Abstract
Microbe-associated molecular patterns (MAMPs) lead to the activation of the first line of plant defence. Few fungal molecules are universally qualified as MAMPs, and proteins belonging to the cerato-platanin protein (CPP) family seem to possess these features. Cerato-platanin (CP) is the name-giving protein of the CPP family and is produced by Ceratocystis platani, the causal agent of the canker stain disease of plane trees (Platanus spp.). On plane tree leaves, the biological activity of CP has been widely studied. Once applied on the leaf surface, CP acts as an elicitor of defence responses. The molecular mechanism by which CP elicits leaves is still unknown, and the protective effect of CP against virulent pathogens has not been clearly demonstrated. In the present study, we tried to address these questions in the model plant Arabidopsis thaliana. Our results suggest that stomata rapidly sense CP since they responded to the treatment with ROS signalling and stomatal closure, and that CP triggers salicylic acid (SA)- and ethylene (ET)-signalling pathways, but not the jasmonic acid (JA)-signalling pathway, as revealed by the expression pattern of 20 marker genes. Among these, EDS1, PAD4, NPR1, GRX480, WRKY70, ACS6, ERF1a/b, COI1, MYC2, PDF1.2a and the pathogenesis-related (PR) genes 1–5. CP rapidly induced MAPK phosphorylation and induced the biosynthesis of camalexin within 12 hours following treatment. The induction of localised resistance was shown by a reduced susceptibility of the leaves to the infection with Botrytis cinerea and Pseudomonas syringae pv. tomato. These results contribute to elucidate the key steps of the signalling process underlying the resistance induction in plants by CP and point out the central role played by the stomata in this process.
Introduction
Microbe- or pathogen-associated molecular patterns (MAMPs/PAMPs) are microbe-derived molecules with a key role in the plant immune system: their perception by pattern recognition receptors (PRRs) leads to the activation of PAMP-triggered immunity (PTI), which is the first line of defence against pathogens [1].
In fungi, relatively few molecules are universally considered as MAMPs, such as chitin, with its variants like chitosan, ethylene-inducing xylanase, β-glucans, necrosis- and ethylene-inducing peptide 1 (Nep1)-like proteins and ergosterol [2]–[4]. Some of these molecules are not only produced by fungi, such as β-glucans and Nep1-lke proteins, which can be found in oomycetes and bacteria [5], [6]. Recently, proteins belonging to a new fungal protein family, the “cerato-platanin family”, have provided more and more experimental evidence of their MAMP activity.
Cerato-platanin proteins (CPPs) are produced by plant pathogenic and non-pathogenic fungi, both ascomycetes and basidiomycetes [7]. Concerning the primary role of these proteins, recent results suggest that they are mono-domain expansin-like proteins localised in the cell wall and involved in the hyphal growth and development [8]–[11]. Moreover, a role in the fungus-plant interaction has also been reported [12]. When CPPs are applied on host and non-host plants, they induce defence-related responses and resistance against pathogens [13]–[16].
Cerato-platanin (CP) is the name-giving protein of this family and is produced by Ceratocystis platani, the causal agent of the canker stain disease of plane trees (Platanus spp.) [17]. After surface treatment of plane tree leaves, CP induces defence-related responses and programmed cell death [18], [19]. In addition, CP has been reported to induce the production of phenolic compounds in various plants [17], [20], [21].
The mechanism by which CP elicits host and non-host plants after foliar treatment is still poorly understood, because the protein is unable to penetrate the leaf cuticle and a receptor has not been identified yet [22]. The data present in the literature about other CPPs do not help to unravel the question because these proteins are usually injected into the tissues by infiltration. Also the signalling pathways activated in the plants by CP or CPPs are unknown, although an involvement of the plant hormone salicylic acid has been recently reported in tobacco after treatment with BcSpl1 from Botrytis cinerea [13].
In the present study we aimed to clarify the questions concerning the resistance-inducing mechanism triggered by CP in the model plant Arabidopsis. In particular, we investigated the role played by the stomata, the activation of the signalling pathways and the biosynthesis of the phytoalexin camalexin following foliar treatment with CP. In addition, the ability of CP to induce localised resistance against Botrytis cinerea and Pseudomonas syringae pv. tomato was analysed.
Results
The production of H2O2 after treatment with CP spreads from stomata to the neighbouring cells
As the oxidative burst is one of the first responses occurring in plants after MAMP recognition [2], we assessed the ability of CP to elicit Arabidopsis leaves by analysing the production of hydrogen peroxide. H2O2 was first analysed in situ by treating the lower (abaxial) leaf surface according to a previous work performed on P. acerifolia [19]. H2O2 was visualised by the fluorescent probe 2,7-dichlorofluorescin diacetate (H2DCF-DA), a compound that easily enters the cells [19]. Fluorescence was already visible after 15 min of treatment (Fig. S1), and some stomata were more fluorescent than the background. After 1 h the fluorescence was already very intense, differently from what occurred when we treated the upper (adaxial) surface of the leaf, which developed a weak fluorescence at this treatment time (data not shown). Leaves remained fluorescent for 24 h. The production of H2O2 in situ was also analysed by treating leaves with serial dilutions of CP, for 15 and 60 min, and CP was able to induce H2O2 production even at a lower concentration (75 µM) (data not shown).
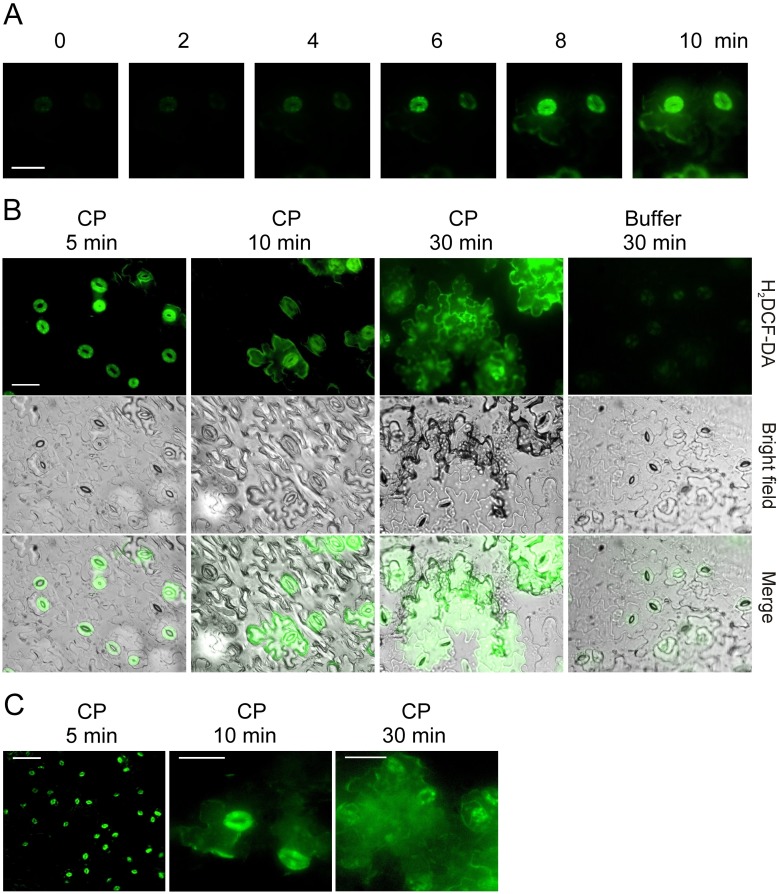
Subsequently, we investigated stomata as a signalling source of H2O2 and we used epidermal peels to analyse the evolution of the H2O2 production on the epidermis. Epidermal peels were first loaded with the fluorescent probe, washed, and then treated on the cuticle side with CP. This procedure allowed the probe to be present in all the cells before the treatment, so that the fluorescence evolution only depended on the H2O2 production over time. H2O2 production by single stomata over time and on the same peel is shown in Fig. 1A. Guard cells turned out to be the first cells on the epidermis to become fluorescent, starting to synthesise H2O2 after 4–6 min of treatment. Before that time, the fluorescence was only due to the oxidizing environment of the chloroplast [23]. After 8–10 min the epidermal cells surrounding the stomata also became fluorescent. Because of the impossibility of analysing a single peel under the microscope for a longer period, we decided to repeat the experiment with different epidermal peels. Peels were treated for 5, 10 and 30 min. As shown in Fig. 1B, once again the first fluorescence signals occurred in the stomata, while after 30 min the fluorescence had spread to most epidermal cells. In the same experiment, closed stomata did not produce H2O2 (Fig. S2A). In addition, no fluorescence was observed in a 2-h period when we treated epidermal peels obtained from plants with stomata still closed (Fig. S2B).
