Drosha is required for the nuclear processing of pri- to pre-miRNAs. Drosha itself is translationally regulated during oocyte-to-egg maturation. Upon completion of meiosis, Drosha mRNA is translationally activated. This leads to a subsequent increase of endogenous miRNAs.
Abstract
MicroRNAs (miRNAs) are ∼21-nucleotide-long, single-stranded noncoding RNAs that regulate gene expression. Biogenesis of miRNAs is mediated by the two RNase III-like enzymes, Drosha and Dicer. Here we study miRNA biogenesis during maturation of Xenopus oocytes to eggs using microinjection of pri-miRNAs. We show that processing of exogenous and endogenous primary miRNAs (pri-miRNAs) is strongly enhanced upon maturation of oocytes to eggs. Overexpression of cloned Xenopus Drosha in oocytes, however, boosts pri-miRNA processing dramatically, indicating that Drosha is a rate-limiting factor in Xenopus oocytes. This developmental regulation of Drosha is controlled by poly(A) length addition to the Drosha mRNA, which boosts translation upon transition from oocytes to eggs. Processing of pri-miRNAs by Drosha and Dicer has been shown to be affected by adenosine-to-inosine deamination–type RNA editing. Using activated Xenopus eggs for microinjection experiments, we demonstrate that RNA editing can reduce pri-miRNA processing in vivo. This processing block is determined by the structural but not sequence changes introduced by RNA editing.
INTRODUCTION
MicroRNAs (miRNAs) are key regulators of gene expression in metazoans and plants. The biogenesis of miRNAs follows a rather conserved pathway in most metazoans (Kim et al., 2009). miRNA genes are transcribed by RNA polymerases II or III into primary transcripts (pri-miRNAs). The pri-miRNA is subsequently processed by the nuclear microprocessor complex (consisting of the double-stranded RNA (dsRNA)–binding protein DGCR8 and the RNase III enzyme Drosha). Cleavage releases a short, ∼60- to 70-nucleotide (nt) hairpin termed pre-miRNA with a 2-nt overhang at the 3′ end. After export from the nucleus via exportin-5, pre-miRNAs are further cleaved in the cytoplasm by the ∼200-kDa RNase III–like enzyme Dicer complexed with TAR-RNA–binding protein (TRBP; Yi et al., 2003; Lund et al., 2004; Lund and Dahlberg, 2006; Chendrimada et al., 2005; Haase et al., 2005). The resulting ∼21-nt miRNAs and Argonaute (Ago) form the RNA-induced silencing complex, mediating posttranscriptional gene silencing, mostly by imperfect base pairing with target mRNAs, usually in the 3′ untranslated region (3′-UTR; Wilson and Doudna, 2013).
Biogenesis of miRNAs can be controlled at several levels (Finnegan and Pasquinelli, 2013). First, transcription of primary miRNAs can be regulated and subsequent processing steps can be modulated by specific binding of RNA-binding proteins to miRNA precursors (Michlewski and Caceres, 2010; Michlewski et al., 2010; Piskounova et al., 2011; Van Wynsberghe et al., 2011). Processing of miRNAs can also be affected by posttranscriptional nucleotide modifications in the pri-miRNA as induced by adenosine deaminases acting on RNA (ADARs; Yang et al., 2006; Wulff and Nishikura, 2012). Moreover, the key enzymes Drosha and Dicer can themselves be subject to posttranslational regulation (Krol et al., 2010). Finally, miRNA stability can be regulated by controlling nucleotide addition at the 3′ end (Liu et al., 2011; Thornton et al., 2012).
Gene regulation by miRNAs plays a pivotal role in animal development and differentiation (Bushati and Cohen, 2007). In early Xenopus development, miRNA expression is regulated in a stage- and tissue-specific manner (Watanabe et al., 2005; Tang and Maxwell, 2008). The maturation of Xenopus oocytes to eggs is accompanied by cessation of transcription and a complex network of translational activation and repression of stored maternal mRNAs. Translation of stored maternal mRNAs can be controlled by the length of the poly(A) tail. Elongation of the poly(A) tail mediated by cytoplasmic polyadenylation element–binding protein facilitates translation of dormant mRNAs during oocyte development (Cooke et al., 2010; Richter and Lasko, 2011; Villalba et al., 2011).
Oocyte maturation can boost Dicer activity, and miRNAs are expressed in a stage-specific manner in Xenopus development (Watanabe et al., 2005; Lund and Dahlberg, 2006; Armisen et al., 2009; Lund et al., 2009). Moreover, Ago protein is a rate-limiting step in RNA interference (RNAi) and processing of pre-miRNAs (Lund et al., 2011).
Here we show that Drosha activity is almost absent in Xenopus stage VI oocytes but is dramatically boosted upon maturation of oocytes to eggs. The boost in Drosha activity occurs independently of ongoing transcription but is concomitant with poly(A) tail extension of Drosha mRNA and increase in Drosha protein. In addition, quantitative PCR (qPCR) indicates that Drosha mRNA levels do not change during oocyte-to-egg maturation. Together the results indicate translational control of Drosha protein during oocyte-to-egg maturation.
Using processing-competent Xenopus eggs, we determine the effect of structural changes induced by ADAR-mediated RNA editing on the processing of hsa-pri-miRNA-142. Previous studies show that the presence of inosines can target pre-miRNAs for destruction by the Tudor staphylococcal nuclease (Yang et al., 2006). Alternatively, structural or sequence changes in RNA induced by RNA editing may impair cleavage reactions by Drosha or Dicer (Yang et al., 2006; Kawahara et al., 2007). By following the fate of a microinjected, radiolabeled pri-miRNA-142, we show that structural changes at the Drosha cleavage site can impair cleavage of pri-miRNA-142 in vivo and that these changes can be suppressed by compensatory mutations that restore the structure of the pri-miRNA.
RESULTS
Drosha cleavage of microinjected pri-miRNAs is enhanced by maturation of Xenopus laevis oocytes to eggs
Pri-miRNAs are processed to pre-miRNAs in the cell nucleus by the Drosha-DGCR8 microprocessor complex (Gregory et al., 2004). pre-miRNAs are subsequently exported to the cytoplasm to be further processed by Dicer both in somatic cells and oocytes (Yi et al., 2003; Lund et al., 2004).
To study the processing of pri-miRNAs and the potential effect of structural changes induced by ADAR-mediated RNA editing in oocytes, constructs mimicking pri-miR-21, pri-miR-29b-1, and pri-miR-142 were cloned and transcribed in vitro. To allow proper folding and processing of the sequence, 40–50 nucleotides were added on either side of the predicted Drosha cleavage sites. Capped, radiolabeled transcripts were injected into stage VI oocyte nuclei (also termed germinal vesicles [GVs]) and eggs at a concentration of 5 ng/GV or cytoplasm. As a control for proper nuclear injection, the pri-miRNA was mixed with either the nuclear-retained U3 small nucleolar RNA (snoRNA) or U6 small nuclear RNA (snRNA).
Of interest, in GVs, only minor processing of the injected pri-miRNA by Drosha could be observed. Instead, the injected pri-miRNA was rather unstable and degraded within 30 min after injection. The control RNA U6, however, was stable. This was true for nuclear-injected pri-miR-21, pri-miR-29b-1, and pri-miR-142 (Figure 1, A and B, and Supplemental Figure S1B). Of interest, injections of pri-miRNAs into the cytoplasm left the pri-miRNAs stable in this cellular compartment (Figure 1C and asterisks in Supplemental Figure 1B). Degradation of the pri-miRNA was unaffected by their capping status, as both capped and uncapped pri-miRNAs were unstable in the nucleus (Supplemental Figure S2).
