 cells/sample were counted. (B) Cell invasion. After transfections/treatments, cells were serum starved for 2 h, lifted, counted, and subjected to Transwell invasion assays using Matrigel. Cells invaded toward 10% FBS for 20 h and were then fixed and counted. (C) Cell migration. After transfections, cells were lifted, counted, and subjected to Transwell migration assays. Means ± SEM from three independent experiments in which 100 cells/sample were counted. Asterisks denote values significantly different from control cells (p < 0.05).
DISCUSSION
In the present study, we report that Syntaxin4 and SNAP23 associate with VAMP7, possibly in a complex, and function to deliver MT1-MMP to invadopodia during degradation of ECM by MDA-MB-231 breast tumor cells. It was previously demonstrated that VAMP7 targets MT1-MMP to invadopodia (Steffen et al., 2008), and our results are consistent with this, as we observed significant reduction in the amount of MT1-MMP at invadopodia upon inhibition of VAMP7 function. We also demonstrate that SNAP23 and Syntaxin4 colocalize with MT1-MMP at invadopodia, and inhibition of either of these SNAREs reduced the amount of both VAMP7 and MT1-MMP at invadopodia. Trafficking of MT1-MMP is important for ECM degradation during tumor progression (Hotary et al., 2003; Sabeh et al., 2004), and our findings highlight the important contribution that multiple SNARE proteins make to this process.
Movement of tumor cells through an ECM is a complex process, and the findings here do not preclude the involvement of other MMPs. MDA-MB-231 cells express several MMPs, including MMP-9 (Sneh et al., 2013), and future work will help to characterize the possible roles of different MMPs in ECM degradation by these cells. Current models suggest that MT1-MMP is a central factor in driving cell invasion in some tumor types (Nakahara et al., 1997; Sabeh et al., 2004, 2009; Frittoli et al., 2011); however, it is possible that other MMPs contribute to ECM remodeling at different stages of tumor progression. The system used here will also be useful for examining the link between estrogen receptor expression and MMP function during tumor invasion. MDA-MB-231 cells are estrogen receptor negative and form prominent invadopodia in an MT1-MMP–dependent manner. It will be informative to determine whether forced expression of estrogen receptor in these cells, as described previously (Li et al., 2013), can reduce their MT1-MMP–mediated invasiveness, producing a phenotype similar to the estrogen receptor–positive, MT1-MMP–negative MCF-7 cell line (Lafleur et al., 2006; Chernov et al., 2009).
The results here suggest that the interaction of Syntaxin4, SNAP23, and VAMP7 contributes to the development of invadopodia into ECM-degrading structures and that changes to SNARE-mediated exocytosis might contribute to an invasive phenotype. The association of Syntaxin4, SNAP23, and VAMP7 was increased when MDA-MB-231 cells were plated on gelatin, and this represents a different SNARE complex from the VAMP3-Syntaxin13-SNAP23 complex induced in Chinese hamster ovary cells plated on fibronectin (Skalski et al., 2010). In addition, the functions of Syntaxin4, SNAP23, and VAMP7 were required for cell invasion, but not two-dimensional cell migration, in MDA-MB-21 cells. We did detect Syntaxin3 in invadopodia, but it was not associated with MT1-MMP, and a possible role for this SNARE in cell invasion will be a subject for future investigation. Together the findings point to potential differences in regulated SNARE interactions that are dependent on cell type, ECM substrate, and mode of cell motility.
Regulation of exocytosis through alteration of SNARE interactions has been described in other systems (Foster et al., 1998; Polgar et al., 2003; Sander et al., 2008), and it is reasonable to propose that SNARE complex formation is modulated during the invasion process. This notion is supported by our finding that an increase in Syntaxin4–SNAP23 interaction correlates with a decrease in Syntaxin4 phosphorylation. We thus speculate that dephosphorylation of Syntaxin4 might facilitate its association with SNAP23 and contribute to the regulation of MT1-MMP trafficking during invadopodium formation. Previous studies showed that phosphorylation of Syntaxin4 can regulate exocytic events (Fu et al., 2005) and decrease the interaction of Syntaxin4 with SNAP23 in platelets (Chung et al., 2000). The findings here now suggest a role for phosphorylation of Syntaxin4 in regulating similar interactions during MT1-MMP–mediated degradation of ECM by tumor cells. Identification of the specific sites of phosphorylation on Syntaxin4 in these conditions will provide important clues for further investigation.