Figure 1. H2O2 production at the level of stomata.
Epidermal peels from Arabidopsis leaves were first loaded with the fluorescent probe H2DCF-DA and then treated either on the cuticle side (A and B) or on the underside (C) with 150 µM CP to analyse the origin of the ROS signalling. (A) Single guard cells photographed at 2-minute intervals after treatment with CP (same peel). Bar = 30 µm. (B) Photographs of representative regions of three different peels treated with CP for 5, 10 or 30 min respectively, or 30 min with MES buffer (control). Fluorescence microscopy (H2DCF-DA), light microscopy (bright field) and merged pictures (merge) are shown in order to facilitate the localisation of stomata. The bar is 40 µm and applies to all photographs. (C) Peels treated on the underside (the side devoid of cuticle) with CP. Bars = 100 µm (5 min) and 30 µm (10 and 30 min).
In order to exclude the possibility that the guard cells responded faster to CP because devoid of cuticle, we treated epidermal peels on the underside (where the cuticle is not present). Once again, the H2O2 production started in the stomata and only subsequently spread to the neighbouring epidermal cells (Fig. 1C).
CP induces stomatal closure
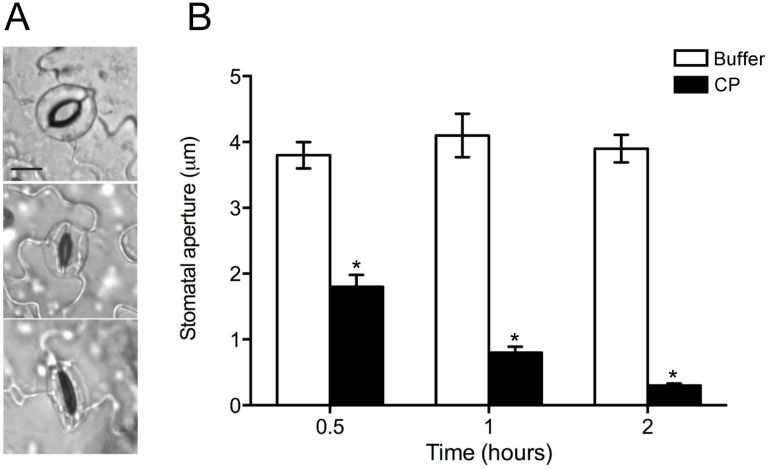
According to previous studies performed with elicitors different from CPPs [24], [25], we used epidermal peels, which allow better microscopic recording, to investigate the ability of CP to influence the stomatal movement (Fig. 2A). As shown in Fig. 2B, CP induced a progressive reduction in the average width of the stomatal aperture. The reduction was already significant after 30 min, and after 2 h the stomata were almost completely closed. The treatment with MES buffer, which was used as a control, did not influence the stomatal movement.
Figure 2. Stomatal closure induced by CP.
(A) Open and closed stomata in Arabidopsis epidermal peels. Bar = 10 µm. (B) Measurement of stomatal aperture after 30 min, 1 and 2 h of treatment with 150 µM CP or MES buffer (control). Results are the mean of 100 measurements ± SEM. Statistical analysis was performed by unpaired t-test (treated vs. control). Asterisks indicate statistically significant difference at P<0.05. The experiment was repeated with similar results.
Phosphorylation of MAP kinases
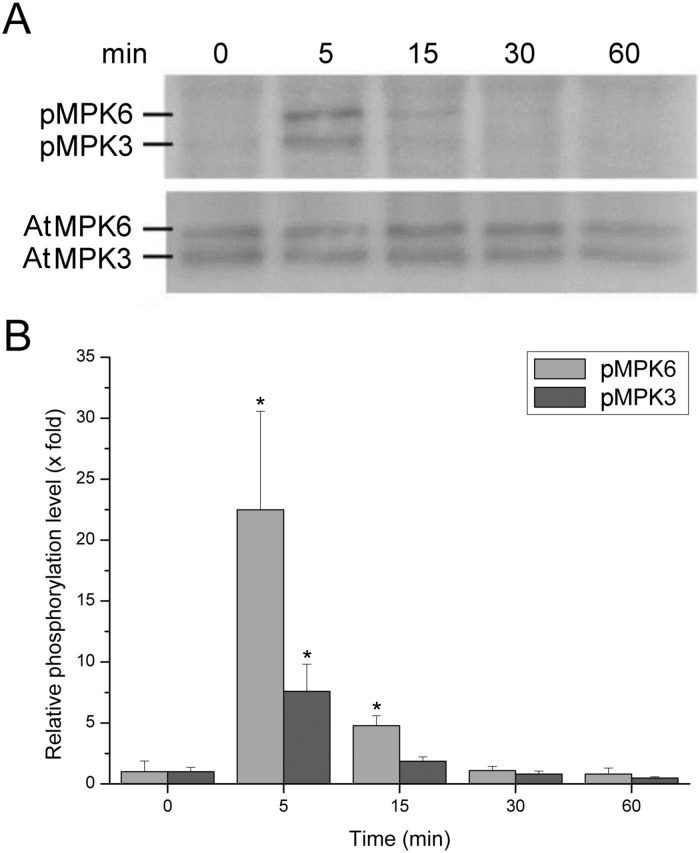
The activation of the MAPK cascade following treatment with CP was studied in Arabidopsis by analysing the phosphorylation of MAPK3 and MAPK6, according to the recent evidence that indicates a key role of the kinase induction in Arabidopsis defence responses [26]–[28]. For example, MPK3 and MPK6 play roles in camalexin induction, the major phytoalexin in Arabidopsis, and posses a critical role in priming Arabidopsis plants for full induction of defence responses during induced resistance [29], [30].
The analysis was carried out with specific phospho-p44/42 (pERK1/2) antibodies on protein extracts obtained after 0, 5, 15, 30 and 60 min of treatment with CP. As shown by immuno-blot analysis, both MPK3 and MPK6 turned out to be rapidly phosphorylated (Fig. 3A). After only 5 min of treatment MPK6 was phosphorylated up to 20-fold the control value (Fig. 3B). As expected, MAPKs were not phosphorylated when leaves were treated with sterile distilled water (data not shown).
Figure 3. Phosphorylation of Arabidopsis MPK6 and MPK3 after treatment with 150 µM CP.
(A) Analysis carried out with human phospho-p44/42 antibodies (pERK1/2) on protein extracts obtained after 0, 5, 15, 30 and 60 min of treatment. pMPK6 and pMPK3 indicate phosphorylation. (B) Image analysis showing the phosphorylation level normalized to the respective protein amount. Error bars indicate SD of three biological replicates. Statistical analysis was performed by unpaired t-test by comparing the phosphorylation level to the appropriate background level (0 h). Asterisks indicate statistically significant difference at P≤0.05.
CP up-regulates SA- and ET-signalling genes, but not JA-signalling genes
As a first approach, we used a primer library purchased from Sigma-Aldrich containing gene-specific primers for pathogen-inducible genes. The study was performed after 6 h of treatment, which was the time in which the greatest number of genes was expected to be modulated (preliminary data not shown). By this analysis, we identified 56 up-regulated genes out of 67 genes analysed (Table S1). It is worth mentioning genes involved at different levels with the oxidative stress, such as glutathione S-transferases (GSTF3, GSTF7), peroxidase 50, late embryogenesis abundant-like protein 5 (LEA5), blue copper protein (BCB) and respiratory burst oxidase homologue D (RBOHD) [31]–[34]; or genes codifying for various receptor kinases, suchs as cysteine-rich receptor-like protein kinases (CRK5, CRK7, CRK10, CRK36), lectin-receptor kinase, receptor-like protein kinase 1 (RLK1), wall-associated receptor kinase 1 (WAK1) and somatic embryogenesis receptor kinase 4 (SERK4) [35]. Many other up-regulated genes could instead be related to defence-signalling pathways, mainly SA and ET. As the signalling pathways activated by CPPs in Arabidopsis are unknown, we focused our attention on the expression of SA-, ET- and JA-dependent genes.
