Abstract
High-confidence prediction of complex traits such as disease risk or drug response is an ultimate goal of personalized medicine. Although genome-wide association studies have discovered thousands of well-replicated polymorphisms associated with a broad spectrum of complex traits, the combined predictive power of these associations for any given trait is generally too low to be of clinical relevance. We propose a novel systems approach to complex trait prediction, which leverages and integrates similarity in genetic, transcriptomic, or other omics-level data. We translate the omic similarity into phenotypic similarity using a method called Kriging, commonly used in geostatistics and machine learning. Our method called OmicKriging emphasizes the use of a wide variety of systems-level data, such as those increasingly made available by comprehensive surveys of the genome, transcriptome, and epigenome, for complex trait prediction. Furthermore, our OmicKriging framework allows easy integration of prior information on the function of subsets of omics-level data from heterogeneous sources without the sometimes heavy computational burden of Bayesian approaches. Using seven disease datasets from the Wellcome Trust Case Control Consortium (WTCCC), we show that OmicKriging allows simple integration of sparse and highly polygenic components yielding comparable performance at a fraction of the computing time of a recently published Bayesian sparse linear mixed model method. Using a cellular growth phenotype, we show that integrating mRNA and microRNA expression data substantially increases performance over either dataset alone. Using clinical statin response, we show improved prediction over existing methods.
Keywords: complex trait prediction, polygenic modeling, systems biology, polygenic prediction, Kriging
Introduction
High-confidence prediction of complex traits is an ultimate goal of personalized medicine. For heritable traits, genotypic similarity contributes to phenotypic similarity. While genome-wide association studies (GWAS) have revealed many statistically significant loci associated with complex traits, the small effect sizes of most loci limit their prediction utility (de los Campos et al., 2010a). For most complex traits, an appreciable proportion of phenotypic variance is explained only when polygenic models, rather than single-marker tests, are used (Purcell et al., 2009; Yang et al., 2010). For example, 45% of the variance in human height can be explained using a mixed linear modeling approach (GCTA) that simultaneously considers all 300 K common SNPs genotyped (Yang et al., 2010, 2011). Because this approach does not rely on selection of loci, it is thought to be appropriate for traits with a highly polygenic or infinitesimal genetic architecture (Makowsky et al., 2011; Ober et al., 2012).
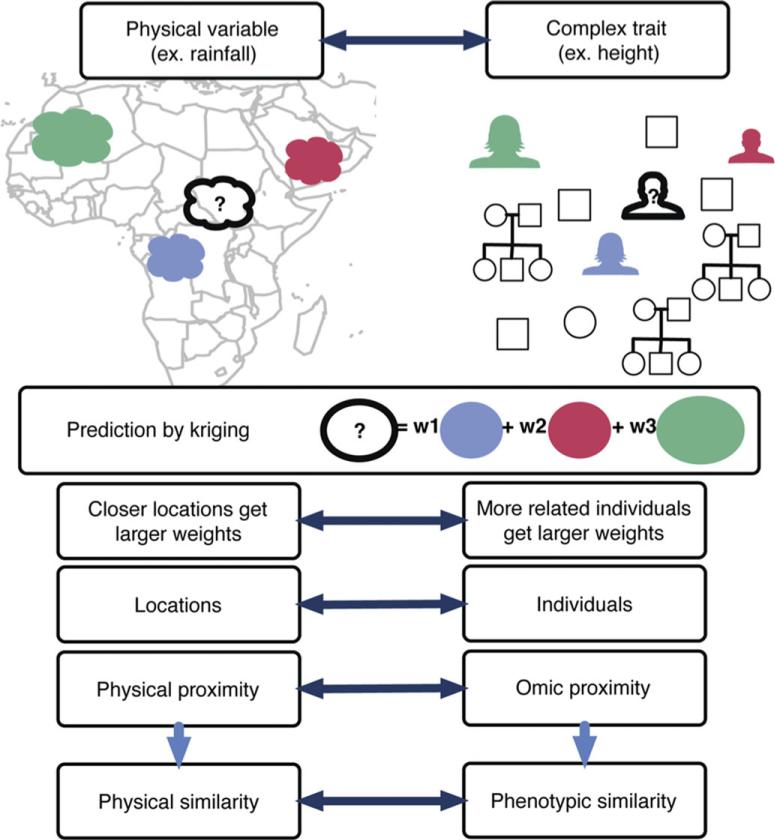
In order to intuitively understand our approach to phenotype prediction from genomic data, an analogy from the field of geostatistics is useful. Kriging interpolates the value of some geographical measurement such as yearly rainfall at an unobserved location (Cressie, 1993; Im et al., 2007; Stein, 1999). The approach assumes the rainfall measured at nearby locations will be more similar to that at the unmeasured location than rainfall at sites farther away. Kriging uses individual weights based on the correlation matrix of these locations along with the observed rainfall amounts to predict the rainfall at the unmeasured location. The locations are analogous to individuals and the rainfall amounts are analogous to the phenotype of interest (Fig. 1). In our application to complex trait prediction, the notion of distance can be replaced with the degree of genetic similarity or dissimilarity. Close ties between genetic distance and geographic distance have been demonstrated in studies of human population structure. For example, in the analysis of genome-wide genotype data from 3000 Europeans, a geographic map of Europe arose naturally when the first two principal components of the data were plotted (Novembre et al., 2008). A diagram of the analogy between geostatistical Kriging and complex trait prediction is shown in Figure 1.