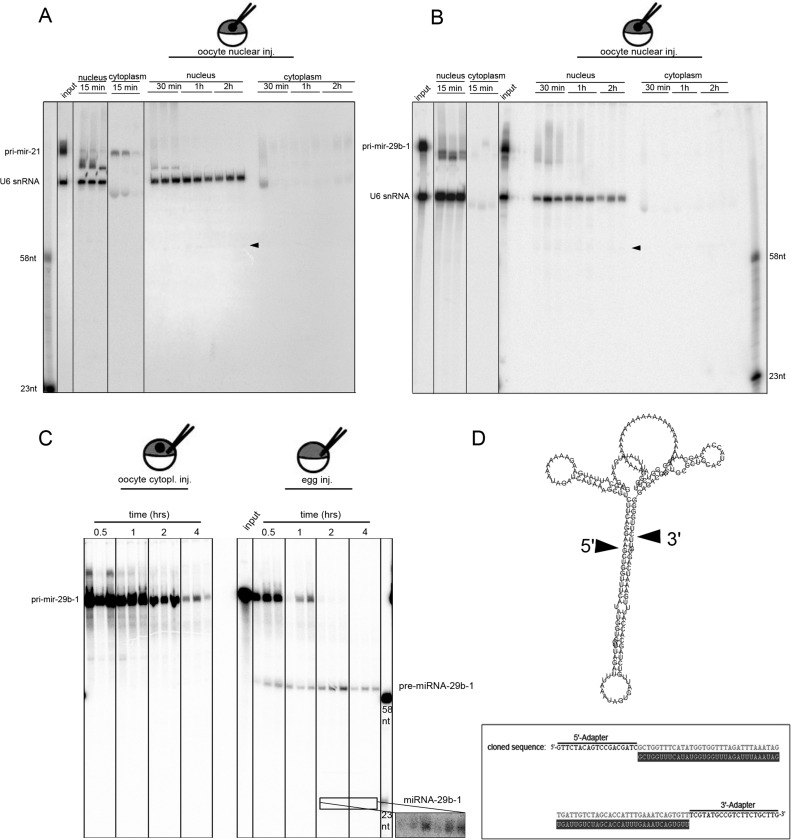
FIGURE 1:
Microinjected pri-miRNAs are not processed in oocyte nuclei but are in matured eggs. Radiolabeled pri-miRNA-21 (A) and pri-miRNA-29b-1 (B) were injected into individual nuclei of oocytes. To control for proper nuclear injection, the nuclear-retained U6 snRNA was coinjected. After incubation for up to 2 h, the individual oocytes were manually dissected into nuclei and cytoplasm. The corresponding nuclear and cytoplasmic RNAs of individual oocytes were extracted and separated on denaturing urea PAGE. An aliquot of the injected mixture is loaded (input), as well as markers corresponding to processed pre- and mature miRNAs (marker). pri-miRNAs injected into oocyte nuclei are processed poorly and get mostly degraded, whereas U6 snoRNA is stable (arrowheads mark weak, processed pre-miRNA bands). (C) pri-miRNA-29b-1 injected into eggs becomes processed into pre- and mature (insert) miRNAs. (D) RNA folding prediction of synthetic pri-miRNA-29b-1 used for injection. The processed band was cloned, and three independent clones were sequenced. Alignment with the predicted pre-miR29b-1 (gray) shows perfect alignment with the cloned pre-miRNA-29b-1 (top). This proves proper Drosha processing of microinjected pri-miRNA-29b-1 in Xenopus eggs. The processing sites are indicated by arrowheads.
Microinjection of in vitro–synthesized pri-miRNAs might saturate the oocyte microprocessor complex, leading to degradation of excess microinjected pri-miRNAs. To test for this possibility, we performed nuclear injection of pri-miR-21 at concentrations of 10 and 2 ng/nucleus, respectively (Supplemental Figure S1A). In these experiments the injected pri-miRNA also was unstable. However, minor processing of pri- to pre-miRNA was observed in GVs injected with 10 ng pri-mRNA/ nucleus. Together these results show that Drosha activity is low in oocytes and that pri-miRNAs are unstable in oocyte germinal vesicles but stable in the oocyte cytoplasm.
Previous studies showed that maturation of oocytes to eggs leads to activation of Dicer (Lund and Dahlberg, 2006). We therefore tested whether matured eggs would also show increased Drosha activity. We matured oocytes to eggs by treating stage VI oocytes with progesterone. Matured eggs were selected by occurrence of a pigmentation spot in the animal hemisphere and manually collected for further experiments. Maturation of oocytes to eggs leads to nuclear envelope breakdown. Therefore microinjection experiments in eggs were done into the mixed cytoplasm. Microinjection of pri-miR-29b-1 or pri-miR-21 in eggs clearly showed that Drosha mediated processing of pri-miRs to pre-miRs and subsequent processing to mature miRNAs (Figure 1C and Supplemental Figure S3). This suggests that Drosha activity is enhanced in eggs.
To confirm proper processing of pri-miRNAs to pre-miRNAs by Drosha in Xenopus eggs, the obtained putative pre-miRNA band was gel purified, ligated to 3′- and 5′-adapters, and amplified by reverse transcription PCR. The resulting cDNA was cloned and sequenced. To avoid contamination with endogenous pre-miRNA molecules, human pri-miR-29b-1 was used for microinjection. The resulting pre-miRNA has no identified homologue in Xenopus. Sequence analysis and comparison to the corresponding miRBase entry confirmed appropriate processing of the microinjected pri-miR-29b-1 to pre-miR-29b-1 in Xenopus eggs (Figure 1D).
Processing of Xenopus miRNAs is enhanced in eggs
Our data show that maturation of oocytes to eggs boosts the processing of injected pri-miRNAs to pre-miRNAs. Similarly, stimulation of processing of pre-miRNAs to mature miRNAs by Dicer was shown to be enhanced upon oocyte-to-egg transition (Lund and Dahlberg, 2006). Moreover, several miRNAs were reported to increase upon oocyte-to-egg maturation in Xenopus tropicalis, consistent with an increase in their processing (Armisen et al., 2009). We therefore asked whether endogenous miRNA levels are also affected upon oocyte maturation in Xenopus laevis. Northern blot experiments were performed to compare levels of miRNAs known to be expressed in oocytes. We chose xtr-miR-101 and xtr-miR-148a, two miRNAs that were shown to be expressed in X. tropicalis germline cells, for these experiments (Armisen et al., 2009). Three Northern blots were made from biological replicates and normalized for U7 snRNA. Blots were exposed to phosphoimager screens and detected on a Bio-Rad FXPro phosphoimager. Quantification of the blots confirmed a twofold increase of these two endogenous miRNAs upon oocyte-to-egg maturation in X. laevis (Supplemental Figure S4).
To confirm that processing of endogenous pri- and pre-xtr-miR-148a can be increased upon maturation of oocytes to eggs, a fragment of pri-xtr-miR-148a and the predicted pre-xtr-miR-148 were cloned and in vitro transcribed. Nuclear injection of pri-xtr-miR-148a precursors in oocytes again confirms only minor processing but prominent degradation of the injected pri-miRNA. In contrast, injection of pri-xtr-miR-148a into eggs leads to efficient accumulation of the corresponding pre- and mature xtr-miRNA, consistent with activation of Drosha in eggs (Figure 2A).
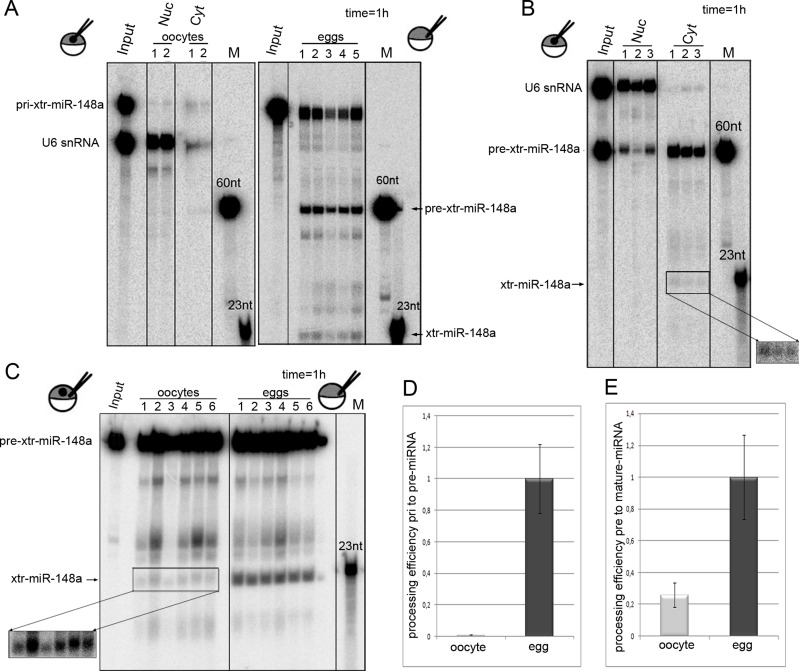
FIGURE 2:
Drosha and Dicer processing are enhanced upon maturation of oocytes to eggs. (A) pri-xtr-miR-148a is not processed when injected into oocyte nuclei (oocyte) but gets processed in eggs. RNA was injected in individual oocyte nuclei or eggs with U6 as a nuclear retention marker. RNAs were reextracted from corresponding single nuclei (Nuc) and cytoplasm (Cyt) of oocytes or from whole eggs. Minor processing of the miRNA could be detected in oocytes, whereas efficient processing to pre- and mature xtr-miR-148a was seen in eggs. (B) Nuclear export and processing of pre-xtr-miR148a in oocytes. Pre-xtr-miR-148a injected into oocyte nuclei is exported to the cytoplasm and processed to mature miR-148a. (C) Dicer processing is enhanced in eggs. Cytoplasmic injection of pre-xtr-miR-148a into oocytes shows moderate processing to mature miR-148a. The same amount of pre-xtr-miR148a injected into eggs is more efficiently processed. (D) Quantification of blot shown in A. The amount of processed RNA was quantified over the amount of input RNA. This shows a strong increase in Drosha cleavage in eggs. (E) Quantification of blot shown in C confirms a fivefold increase in Dicer processing. For A–C, incubation time was 1 h.