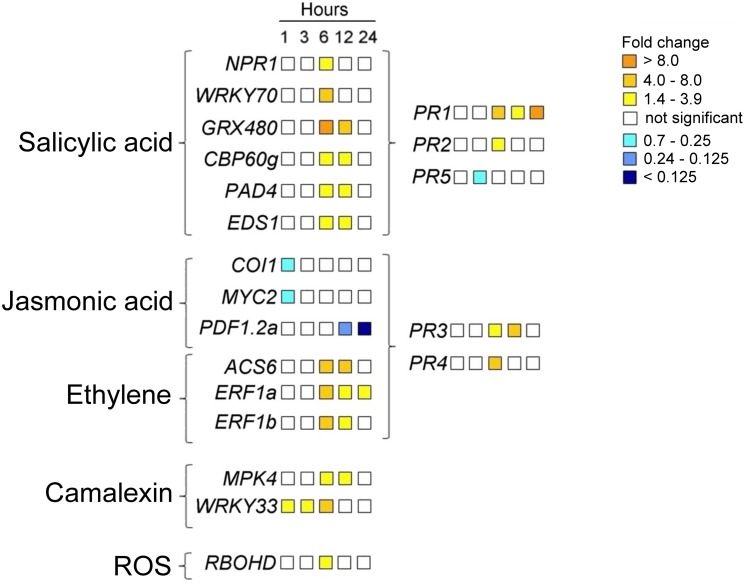
In order to perform a detailed study, we designed 13 more primer pairs to include, in the analysis, some of the most studied genes that we found in the literature. The study was performed at various treatment times (1, 3, 6, 12 and 24 h) on a total of 20 genes (7 were from the Sigma-Aldrich primer library). Among these, we analysed the expression of MAPK4 and the transcription factor WRKY33, which act upstream of the synthesis of camalexin [36], and RBOHD (respiratory burst oxidase homologue D), which is implicated in the production of reactive oxygen species (ROS) [33]. The expression pattern of the analysed genes is shown in Fig. 4. CP induced the expression of SA- and ET-dependent genes starting from 6 h after the treatment, and this was also the treatment time in which most genes were found modulated following application of CP. This did not occur for JA-dependent genes, which were instead down-regulated by CP. COI1 (coronatine insensitive 1) and MYC2 (jasmonate insensitive 1, JIN1), two key regulators of JA signalling [37], were down-regulated after only 1 h of treatment, while PDF1.2a (plant defensin) was strongly down-regulated after 12 and 24 h.
Figure 4. Expression analysis of genes related to defence-signalling pathways, camalexin synthesis and ROS production.
Significant variations (P≤0.05) in the level of expression are reported in the figure with a colour scale (up-regulated genes = yellow; down-regulated genes = blue). Arabidopsis leaves were treated on the lower surface with 150 µM CP (treated sample) or sterile distilled water (calibrator sample) for 1, 3, 6, 12 or 24 h. Actin-2 was used as the endogenous reference gene. Relative gene expression values (2−ΔΔCt or fold change values) from three biological replicates and two technical replicates are shown. Statistical analysis was performed by unpaired t-test (treated sample vs. calibrator sample per each time point). The qPCR results and the locus ID of each gene can be seen in Table S1.
The expression of pathogenesis-related (PR) genes was analysed taking into account their differential expression in SA- and JA-/ET-signalling pathways [38]. As shown in Fig. 4, PR1, PR2 (β-1, 3-glucanase), PR3 (basic chitinase) and PR4 (hevein-like protein) turned out to be all up-regulated by CP, starting from 6 h after the treatment. However, their up-regulated status was differently maintained. In particular, PR1 was up-regulated until 24 h. PR5 (thaumatin-like) was instead down-regulated.
WRKY33 turned out to be the earliest up-regulated gene (1 h) by CP and its status was maintained until 6 h; MAPK4 was up-regulated after 6 h and its status was maintained until 12 h. Finally, RBOHD was up-regulated after 6 h of treatment.
CP induces the biosynthesis of camalexin
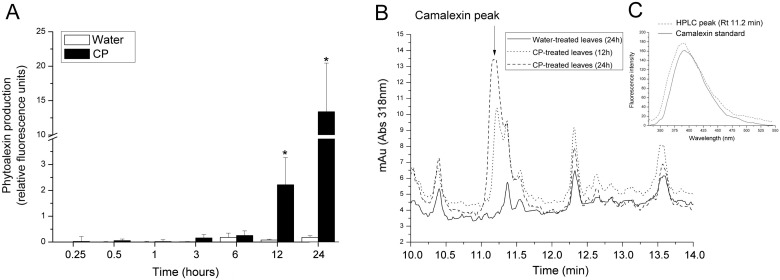
Phytoalexins are phenolic compounds whose production after treatment with CP has been reported in different plants as a release of fluorescence substances [17], [20], [21]. Nevertheless, the biochemical nature of these substances has never been investigated in detail. In the present study, the synthesis of phytoalexins was first determined in Arabidopsis leaves by measuring the fluorescence of the droplets recovered from the leaves. As shown in Fig. 5A, the fluorescence in the droplets significantly increased starting from 12 h following treatment and became about six times higher after 24 h. In order to determine whether the fluorescence was due to camalexin biosynthesis, the emission peak of a camalexin standard was compared to the peak obtained from the recovered droplets. As shown in Fig. S3, the two spectra almost overlapped, thus suggesting that the fluorescence emitted after treatment with CP was due to camalexin production. To confirm this observation and to investigate the presence of camalexin in the tissues, treated leaves were extracted with methanol and analysed by reverse-phase chromatography (Fig. 5B). The analysis revealed the presence of a main peak (with a retention time of 11.2 min) that matches the retention time of a camalexin standard analysed with the same method (Fig. S4). To further confirm the result, the eluted peak at 11.2 min was recovered and its fluorescence emission compared to the standard (Fig. 5C). The overall results indicate that camalexin was produced by Arabidopsis leaves following treatment with CP.
Figure 5. Phytoalexin production by Arabidopsis leaves treated with CP.
(A) Leaves treated on the lower surface with 150 µM CP or sterile distilled water (control). The drops were collected after 15 and 30 min, 1, 3, 6, 9, 12 or 24 h of treatment and the phytoalexin release measured by fluorescence analysis (λex = 320 nm, λem = 386 nm). The fluorescence value was normalized to the number of droplets analysed and expressed as relative fluorescence units. Error bars indicate SD of three biological replicates. Statistical analysis was performed by unpaired t-test (treated vs. control). Asterisks indicate statistically significant difference at P<0.05. (B) Reverse Phase-High Performance Liquid Chromatography (RP-HPLC) of camalexin extracted from the leaves treated for 12 and 24 h. The retention time (Rt) of the peak indicated by the arrow is the same as the camalexin standard, as experimentally determined. (C) Fluorescence emission spectrum of the peaks eluted at 11.2 min compared to a camalexin standard.
As these results were obtained by treating the lower leaf surface, we compared the two leaf surfaces in their ability to release phytoalexins after treatment with CP. After 48 h of treatment, the fluorescence emitted by the droplets recovered from the lower surface was about double than that emitted by the droplets recovered from the upper surface (Fig. S5).
CP protects Arabidopsis leaves from Botrytis cinerea and Pseudomonas syringae pv. tomato
In order to assess the effectiveness of CP in protecting the plant from pathogens, Arabidopsis leaves were treated on the lower surface with 10-µl drops of CP, sterile distilled water (negative control) or 0.1% chitosan (positive control). After 24 h of treatment, the drops were recovered and the leaves were infected on the same foliar surface with virulent pathogens.
