Abstract
Long non-coding RNAs (lncRNAs) have been shown to be implicated in the complex network of cancer including malignant melanoma and play important roles in tumorigenesis and progression. However, their functions and downstream mechanisms are largely unknown. This study aimed to investigate whether BRAF-activated non-coding RNA (BANCR), a novel and potential regulator of melanoma cell, participates in the proliferation of malignant melanoma and elucidate the underlying mechanism in this process. We found that BANCR was abnormally overexpressed in human malignant melanoma cell lines and tissues, and increased with tumor stages by quantitative PCR. BANCR knockdown induced by shRNA transfection significantly inhibited proliferation of tumor cells and inactivated MAPK pathway, especially by silencing the ERK1/2 and JNK component. Moreover, combination treatment of BANCR knockdown and suppression ERK1/2 or JNK (induced by specific inhibitors U0126 or SP600125 respectively) produced synergistic inhibitory effects in vitro. And the inhibitory effects induced by ERK1/2 or JNK could be rescued by BANCR overexpression. By tumorigenicity assay in BALB/c nude mice, we further found that BANCR knockdown inhibited tumor growth in vivo. In addition, patients with high expression of BANCR had a lower survival rate. Taken together, we confirmed the abnormal upregulation of a novel lncRNA, BANCR, in human malignant melanoma. BANCR was involved in melanoma cell proliferation both in vitro and in vivo. The linkage between BANCR and MAPK pathway may provide a novel interpretation for the mechanism of proliferation regulation in malignant melanoma.
Introduction
Melanoma is the most aggressive type of skin cancer, characterized by a rapid progression, metastasis to regional lymph nodes and distant organs as well as a limited efficiency of therapeutics [1]. Although it accounts for only about 4% of all dermatological cancers, it contributes to more than 80% death of skin cancer patients [2]. Therefore, it is necessary to understand thoroughly which factors involved in the development and progression of melanoma. In addition, though many molecules contributing to invasion and migration have been reported [3], [4], the precise mechanisms underlying proliferation, the first step of tumor pathogenesis, remain to be fully elucidated.
Recently, growing attention has been given to a class of non-protein-coding RNAs (ncRNAs), known as long non-coding RNAs (lncRNAs), which participates in cell fate determination and disease pathogenesis [5], [6]. LncRNAs are non-coding RNAs greater than 200 nucleotides in length and can regulate gene expression in different biological processes transcriptionally or post-transcriptionally. And its dysregulation has been observed under many pathological conditions including cancers, heart diseases and Alzheimer disease [7]. For example, HOTAIR seems to modulate tumor invasiveness by enhancing PRC2-mediated suppression of metastasis [8]. PRNCR1 and PCGEM1 can bind successively to androgen receptor and strongly enhance androgen receptor-mediated gene activation and proliferation in prostate cancer [9]. And dysregulation of HNF1A-AS1 participates in oesophageal tumorigenesis by modulation of chromatin and nucleosome assembly as well as by induction of cancer-related H19 [10].
In previous study, Flockhart RJ et al. [11] and McCarthy N [12] identified a previously unstudied but widely expressed lncRNA, BRAF-activated non-coding RNA (BANCR), as playing a potentially functional role in melanoma cell migration by RNA-sequencing. In the present study, we confirmed the role of BANCR in the proliferation of malignant melanoma, and aimed to elucidate the contribution of MAPK pathway in this process. Our data demonstrated that BANCR can promote melanoma proliferation via activating ERK1/2 and JNK MAPK pathway both in vitro and in vivo. The linkage of BANCR and the MAPK pathway may unravel a novel mechanism in the regulation of melanoma proliferation.
Materials and Methods
Tissue samples
Tissues were obtained from patients (62 male and 41 female) who underwent surgery at Department of Dermatology, Inner Mongolia People's Hospital from 2005 to 2010. The study was carried out in accordance with the institutional ethical guidelines and the use of human skin tissues was approved by the Medical Ethics Committee of Inner Mongolia People's Hospital (IMP study ID H05-561321). Every patient involved in the study was asked to sign a piece of written informed consent which has been approved by the ethics committee of Inner Mongolia People's Hospital, and all the consents were saved by the ethics committee. The study was conducted according to the principles expressed in the Declaration of Helsinki. Skin tissues from 12 patients with melanocytic nevus (matched by gender and age) were collected as controls. All samples were snap-frozen in liquid nitrogen, then stored at −80°C for further use. Patient descriptions are detailed in table 1. Clinical follow-up data were available for 72 patients, while 31 patients were excluded for lack of information.