To test for Dicer-meditated processing, we injected xtr-pre-miR-148a into oocyte nuclei (Figure 2B). One hour after injection, a large fraction of the nuclear-injected pre-miRNA was exported to the cytoplasm, but only a small fraction was further processed to mature xtr-miR-148a by Dicer (Figure 2B).
To compare Dicer processing in oocytes and eggs, we injected xtr-pri-miR-148a into the cytoplasm of oocytes or the mixed cell contents of activated eggs (Figure 2C). Dicer processing activity was almost fourfold increased in Xenopus eggs compared with oocytes (Figure 2, C and D). This increase is consistent with the previously reported increase in Dicer activity in Xenopus eggs (Lund et al., 2011).
Ectopically expressed Drosha is active in Xenopus oocyte nuclei
To test whether Drosha activity might be inhibited in Xenopus oocytes, we expressed recombinant Drosha in oocytes and eggs and tested for its enzymatic activity. We cloned the cDNA encoding X. laevis Drosha using primers against highly conserved regions in the protein followed by 5′ and 3′ rapid amplification of cDNA ends (RACE). The cDNA clone isolated in this way was fused to an N-terminal 7x myc tag and to the 3′ UTR of Xenopus NO38 (Peculis and Gall, 1992).
Capped, in vitro–transcribed RNA of this construct was injected into Xenopus oocytes. Translation was allowed to occur overnight. Subsequently, half of the cells were matured to eggs. Injected or control oocytes and eggs were homogenized, and the translated protein was immunoprecipitated with an anti-myc antibody coupled to protein A–Sepharose beads. The proteins bound to the beads were directly used for in vitro processing assays using radiolabeled pri-miR-29b-1 (Supplemental Figure S5). Distinct processing patterns were observed in immunoprecipitates of both oocytes and eggs expressing recombinant Drosha. These bands were not observed in MOCK immunoprecipitates (Supplemental Figure S5).
On quantification, no differences in Drosha processing patterns could be detected between precipitates derived from oocytes or eggs expressing recombinant Drosha (Supplemental Figure S5). This indicates that Drosha protein purified from oocytes or eggs is equally active and argues against posttranslational modification of the protein that would lead to inactivation/activation of the protein in oocytes or eggs, respectively.
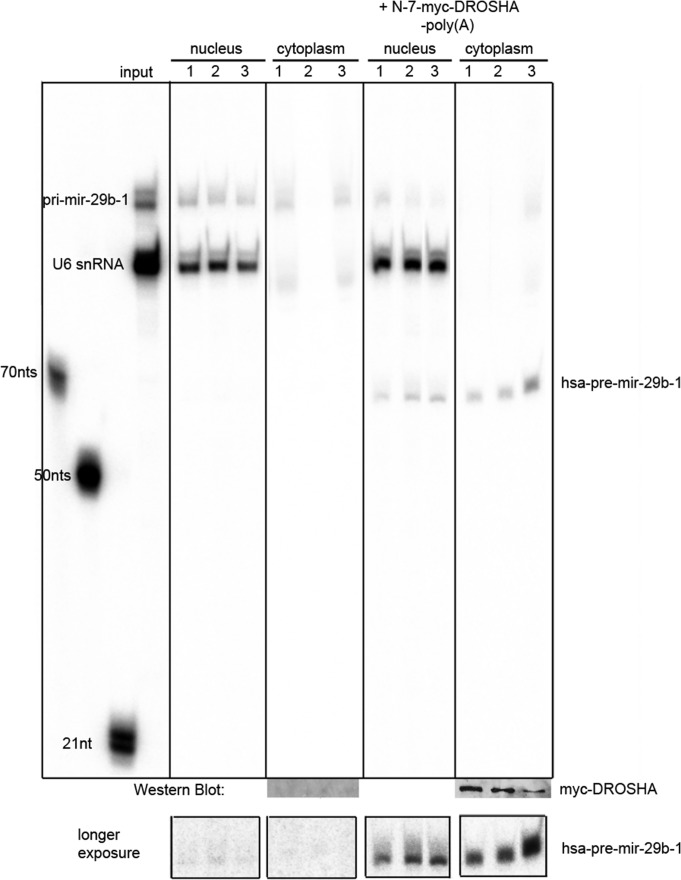
Next we tested whether ectopically expressed Drosha is also active in live oocytes. Again, Drosha was ectopically expressed in oocytes. Subsequently, pri-miR-29b-1 was microinjected into oocyte nuclei or matured eggs, and processing was monitored 30–60 min after injection. No processing was observed in nuclei of untreated oocytes, whereas efficient Drosha cleavage was observed in nuclei of oocytes ectopically expressing X. laevis Drosha (Figure 3). The Drosha cleavage product was also efficiently exported to the cytoplasm of the corresponding oocytes (Figure 3). This experiment clearly demonstrates that ectopically expressed Drosha is active in oocyte nuclei, arguing against the existence of an inhibitory factor in oocytes.
FIGURE 3:
Ectopic expression of myc-tagged Drosha stimulates miRNA processing in oocytes. Oocytes were first injected with RNA encoding N-terminally myc-tagged Drosha (+N-myc-Drosha-polyA) or left uninjected (control). After overnight incubation to allow for translation of Drosha, radiolabeled pri-miRNA-29b-1 was injected into oocyte nuclei. After 30–60 min, nuclei and corresponding cytoplasm were isolated. RNA was reextracted from individual nuclei, whereas only half of the corresponding cytoplasm was used. The remaining cytoplasm was prepared for SDS protein electrophoresis and subsequent Western blotting. RNAs isolated from three nuclei and cytoplasm from control and Drosha-expressing oocytes were separated on urea-polyacrylamide gels. Nuclear processing of pri- to pre-miRNA and subsequent export to the cytoplasm was well observed in oocytes ectopically expressing Drosha, whereas only minor processing was observed in control oocytes (see longer exposure of pre-miRNA region). Cytoplasmic Western blots for the presence of myc-Drosha were generated from half of the cytoplasm used for RNA extraction with an anti-myc monoclonal antibody. Nuclear accumulation of myc-Drosha was tested on separate Western blots (not shown). Individual lanes showing good nuclear injection of pri-miR-29b-1 were cut and pasted together.
Transcription-independent regulation of Drosha activity
So far, we could show that oocyte maturation led to increased pri-miRNA processing in eggs. This raised the question of how Drosha processing could be activated during oocyte maturation. Conceptually, Drosha activity could be regulated at the transcriptional, translational, or posttranslational level either directly or via a factor that interferes with Drosha activity.
To test for transcriptional activation of Drosha during progesterone-mediated maturation, we pretreated oocytes with the transcriptional inhibitor actinomycin D (AMD). However, upon maturation, processing patterns of microinjected pri-miRNA-29b-1 looked identical in AMD-treated and untreated cells, indicating that transcription is not required for stimulation of Drosha activity (Supplemental Figure S6).