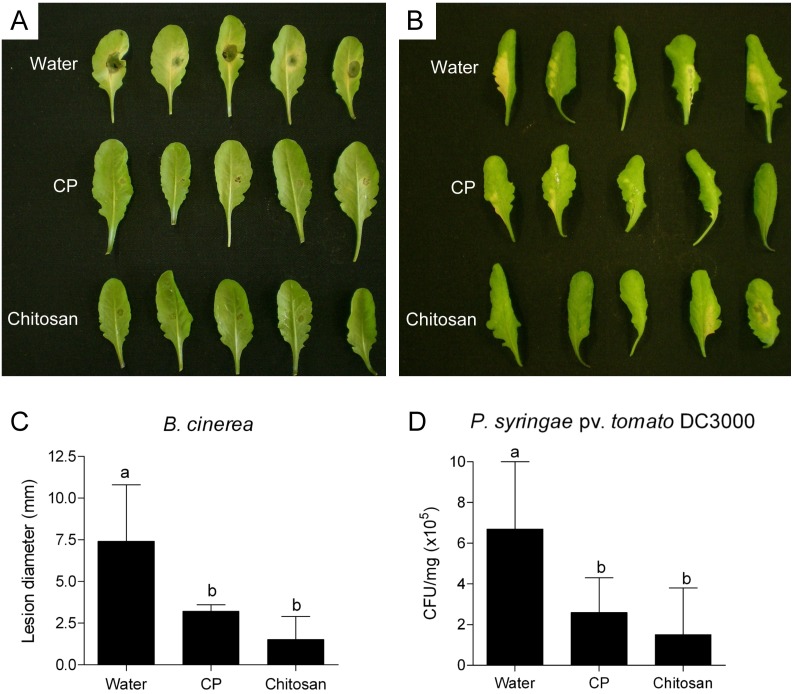
B. cinerea strain PM10 was inoculated by placing a single 10-µl drop of a suspension 2×105 conidia ml−1 on a side of the middle vein. After 3 days of incubation at 22°C, CP-treated leaves showed a consistent reduction of the disease symptoms (Fig. 6A, C). The lesion size in CP-treated leaves was statistically similar to what occurred after the treatment with chitosan. The lesions remained basically limited to the point of application of the conidial suspension, although after the treatment with chitosan some leaves showed a complete absence of lesions.
Figure 6. Resistance induction assays against Botrytis cinerea and Pseudomonas syringae pv. tomato (Pst).
Arabidopsis detached leaves were treated on the lower surface with 150 µM CP, sterile distilled water (control) or 0.1% chitosan for 24 h before pathogen inoculation. (A) B. cinerea strain PM10 was inoculated by placing a single 10-µl drop of a suspension 2×105 conidia ml−1 in 1% Sabouraud Maltose Broth on a side of the middle vein. Infected leaves were incubated for 3 days at 22°C before taking photos and measuring the lesion size. (B) Pst strain DC3000 was inoculated by placing a single 10-µl drop of a suspension 108 colony-forming units (CFU) ml−1 (OD600 = 0.2) in sterile distilled water containing 0.02% Silwet L-77 on a side of the middle vein. Infected leaves were incubated for 3 days at 28°C before taking photos and determining bacterial titer. (C) Measurement of the lesion diameter (mm) ± SD after 3 days of incubation with B. cinerea (n = 8). (D) Counting of CFU mg−1±SD after 3 days of incubation with Pst DC3000 (n = 8). Statistically significant differences among treatments are indicated at P≤0.05.
P. syringae pv. tomato (Pst) strain DC3000 was similarly inoculated by placing on the leaf surface a single 10-µl drop of a suspension 108 colony-forming units (CFU) ml−1. After 3 days of incubation at 28°C, the leaves were photographed and the bacterial titer was determined. As shown in Fig. 6B, the treatments with CP and chitosan reduced both symptom development and bacterial growth (Fig. 6D) compared to water-treated leaves. Chitosan appeared more effective than CP in limiting the growth of Pst DC3000, but the difference compared to CP was again not significant.
No direct inhibiting effect of CP against the germination of B. cinerea conidia or the growth of Pst DC3000 was found when the pathogens were grown in vitro in the presence of 150 µM CP (data not shown).
Discussion
ROS are central players in the complex signalling network of cells and it is well known that MAMPs cause an oxidative burst upon their recognition by the plant [2]. An initial burst of ROS production can trigger a cascade of cell-to-cell communication events that propagate the signal over long distances like a wave [39]. In the present study, we analysed the production of H2O2 after treatment with CP at the level of stomata. We treated Arabidopsis leaves on the external epidermal surface for two reasons: the external application mimics the first natural contact between a MAMP and the plant tissue, and above all it can be representative of a possible use of CP in plant protection.
Hypothesizing a role for stomata in the perception mechanism of CP, we assumed that H2O2 had to be produced first by the stomata. Accordingly, we designed the experiment to monitor the H2O2 evolution on the epidermis and we started the analysis almost instantaneously (2 min). To our knowledge, there are no other data in the literature that show the initial production of H2O2 at the level of stomata and the subsequent spreading on the epidermis after surface treatment with microbial-derived elicitors. Among the CPPs, the H2O2 production in Arabidopsis leaves has been reported for MgSM1 and BcSpl1 [16], [40]. However, MgSM1 had been assessed 24 h after the ectopic expression in transgenic plants, whereas BcSpl1 had been assessed 4 h after the infiltration.
Our results suggest that CP elicited defence responses by entering through the stomata, and perhaps its perception occurred at the level of the inner surface of the guard cells. This action model was suggested by some clear observations: the H2O2 production after the treatment with CP was initiated by guard cells (4–6 min) and only subsequently spread from the stomata to the neighbouring epidermal cells (not vice versa); this occurred even when the peels were treated on the underside, which is devoid of cuticle; the treatment of the lower leaf surface, which normally presents a higher stomatal density [41], [42], resulted in a higher production of H2O2 and phytoalexins; the treatment performed with closed stomata did not result in H2O2 production. Accordingly, we can deduce that (a) stomata have a key role in the Arabidopsis immunity triggered by CP; (b) the guard cells probably possess a receptor for this MAMP. This assumption is in accordance with the role in MAMP sensing attributed to guard cells by Melotto et al. [25], who also hypothesized the presence in these cells of several receptors for multiple MAMPs, and with the actual presence in these cells of the flagellin receptor [43]. At the same time our results do not conflict with the presence of a receptor also in the plasma membrane of epidermal or mesophyll cells, as recently suggested by Frías et al. [44] for the CPP BcSpl1. However, at the level of epidermal cells our results strongly suggest that the stomata are the most responsive to CP. In fact, even when the peels were treated on the underside, i.e. the side devoid of cuticle, the H2O2 production showed the same evolution, namely from guard cells to neighbouring cells, and thus the presence of cuticle cannot be considered the only determinant of the obtained results.
Stomata represent a natural entry site for potentially harmful microbes, and the stomatal closure in response to MAMPs is considered as an innate immune response active at the pre-invasive level [25], [45]. CP induced a progressive reduction of the stomatal aperture similarly to flagellin-22 (flg22) and lipopolysaccharides [25]. The generation of H2O2 is crucial to initiate this process and MAMPs rely on RBOHD as the primary NADPH oxidase for ROS production [45], [46]. As found by qPCR, RBOHD was effectively up-regulated by CP but the timing (6 h) could not explain the early oxidative burst observed. Therefore, we can assume that the rapid generation of H2O2 was caused by this enzyme in the form already present in the cells.
The activation of MAPK cascades is an early response to MAMP signals [2]. CP rapidly induced the phosphorylation of MAPK3 and MAPK6. This result was expected from previous results obtained in P. acerifolia leaves [19] and confirms that the activation of the kinase cascade is one of the first events in plant defence signalling.
In order to establish the signalling events triggered by CP, we performed an extensive gene expression analysis never performed before for CPPs. We analysed 17 genes related to SA-, JA- and ET-signalling pathways, which are basically new in relation to the CPPs.
We found that the SA-dependent and the ET-dependent genes, whose roles and reciprocal interactions in the signalling pathways have been reported in the literature [47]–[49], showed the same activation time (6 h). However, the intensity and the duration of the overexpression varied depending on the gene. On the contrary, the down-regulation of the JA-dependent genes occurred almost instantaneously (1 h), as revealed by COI1, the JA receptor [50], and MYC2, a transcription factor involved in the transcriptional regulation of some JA-responsive genes [51]. The lack of activation of the JA signalling could also be confirmed by the subsequent down-regulation of the plant defensin PDF1.2a [52]. Confirming the well-known antagonism between SA and JA signalling [53], our result leads us to predict a role for SA and ET, rather than JA, in CP-induced defence signalling in Arabidopsis. In tobacco, Frías et al. [13] have recently reported a role for SA after treatment with the CPP BcSpl1. However, this is the first time that a role for ET signalling has been reported.