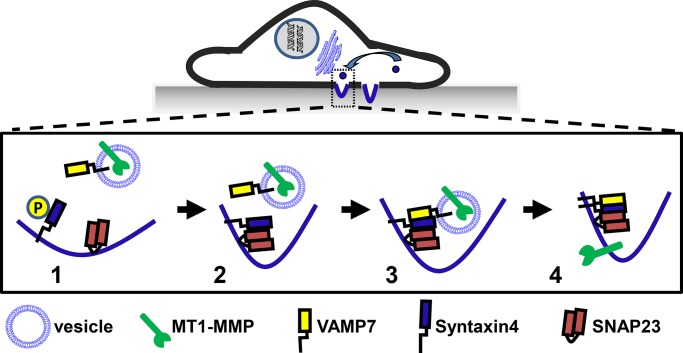
Previously we (Williams and Coppolino, 2011) and others (Steffen et al., 2008) showed that MT1-MMP localizes to a late endosomal compartment, marked by VAMP7, which contributes to the recycling of MT1-MMP to membrane protrusions rather than its down-regulation. This pathway involves a novel role for Rab7 in the recycling of MT1-MMP (Williams and Coppolino, 2011). Here we propose a possible mechanism for the delivery of MT1-MMP to invadopodia that is mediated through interaction of SNAP23-Syntaxin4-VAMP7 (Figure 8), and future studies are planned to address the role of Rab7 in this MT1-MMP trafficking pathway.
FIGURE 8:
A model for MT1-MMP trafficking to invadopodia. MT1-MMP localizes to the late endosomal compartment, characterized by VAMP7, and is targeted to invadopodia, where it functions to degrade the extracellular matrix. 1) VAMP7 is located on MT1-MMP–containing vesicles, and Syntaxin4 and SNAP23 are on the target membrane (site of invadopodium formation). 2) Syntaxin4 associates with SNAP23; this correlates with dephosphorylation of Syntaxin4. 3) The SNAP23-Syntaxin4 complex can bind VAMP7, facilitating membrane fusion and 4) delivery of MT1-MMP to forming invadopodia.
It was also reported that MT1-MMP localizes to lipid raft domains at invadopodia (Grass et al., 2012). In addition, it has been demonstrated that during mast cell exocytosis, Syntaxin4 is able to associate with lipid rafts by binding to SNAP23 (Puri and Roche, 2006). It is possible that MT1-MMP is targeted to lipid raft domains at invadopodia through the formation of a SNAP23-Syntaxin4–containing complex. Dephosphorylation of Syntaxin4, leading to enhanced SNAP23–Syntaxin4 interaction, may therefore represent an important regulatory step in the formation of invadopodia. It has been suggested that phosphatases have important roles in invadopodium formation (Murphy and Courtneidge, 2011), but few have been well characterized in this context, and this is an important area for future investigation. Collectively the work presented here identifies important SNARE interactions that are regulated during cell invasion and elucidates roles for the involved SNAREs in trafficking of MT1-MMP during degradation of ECM.
MATERIALS AND METHODS
Reagents and cDNA constructs
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) or Fisher Scientific (Nepean, ON, Canada) unless otherwise indicated. The MMP inhibitor SB-3CT pMS was purchased from EMD Chemicals (Gibbstown, NJ). Antibodies to the following proteins were obtained from the indicated suppliers: MT1-MMP, SNAP23, VAMP7, SNAP29 and actin (ab3644, ab3340, ab36195, ab138500, ab3280; Abcam, Toronto, Canada); phospho-Ser/Thr, Syntaxin4, and Giantin (610281, 610439, 610960; BD Biosciences, Mississauga, Canada); β1 (P4C10; Developmental Studies Hybridoma Bank, Iowa City, IA); GS15 (PRB114C; Cederlane, Burlington, Canada); VAMP3 (PA1-767A; Fisher Scientific); and Syntaxin2 and Syntaxin3 (ANR-008, ANR-005; Alomone Labs, Jerusalem, Israel). All secondary antibodies and Alexa Fluor 647–labeled phalloidin were purchased from Invitrogen (Burlington, Canada). Generation of MT1-MMP-GFP, GFP-SNAP23FL, GFP-SNAP23C∆9, GFP-Syntaxin4FL, GFP-Syntaxin4cyto, GFP-VAMP7FL, and GFP-VAMP7cyto is described elsewhere (Kean et al., 2009; Skalski et al., 2010; Williams and Coppolino, 2011). MT1-MMP-FLAG construct was generated through PCR and cloned into pcDNA3.1(-). Primers: forward, 5′-TATAAAGAATTCGGTGGTCTCGGACCA TGTC-3′; reverse, 5′-ATAAAAAAGCTTTTATTATTTATCATCATCATCTTTATAATCCCGC GGAACCTTGTCCAGCAGGGAAC-3′. shRNAs against SNAP23 (144931 and 144789) and VAMP7 and siRNAs against Syntaxin4 (ON-TARGETplus SMARTpool L-016256-00 and ON-TARGETplus siRNA J-016256-06) were purchased from Fisher Scientific.