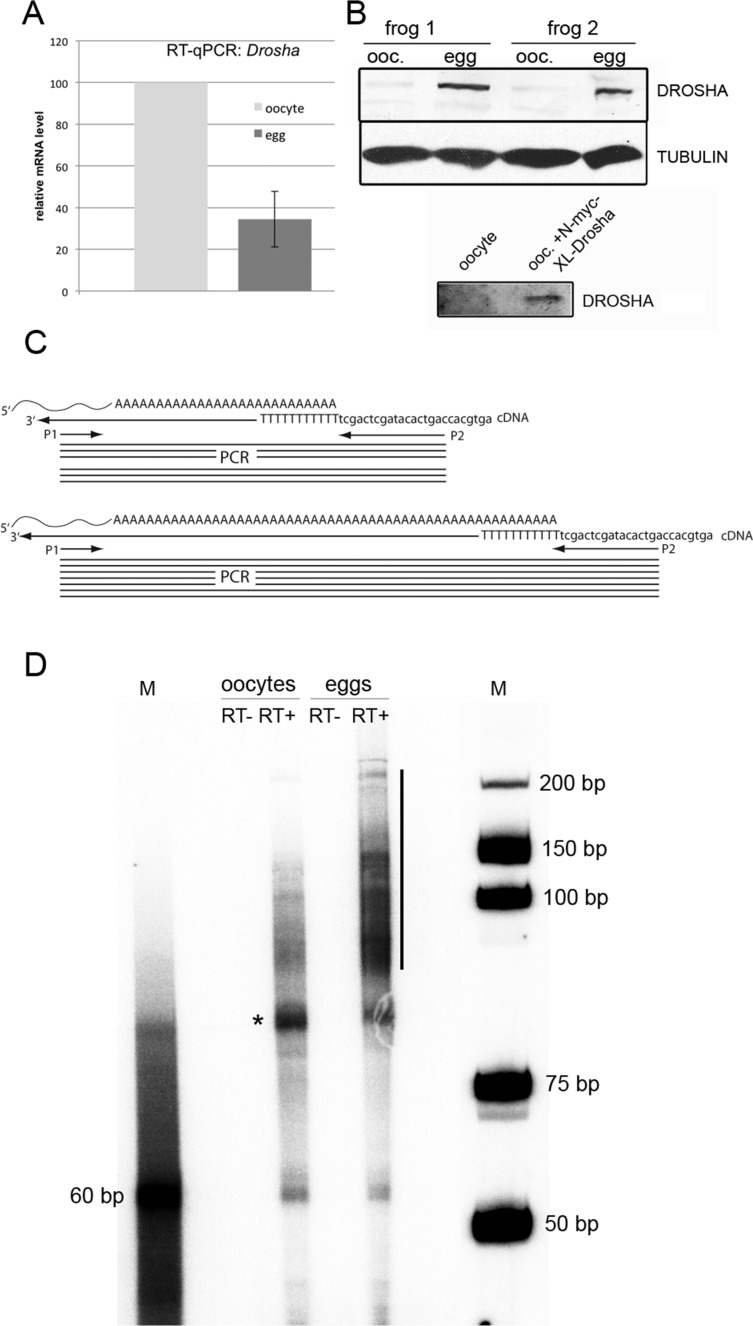
We also compared Drosha RNA levels during oocyte-to-egg maturation via qPCR on cDNAs prepared from oocytes and eggs. However, when compared with Smn2, a validated qPCR standard during Xenopus development, Drosha mRNA levels even dropped about threefold during oocyte-to-egg maturation (Dhorne-Pollet et al., 2013; Figure 4A).
FIGURE 4:
Maturation of oocytes to eggs leads to an increase of Xenopus Drosha protein and poly A+ extension of Drosha mRNA. (A) qPCR of oocyte and egg cDNA shows threefold decrease in Drosha mRNA relative to Smn-2 mRNA. (B) Western blot of individual oocytes and eggs of two different frogs with a monoclonal anti-Drosha antibody. To verify that the antibody is recognizing the correct proteins, oocytes were also injected with mRNA encoding myc-xlDrosha. Drosha is well detected in eggs but barely visible in oocytes unless they were previously injected with RNA encoding myc-Drosha. (C) Scheme of poly(A) tail determination. cDNA was prepared with an anchoring primer. Depending on the length of the poly(A) tail, a short (top) or long (bottom) PCR product is obtained between a specific primer (P1) and an anchored primer (P2). (D) Whereas the poly(A) tail extends up to 90 nucleotides in oocytes (asterisk), an extension of up to 200 nucleotides can be observed in eggs (black bar). M, marker bands. The 60–base pair band in the poly(A) tail PCR lane most likely originates from priming of the oligo(dT) primer used for cDNA synthesis close to the poly(A) site.
Drosha is translationally regulated
Based on the described experiments, a transcriptional or posttranslational regulation of Drosha appeared rather unlikely. Xenopus oocytes store a plethora of maternal RNAs, which are only translated upon maturation, fertilization, or later during development (Kuge and Inoue, 1992). Therefore, to test for possible translational control of Drosha during oocyte-to-egg maturation, we compared protein levels in oocytes and eggs.
Several commercially available antibodies were tested for their cross-reactivity with X. laevis Drosha protein. One rabbit monoclonal antibody, raised against a conserved peptide around Gly-953 of human Drosha, showed good cross-reactivity with Xenopus Drosha. The cross-reactivity was tested on oocytes ectopically expressing myc-tagged Xenopus Drosha (Figure 4B). Using this antibody on defolliculated oocyte and egg lysates of two different frogs, we showed that Drosha protein is clearly detectable in lysates of eggs, whereas only a faint band is observed in oocytes (Figure 4B). Thus it seems that the amount of Drosha protein increases during oocyte-to-egg maturation.
We also tried to inhibit translation using cycloheximide. However, this treatment precludes maturation of oocytes to eggs, as previously reported, making this experimental strategy uninformative (Wasserman and Masui, 1975; Schuetz and Samson, 1979).
Storage of maternally expressed mRNAs in oocytes and their translational activation in eggs is frequently mediated by extending the poly(A) tail of mRNAs in the cytoplasm (McGrew and Richter, 1990; Paillard and Osborne, 2003). We therefore set out to determine the length of the poly(A) tail of endogenous Drosha mRNA via an anchored 3′ RACE with an oligo(dT) primer at the 3′ end (Figure 4C; Murray and Schoenberg, 2008). As expected, an extension of the poly(A) tail was clearly visible upon maturation of oocytes to eggs. Whereas Drosha mRNA had a poly(A) tail of ∼80 nucleotides in oocytes, this tail was extended to more than 200 adenosine residues in eggs (Figure 4D). Consistent with a poly(A)-tail extension during oocyte-to-egg maturation, the X. tropicalis cDNA encoding the Drosha protein contains several cytoplasmic polyadenylation signals (Supplemental Figure S7; Villalba et al., 2011).
Effect of RNA editing on pri-miRNA processing in vivo
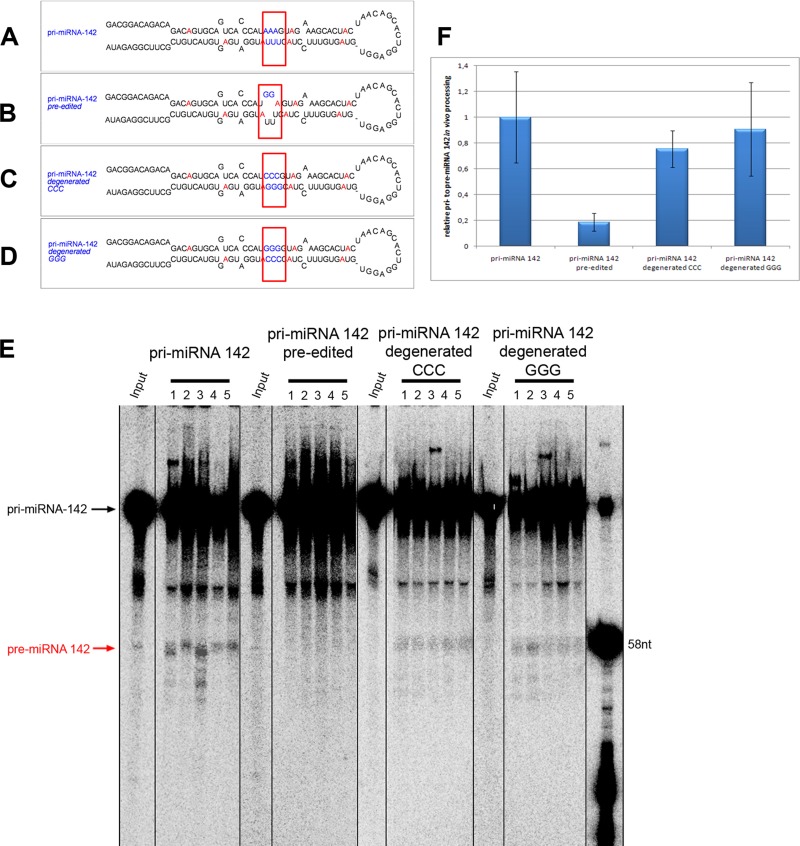
RNA editing of pri- and pre-miRNAs can alter miRNA processing patterns or miRNA target specificity. RNA editing of mouse pri-miR-142 at positions +4 and +5 efficiently blocks pri-miRNA processing in vivo and may lead to efficient degradation of the precursor RNA by staphylococcal nuclease (Yang et al., 2006). Inhibition of processing most likely occurs through folding differences near the Drosha cleavage sites induced by the edited nucleotides (Yang et al., 2006). We tested this hypothesis by introducing guanosines that mimic the edited inosines at positions +4 and +5 of pri-miR-148. Introduction of guanosines instead of edited inosines was already shown to be sufficient to block efficient processing of pri-miR-142 (Yang et al., 2006). To correct for structural changes, we also introduced compensatory mutations—cytosines opposite the “edited” guanosines. Moreover, the positions of “edited” guanosines and compensatory cytosines were exchanged to control for any sequence-specific differences (Figure 5, A–D).