MAPK4 and WRKY33 were analysed for their involvement in the biosynthesis of camalexin, the major phytoalexin in Arabidopsis [36], [54]. Phytoalexins are secondary metabolites that show biological activity against a variety of pathogens and are considered molecular markers of disease resistance [54]. Interestingly, the resistance induced by elicitors in Arabidopsis to B. cinerea is independent of SA, ET, or JA signalling, whereas the biosynthesis of camalexin and other secondary metabolites seems to have a prominent role [55], [56]. CP up-regulated MAPK4 and WRKY33, and induced the biosynthesis of fluorescent phenolic compounds identified as camalexin. The camalexin level was increased both inside the leaves treated with CP and in the droplets recovered from the leaf surface, starting from 12 h after treatment.
The present study was concluded by demonstrating the ability of CP to induce resistance against pathogens. To date the effectiveness of CP against pathogens had only been demonstrated against C. platani [57]. In that report, plane tree leaves treated with CP hampered C. platani germination and growth when inoculated with conidia. However C. platani is not a foliar pathogen. In this study we demonstrated that CP effectively enhances resistance against virulent foliar pathogens on the non-host plant A. thaliana. CP induced disease resistance to the necrotrophic fungus B. cinerea and the hemibiotrophic bacterium P. syringae pv. tomato.
In conclusion, with the results obtained in the present study we can assume that CP induces resistance in Arabidopsis leaves through stomata, which very rapidly sense CP and respond to the treatment with ROS signalling and stomatal closure. The signalling pathways triggered by CP certainly involve both SA and ET, as revealed by the expression pattern of marker genes, while JA does not seem to be involved. Biotrophic and hemibiotrophic pathogens are generally sensitive to defence responses that are regulated by SA, whereas pathogens with a necrotrophic lifestyle are commonly deterred by defences that are controlled by JA and ET [37], [47]. Accordingly, P. syringae pv. tomato was likely hampered by the activation of the SA-signalling pathway, whereas the activation of the ET-signalling pathway combined with the camalexin production were probably crucial for the outcome of the protection against B. cinerea. The results shown in the present study not only contribute to elucidate the key steps of the signalling process underlying the resistance induction in plants by CP but also point out the crucial role played by the stomata in the early signalling of microbial-derived molecules on the epidermis.
Materials and Methods
Production and purification of cerato-platanin
Cerato-platanin (CP) was produced by heterologous expression in the yeast Pichia pastoris [58], purified by C-18 Reverse Phase-High Performance Liquid Chromatography (RP-HPLC), quantified by the bicinchoninic acid (BCA) method (Pierce, Rockford, IL) and checked for purity by SDS-PAGE and mass spectrometry [59].
Plant material and growth conditions
Arabidopsis thaliana plants, ecotype Col-0, were grown in soil in a growth chamber at 22°C with a 12-h photoperiod under light intensity of 100 µE m−2 s−1 for 4–5 weeks. The phosphorylation assay for MAP kinases was performed on 12-day-old seedlings as described below. Unless differently specified, the experiments were performed on intact detached leaves.
Detection of hydrogen peroxide
The production of H2O2 in intact leaves after the treatment with CP was visualized in situ with the probe H2DCF-DA 10 µM (Sigma-Aldrich, St Louis, MO, USA) according to Lombardi et al. [19]. Leaves from 4-week-old plants were detached and treated on the leaf surface with 10-µl drops (12–15 drops per leaf) of 150 µM CP and incubated in a moist chamber for 5, 15, 30 min, 1, 3, 6, 12, 24 h. At the end of the incubation period the leaves were incubated for 30 min with the probe, washed twice with buffer to remove the excess of fluorophore, and mounted on slides. Controls were prepared by treating leaves with both sterile distilled water and the protein bovine serum albumin (BSA) 150 µM. Photographs were taken with a QICAM digital CCD camera (QImaging, CA, USA) attached to a Nikon Eclipse Ti-S Microscope, equipped with filter block FITC (λex 488, λem 502–540 nm).
In order to find the minimum active concentration of CP, leaves were treated with 1∶2 serial dilutions of CP (starting from 300 µM) and the production of H2O2 in situ was analysed after 15 and 60 min.
In order to analyse the H2O2 production on the epidermis around the stomata, epidermal peels were used. Arabidopsis plants were kept for at least 3 h under the light to ensure that most stomata were open before beginning the experiments, then epidermis was peeled off and placed on glass slides with the cuticle side in contact with MES buffer. Peels were pre-incubated for 2 h in MES buffer (25 mM MES-KOH pH 6.15, 10 mM KCl) under the light, soaked for 20 min in 10 µM H2DCF-DA diluted in MES buffer, washed three times in MES buffer, and then incubated with 150 µM CP for 5, 10 or 30 min. MES buffer was used as a negative control. The experiment was repeated by treating peels with closed stomata. As a further analysis, epidermal peels were treated with CP on the side devoid of cuticle.
In order to analyse H2O2 production at the level of single guard cells, an epidermal peel was placed on a glass slide with the cuticle side in contact with CP and was visualized in continuum under the microscope for 10 min; single stomata were photographed at 2-minute intervals.
Stomatal closure assay
Arabidopsis plants were kept for at least 3 h under the light to ensure that most stomata were open before beginning the experiments. Epidermal peels were prepared from three different fully expanded leaves as described in the previous paragraph. In those conditions, peels have about 70% open stomata [25]. The tissues were placed on glass slides with the cuticle side in contact with MES buffer or with 150 µM CP dissolved in MES buffer. MES buffer was used as a negative control. At various time points (30, 60 and 120 min) pictures were taken of random regions and the width of the stomatal aperture was measured by using Image-Pro Plus 6.0 (Media Cybernetics, Bethesda, MD, USA).
MAPK phosphorylation assay
Arabidopsis seeds were sterilized by treating them for 3 min in 70% ethanol, 3 min in 20% bleach with 0.1% Tween 20 and extensively washed with sterile distilled water. Subsequently, the seeds were incubated in the dark at 4°C for 1 day and then dispensed into a 24-well tissue culture plate (10 seeds per well) containing 1 ml of Murashige and Skoog Basal medium supplemented with 0.5% sucrose, pH 5.7. The plates were incubated at 22°C with a 12-h-light photoperiod (100 µE m−2 s−1 intensity). After 8 days, the medium was replaced and the treatments were performed after four additional days. Seedlings were treated for 5, 15, 30 and 60 min by adding to the medium either CP to a final concentration of 150 µM or an equal volume of sterile distilled water as a control.
Protein extraction and immunoblot analysis were performed according to Galletti et al. [26]. In summary, proteins were extracted with a buffer containing 50 mM Tris-HCl (pH 7.5), 200 mM NaCl, 1 mM EDTA, 10 mM NaF, 2 mM sodium orthovanadate, 1 mM sodium molybdate, 10% (v/v) glycerol, 0.1% Tween 20, 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol, and 1X protease inhibitor cocktail P9599 (Sigma-Aldrich). Fifteen µg of proteins per each sample were resolved on 12% polyacrylamide gel and transferred onto a PVDF membrane (BioRad, Hercules, CA, USA). Primary antibodies against MPK3 and MPK6 (Sigma-Aldrich), and against phospho-p44/42 MAP kinase (Cell Signaling Technologies), were used with horseradish peroxidase-conjugated anti-rabbit as a secondary antibody. Signal detection was performed by ECL western detection kit (GE Healthcare). Image analysis was performed with the software KODAK MI v.4.0.4 (KODAK Rochester, New York, USA).