Cell culture and transfection
MDA-MB-231 cells were cultured in DMEM (Sigma-Aldrich) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich) under 5% CO2 at 37°C. Cells were transfected with PolyPlus transfection reagent (VWR Internationa, Mississauga, Canada) as described by the manufacturer's protocol. All transfected constructs were expressed for 18–24 h.
Immunofluorescence microscopy
Cells were grown on glass coverslips coated as described under Invadopodia formation assay and subsequently fixed (4% [wt/vol] paraformaldehyde/phosphate-buffered saline [PBS]) and permeabilized with 0.2% Triton X-100/PBS. Samples were then blocked with 5% bovine serum albumin (BSA; wt/vol) powder/PBS before staining with primary and secondary antibody, followed by washing and mounting. Samples were imaged using a 63×/numerical aperture 1.4 lens on a Leica DM-IRE2 inverted microscope with a Leica TCS SP2 system (Leica, Heidelberg, Germany). Images were captured using Leica Confocal Software.
ImageJ analysis
Pearson's correlation analysis was performed using the Colocalization Analysis plug-in. All images were processed to remove noise and background, and region of interest was manually selected for each image. Pearson's correlation for pixels where intensity is greater than the threshold for both channels is represented, and values >0.5 represent a biological correlation and an observed colocalization.
Invadopodia formation assay
Invadopodia formation was performed as previously described (Artym et al., 2009). Briefly, coverslips were coated with 50 μg/ml PLL (Sigma-Aldrich), followed by 0.5% glutaraldehyde (Sigma-Aldrich), and inverted on an 80-μl drop of Alexa 594–labeled gelatin, incubated with 5 mg/ml Na borohydride (Sigma-Aldrich), and washed extensively with PBS. For immunoprecipitations, cell culture plates were coated in a similar manner, with the exception that plates were overlaid with unlabeled 0.2% gelatin. Control plates were coated with 50 μg/ml PLL.
Cell invasion assay
Cell culture inserts, in 24-well dishes (Costar), were prepared with and without (control) Matrigel. The bottom chamber was coated with 20 μg/ml fibronectin and the upper chamber with 0.125 mg/ml Growth Factor Reduced Matrigel (BD Biosciences). MDA-MB-231 cells were transfected for 8 h, at which point they were lifted and seeded onto Matrigel-coated and control (without Matrigel) upper surface in serum-free media (80,000 cells/well). The cells that invaded toward chemoattractant (10% FBS/0.1%BSA) in the lower chamber and penetrated the Matrigel were fixed with 4% paraformaldehyde, stained with 4′,6-diamidino-2-phenylindole (DAPI), and counted. Cells that did not invade were removed with a cotton swab before fixation of sample. Ten fields of cells per membrane were counted. The data are presented as the number of cells that invaded through the Matrigel divided by the number of cells that migrated through the control insert (setting mock-treated, GFP-transfected cells at 100%).
Cell migration assays
Boyden Transwell migration chambers (Costar) were coated with 20 μg/ml fibronectin on the bottom chamber. Transfected cells were counted, and 20,000 cells/well in serum-free media were added to the top well. The lower wells were filled with DMEM/10% FBS, and cells were allowed to migrate for 6 h. The top and bottom of the membrane was fixed in 4% paraformaldehyde/PBS, stained with DAPI, and mounted on coverslips. Ten fields of cells on the membrane were counted per experiment using fluorescence microscopy. The data are presented as number of transfected cells that migrated to the bottom membrane divided by number of cells that remained on the top membrane.