FIGURE 5:
Structural changes induced by RNA editing lead to reduced processing of pri-miRNA-142. Constructs mimicking wild-type, preedited, and constructs carrying compensatory mutations that revert secondary structure changes induced by RNA editing were injected into eggs, and their processing was monitored. (A–D) Constructs used for injection. (A) Wild-type pri-miRNA-142, (B) pri-miRNA-142 preedited at positions +4 and +5, (C) editing incompetent pri-miRNA-142, and (D) compensatory mutation closing the loop induced upon editing. Red nucleotides mark adenosines reported to be edited (Yang et al., 2006). Blue nucleotides mark the position of introduced mutations. (E) Processing of pri-miRNA-142 variants in Xenopus eggs shows that processing is strongly inhibited in the preedited pri-miRNA-142. RNAs were reextracted 30 min after injection from individual microinjected eggs and separated on denaturing polyacrylamide gels. Processing can be restored by mutations that prevent editing or a compensatory mutation that restores proper folding. (F) Quantification of processing levels (ratio of pre-miRNA to pri-miRNAs) shows a strong reduction in processing in preedited pri-miRNA 142. Processing ratio of wild-type pri-miRNA-142 to pre-miRNA-142 was set to 1, and all other processing ratios were normalized to this. For quantification, different exposures were chosen to prevent saturated pixels in the pri-miRNA region and obtain sharp bands in this region.
Drosha processing of wild-type pri-miR-142, preedited pri-miR-142, and the two constructs in which the editing-induced structural changes were corrected was then tested in Xenopus eggs. Radiolabeled pri-miRNAs were microinjected into eggs. After a 30-min incubation, the RNAs were reextracted from individual eggs, and processing products were analyzed by PAGE.
Processing rates were analyzed and quantified (Figure 5, E and F). Compared to wild type, the preedited pri-miRNA-142 showed a fivefold-reduced processing level. Of interest, both mutations that compensate for the editing event by closing the bulge structure formed upon editing showed almost-normal processing rates, demonstrating that structural changes introduced by RNA editing but not sequence changes lead to reduced Drosha processing.
Cloning and sequence analysis of microinjected pri-miRNA-142 variants did not reveal de novo A-to-I RNA editing in eggs. Hence, pri-miRNA-142 variants remain stable and are not targeted by Tudor-SN, which degrades hyperedited, inosine-containing RNA molecules (Scadden, 2005).
DISCUSSION
Developmental activation of pri-miRNA processing
We show that pri-miRNA processing is activated upon maturation of oocytes to eggs. Stage VI oocytes are transcriptionally rather inactive. However, this does not rule out that miRNA gene transcription and processing occur earlier in development. The fact that mature miRNAs can also be detected in stage VI oocytes by Northern blotting indicates that transcription and processing of miRNAs must occur at low levels during the many months of oogenesis. However, in contrast to somatic cells, oocytes at this stage of development have barely detectable levels of nuclear Drosha.
In contrast, a strong increase in Drosha levels is accompanied by increased pri-miRNA processing in eggs. Northern blot experiments show that Drosha activation also leads to a strong increase of endogenous xtr-miRNA-101 and xtr-miRNA-148a, demonstrating that Drosha activation may also act on endogenous pri-miRNAs. Together with the reported increase of Dicer activity upon oocyte maturation and the finding that Ago is a limiting factor during Xenopus oogenesis and early development, our findings support the notion that miRNA biogenesis is strongly regulated during oogenesis and meiosis (Lund and Dahlberg, 2006; Lund et al., 2011; Armisen et al., 2009).
The fact that all microinjected pri-miRNAs used in this study were rather unstable upon injection in the GV raises the question of whether many pri-miRNAs are stored in oocytes to become processed upon completion of meiosis I. However, the artificial constructs we used do not resemble the full-length primary transcripts, which may be stabilized by association with proteins in vivo. In addition, the poly(A) tails added to the constructs we used may be too short to stabilize the injected RNAs. Moreover, in vivo, processing factors, including the microprocessor complex, may normally be deposited cotranscriptionally, leading to greater stability of in vivo–transcribed RNAs than observed for the in vitro–synthesized, microinjected RNAs used here. Whether pri-miRNAs can be exported to the oocyte cytoplasm, where they appear stable, remains to be determined. Because oocyte-to-egg maturation is accompanied by nuclear envelope breakdown, any subsequent Drosha processing event would then occur in the mixed cytosol.
A possible source of pri-miRNAs might originate from intronic sequences that have been shown to be enriched and stable in the oocyte GV (Gardner et al., 2012). Consistent with this idea is the finding that in Xenopus, many miRNAs are intronically located (Tang and Maxwell, 2008). Such stable, intron-located pri-miRNAs may therefore become processed by the increased Drosha levels observed upon oocyte-to-egg maturation. Moreover, transcriptional stimulation at the mid blastula transition will strongly contribute to an increase in the pri-miRNA pool.
In the course of our experiments, we also observed some variability in the speed by which nuclear-injected pri-miRNAs were degraded. Degradation rates did not depend on the amount of RNA injected, but instead varied between experiments, suggesting that the source of the oocyte may contribute to the variable degradation rates observed.
We also observed variability in Drosha activity on different substrates. Whereas pri-xtr-miR-148a was very efficiently processed in eggs, mouse pri-miR-142 was processed rather poorly. This difference may well reflect the folding and thus suitability of various substrates for Drosha processing.
In contrast to Drosha activation, xtr-pre-miRNA-148a Dicer processing can already be detected in stage VI oocytes. Oocyte maturation then further intensifies Dicer processing of xtr-pre-miRNA-148a. Together these results demonstrate a developmentally regulated increase in Drosha protein and an increase of Dicer activity that jointly will lead to enhanced miRNA biogenesis. Accordingly, our data suggest that Drosha processing is the rate-limiting step during miRNA maturation. This may have important implications for miRNA-mediated gene regulation during oogenesis and fertilization. Previous work showed that miRNAs play an important role in the turnover of maternal mRNAs during the switch from maternal to zygotic gene expression by mediating deadenylation of target mRNAs (Giraldez et al., 2006; Bushati et al., 2008; Lund et al., 2009). Thus miRNA up-regulation and changes in affected targets may play an important role in regulating maternally inherited RNAs.
Translational regulation of Drosha activity
Our data show that Drosha protein increases during oocyte-to-egg maturation. This increase is accompanied by an extension of the poly(A) tail found on the Drosha mRNA, whereas Drosha mRNA levels even show a slight decrease compared with Smn2 mRNA. Moreover, activation of Drosha activity does not require ongoing transcription. This, together with the fact that ectopically expressed Drosha is enzymatically active in the germinal vesicle, indicates that Drosha mRNA is translationally regulated via the length of its poly(A) tail, a phenomenon shown for many maternally stored RNAs (Wormington, 1993; Meric et al., 1996; Radford et al., 2008).