Table 1. Association between BANCR and clinicopathological parameters of patients.
| Characteristic | N (%) | BANCR | P value |
| All cases | 103 (100) | ||
| Age | 0.092 | ||
| ≤60 y | 48 (46.6) | 4.8(3.4–7.6) | |
| >60 y | 55 (53.4) | 5.1(3.0–8.2) | |
| Histologic type | 0.316 | ||
| Superficial spreading | 33 (32.0) | 5.2(2.8–7.7) | |
| Nodular | 25 (24.3) | 4.9(2.5–6.5) | |
| Acral lentiginous | 21 (20.4) | 5.4(3.0–7.1) | |
| Lentigo maligna | 7 (6.8) | 4.6(3.1–6.9) | |
| Other | 17(16.5) | 5.0(2.2–8.3) | |
| Location | 0.081 | ||
| Head and neck | 53 (51.5) | 4.2(2.3–6.6) | |
| Trunk | 27 (26.2) | 3.9(2.5–7.3) | |
| Extremities | 23 (22.3) | 4.3(3.1–8.6) | |
| TNM classification | 0.017 | ||
| I–II | 20 (19.4) | 2.2(1.8–3.5) | |
| III | 70 (68.0) | 4.2(3.9–6.1) | |
| IV | 13 (12.6) | 6.8(6.3–8.3) |
Cell culture and treatment
Five human melanoma cells lines, including A-375, 1205Lu, UACC903, CHL-1 and sk-mel-5, were purchased from American Type Culture Collection. Cells were maintained in RPMI-1640 medium (A-375, 1205Lu) or DMEM medium (UACC903, CHL-1 and sk-mel-5) respectively supplemented with 10% fetal bovine serum (Gibco, CA, USA) in a 37°C humidified atmosphere of 5% CO2.
Knockdown of BANCR was induced by transfection with BANCR shRNA (Genepharma, Shanghai, China) using Lipofectamine2000 (Invitrogen, CA, USA). The target sequences were as follows: shRNA #1: 5′-CGGAAATAGACTGCAGCAC CAA-3′; shRNA #2: 5′-CCTTTATGGATTCAACTGTAAT-3′. Ectopic expression of BANCR was achieved through pCDNA3.1-BANCR transfection using Lipofectamine2000. Oligonucleotides for amplification of BANCR were as follows: sense: 5′- CAGGAAGGGGTGAATGAAGA-3′; antisense: 5′- CCAGTGCAGGGTAATGTGTG-3′.
Stable transfectants were selected in medium containing 600 µg/mL G418 (Invitrogen, CA, USA) and used in further assays or RNA/protein extraction.
For treatment with pharmacological inhibitors, the overnight starved cells were kept in complete medium containing 10 µM U0126 (inhibitor of the upstream ERK1/2 activator MEK1/2, Sigma-Aldrich, MO, USA) or 20 µM SP600125 (JNK inhibitor, Sigma-Aldrich, MO, USA). DMSO was used as a control. Medium containing inhibitor was renewed every day.
RNA extraction and quantitative real-time PCR analysis
Total RNA was extracted from tissues or cells using Trizol reagent (Invitrogen, CA, USA). BANCR expressions were measured by SYBR green qPCR assay (Takara, Dalian, China) in triplicate. Primer sequences were as follows: BANCR, forward: ACAGGACTCCATGGCAAACG, reverse: ATGAAGAAAGCCTGGTGCAGT; GAPDH, forward: ACCACAGTCCATGCCATCAC, reverse: TCCACCACCCTGTTGCTGTA. Data were processed using 2-ΔΔCT method and normalized to GAPDH expression.
Protein extraction and western blot analysis
Cells were lysed in RIPA buffer with protease inhibitors and phosphatase inhibitors. Protein was loaded onto a SDS-PAGE minigel and transferred onto PVDF membrane. The blots were probed with primary antibodies (Cell signal technology, MA, USA) followed by the HRP-conjugated secondary antibody. Signals were visualized using ECL Substrates (Millipore, MA, USA). GAPDH served as the loading control.
CCK-8 cell proliferation assay
Cell proliferation rates were measured using Cell Counting Kit-8 (CCK-8, Beyotime, Hangzhou, China). 0.5×104 cells were seeded in each 96-well plate for 24 h, treated with or without U0126 or SP600125, and further incubated for 24 h, 48 h and 72 h respectively. After 10 µl CCK-8 reagent was added to each well, the plate was returned to the incubator for 1 h, the absorbance at 450 nm was measured using an enzyme-linked immunosorbent assay plate reader.