Coimmunoprecipitation
Cyanogen bromide–activated Sepharose beads (Sigma-Aldrich) were coated with antibody as per manufacturer's instructions. Cells were lysed in 1% NP-40, 0.5% NaDOC, 2 mM EDTA, 10% glycerol, 137 mM NaCl, 20 mM Tris-HCl, pH 8.0, 10 mM NaF, 10 mM Na4P2O7, 0.2 mM Na3VO4, and protease inhibitor cocktail (Sigma-Aldrich). Lysate was incubated with antibody bound beads overnight at 4°C and washed three times with lysis buffer followed by four washes with 0.1% Tween/PBS. Bound proteins were eluted using 2.5× SDS running buffer heated to 100°C. Alternatively, cell lysates were incubated with antibody for 4 h, followed by protein G magnetic bead (New England BioLabs) addition for 2 h, extensively washed, and eluted using 2.5× SDS running buffer heated to 50°C. Proteins were separated using SDS–PAGE and analyzed by Western blot.
Subcellular fractionation and invadopodia isolation
Purification of an enriched invadopodia cell fraction was performed based on a previous protocol with modifications (Mueller et al., 1992). Briefly, cells were cultured for 2.5 h on coverslips or plates prepared as described under Invadopodia formation assay. Cell membranes and cytoskeleton were stabilized with a buffer containing 10 mM 3-(N-morpholino)propanesulfonic acid, pH 6.8, 100 mM KCl, 2.5 mM MgCl2, 0.3 M sucrose, 1 mM Na3VO4, 10 mM NaF, and 10 mM Na4P2O7 (shearing buffer). Cells were placed on ice and then sheared in the same buffer with the addition of protease inhibitor cocktail (Sigma-Aldrich). Cell bodies were sheared by gently sweeping an L-shaped glass pipette rod across the entire surface. Conformation of cell body removal was determined by phase-contrast microscopy. Cell bodies were immediately pelleted, and the supernatant fraction was obtained with the cell body fraction and lysed in 1% NP-40, 0.5% NaDOC, 2 mM EDTA, 10% glycerol, 137 mM NaCl, 20 mM Tris-HCl, pH 8.0, 10 mM NaF, 10 mM Na4P2O7, 0.2 mM Na3VO4, and protease inhibitor cocktail. Sheared plates containing invadopodia embedded into the matrix were washed three times with shearing buffer followed by the addition of lysis buffer and vigorously scraped with a cell scraper. Whole-cell lysate was obtained from cells cultured on gelatin plates and lysed on the plate.
Supplementary Material
Acknowledgments
This work was supported by the Natural Sciences and Engineering Research Council of Canada. K.C.W. holds a Natural Sciences and Engineering Research Council of Canada scholarship.
Abbreviations used:
- ECM
extracellular matrix
- MT1-MMP
membrane type 1–matrix metalloproteinase
- VAMP
vesicle-associated membrane protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-10-0582) on May 7, 2014.