Effect of RNA editing on pri-miRNA processing in vivo
ADAR proteins and proteins of the RNAi machinery, such as the microprocessor complex (Drosha-DGCR8), Dicer, and TRBP, contain dsRNA-binding domains (dsRBDs). dsRBDs show little sequence specificity, which makes them potential competitors for RNA binding (Saunders and Barber, 2003; Nishikura, 2006). Consistently, ADARs were shown to interfere with miRNA maturation even in the absence of RNA-editing activity (Heale et al., 2009; Vesely et al., 2012). Moreover, ADAR has also been shown to stimulate processing of miRNAs (Ota et al., 2013). In vitro experiments using Drosha/DGCR8 complexes revealed that editing of positions +4 and +5 located in the dsRNA stem near the Drosha cleavage site is both necessary and sufficient to inhibit Drosha cleavage (Yang et al., 2006). It was also shown that expressed pri-miRNAs that mimic the edited state fail to produce mature miRNAs, suggesting that the Drosha cleavage step might also be affected in vivo (Yang et al., 2006).
To address this point more clearly, we tested the processing of preedited pri-miRNA-142 using our established Xenopus egg microinjection system. This system has the advantage that body-labeled RNAs can be followed along each processing step and all cleavage products can be visualized. In this study we showed that processing of preedited pri-miRNA-142 is strongly reduced in vivo. Reduction in processing levels is most likely caused by the introduced structural change in the hairpin of the preedited pri-miRNA-142 variant. Compensatory mutations that correct this structural change show normal processing.
Of interest, sequencing of cDNAs generated from microinjected pri-miRNA-142 failed to detect de novo RNA editing in vivo, leaving unprocessed pri-miRNA-142 stable in the egg. This finding is in contrast to the situation in tissue culture cells, where unprocessed pri-miRNA-142 was edited at multiple positions and then cleaved by Tudor-SN (Yang et al., 2006). The finding is even more surprising when one considers that RNA-editing activity was first identified in Xenopus oocytes and eggs (Rebagliati and Melton, 1987; Bass and Weintraub, 1988). However, it is possible that either the incubation times chosen in our experiments are too short to detect efficient editing or the construct we used may not reflect a perfect editing substrate for editing in Xenopus oocytes or eggs.
Nonetheless, reduction of preedited pri-miRNA-142 in vivo agrees well with results published for inhibition of pri-miRNA-142 in vitro and pre-miRNA-151 in vivo processing (Yang et al., 2006; Kawahara et al., 2007). This confirms the importance of RNA editing as a regulator of miRNA biogenesis and consequently miRNA-mediated gene silencing.
MATERIALS AND METHODS
Handling of Xenopus oocytes and microinjection experiments
X. laevis oocytes and eggs were stored in OR2-buffer (82.5 mM NaCl, 2.5 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 1 mM Na2HPO4, 5 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 8.3) at 16°C (Wallace et al., 1973). Oocytes were matured to eggs by adding 20 μg/ml progesterone and incubating at 16°C overnight. X. laevis eggs were selected for visible germinal vesicle breakdown. Nuclear and cytoplasmic injections were performed using an Eppendorf microinjector. Up to 50 nl (500 ng/μl) of Drosha-encoding mRNA was injected in cytoplasms. Pri- and pre-miRNAs were injected at 5–10 nl per nucleus or cytoplasm. RNAs were injected at concentrations of 200–500 ng/μl.
For nuclear injections, oocytes were centrifuged with the animal pole facing upward at 800 × g for 20 min. This results in the germinal vesicle becoming visible as a bright spot in the animal hemisphere.
Extraction of total RNA was carried out using Invitrogen (Carlsbad, CA) TRIzol reagent according to the manufacturer's protocol. RNA was separated on 10% 29:1 acrylamide:bis-acrylamide gels containing 8 M urea in 1× Tris/borate/EDTA (TBE).
Quantifications of processed versus unprocessed bands were done on a phosphoimager in three independent experiments using Bio-Rad Quantity One software. Care was taken that no saturated pixels were contained in the exposure.
Cloning of miRNA variants containing a T7 promoter
To produce DNA constructs that can be in vitro transcribed by T7 RNA polymerase, we PCR amplified miRNA sequences from 1 μg of human genomic DNA. To stabilize the transcript, we inserted an artificial 40-nt-long poly(A) tail at the 3′ end of the pri-miRNA sequence.
PCR products were cloned in Promega (Madison, WI) pGEM-Teasy vector and verified by sequencing.
The following sequences were cloned (T7 promoter sequences are shown in bold):
hsPri-miRNA-21
TAATACGACTCACTATAGGACATCTCCATGGCTGTACCACCTTGTCGGGTAGCTTATCAGACTGATGTTGACTGTTGAATCTCATGGCAACACCAGTCGATGGGCTGTCTGACATTTTGGTATCTTTCATCTGACCATAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA
hsPri-miRNA-29
TAATACGACTCACTATAGGGTCTTGGGTTTATTGTAAGAGAGCATTATGAAGAAAAAAATAGATCATAAAGCTTCTTCAGGAAGCTGGTTTCATATGGTGGTTTAGATTTAAATAGTGATTGTCTAGCACCATTTGAAATCAGTGTTCTTGGGGGAGACCAGCTGCGCTGCACTACCAACAGCAAAAGAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA
xtrPre-miR148a
TAATACGACTCACTATAGGAGTTCTGTGACACTTAGACTCTGAATATGATAGCAGTCAGTGCACTACAGAACTTTGT
xtrPri-miR148a
TAATACGACTCACTATAGGGTCTTGGGTTTATTGTAAGAGAGCATTATGAATAGTCTTTTAAATCAAAGTTCTGTGACACTTAGACTCTGAATATGATAGCAGTCAGTGCACTACAGAACTTTGTTTTGGGAGTCTGGCTGCGCTGCACTACCAACAGCAAAAGAA
Human U3
TAATACGACTCACTATAGAAGACTATATTTTCAGGGATCATTTCTACAGTGCACTACTAGAGAAGTTTCTGTGAACTTGTAGAGCACCGGAAACCATGAGCAGGAAGTGCAGCGTTCTCTCCTGAGCATGAAGCCGGCTCTTGGTGTGGCTTCGCTGCAACTGCCATTGGCCATTGATGATCGTTCTTCTCTTCTCTGGGAGAGTAAGAGAGAGAGGACACAGTCTGAGTGG
Human U6
TAATACGACTCACTATAGGTGCTCATTTTGGCAGCACATATACTAAAATTAGAACACTGCAGAGAAGATTAGCATGGCCCCTGCACAAGGATGACAATAAAAATTAAAAAATGAATTT
T7 in vitro transcription
For in vitro transcription, the plasmid containing the Drosha template was linearized at the 3′ end. T7 in vitro transcription using 500 ng of template DNA was performed in 10 mM dithiothreitol, 1× Stratagene (La Jolla, CA) transcription buffer, 0.3 mM GpppGTP (cap), 0.5 mM CTP, 0.5 mM UTP, 0.5 mM ATP, 1 μl (40 U) RNase inhibitor, and T7 RNA polymerase. For capped transcripts, 0.1 mM GTP was added after a 10-min preincubation with the cap analogue. The reaction was incubated for 2 h at 37°C, purified using Sephadex G-25 columns, and precipitated. For the production of radiolabeled RNA, 0.1 mM ATP and 30 μCi of [α-32P]ATP were added to the nucleotide mix while all other nucleotides were left at 0.3 mM.
The in vitro transcripts were separated on 10 or 15% 29:1 AA:bis-AA gels (8 M urea in 1× TBE). The wet gel was exposed to x-ray films, and appropriate bands were cut out and eluted in 400 μl of elution buffer (500 mM NH4OAc, 0.2% SDS, 100 mM EDTA) overnight. Eluted samples were purified over Sephadex G-25 columns and precipitated.