Tumorigenicity assay in nude mice
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of Inner Mongolia People's Hospital. The protocol was approved by the Committee on the Ethics of Animal Experiments of Inner Mongolia People's Hospital (IMP study ID H05-561321).
A total of 24 male BALB/c nude mice (18–20 g, 5 weeks old, Animal Center of the Chinese Academy of Science, Shanghai, China) were randomized into three groups of eight each and housed 4 per cage under specific-pathogen-free conditions (room temperature-controlled at 24±2°C with a relative humidity of 60±5%, on a 12 h light/dark cycles) in the Animal Care Facility Service (Inner Mongolia People's Hospital). Standard rodent chow and fresh distilled water were supplemented every two days. Mice were anesthetized with intraperitoneal injection of sodium pentobarbital (0.3%, 50–60 mg/Kg) to relieve pain and then injected subcutaneously with 4×106 cells into the flanks. Physiological parameters, including activity, food/water intake, skin, hair, excrement and mental state were monitored every day. And tumor volume and weight were monitored once every 5 days. Tumor volume was calculated according to the formula: tumor volume = length × (width) 2/2. After five weeks, mice (0/8 in group NC; 5/8 in group shRNA #1; 6/8 in group shRNA #2) presented symptoms including extremely slow activity, lethargy, hair loss and obvious weight loss, thus all the mice were sacrificed by pentobarbital overdose (1%) and exsanguination according to American Veterinary Medical Association guidelines for euthanasia.
Statistical analysis
All values were expressed as mean ± SD and processed using the SPSS 13.0 software. Statistical significance was noted at P<0.05. Three independent experiments were performed. The differences among the groups in proliferation assays, western blot and PCR assays were estimated by Student's t-test or one-way ANOVA. The survival curves were plotted according to the Kaplan-Meier method and compared using the Log-rank test.
Results
BANCR is frequently upregulated in malignant melanoma tissues and cell lines
Expression of BANCR in a total of 103 human tissue specimens of malignant melanoma with various cancer stages and melanoma cell lines was detected using SYBR green quantitative PCR analysis. A significantly increased level of BANCR was seen in patients with malignant melanoma (Figure 1A, P = 0.007), compared with the levels detected in age/gender-matched controls with melanocytic nevus. Patients were then grouped by clinical cancer stages. Differences between controls and patients with stages I/II, stage III, and stage IV tumors were also statistically significant (Figure 1B, P<0.001). A direct correlation between clinical cancer stages and BANCR was noted. There was a significantly positive correlation between BANCR expression and tumor stages (P<0.05), while no correlation was observed between BANCR expression and age, gender, localization or tumor size. Then we extended our test to five human melanoma cell lines. The total five cell lines showed a notable overexpression of BANCR compared to the control skin tissue pooled from 3 controls with melanocytic nevus (Figure 1C, P<0.001).
Figure 1. Increased expression of BANCR in both malignant melanoma tissues and cell lines.
(A) Increased BANCR expression in malignant melanoma tissues compared to control skin tissues. (B) BANCR expression increased with clinical stages of malignant melanoma. (C) Significant high expression of BANCR in five melanoma cell lines in comparison with control skin tissues pooled from 3 controls with melanocytic nevus. (** P<0.01).