REFERENCES
- Aquino G, et al. pEGFR-Tyr 845 expression as prognostic factors in oral squamous cell carcinoma: a tissue-microarray study with clinic-pathological correlations. Cancer Biol Ther. 2012;13:967–977. doi: 10.4161/cbt.20991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artym VV, Yamada KM, Mueller SC. ECM degradation assays for analyzing local cell invasion. Methods Mol Biol. 2009;522:211–219. doi: 10.1007/978-1-59745-413-1_15. [DOI] [PubMed] [Google Scholar]
- Artym VV, Zhang Y, Seillier-Moiseiwitsch F, Yamada KM, Mueller SC. Dynamic interactions of cortactin and membrane type 1 matrix metalloproteinase at invadopodia: defining the stages of invadopodia formation and function. Cancer Res. 2006;66:3034–3043. doi: 10.1158/0008-5472.CAN-05-2177. [DOI] [PubMed] [Google Scholar]
- Blouw B, Seals DF, Pass I, Diaz B, Courtneidge SA. A role for the podosome/invadopodia scaffold protein Tks5 in tumor growth in vivo. Eur J Cell Biol. 2008;87:555–567. doi: 10.1016/j.ejcb.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden ET, Barth M, Thomas D, Glazer RI, Mueller SC. An invasion-related complex of cortactin, paxillin and PKCmu associates with invadopodia at sites of extracellular matrix degradation. Oncogene. 1999;18:4440–4449. doi: 10.1038/sj.onc.1202827. [DOI] [PubMed] [Google Scholar]
- Bowden ET, Onikoyi E, Slack R, Myoui A, Yoneda T, Yamada KM, Mueller SC. Co-localization of cortactin and phosphotyrosine identifies active invadopodia in human breast cancer cells. Exp Cell Res. 2006;312:1240–1253. doi: 10.1016/j.yexcr.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Chernov AV, Sounni NE, Remacle AG, Strongin AY. Epigenetic control of the invasion-promoting MT1-MMP/MMP-2/TIMP-2 axis in cancer cells. J Biol Chem. 2009;284:12727–12734. doi: 10.1074/jbc.M900273200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung SH, Polgar J, Reed GL. Protein kinase C phosphorylation of syntaxin 4 in thrombin-activated human platelets. J Biol Chem. 2000;275:25286–25291. doi: 10.1074/jbc.M004204200. [DOI] [PubMed] [Google Scholar]
- Clark ES, Brown B, Whigham AS, Kochaishvili A, Yarbrough WG, Weaver AM. Aggressiveness of HNSCC tumors depends on expression levels of cortactin, a gene in the 11q13 amplicon. Oncogene. 2009;28:431–444. doi: 10.1038/onc.2008.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins RF, Schreiber AD, Grinstein S, Trimble WS. Syntaxins 13 and 7 function at distinct steps during phagocytosis. J Immunol. 2002;169:3250–3256. doi: 10.4049/jimmunol.169.6.3250. [DOI] [PubMed] [Google Scholar]
- Foster LJ, Yeung B, Mohtashami M, Ross K, Trimble WS, Klip A. Binary interactions of the SNARE proteins syntaxin-4, SNAP23, and VAMP-2 and their regulation by phosphorylation. Biochemistry. 1998;37:11089–11096. doi: 10.1021/bi980253t. [DOI] [PubMed] [Google Scholar]
- Frittoli E, Palamidessi A, Disanza A, Scita G. Secretory and endo/exocytic trafficking in invadopodia formation: the MT1-MMP paradigm. Eur J Cell Biol. 2011;90:108–114. doi: 10.1016/j.ejcb.2010.04.007. [DOI] [PubMed] [Google Scholar]
- Fu J, Naren AP, Gao X, Ahmmed GU, Malik AB. Protease-activated receptor-1 activation of endothelial cells induces protein kinase Calpha-dependent phosphorylation of syntaxin 4 and Munc18c: role in signaling p-selectin expression. J Biol Chem. 2005;280:3178–3184. doi: 10.1074/jbc.M410044200. [DOI] [PubMed] [Google Scholar]
- Grass GD, Bratoeva M, Toole BP. Regulation of invadopodia formation and activity by CD147. J Cell Sci. 2012;125:777–788. doi: 10.1242/jcs.097956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hirling H, Steiner P, Chaperon C, Marsault R, Regazzi R, Catsicas S. Syntaxin 13 is a developmentally regulated SNARE involved in neurite outgrowth and endosomal trafficking. Eur J Neurosci. 2000;12:1913–1923. doi: 10.1046/j.1460-9568.2000.00076.x. [DOI] [PubMed] [Google Scholar]