Northern blotting
Northern blots were done as described (Pall and Hamilton, 2008). Briefly, total RNA was isolated from 35 oocytes or eggs. The equal amount of oocyte and egg total RNA was confirmed by RiboGreen fluorometric quantification. Isolated RNAs were separated on a 15% polyacrylamide gel. Gels were blotted onto Hybond N+ membrane (GE Healthcare, Freiburg, Germany) using a semidry blotting technique. For hybridization, oligonucleotides xtr-miRNA-101, 5′-CTTCAGTTATCACAGTACTGTA-3′, and xtr-miRNA-148a, 5′-ACAAAGTTCTGTAATGCACTGA-3′, were 5′-end labeled with 20 μCi of [γ-32P]ATP using T4 polynucleotide kinase according to manufacturer's protocol. Hybridization was carried out as described (Pall and Hamilton, 2008). Blots were exposed to phosphoimager screens, and signals were detected using an FX Pro phosphoimager. Quantification and integration of signals of equally sized squares were done using Quantity One Software.
Immunoprecipitation of recombinant Drosha
Oocytes and eggs (10 cells) injected with N-terminally tagged 7xmyc-Drosha-poly(A) RNA were homogenized, and 100 μl of NET-2 (150 mM NaCl, 50 mM Tris, pH 7.4, 0.05% NP-40) buffer was added. The mixture was spun for 5 min at 14,000 rpm, and the supernatant was transferred to a fresh tube. An aliquot of one cell (10 μl) was transferred to a fresh tube and saved for Western blotting (input control).
The α-myc antibody 9E10 was coupled to Sepharose A beads (4 mg of beads in 5 ml of 9E10) overnight at 4°C. The coupled beads were washed four times in NET-2 buffer and resuspended in 500 μl of NET-2 buffer. Supernatants of cell extracts were added to the resuspended antibody-coupled beads and incubated for 1 h at 4°C on a rotating wheel. The beads were washed four times with NET-2 buffer and resuspended in 500 μl of NET-2 buffer. An aliquot of one cell (50 μl) was transferred to a fresh tube for Western blotting (purification control). The remaining immunoprecipitate was washed in 500 μl of 6.4 mM MgCl2 and used for Drosha in vitro processing assay.
Drosha in vitro processing assay
Capped, in vitro–transcribed RNA encoding myc-tagged Drosha was injected in oocytes, and half of the cells were matured to eggs. Cells were lysed, and lysates were used to immunoprecipitate the translated Drosha protein using α-myc antibody coupled to protein A– Sepharose following the protocol described by Steitz (1989). The precipitated material was used for an in vitro processing assay with protein still coupled to beads as described at www.narrykim.org/in_vitro_Drosha_processing.pdf.
After processing, the cleaved RNA was boiled in 8 M urea and loaded on a 10% urea polyacrylamide gel.
Cloning of processed pre-miRNAs
Nuclear or cytoplasmic RNA isolations were run on polyacrylamide gels next to radioactively labeled markers. After a short exposure of the wet gel to film, the region of interest was excised, and RNAs were extracted by macerating the gel in 500 mM NH4Ac and 0.1% SDS with overnight incubation. Extracted RNAs were precipitated, and cDNA synthesis was done with a primer overlapping the template by eight nucleotides. cDNA synthesis was first performed at 16°C and then extended at room temperature. After cDNA synthesis, PCR was performed with the primer used for cDNA synthesis and a 5′ primer overlapping the cDNA again by eight nucleotides. PCR products were gel purified, cloned, and sequenced. Of 24 clones sequenced, 3 contained the correct and expected sequence.
Reverse transcription quantitative PCR
For the quantification of Drosha, total RNA prepared from stage VI oocytes and unfertilized eggs was DNase I digested (1 U/μl, 10 U; Fermentas) and reverse transcribed using M-MLV Reverse Transcriptase (Thermo Scientific, Waltham, MA) and random hexamers in a total volume of 20 μl. Real-time PCR was performed on a Bio-Rad iQ5 Mastercycler, using GoTaq SYBR Green qPCR master mix (Promega, Madison, WI) and the following primers. Drosha: sense, 5′-CCTTTATCGCTGCCCTTTAT-3′; antisense, 5′-CCATCTGGGGGAAGTTATAT-3′. Smn2: sense, 5′-ATAGGAGACACGTGTAATGC-3′; antisense 5′-GAGGATCTTTGCTTTGATGC-3′. The PCR program was 40 cycles of 95°C/15 s, 57.5°C/30 s, and 72°C/1 min, preceded by denaturation at 95°C/3 min. The relative difference in expression of Drosha was calculated by the ΔΔCt method using Smn2 as a reference gene.
Poly(A) length assay
This assay was performed according to Murray and Schoenberg (2008). We used the oligo(dT) adapter 5′-GGCCACGCGTCGACTAGTACTTTTTTTTTTTTTTTTT in combination with the mRNA-specific primer 5′-CCTTTATCGCTGCCCTTTAT.
Cloning of Xenopus Drosha
To clone full-length Xenopus Drosha, a 5′ and 3′ RACE protocol was applied using primers in a region that was found conserved from fish to mammals. The forward primer taggccacaatcagagaat and the reverse primer ccatctgggggaagttatat were used for the anchored PCR on cDNA isolated from X. laevis poly(A) RNA (Zhang and Frohman, 1997). The ends of the Drosha cDNA were subsequently cloned using 5′ and 3′ RACE protocols as published (Zhang and Frohman, 1997). The resulting fragments were assembled and verified by sequencing.
Western blotting
The same numbers of Xenopus oocytes and eggs from two different frogs were manually defolliculated and homogenized, and the insoluble fraction was removed by centrifugation. The soluble fraction was denatured by addition of 2× Laemmli buffer and heating to 95°C for 5 min (Laemmli, 1970). The equivalent of 1.5 oocytes or eggs was run on 6% SDS polyacrylamide gels, and proteins were blotted to nitrocellulose and detected as follows: detection of Drosha was achieved with a human- and mouse-specific antibody that cross-reacts with Xenopus Drosha (D28B1 rabbit monoclonal antibody 3364; Cell Signaling Technology, Beverly, MA). To control for equal loading, the same blot was also incubated with α-tubulin antibody (Sigma-Aldrich, St. Louis, MO). As secondary, an anti-rabbit AP antibody (31340; Pierce) was used. Myc-tagged Drosha was detected with the anti-myc monoclonal antibody 9E10 and a labeled, secondary anti-mouse antibody.
Supplementary Material
Acknowledgments
This work was supported by Austrian Science Foundation Special Research Program SFB 4313 to M.F.J. and National Natural Science Foundation of China Grant 30770469 to Y.F.J. C.V. was funded by Austrian Science Foundation Doctoral Program W1207.
Abbreviations used:
- Ago
Agonaute
- DGCR8
DiGeorge critical region 8
- hsa-pri-miRNA
Homo sapiens primary microRNA
- miRNA
micro RNA
- UTR
untranslated region
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-07-0386) on May 14, 2014.
*These authors contributed equally to this work.
The authors declare no conflict of interest.