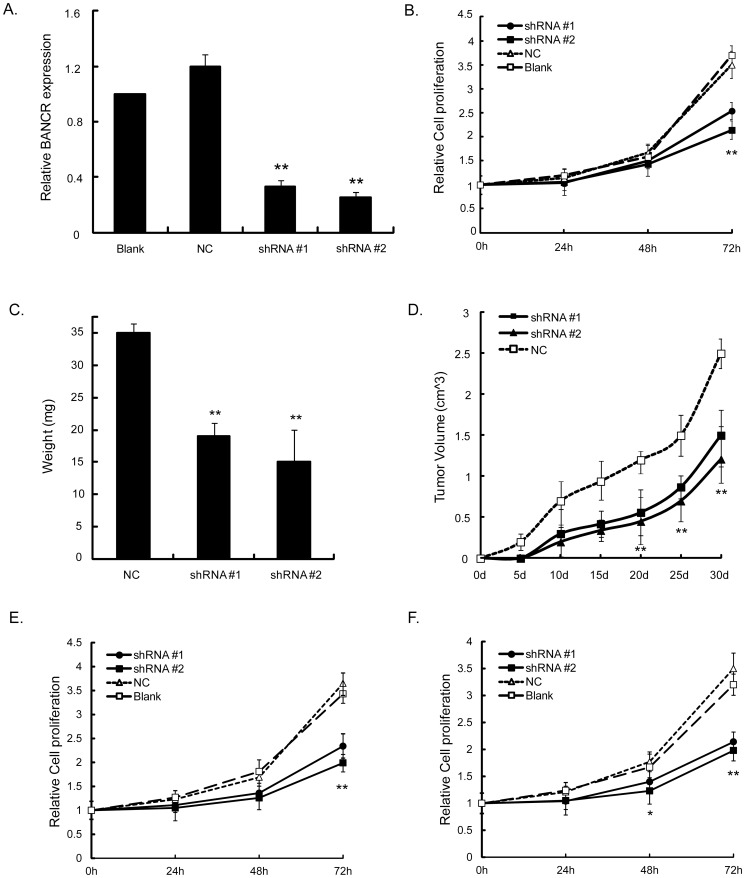
Knockdown of BANCR inhibited melanoma proliferation in vitro and in vivo
We selected sk-mel-5 cells to perform further assays to test whether BANCR was functionally involved in malignant melanoma tumorigenesis. BANCR shRNA was stably transfected into the sk-mel-5 cells and expression of BANCR in these clones was confirmed by PCR analysis (Figure 2A). shRNA transfectant cells exhibited significantly decreased proliferation compared with either NC shRNA transfectants cells or parental cells (P<0.01, Figure 2B). To examine whether downregulation of BANCR could inhibit tumorigenicity of sk-mel-5 cells in vivo, a xenograft tumor model was applied. As shown in Figure 2C and 2D, the shRNA transfectant cells developed tumors with much smaller volumes (1.27±0.13 for shRNA #1 and 1.01±0.14 for shRNA #2 vs. 2.51±0.17 at the fifth week, P<0.01) and lighter weight (15±2.1 for shRNA #1 and 13±3.5 for shRNA #2 vs. 35±1.5 at the fifth week, P<0.01) than controls.
Figure 2. Effects of BANCR on proliferation melanoma cells.
(A) Expression of BANCR was significantly silenced by transfecting sk-mel-5 cells with shRNA. Loss of BANCR expression significantly inhibited (B) proliferation of sk-mel-5 cells, tumor growth, including (C) tumor weight and (D) volume in nude mice. Loss of BANCR expression significantly inhibited proliferation of UACC903 (E) and CHL-1 (F) cells, (**P<0.01, Figure is representative of 3 experiments with similar results.)
Furthermore, we performed the same proliferation assays in UACC903 and CHL-1 cells which also exhibit relatively high expression of BANCR to explore whether the above effect on proliferation seen in sk-mel-5 cells was cell line specific. As shown in Figure 2E and 2F, the inhibitory effects of sh-BANCR transfection on cell proliferation were observed as well. Taken together, it indicated that BANCR regulated cell proliferation, and BANCR depletion impaired proliferation of melanoma cells with high expression of BANCR.
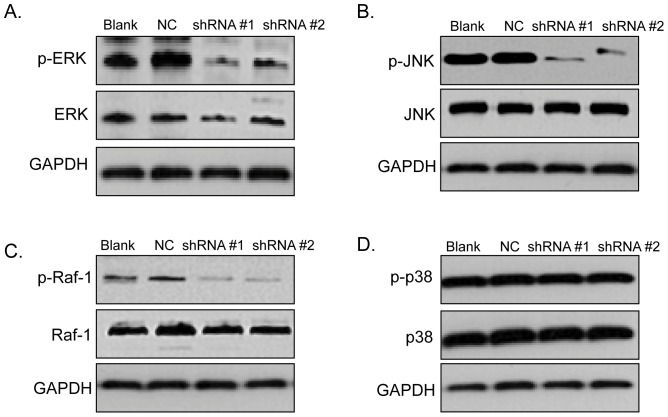
Knockdown of BANCR inactivated ERK1/2, Raf-1 and JNK, but had no effect on p38 MAPK
We tested the activation of MAPK pathway to explore the potential underlying mechanisms of BANCR-related low cell growth by western blot. The results showed that ERK1/2 and JNK were inactivated in shRNA transfectant cells compared with NC shRNA transfectant cells (Figure 3A and B). However, no differences of p38 MAPK were observed (Figure 3D). To determine the role of BANCR in the Raf-ERK pathway, we further detected expression of Raf-1, the upstream ERK1/2 activator. And Raf-1 protein levels decreased as well after BANCR was downregulated (Figure 3C).