Gene expression analysis
A primer library containing 93 gene-specific primers for pathogen-inducible genes (Primer Library for Arabidopsis Pathogen-inducible Genes, Sigma-Aldrich) was employed for a large scale analysis that we performed by qPCR after 6 h of treatment. Thirteen more primer pairs (Table S2) were designed with Primer Express Software 3.0 (Applied Biosystems) for a detailed analysis of defence-signalling pathways that we performed at 1, 3, 6, 12 or 24 h. Leaves from 4-week-old plants were detached and treated on the lower surface with 10-µl drops of CP 150 µM (treated sample) or sterile distilled water (control sample) as previously described and incubated in a moist chamber for 1, 3, 6, 12 or 24 h. Three treated leaves and three control leaves were incubated per each time.
Total RNA was extracted by using GenElute Mammalian Total RNA Miniprep Kit (Sigma-Aldrich), treated with Amplification Grade DNase I (Sigma-Aldrich) and reverse-transcribed into cDNA (400 ng per sample) by using iScript cDNA synthesis kit (BioRad, Hercules, CA, USA).
Real-time reactions (20 µl) were carried out with 10 ng of cDNA, 250 nM primers, and 1×Fast SYBR Green Master Mix (Applied Biosystems, Foster City, CA, USA) following the manufacturer’s instructions. PCRs were run in a StepOne real-time PCR System (Applied Biosystems) by using the recommended thermal-cycling conditions (hold 95°C, 20 s; 40 cycles 95°C, 3 s; 60°C, 30 s). Before performing the relative expression calculation the performance of each amplification was checked and some primers were excluded from the study.
The relative gene expression values (2−ΔΔCt) were calculated by using Actin-2 as the endogenous reference gene and water-treated leaves as the calibrator sample (control sample), following the calculation described by Livak and Schmittgen [60]. Actin-2 was used as a reference gene after confirmation of its transcriptional stability in our experimental conditions (CP- vs. water-treated leaves) (data not shown). Actin-2 primers were included in the primer library. Statistical analysis (treated vs. control) was performed by unpaired t-test by using GraphPad InStat v. 3.05 (GraphPad Software, San Diego, CA, USA).
Phytoalexin synthesis assay and camalexin detection
Drops were collected from the lower leaf surface of CP- or water-treated leaves after 15, 30, 60 min, 3, 6, 9, 12 and 24 h of incubation and the phytoalexin measurement was performed by fluorescence analysis (λex = 320 nm, λem = 386 nm, slit 5) by using a Perkin Elmer spectrofluorimeter 650-10 S (Perkin Elmer, Wellesley, MA). In order to confirm the release of camalexin from the leaves, the fluorescence spectrum (λex = 320 nm, λem = 330–550 nm, slit 5) of the droplets collected after 24 h of incubation was compared to the spectrum obtained by using a pure camalexin standard (kindly provided by Dr. E. Glawischnig, University of Technology, Munich, Germany). The experiment was also repeated to compare the two leaf surfaces in their ability to release phytoalexins after 48 h of treatment.
Camalexin was extracted with methanol from CP-treated leaves (12 and 24 h) according to Beets and Dubery [61].
Ten µl of a standard stock solution of camalexin (15 µg ml−1 in methanol) and 40 µl of extracted sample were analysed with RP-HPLC using a C18 column, 3 µm, 15×4.6 cm (Supelco, Bellefonte, Pennsylvania, USA). Elution gradient was performed at a flow rate of 0.8 ml min−1 with the following solvent system: 10 mM trifluoroacetic acid (TFA) in acetonitrile (solvent A); 10 mM TFA in water (solvent B). The gradient used was 10% A for 2 min, from 10% to 98% A for 13 min, holding at 98% A for 10 min, from 98% to 100% A for 5 min. Detection was based on UV absorbance at 318 nm.
Resistance induction assays
Arabidopsis leaves were detached from 5-week-old plants and treated on the lower surface with 10-µl drops of 150 µM CP, sterile distilled water (control) or 0.1% chitosan (about 20 µM in 10 mM acetic acid; molecular weight 50–60 kDa, deacetylation degree 85%). The leaves were incubated for 24 h in Petri dishes containing 0.8% agar, after that the drops were recovered and the leaves were infected on the same foliar surface with pathogens.
Botrytis cinerea strain PM10 [62] was grown on potato dextrose agar (PDA, Difco, Detroit, MI) at 22°C for 8 days before collecting conidia. Inoculations were performed by placing a single 10-µl drop per leaf of a suspension 2×105 conidia ml−1 in 1% Sabouraud Maltose Broth (SMB) (maltose 8 g l−1, peptone 2 g l−1) on a side of the middle vein. Leaves were incubated at 22°C for 3 days before measuring the lesion diameter and taking photographs. In order to test for a direct inhibiting effect of CP against the germination of B. cinerea conidia, two drops of conidial suspension were placed on a glass slide and one of these was added with CP, whereas the other one was added with sterile distilled water for an equal volume. Germination and growth were monitored.
Pseudomonas syringae pv. tomato (Pst) strain DC3000 was cultured at 28°C, 300 rpm, in Luria-Bertani (LB; tryptone 10 g l−1, yeast extract 5 g l−1, NaCl 5 g L−1) medium supplemented with 50 mg l−1 rifampicin (Sigma-Aldrich) until an OD600 of 0.8 was reached (24–26 h). Bacteria were collected by centrifugation and resuspended in sterile water containing the surfactant Silwet L-77 0.02% (Momentive, Columbus, OH, USA) to the final concentration of 108 colony-forming units (CFU) ml−1 (OD600 = 0.2). Inoculations were performed by placing a single 10-µl drop per leaf on a side of the middle vein. Leaves were incubated at 28°C for 3 days after that photographs were taken and the leaf bacterial titer was determined. The titer was determined by grounding the infected side of the leaf in 1 ml of 10 mM MgCl2 and by overspreading serial dilutions on LB agar (10 g l−1 agar) plates containing 50 mg l−1 rifampicin. CFU were counted after 2 days of incubation at 28°C. In order to test for a direct inhibiting effect of CP on the bacterial growth, Pst DC3000 was grown with 150 µM CP for 3 days.
Statistically significant differences among treatments were calculated by one-way ANOVA with Tukey-Kramer post test (P≤0.05) with GraphPad InStat v. 3.05 (GraphPad Software, San Diego, CA, USA).
Supporting Information
H2O2 production in Arabidopsis leaves treated with CP. Leaves were treated on the lower (abaxial) surface with 10-µl drops of 150 µM CP or water (control) for 15 and 30 min, 1, 3, 6, 12 and 24 h. H2O2 was visualized in situ by the fluorescent probe 2,7-dichlorofluorescin diacetate (H2DCF-DA). The bar is 100 µm and applies to all photographs.
(TIF)
The production of H2O2 only origins at the level of open stomata. (A) Treatment of epidermal peels obtained from leaves with open stomata. Some closed stomata are indicated by the arrows. The peels were first loaded with the fluorescent probe H2DCF-DA and then treated on the cuticle side with 150 µM CP. (B) Treatment of epidermal peels obtained from leaves with closed stomata. Fluorescence microscopy (H2DCF-DA), light microscopy (bright field) and merged pictures (merge) are shown. The bar is 40 µm and applies to all photographs.
(TIF)
Fluorescence spectrum of a pure camalexin standard and of sample of droplets collected from the foliar surface after 24 h of treatment. Leaves were treated on the lower surface with 10-µl drops of 150 µM CP. The droplets were recovered and the fluorescence analysis was performed at λex = 320 nm.
(TIF)
Retention time (Rt) of camalexin as revealed by RP-HPLC. Standard stock solution of camalexin analysed by Reverse Phase-High Performance Liquid Chromatography (RP-HPLC). Rt = 11.2 min.
(TIF)
Phytoalexin production by Arabidopsis leaves treated on the adaxial or abaxial surface with CP. Leaves were treated with 10-µl drops of 150 µM CP or sterile distilled water (control) for 48 h. The phytoalexin release was measured by fluorescence analysis (λex = 320 nm, λem = 386 nm). The fluorescence value was normalized to the number of droplets analysed and was expressed as relative fluorescence units. Error bars indicate SD of three measurements. Statistical analysis was performed by unpaired t-test (treated vs. control). Asterisks indicate statistically significant difference at P<0.05.
(TIF)
Real-time RT-qPCR results. Pathogen library (6 h) and time-course analysis (1, 3, 6, 12, 24 h) for selected genes.
(XLS)
Primers designed in the present study and used for the RT-qPCR analysis.