- Hotary KB, Allen ED, Brooks PC, Datta NS, Long MW, Weiss SJ. Membrane type I matrix metalloproteinase usurps tumor growth control imposed by the three-dimensional extracellular matrix. Cell. 2003;114:33–45. doi: 10.1016/s0092-8674(03)00513-0. [DOI] [PubMed] [Google Scholar]
- Huang X, Sheu L, Tamori Y, Trimble WS, Gaisano HY. Cholecystokinin-regulated exocytosis in rat pancreatic acinar cells is inhibited by a C-terminus truncated mutant of SNAP-23. Pancreas. 2001;23:125–133. doi: 10.1097/00006676-200108000-00002. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Seiki M. MT1-MMP: A potent modifier of pericellular microenvironment. J Cell Physiol. 2006;206:1–8. doi: 10.1002/jcp.20431. [DOI] [PubMed] [Google Scholar]
- Kalwat MA, Wiseman DA, Luo W, Wang Z, Thurmond DC. Gelsolin associates with the N terminus of syntaxin 4 to regulate insulin granule exocytosis. Mol Endocrinol. 2012;26:128–141. doi: 10.1210/me.2011-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kean MJ, Williams KC, Skalski M, Myers D, Burtnik A, Foster D, Coppolino MG. VAMP3, syntaxin-13 and SNAP23 are involved in secretion of matrix metalloproteinases, degradation of the extracellular matrix and cell invasion. J Cell Sci. 2009;122:4089–4098. doi: 10.1242/jcs.052761. [DOI] [PubMed] [Google Scholar]
- Kondratiev S, Gnepp DR, Yakirevich E, Sabo E, Annino DJ, Rebeiz E, Laver NV. Expression and prognostic role of MMP2, MMP9, MMP13, and MMP14 matrix metalloproteinases in sinonasal and oral malignant melanomas. Hum Pathol. 2008;39:337–343. doi: 10.1016/j.humpath.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Lafleur MA, Mercuri FA, Ruangpanit N, Seiki M, Sato H, Thompson EW. Type I collagen abrogates the clathrin-mediated internalization of membrane type 1 matrix metalloproteinase (MT1-MMP) via the MT1-MMP hemopexin domain. J Biol Chem. 2006;281:6826–6840. doi: 10.1074/jbc.M513084200. [DOI] [PubMed] [Google Scholar]
- Li Q, Sun H, Zou J, Ge C, Yu K, Cao Y, Hong Q. Increased expression of estrogen receptor alpha-36 by breast cancer oncogene IKKepsilon promotes growth of ER-negative breast cancer cells. Cell Physiol Biochem. 2013;31:833–841. doi: 10.1159/000350101. [DOI] [PubMed] [Google Scholar]
- Miyata T, et al. Involvement of syntaxin 4 in the transport of membrane-type 1 matrix metalloproteinase to the plasma membrane in human gastric epithelial cells. Biochem Biophys Res Commun. 2004;323:118–124. doi: 10.1016/j.bbrc.2004.08.064. [DOI] [PubMed] [Google Scholar]
- Mori H, Tomari T, Koshikawa N, Kajita M, Itoh Y, Sato H, Tojo H, Yana I, Seiki M. CD44 directs membrane-type 1 matrix metalloproteinase to lamellipodia by associating with its hemopexin-like domain. EMBO J. 2002;21:3949–3959. doi: 10.1093/emboj/cdf411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SC, Ghersi G, Akiyama SK, Sang QX, Howard L, Pineiro-Sanchez M, Nakahara H, Yeh Y, Chen WT. A novel protease-docking function of integrin at invadopodia. J Biol Chem. 1999;274:24947–24952. doi: 10.1074/jbc.274.35.24947. [DOI] [PubMed] [Google Scholar]
- Mueller SC, Yeh Y, Chen WT. Tyrosine phosphorylation of membrane proteins mediates cellular invasion by transformed cells. J Cell Biol. 1992;119:1309–1325. doi: 10.1083/jcb.119.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DA, Courtneidge SA. The “ins” and “outs” of podosomes and invadopodia: characteristics, formation and function. Nat Rev Mol Cell Biol. 2011;12:413–426. doi: 10.1038/nrm3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara H, Howard L, Thompson EW, Sato H, Seiki M, Yeh Y, Chen WT. Transmembrane/cytoplasmic domain-mediated membrane type 1-matrix metalloprotease docking to invadopodia is required for cell invasion. Proc Natl Acad Sci USA. 1997;94:7959–7964. doi: 10.1073/pnas.94.15.7959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perentes JY, Kirkpatrick ND, Nagano S, Smith EY, Shaver CM, Sgroi D, Garkavtsev I, Munn LL, Jain RK, Boucher Y. Cancer cell-associated MT1-MMP promotes blood vessel invasion and distant metastasis in triple-negative mammary tumors. Cancer Res. 2011;71:4527–4538. doi: 10.1158/0008-5472.CAN-10-4376. [DOI] [PubMed] [Google Scholar]