Figure 3. The activities of ERK1/2, Raf-1 and JNK in BANCR silencing sk-mel-5 cells were significantly repressed.
Expressions of ERK1/1, Raf-1, p38 and JNK in BANCR silencing sk-mel-5 cells were detected by western blot. Loss of BANCR induced the inactivation of (A) ERK1/2, (B) JNK and (C) the upstream molecule of ERK1/2, Raf-1. (D) However, no activation was observed in p38 MAPK. GAPDH expression was used to normalize for equal loading. Figure is representative of 3 experiments with similar results.
BANCR regulated melanoma proliferation synergistically with ERK1/2 and JNK inactivation
We treated shRNA transfectant cells with specific pharmacological inhibitors U0126 and SP600125 to investigate the effects of ERK1/2 and JNK on BANCR-related proliferation. Western blot was performed to detect total and phosphorylation changes of ERK1/2 and JNK after cells were treated with U0126 and SP600125. As shown in Figure 4, the indicated inhibitors induced significant inactivation of ERK1/2 (Figure 4A) and JNK (Figure 4C). Consistently, cell proliferation decreased significantly (Figure 4B and 4D) after 72 h. To further investigate the interaction of BANCR and ERK1/2 or JNK, we performed rescue experiment by treating BANCR-plasmid transfected cells with U0126 and SP600125. When BANCR was upregulated, ERK1/2 and JNK pathways were both activated. And more importantly, when ERK1/2 and JNK were inactivated by the indicated inhibitors, overexpression of BANCR rescued the inactivation (Figure 4E and 4G). Proliferation assays showed that the inhibitory proliferation induced by ERK1/2 and JNK inactivation could also be ameliorated by BANCR (Figure 4F and 4H).
Figure 4. Inactivation of ERK1/2 and JNK participated in BANCR-regulated proliferation.
Cells transfected with sh-BANCR or BANCR plasmid were treated with U0126 or SP600125 respectively. Western blot was performed to detect total and phosphorylation changes of ERK1/2 and JNK. U0126 or SP600125 induced significant inactivation of ERK1/2 (A) and JNK (C). Cell proliferation decreased significantly after 72 h (B, D). Note the notable effects induced by combined treatment with shRNA and inhibitors. BANCR activated ERK1/2 (E) and JNK (G) pathways. Inactivation of ERK1/2 and JNK were rescued by overexpression of BANCR. Inhibitory proliferation induced by ERK1/2 and JNK inactivation was ameliorated by BANCR (F, H). Figure is representative of 3 experiments with similar results.
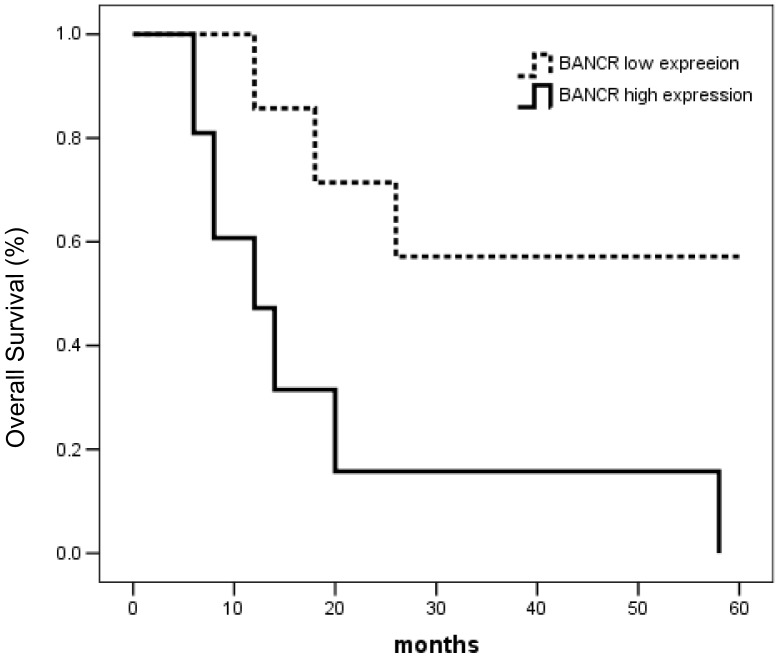
Expression of BANCR was associated with the poor prognosis of malignant melanoma patients
To investigate the prognostic value of BANCR, the association of BANCR with an overall survival was evaluated using Kaplan-Meier survival curves with the log-rank test. Seventy-two patients were enrolled for this analysis. The follow-up time ranged from 1 to 60 months. The median survival time of the group with higher expression of BANCR was 13.055 months, and the cumulative 1-, 3- and 5-year survival rates were 61%, 40% and 29%, respectively. The median survival time of the lower expression group was 55.021 months, and the 1-, 3- and 5-year survival rates were 86%, 71% and 57%, respectively. The difference between the groups was significant (P<0.01). The univariate survival analysis indicated that the survival rates of patients with higher expression of BANCR was lower than that of patients with lower expression (Figure 5, χ2 = 12.826, P = 0.000).