REFERENCES
- Armisen J, Gilchrist MJ, Wilczynska A, Standart N, Miska EA. Abundant and dynamically expressed miRNAs, piRNAs, and other small RNAs in the vertebrate Xenopus tropicalis. Genome Res. 2009;19:1766–1775. doi: 10.1101/gr.093054.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass BL, Weintraub H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell. 1988;55:1089–1098. doi: 10.1016/0092-8674(88)90253-x. [DOI] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Bushati N, Stark A, Brennecke J, Cohen SM. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr Biol. 2008;18:501–506. doi: 10.1016/j.cub.2008.02.081. [DOI] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke A, Prigge A, Wickens M. Translational repression by deadenylases. J Biol Chem. 2010;285:28506–28513. doi: 10.1074/jbc.M110.150763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhorne-Pollet S, Thelie A, Pollet N. Validation of novel reference genes for RT-qPCR studies of gene expression in Xenopus tropicalis during embryonic and post-embryonic development. Dev Dyn. 2013;242:709–717. doi: 10.1002/dvdy.23972. [DOI] [PubMed] [Google Scholar]
- Finnegan EF, Pasquinelli AE. MicroRNA biogenesis: regulating the regulators. Crit Rev Biochem Mol Biol. 2013;48:51–68. doi: 10.3109/10409238.2012.738643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner EJ, Nizami ZF, Talbot CC, Jr, Gall JG. Stable intronic sequence RNA (sisRNA), a new class of noncoding RNA from the oocyte nucleus of Xenopus tropicalis. Genes Dev. 2012;26:2550–2559. doi: 10.1101/gad.202184.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heale BS, Keegan LP, McGurk L, Michlewski G, Brindle J, Stanton CM, Caceres JF, O'Connell MA. Editing independent effects of ADARs on the miRNA/siRNA pathways. EMBO J. 2009;28:3145–3156. doi: 10.1038/emboj.2009.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 2007;8:763–769. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Kuge H, Inoue A. Maturation of Xenopus laevis oocyte by progesterone requires poly(A) tail elongation of mRNA. Exp Cell Res. 1992;202:52–58. doi: 10.1016/0014-4827(92)90403-u. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Liu N, Abe M, Sabin LR, Hendriks GJ, Naqvi AS, Yu Z, Cherry S, Bonini NM. The exoribonuclease Nibbler controls 3’ end processing of microRNAs in Drosophila. Curr Biol. 2011;21:1888–1893. doi: 10.1016/j.cub.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Dahlberg JE. Substrate selectivity of exportin 5 and Dicer in the biogenesis of microRNAs. Cold Spring Harb Symp Quant Biol. 2006;71:59–66. doi: 10.1101/sqb.2006.71.050. [DOI] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Lund E, Liu M, Hartley RS, Sheets MD, Dahlberg JE. Deadenylation of maternal mRNAs mediated by miR-427 in Xenopus laevis embryos. RNA. 2009;15:2351–2363. doi: 10.1261/rna.1882009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Sheets MD, Imboden SB, Dahlberg JE. Limiting Ago protein restricts RNAi and microRNA biogenesis during early development in Xenopus laevis. Genes Dev. 2011;25:1121–1131. doi: 10.1101/gad.2038811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrew LL, Richter JD. Translational control by cytoplasmic polyadenylation during Xenopus oocyte maturation: characterization of cis and trans elements and regulation by cyclin/MPF. EMBO J. 1990;9:3743–3751. doi: 10.1002/j.1460-2075.1990.tb07587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meric F, Searfoss AM, Wormington M, Wolffe AP. Masking and unmasking maternal mRNA. The role of polyadenylation, transcription, splicing, and nuclear history. J Biol Chem. 1996;271:30804–30810. doi: 10.1074/jbc.271.48.30804. [DOI] [PubMed] [Google Scholar]
- Michlewski G, Caceres JF. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat Struct Mol Biol. 2010;17:1011–1018. doi: 10.1038/nsmb.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michlewski G, Guil S, Caceres JF. Stimulation of pri-miR-18a Processing by hnRNP A1. Adv Exp Med Biol. 2010;700:28–35. doi: 10.1007/978-1-4419-7823-3_3. [DOI] [PubMed] [Google Scholar]
- Murray EL, Schoenberg DR. Assays for determining poly(A) tail length and the polarity of mRNA decay in mammalian cells. Methods Enzymol. 2008;448:483–504. doi: 10.1016/S0076-6879(08)02624-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K. Editor meets silencer: crosstalk between RNA editing and RNA interference. Nat Rev Mol Cell Biol. 2006;7:919–931. doi: 10.1038/nrm2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota H, Sakurai M, Gupta R, Valente L, Wulff BE, Ariyoshi K, Iizasa H, Davuluri RV, Nishikura K. ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell. 2013;153:575–589. doi: 10.1016/j.cell.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillard L, Osborne HB. East of EDEN was a poly(A) tail. Biol Cell. 2003;95:211–219. doi: 10.1016/s0248-4900(03)00038-8. [DOI] [PubMed] [Google Scholar]
- Pall GS, Hamilton AJ. Improved Northern blot method for enhanced detection of small RNA. Nat Protoc. 2008;3:1077–1084. doi: 10.1038/nprot.2008.67. [DOI] [PubMed] [Google Scholar]
- Peculis BA, Gall JG. Localization of the nucleolar protein NO38 in amphibian oocytes. J Cell Biol. 1992;116:1–14. doi: 10.1083/jcb.116.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskounova E, Polytarchou C, Thornton JE, LaPierre RJ, Pothoulakis C, Hagan JP, Iliopoulos D, Gregory RI. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell. 2011;147:1066–1079. doi: 10.1016/j.cell.2011.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford HE, Meijer HA, de Moor CH. Translational control by cytoplasmic polyadenylation in Xenopus oocytes. Biochim Biophys Acta. 2008;1779:217–229. doi: 10.1016/j.bbagrm.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebagliati MR, Melton DA. Antisense RNA injections in fertilized frog eggs reveal an RNA duplex unwinding activity. Cell. 1987;48:599–605. doi: 10.1016/0092-8674(87)90238-8. [DOI] [PubMed] [Google Scholar]
- Richter JD, Lasko P. Translational control in oocyte development. Cold Spring Harb Perspect Biol. 2011;3:a002758. doi: 10.1101/cshperspect.a002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders LR, Barber GN. The dsRNA binding protein family: critical roles, diverse cellular functions. FASEB J. 2003;17:961–983. doi: 10.1096/fj.02-0958rev. [DOI] [PubMed] [Google Scholar]
- Scadden AD. The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat Struct Mol Biol. 2005;12:489–496. doi: 10.1038/nsmb936. [DOI] [PubMed] [Google Scholar]
- Schuetz AW, Samson D. Nuclear requirement of post-maturational cortical differentiation of amphibian oocytes: effects of cycloheximide. J Exp Zool. 1979;210:307–319. doi: 10.1002/jez.1402100214. [DOI] [PubMed] [Google Scholar]
- Steitz JA. Immunoprecipitation of ribonucleoproteins using autoantibodies. Methods Enzymol. 1989;180:468–481. doi: 10.1016/0076-6879(89)80118-1. [DOI] [PubMed] [Google Scholar]
- Tang GQ, Maxwell ES. Xenopus microRNA genes are predominantly located within introns and are differentially expressed in adult frog tissues via post-transcriptional regulation. Genome Res. 2008;18:104–112. doi: 10.1101/gr.6539108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton JE, Chang HM, Piskounova E, Gregory RI. Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7) RNA. 2012;18:1875–1885. doi: 10.1261/rna.034538.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wynsberghe PM, Kai ZS, Massirer KB, Burton VH, Yeo GW, Pasquinelli AE. LIN-28 co-transcriptionally binds primary let-7 to regulate miRNA maturation in Caenorhabditis elegans. Nat Struct Mol Biol. 2011;18:302–308. doi: 10.1038/nsmb.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesely C, Tauber S, Sedlazeck FJ, von Haeseler A, Jantsch MF. Adenosine deaminases that act on RNA induce reproducible changes in abundance and sequence of embryonic miRNAs. Genome Res. 2012;22:1468–1476. doi: 10.1101/gr.133025.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalba A, Coll O, Gebauer F. Cytoplasmic polyadenylation and translational control. Curr Opin Genet Dev. 2011;21:452–457. doi: 10.1016/j.gde.2011.04.006. [DOI] [PubMed] [Google Scholar]
- Wallace RA, Jared DW, Dumont JN, Sega MW. Protein incorporation by isolated amphibian oocytes. 3. Optimum incubation conditions. J Exp Zool. 1973;184:321–333. doi: 10.1002/jez.1401840305. [DOI] [PubMed] [Google Scholar]
- Wasserman WJ, Masui Y. Effects of cyclohexamide on a cytoplasmic factor initiating meiotic maturation in Xenopus oocytes. Exp Cell Res. 1975;91:381–388. doi: 10.1016/0014-4827(75)90118-4. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Takeda A, Mise K, Okuno T, Suzuki T, Minami N, Imai H. Stage-specific expression of microRNAs during Xenopus development. FEBS Lett. 2005;579:318–324. doi: 10.1016/j.febslet.2004.11.067. [DOI] [PubMed] [Google Scholar]
- Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annu Rev Biophys. 2013;42:217–239. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wormington M. Poly(A) and translation: development control. Curr Opin Cell Biol. 1993;5:950–954. doi: 10.1016/0955-0674(93)90075-2. [DOI] [PubMed] [Google Scholar]
- Wulff BE, Nishikura K. Modulation of microRNA expression and function by ADARs. Curr Top Microbiol Immunol. 2012;353:91–109. doi: 10.1007/82_2011_151. [DOI] [PubMed] [Google Scholar]
- Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, Nishikura K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Frohman MA. Using rapid amplification of cDNA ends (RACE) to obtain full-length cDNAs. Methods Mol Biol. 1997;69:61–87. doi: 10.1385/0-89603-383-x:61. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.