(DOC)
Acknowledgments
We thank Dr. E. Glawischnig, University of Technology, Munich, Germany, for kindly providing the camalexin standard; Prof. F. Faoro, University of Milan, Italy, for kindly providing chitosan; Prof. R. Buonaurio, University of Perugia, Italy, for kindly providing Pst DC3000 and Prof. F. Favaron, University of Padua, Italy, for kindly providing the B. cinerea strain PM10.
Funding Statement
This work was supported by Ministero Italiano dell’Università e della Ricerca Scientifica, Progetti di Ricerca di Interesse Nazionale (PRIN) 2009 (http://prin.miur.it/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jones JDG, Dangl JL (2006) The plant immune system. Nature 444: 323–332. [DOI] [PubMed] [Google Scholar]
- 2. Boller T, Felix G (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406. [DOI] [PubMed] [Google Scholar]
- 3. Iriti M, Faoro F (2009) Chitosan as a MAMP, searching for a PRR. Plant Signal Behav 4: 66–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Thomma BP, Nürnberger T, Joosten MH (2011) Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23: 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gijzen M, Nürnberger T (2006) Nep1-like proteins from plant pathogens: Recruitment and diversification of the NPP1 domain across taxa. Phytochemistry 67: 1800–1807. [DOI] [PubMed] [Google Scholar]
- 6. Klarzynski O, Plesse B, Joubert JM, Yvin JC, Kopp M, et al. (2000) Linear β-1, 3 glucans are elicitors of defense responses in tobacco. Plant Physiol 124: 1027–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen H, Kovalchuk A, Keriö S, Asiegbu FO (2013) Distribution and bioinformatic analysis of the cerato-platanin protein family in Dikarya. Mycologia 105: 1479–1488. [DOI] [PubMed] [Google Scholar]
- 8. Baccelli I, Luti S, Bernardi R, Scala A, Pazzagli L (2014) Cerato-platanin shows expansin-like activity on cellulosic materials. Appl Microbiol Biotechnol 98: 175–184. [DOI] [PubMed] [Google Scholar]
- 9. de O Barsottini MR, de Oliveira JF, Adamoski D, Teixeira PJ, do Prado PF, et al. (2013) Functional diversification of cerato-platanins in Moniliophthora perniciosa as seen by differential expression and protein function specialization. Mol Plant Microbe Interact 26: 1281–1293. [DOI] [PubMed] [Google Scholar]
- 10. de Oliveira AL, Gallo M, Pazzagli L, Benedetti CE, Cappugi G, et al. (2011) The structure of the elicitor cerato-platanin (CP), the first member of the CP fungal protein family, reveals a double ψβ-barrel fold and carbohydrate binding. J Biol Chem 286: 17560–17568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Frischmann A, Neudl S, Gaderer R, Bonazza K, Zach S, et al. (2013) Self-assembly at air/water interfaces and carbohydrate binding properties of the small secreted protein EPL1 from the fungus Trichoderma atroviride. . J Biol Chem 288: 4278–4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pazzagli L, Seidl-Seiboth V, Barsottini M, Vargas WA, Scala A, et al. (2014) Cerato-platanins: Elicitors and effectors. Plant Sci in press http://dx.doi.org/10.1016/j.plantsci.2014.02.009. [DOI] [PubMed]
- 13. Frías M, Brito N, González C (2013) The Botrytis cinerea cerato-platanin BcSpl1 is a potent inducer of systemic acquired resistance (SAR) in tobacco and generates a wave of salicylic acid expanding from the site of application. Mol Plant Pathol 14: 191–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vargas WA, Djonović S, Sukno SA, Kenerley CM (2008) Dimerization controls the activity of fungal elicitors that trigger systemic resistance in plants. J Biol Chem 283: 19804–19815. [DOI] [PubMed] [Google Scholar]
- 15. Djonović S, Pozo MJ, Dangott LJ, Howell CR, Kenerley CM (2006) Sm1, a proteinaceous elicitor secreted by the biocontrol fungus Trichoderma virens induces plant defense responses and systemic resistance. Mol Plant Microbe Interact 19: 838–853. [DOI] [PubMed] [Google Scholar]
- 16. Yang Y, Zhang H, Li G, Li W, Wang X, et al. (2009) Ectopic expression of MgSM1, a cerato-platanin family protein from Magnaporthe grisea, confers broad-spectrum disease resistance in Arabidopsis . Plant Biotechnol J 7: 763–777. [DOI] [PubMed] [Google Scholar]
- 17. Pazzagli L, Cappugi G, Manao G, Camici G, Santini A, et al. (1999) Purification, characterization, and amino acid sequence of cerato-platanin, a new phytotoxic protein from Ceratocystis fimbriata f. sp. platani . J Biol Chem 274: 24959–24964. [DOI] [PubMed] [Google Scholar]
- 18. Baccelli I, Scala A, Pazzagli L, Bernardi R (2013b) Early transcription of defence-related genes in Platanus x acerifolia leaves following treatment with cerato-platanin. Biol Plant 57: 571–575. [Google Scholar]
- 19. Lombardi L, Faoro F, Luti S, Baccelli I, Martellini F, et al. (2013) Differential timing of defense-related responses induced by cerato-platanin and cerato-populin, two non-catalytic fungal elicitors. Physiol Plant 149: 408–421. [DOI] [PubMed] [Google Scholar]
- 20. Comparini C, Carresi L, Pagni E, Sbrana F, Sebastiani F, et al. (2009) New proteins orthologous to cerato-platanin in various Ceratocystis species and the purification and characterization of cerato-populin from Ceratocystis populicola. . Appl Microbiol Biotechnol 84: 309–322. [DOI] [PubMed] [Google Scholar]
- 21. Scala A, Pazzagli L, Comparini C, Santini A, Tegli S, et al. (2004) Cerato-platanin an early-produced protein by Ceratocystis fimbriata f.sp. platani elicits phytoalexin synthesis in host and non-host plants. J Plant Pathol 86: 23–29. [Google Scholar]
- 22. Martellini F, Faoro F, Carresi L, Pantera B, Baccelli I, et al. (2013) Cerato-populin and cerato-platanin, two non-catalytic proteins from phytopathogenic fungi, interact with hydrophobic inanimate surfaces and leaves. Mol Biotechnol 55: 27–42. [DOI] [PubMed] [Google Scholar]
- 23. Allan AC, Fluhr R (1997) Two distinct sources of elicited reactive oxygen species in tobacco epidermal cells. Plant Cell 9: 1559–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee S, Choi H, Suh S, Doo IS, Oh KY, et al. (1999) Oligogalacturonic acid and chitosan reduce stomatal aperture by inducing the evolution of reactive oxygen species from guard cells of tomato and Commelina communis . Plant Physiol 121: 147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Melotto M, Underwood W, Koczan J, Nomura K, He SY (2006) Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980. [DOI] [PubMed] [Google Scholar]
- 26. Galletti R, Ferrari S, De Lorenzo G (2011) Arabidopsis MPK3 and MPK6 play different roles in basal and oligogalacturonide or flagellin-induced resistance against Botrytis cinerea . Plant Physiol 157: 804–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ren D, Liu Y, Yang KY, Han L, Mao G, et al. (2008) A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis . Proc Natl Acad Sci USA 105: 5638–5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wan J, Zhang S, Stacey G (2004) Activation of a mitogen-activated protein kinase pathway in Arabidopsis by chitin. Mol Plant Pathol 5: 125–135. [DOI] [PubMed] [Google Scholar]
- 29. Beckers GJ, Jaskiewicz M, Liu Y, Underwood WR, He SY, et al. (2009) Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana . Plant Cell 21: 944–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mao G, Meng X, Liu Y, Zheng Z, Chen Z, et al. (2011) Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis . Plant Cell 23: 1639–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ezaki B, Gardner RC, Ezaki Y, Matsumoto H (2000) Expression of aluminum-induced genes in transgenic arabidopsis plants can ameliorate aluminum stress and/or oxidative stress. Plant Physiol 122: 657–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mowla SB, Cuypers A, Driscoll SP, Kiddle G, Thomson J, et al. (2006) Yeast complementation reveals a role for an Arabidopsis thaliana late embryogenesis abundant (LEA)-like protein in oxidative stress tolerance. Plant J 48: 743–756. [DOI] [PubMed] [Google Scholar]