Figure 5. Kaplan-Meier survival curves of malignant melanoma patients relating to the status of BANCR expression.
Discussion
It has been demonstrated by cDNA cloning studies [13] and genomic microarray analysis [14] that more than 90% of the human genome transcriptional products do not code for proteins [15], which are referred to as non-protein coding RNAs (ncRNAs). Among these, lncRNAs, a newly discovered class of noncoding genes, have gained increasing attention because of their crucial roles in gene regulatory processes, such as drug resistance [16], cellular metabolism [17] and apoptosis [18], especially in skin disease and cancer development [19], [20]. Although many new functions have been ascribed to lncRNAs, their functional roles still remain unclear enough. In particular, the involvement of lncRNAs in malignant melanoma tumorigenesis and progression is not fully studied.
In previous study, Flockhart RJ et al. [11] and McCarthy N [12] indicated the potential regulation of BANCR in melanoma cell migration. In the current study, we confirmed the contribution of BANCR in the proliferation of melanoma cells and the potential mechanisms. Our results showed that BANCR was highly expressed in human malignant melanoma cell lines and tissues, and increased with tumor stages. BANCR could repress proliferation of tumor cells both in vitro and in vivo. Moreover, this inhibitory effect was related with inactivation of MAPK pathway, especially the ERK1/2 and JNK component. Thus BANCR and MAPK pathway can regulate cell proliferation synergistically. Taken together, our finding provided a novel interpretation for the mechanism of BANCR-regulated proliferation in malignant melanoma.
To date, published studies about BANCR are rare and limited. Being overexpressed in melanoma, the oncogenic BRAF-induced BANCR can regulate a set of genes involved in cell migration and is required for full migratory capacity of melanoma cells [20]. However, the underlying mechanism remains obscure. We for the first time demonstrated that BANCR participated in cell proliferation in MAPK pathway-dependent way in detail. The MAPK cascades are key signaling pathways involved in the regulation of cell proliferation, survival and differentiation. Aberrant regulation of MAPK cascades contribute to cancer including malignant melanoma and many other human diseases [21], [22]. The terminal MAPKs are the ERK1/2, JNK, p38 kinases and ERK5. The Raf-ERK1/2 pathway is widely expressed and there has been substantial evidence validating the importance of Raf and ERK in cancer growth and progression [23]. Blocking the pathway by selective inhibitor PD98059 or U0126, inhibits tumor growth in melanoma-bearing mice [24] and induces cell death and abrogates invasive growth of melanoma cells [25]. The JNK pathway is involved in regulating an array of cellular processes, including cell proliferation, migration, and survival, and are thus indispensable for epidermal cancers [26], [27]. Deregulation of the JNK/AP-1 proteins promotes melanoma tumorigenesis [28], and JNK activation is necessary for mitochondrial membrane potential change and apoptosis induced by doxycycline in melanoma cell [29]. Our present results showed that ERK1/2 and JNK were inactivated when BANCR was silenced and vice versa. This inactivation was required for BANCR-regulated proliferation and could be rescued by BANCR upregulation. However, p38 MAPK, which suppresses tumor growth by negatively regulating cell survival and proliferation [30], did not participate in the process.
The RAS/RAF/MAPK pathway is hyperactive in about 30% of human cancers, and activating mutations in key members of this pathway serve as driver mutations in many malignancies [31]. Among which, mutationally activated BRAF has been identified in a variety of cancers recently [32] and it occurs in a non-overlapping occurrence in many cancers such as melanomas, colorectal carcinomas, papillary thyroid carcinomas, serous ovarian carcinomas and lung cancers [21], [33]. Moreover, 90% of melanomas containing the activating mutations in BRAF produce active mutant BRAFV600E protein. BANCR was the mutant BRAFV600E-activated long non-coding RNA identified from samples from BRAF-mutant human melanomas. In our study, high expression of BANCR was correlated significantly to shortened survival of patients with malignant melanoma, suggesting that BANCR may be a predictor of poor clinical outcome.