- Pfeffer SR. Unsolved mysteries in membrane traffic. Annu Rev Biochem. 2007;76:629–645. doi: 10.1146/annurev.biochem.76.061705.130002. [DOI] [PubMed] [Google Scholar]
- Polgar J, Lane WS, Chung SH, Houng AK, Reed GL. Phosphorylation of SNAP-23 in activated human platelets. J Biol Chem. 2003;278:44369–44376. doi: 10.1074/jbc.M307864200. [DOI] [PubMed] [Google Scholar]
- Proux-Gillardeaux V, Gavard J, Irinopoulou T, Mege RM, Galli T. Tetanus neurotoxin-mediated cleavage of cellubrevin impairs epithelial cell migration and integrin-dependent cell adhesion. Proc Natl Acad Sci USA. 2005;102:6362–6367. doi: 10.1073/pnas.0409613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri N, Roche PA. Ternary SNARE complexes are enriched in lipid rafts during mast cell exocytosis. Traffic. 2006;7:1482–1494. doi: 10.1111/j.1600-0854.2006.00490.x. [DOI] [PubMed] [Google Scholar]
- Sabeh F, et al. Tumor cell traffic through the extracellular matrix is controlled by the membrane-anchored collagenase MT1-MMP. J Cell Biol. 2004;167:769–781. doi: 10.1083/jcb.200408028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeh F, Shimizu-Hirota R, Weiss SJ. Protease-dependent versus -independent cancer cell invasion programs: three-dimensional amoeboid movement revisited. J Cell Biol. 2009;185:11–19. doi: 10.1083/jcb.200807195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander LE, Frank SP, Bolat S, Blank U, Galli T, Bigalke H, Bischoff SC, Lorentz A. Vesicle associated membrane protein (VAMP)-7 and VAMP-8, but not VAMP-2 or VAMP-3, are required for activation-induced degranulation of mature human mast cells. Eur J Immunol. 2008;38:855–863. doi: 10.1002/eji.200737634. [DOI] [PubMed] [Google Scholar]
- Sato H, Takino T, Miyamori H. Roles of membrane-type matrix metalloproteinase-1 in tumor invasion and metastasis. Cancer Sci. 2005;96:212–217. doi: 10.1111/j.1349-7006.2005.00039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalski M, Yi Q, Kean MJ, Myers DW, Williams KC, Burtnik A, Coppolino MG. Lamellipodium extension and membrane ruffling require different SNARE-mediated trafficking pathways. BMC Cell Biol. 2010;11:62. doi: 10.1186/1471-2121-11-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneh A, Deol YS, Ganju A, Shilo K, Rosol TJ, Nasser MW, Ganju RK. Differential role of psoriasin (S100A7) in estrogen receptor alpha positive and negative breast cancer cells occur through actin remodeling. Breast Cancer Res Treat. 2013;138:727–739. doi: 10.1007/s10549-013-2491-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodek KL, Ringuette MJ, Brown TJ. MT1-MMP is the critical determinant of matrix degradation and invasion by ovarian cancer cells. Br J Cancer. 2007;97:358–367. doi: 10.1038/sj.bjc.6603863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen A, Le Dez G, Poincloux R, Recchi C, Nassoy P, Rottner K, Galli T, Chavrier P. MT1-MMP-dependent invasion is regulated by TI-VAMP/VAMP7. Curr Biol. 2008;18:926–931. doi: 10.1016/j.cub.2008.05.044. [DOI] [PubMed] [Google Scholar]
- Tayeb MA, Skalski M, Cha MC, Kean MJ, Scaife M, Coppolino MG. Inhibition of SNARE-mediated membrane traffic impairs cell migration. Exp Cell Res. 2005;305:63–73. doi: 10.1016/j.yexcr.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Williams KC, Coppolino MG. Phosphorylation of membrane type 1-matrix metalloproteinase (MT1-MMP) and its vesicle-associated membrane protein 7 (VAMP7)-dependent trafficking facilitate cell invasion and migration. J Biol Chem. 2011;286:43405–43416. doi: 10.1074/jbc.M111.297069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.