- 33. Torres MA, Dangl JL (2005) Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr Opin Plant Biol 8: 397–403. [DOI] [PubMed] [Google Scholar]
- 34. Wagner U, Edwards R, Dixon DP, Mauch F (2002) Probing the diversity of the Arabidopsis glutathione S-transferase gene family. Plant Mol Biol 49: 515–532. [DOI] [PubMed] [Google Scholar]
- 35. Schwessinger B, Ronald PC (2012) Plant innate immunity: perception of conserved microbial signatures. Annu Rev Plant Biol 63: 451–482. [DOI] [PubMed] [Google Scholar]
- 36. Qiu JL, Fiil BK, Petersen K, Nielsen HB, Botanga CJ, et al. (2008) Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 27: 2214–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bari R, Jones JD (2009) Role of plant hormones in plant defence responses. Plant Mol Biol 69: 473–488. [DOI] [PubMed] [Google Scholar]
- 38. Thomma BP, Penninckx IA, Broekaert WF, Cammue BP (2001) The complexity of disease signaling in Arabidopsis . Curr Opin Immunol 13: 63–68. [DOI] [PubMed] [Google Scholar]
- 39. Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, et al. (2011) ROS signaling: the new wave? Trends Plant Sci 16: 300–309. [DOI] [PubMed] [Google Scholar]
- 40. Frías M, González C, Brito N (2011) BcSpl1, a cerato-platanin family protein, contributes to Botrytis cinerea virulence and elicits the hypersensitive response in the host. New Phytol 192: 483–495. [DOI] [PubMed] [Google Scholar]
- 41. Kumar MN, Jane WN, Verslues PE (2013) Role of the putative osmosensor Arabidopsis histidine kinase1 in dehydration avoidance and low-water-potential response. Plant Physiol 161: 942–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lake JA, Woodward FI, Quick WP (2002) Long-distance CO(2) signalling in plants. J Exp Bot 53: 183–193. [DOI] [PubMed] [Google Scholar]
- 43. Robatzek S, Chinchilla D, Boller T (2006) Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis . Genes Dev 20: 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frías M, Brito N, González M, González C (2013) The phytotoxic activity of the cerato-platanin BcSpl1 resides in a two-peptide motif in the protein surface. Mol Plant Pathol in press. doi:10.1111/mpp.12097. [DOI] [PMC free article] [PubMed]
- 45. Sawinski K, Mersmann S, Robatzek S, Böhmer M (2013) Guarding the green: pathways to stomatal immunity. Mol Plant Microbe Interact 26: 626–632. [DOI] [PubMed] [Google Scholar]
- 46. Wang P, Song CP (2008) Guard-cell signalling for hydrogen peroxide and abscisic acid. New Phytol 178: 703–718. [DOI] [PubMed] [Google Scholar]
- 47. Pieterse CM, Leon-Reyes A, Van der Ent S, Van Wees SC (2009) Networking by small-molecule hormones in plant immunity. Nat Chem Biol 5: 308–316. [DOI] [PubMed] [Google Scholar]
- 48. Wang L, Tsuda K, Sato M, Cohen JD, Katagiri F, et al. (2009) Arabidopsis CaM binding protein CBP60g contributes to MAMP-induced SA accumulation and is involved in disease resistance against Pseudomonas syringae . PLoS Pathog 5: e1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu Y, Zhang S (2004) Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis . Plant Cell 16: 3386–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA (2008) COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci USA 105: 7100–7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R (2004) JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis . Plant Cell 16: 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Penninckx IA, Thomma BP, Buchala A, Métraux JP, Broekaert WF (1998) Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis . Plant Cell 10: 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Thaler JS, Humphrey PT, Whiteman NK (2012) Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci 17: 260–270. [DOI] [PubMed] [Google Scholar]
- 54. Ahuja I, Kissen R, Bones AM (2012) Phytoalexins in defense against pathogens. Trends Plant Sci 17: 73–90. [DOI] [PubMed] [Google Scholar]
- 55. Ferrari S, Galletti R, Denoux C, De Lorenzo G, Ausubel FM, et al. (2007) Resistance to Botrytis cinerea induced in Arabidopsis by elicitors is independent of salicylic acid, ethylene, or jasmonate signaling but requires PHYTOALEXIN DEFICIENT3. Plant Physiol 144: 367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kliebenstein DJ, Rowe HC, Denby KJ (2005) Secondary metabolites influence Arabidopsis/Botrytis interactions: variation in host production and pathogen sensitivity. Plant J 44: 25–36. [DOI] [PubMed] [Google Scholar]
- 57. Fontana F, Santini A, Salvini M, Pazzagli L, Cappugi G, et al. (2008) Cerato-platanin pre-treated plane leaves restrict Ceratocystis platani growth and overexpress defense-related genes. J Plant Pathol 90: 295–306. [Google Scholar]
- 58. Carresi L, Pantera B, Zoppi C, Cappugi G, Oliveira AL, et al. (2006) Cerato-platanin, a phytotoxic protein from Ceratocystis fimbriata: expression in Pichia pastoris, purification and characterization. Protein Expr Purif 49: 159–167. [DOI] [PubMed] [Google Scholar]
- 59. Pazzagli L, Zoppi C, Carresi L, Tiribilli B, Sbrana F, et al. (2009) Characterization of ordered aggregates of cerato-platanin and their involvement in fungus–host interactions. Biochim Biophys Acta 1790: 1334–1344. [DOI] [PubMed] [Google Scholar]
- 60. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 61. Beets C, Dubery I (2011) Quantification of camalexin, a phytoalexin from Arabidopsis thaliana: a comparison of five analytical methods. Anal Biochem 419: 260–265. [DOI] [PubMed] [Google Scholar]
- 62. Cettul E, Rekab D, Locci R, Firrao G (2008) Evolutionary analysis of endopolygalacturonase-encoding genes of Botrytis cinerea . Mol Plant Pathol 9: 675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
H2O2 production in Arabidopsis leaves treated with CP. Leaves were treated on the lower (abaxial) surface with 10-µl drops of 150 µM CP or water (control) for 15 and 30 min, 1, 3, 6, 12 and 24 h. H2O2 was visualized in situ by the fluorescent probe 2,7-dichlorofluorescin diacetate (H2DCF-DA). The bar is 100 µm and applies to all photographs.
(TIF)
The production of H2O2 only origins at the level of open stomata. (A) Treatment of epidermal peels obtained from leaves with open stomata. Some closed stomata are indicated by the arrows. The peels were first loaded with the fluorescent probe H2DCF-DA and then treated on the cuticle side with 150 µM CP. (B) Treatment of epidermal peels obtained from leaves with closed stomata. Fluorescence microscopy (H2DCF-DA), light microscopy (bright field) and merged pictures (merge) are shown. The bar is 40 µm and applies to all photographs.
(TIF)
Fluorescence spectrum of a pure camalexin standard and of sample of droplets collected from the foliar surface after 24 h of treatment. Leaves were treated on the lower surface with 10-µl drops of 150 µM CP. The droplets were recovered and the fluorescence analysis was performed at λex = 320 nm.
(TIF)
Retention time (Rt) of camalexin as revealed by RP-HPLC. Standard stock solution of camalexin analysed by Reverse Phase-High Performance Liquid Chromatography (RP-HPLC). Rt = 11.2 min.
(TIF)
Phytoalexin production by Arabidopsis leaves treated on the adaxial or abaxial surface with CP. Leaves were treated with 10-µl drops of 150 µM CP or sterile distilled water (control) for 48 h. The phytoalexin release was measured by fluorescence analysis (λex = 320 nm, λem = 386 nm). The fluorescence value was normalized to the number of droplets analysed and was expressed as relative fluorescence units. Error bars indicate SD of three measurements. Statistical analysis was performed by unpaired t-test (treated vs. control). Asterisks indicate statistically significant difference at P<0.05.
(TIF)
Real-time RT-qPCR results. Pathogen library (6 h) and time-course analysis (1, 3, 6, 12, 24 h) for selected genes.
(XLS)
Primers designed in the present study and used for the RT-qPCR analysis.
(DOC)