It has been previously shown that ERK1/2 or JNK pathway is a potential target for therapy of cancer [34], [35]. However, the interaction between such pathway inhibitors and cancer is complex. Flores LG 2nd [36] found that therapy with U0126 produced only a transient inhibition of tumor glycolytic activity but did not significantly affect the rate of tumor growth in mice nephroblastoma. Our data showed that BANCR-regulated proliferation was not only ERK1/2-dependent but also JNK-dependent. And the inhibitory effect of BANCR silencing was more remarkable when introduced in combination with ERK1/2 and JNK inactivation induced by pharmacological inhibitors U0126 or SP600125. Moreover, we found BANCR is associated with poor prognosis of patients with malignant melanoma. Taken together, BANCR may be both a new potential target and prognostic factor of malignant melanoma. Further investigation will be required to elucidate this question.
Funding Statement
The authors have no support or funding to report.
References
- 1.MacKie RM, Hauschild A, Eggermont AM (2009) Epidemiology of invasive cutaneous melanoma. Ann Oncol (suppl. 6):vi1–7. [DOI] [PMC free article] [PubMed]
- 2. Miller AJ, Mihm MC Jr (2006) Melanoma. N Engl J Med 355(1): 51–65. [DOI] [PubMed] [Google Scholar]
- 3. Margue C, Philippidou D, Reinsbach SE, Schmitt M, Behrmann I, et al. (2013) New Target Genes of MITF-Induced microRNA-211 Contribute to Melanoma Cell Invasion. PLoS One 8(9): e73473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee DJ, Kang DH, Choi M, Choi YJ, Lee JY, et al. (2013) Peroxiredoxin-2 represses melanoma metastasis by increasing E-Cadherin/β-Catenin complexes in adherens junctions. Cancer Res 73(15): 4744–57. [DOI] [PubMed] [Google Scholar]
- 5. Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, et al. (2011) lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477: 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cheetham SW, Gruhl F, Mattick JS, Dinger ME (2013) Long noncoding RNAs and the genetics of cancer. Br J Cancer 108(12): 2419–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wapinski O, Chang HY (2011) Long noncoding RNAs and human disease. Trends Cell Biol 21: 354–361. [DOI] [PubMed] [Google Scholar]
- 8. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, et al. (2010) Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464(7291): 1071–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang L, Lin C, Jin C, Yang JC, Tanasa B, et al. (2013) lncRNA-dependent mechanisms of androgen- receptor-regulated gene activation programs. Nature 500(7464): 598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang X, Song JH, Cheng Y, Wu W, Bhagat T, et al. (2013) Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut 9(2): 305266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Flockhart RJ, Webster DE, Qu K, Mascarenhas N, Kovalski J, et al. (2012) BRAFV600E remodels the melanocyte transcriptome and induces BANCR to regulate melanoma cell migration. Genome Res 22(6): 1006–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McCarthy N (2012) Epigenetics. Going places with BANCR. Nat Rev Cancer 12(7): 451. [DOI] [PubMed] [Google Scholar]
- 13. Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, et al. (2005) The transcriptional landscape of the mammalian genome. Science 309(5740): 1559–63. [DOI] [PubMed] [Google Scholar]
- 14. Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, et al. (2005) Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308(5725): 1149–54. [DOI] [PubMed] [Google Scholar]
- 15. Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, et al. (2011) Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol 18(5): 1243–50. [DOI] [PubMed] [Google Scholar]
- 16.Jiang M, Huang O, Xie Z, Wu S, Zhang X, et al. (2013) A novel long non-coding RNA-ARA: Adriamycin Resistance Associated. Biochem Pharmacol pii: S0006-2952(13)00687-4. [DOI] [PubMed]
- 17.Ellis BC, Graham LD, Molloy PL (2013) CRNDE, a long non-coding RNA responsive to insulin/IGF signaling, regulates genes involved in central metabolism. Biochim Biophys Acta pii: S0167-4889(13)00361-3. [DOI] [PubMed]
- 18. Wu W, Zhang S, Li X, Xue M, Cao S, et al. (2013) Ets-2 regulates cell apoptosis via the Akt pathway, through the regulation of urothelial cancer associated 1, a long non-coding RNA, in bladder cancer cells. PLoS One 8(9): e73920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hombach S, Kretz M (2013) The non-coding skin: Exploring the role of long non-coding RNAs in epidermal homeostasis and disease. Bioessays 35(12): 1093–100. [DOI] [PubMed] [Google Scholar]
- 20. Nagano T, Fraser P (2011) No-nonsense functions for long noncoding RNAs. Cell 145(2): 178–181. [DOI] [PubMed] [Google Scholar]
- 21. Roberts PJ, Der CJ (2007) Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene 26(22): 3291–310. [DOI] [PubMed] [Google Scholar]
- 22. Solus JF, Kraft S (2013) Ras, Raf, and MAP kinase in melanoma. Adv Anat Pathol 20(4): 217–26. [DOI] [PubMed] [Google Scholar]
- 23. Shields JM, Pruitt K, McFall A, Shaub A, Der CJ (2000) Understanding Ras: 'it ain't over ‘til it's over’. Trends Cell Biol 10(4): 147–54. [DOI] [PubMed] [Google Scholar]
- 24. Basu S, Harfouche R, Soni S, Chimote G, Mashelkar RA, et al. (2009) Nanoparticle-mediated targeting of MAPK signaling predisposes tumor to chemotherapy. Proc Natl Acad Sci U S A 106(19): 7957–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lasithiotakis KG, Sinnberg TW, Schittek B, Flaherty KT, Kulms D, et al. Combined inhibition of MAPK and mTOR signaling inhibits growth, induces cell death, and abrogates invasive growth of melanoma cells. J Invest Dermatol 128(8): 2013–23. [DOI] [PubMed] [Google Scholar]
- 26. Ke H, Harris R, Coloff JL, Jin JY, Leshin B, et al. (2010) The c-Jun NH2-terminal kinase 2 plays a dominant role in human epidermal neoplasia. Cancer Res 70(8): 3080–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jin JY, Ke H, Hall RP, Zhang JY (2011) c-Jun promotes whereas JunB inhibits epidermal neoplasia. J Invest Dermatol 131(5): 1149–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ke H, Augustine CK, Gandham VD, Jin JY, Tyler DS, et al. (2013) CYLD inhibits melanoma growth and progression through suppression of the JNK/AP-1 and β1-integrin signaling pathways. J Invest Dermatol 133(1): 221–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shieh JM, Huang TF, Hung CF, Chou KH, Tsai YJ, et al. (2010) Activation of c-Jun N-terminal kinase is essential for mitochondrial membrane potential change and apoptosis induced by doxycycline in melanoma cells. Br J Pharmacol 160(5): 1171–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Han J, Sun P (2007) The pathways to tumor suppression via route p38. Trends Biochem Sci 32(8): 364–71. [DOI] [PubMed] [Google Scholar]
- 31. Dhomen N, Reis-Filho JS, da Rocha Dias S, Hayward R, Savage K, et al. (2009) Oncogenic Braf induces melanocyte senescence and melanoma in mice. Cancer Cell 15(4): 294–303. [DOI] [PubMed] [Google Scholar]
- 32. Davies H, Bignell GR, Cox C, Stephens P, Edkins S, et al. (2002) Mutations of the BRAF gene in human cancer. Nature 417(6892): 949–54. [DOI] [PubMed] [Google Scholar]
- 33. Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, et al. (2002) Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature 418(6901): 934. [DOI] [PubMed] [Google Scholar]
- 34. Wang W, Wang X, Peng L, Deng Q, Liang Y, et al. (2010) CD24-dependent MAPK pathway activation is required for colorectal cancer cell proliferation. Cancer Sci 101(1): 112–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marampon F, Bossi G, Ciccarelli C, Di Rocco A, Sacchi A, et al. (2009) MEK/ERK inhibitor U0126 affects in vitro and in vivo growth of embryonal rhabdomyosarcoma. Mol Cancer Ther 8(3): 543–51. [DOI] [PubMed] [Google Scholar]
- 36. Flores LG 2nd, Yeh HH, Soghomonyan S, Young D, Bankson J, et al. (2013) Monitoring therapy with MEK inhibitor U0126 in a novel Wilms tumor model in Wt1 knockout Igf2 transgenic mice using 18F-FDG PET with dual-contrast enhanced CT and MRI: early metabolic response without inhibition of tumor growth. Mol Imaging Biol 15(2): 175–85. [DOI] [PMC free article] [PubMed] [Google Scholar]