Figure 1.
Kriging and whole-genome prediction connection. This figure shows the analogous relationships between components of the Kriging method used in geostatistics and whole-genome prediction. The prediction at an unobserved location (?) is computed as a weighted average of the variable at observed locations. The weights are functions of the correlation between the rainfall at the new location and the rainfalls at the observed locations. The closer the distance between each observed location and the new location, the higher the weight. In complex trait prediction, locations correspond to individuals, physical proximity corresponds to genetic relatedness. The correlation between two locations or individuals is the key component of this method. In animal breeding approaches, the genetic relatedness matrix or kinship matrix is used. In OmicKriging, a genetic relatedness matrix, a gene expression similarity matrix, or any combination of available high-throughput data similarity measures can be tested for complex trait prediction performance.
Methods of complex trait prediction using whole-genome data were pioneered by Meuwissen et al. [2001] who proposed to predict the genetic effects on phenotypes as the sum of marker effects , where Xil is the genotype of individual i for marker l and is the estimated effect size of marker l. One early application to human disease risk used a polygenic score derived from the logistic regression effect sizes of GWAS SNPs with P-values below a chosen threshold in a training set to predict a significant proportion of the risk of schizophrenia and bipolar disorder in test sets (Purcell et al., 2009). The training set was a large schizophrenia GWAS and the testing sets included multiple additional schizophrenia, as well as bipolar disorder, datasets. A similar polygenic score approach was used to predict risk of the seven diseases within the WTCCC, by dividing each sample into training sets and test sets (Evans et al., 2009). When implementing polygenic score for trait prediction, the single-marker association effect sizes of SNPs that meet a chosen threshold from the training set are multiplied by the genotypes (0,1,2) of each respective SNP in the test set and the sum taken to generate a predicted phenotype value (polygenic score) for each individual. Performance of the polygenic score model is then assessed by comparing predicted to observed values. Principal components or any other known covariates that associate with the phenotype can be included in the polygenic score model to further improve prediction.
Additional whole-genome prediction (WGP) methods, which are reviewed in de los Campos et al. [2010a] and Abraham et al. [2013], include penalized estimation methods such as Least Absolute Shrinkage and Selection Operator (LASSO) (Tibshirani, 1996), Ridge Regression (Hoerl and Kennard, 1970), and Elastic Net (Abraham et al., 2013; Zou and Hastie, 2005) and the Bayesian versions of each method (Park and Casella, 2008; Pérez, et al., 2010). These methods use different penalty functions or different types of priors to overcome the infeasibility of estimating marker effects through ordinary least squares regression in typical GWAS datasets with much larger numbers of markers than individuals. These methods prevent overfitting and may reduce the mean-squared error of estimates and predictions (de los Campos et al., 2010a). As an example, Vazquez et al. used Bayesian LASSO to structure the prior density of marker effects in their Bayesian regression WGP model of skin cancer risk (Vazquez et al., 2012). Their best prediction model, which included 41K genome-wide SNPs, had an area under the receiver operating characteristic curve (AUC) of 0.635, 18.9% higher than that of the baseline model, which just included nongenetic covariates (Vazquez et al., 2012).
The idea of using genetic similarity to generate prediction dates back to Fisher [1918] and Wright [1921]. These ideas were formalized as best linear unbiased prediction (BLUP) approaches based on multivariate normal processes by Henderson, Goldberger, and others (Goldberger, 1962; Henderson, 1950, 1975). Originally, these methods used pedigree-based similarity matrices but with the advent of affordable high throughput genotyping several authors have used similarity measures computed using larger numbers of genetic markers (de los Campos et al., 2009; Habier et al., 2007; Janss et al., 2012; Lynch and Ritland, 1999; Makowsky et al., 2011; Ober et al., 2012; VanRaden, 2008; Yang et al., 2010). These approaches are commonly called G-BLUP for genomic-BLUP. Unlike the polygenic score methods, G-BLUP regresses phenotypes on hundreds of thousands of markers simultaneously using a genetic relationship matrix (GRM) derived from the marker data. While most applications of G-BLUP have assumed all measured genotypes affect the phenotype with normally distributed effect sizes and thus include all markers in the GRM, a newer approach by Zhou et al. combines G-BLUP with a sparse regression model that allows for a small proportion of markers with large effect sizes and estimates the most likely model for a particular phenotype from the data (Zhou et al., 2013).
The equivalence between Kriging and the BLUP methods used in the animal breeding and quantitative genetics fields has been demonstrated (Harville, 1984; Robinson, 1991). Ridge regression is equivalent to standard Kriging/BLUP, but does not have the dimension reduction advantage that the latter offers. Kriging in genomic prediction has been previously used, but it was restricted to simulation studies of genetic similarities (Ober et al., 2011). Based on whole genome simulations, Ober et al. reported that using Matérn functions (class of special functions commonly used in geostatistics) to scale the genetic relatedness measure works better than standard measures of relatedness in the presence of dominance and epistatic effects (Ober et al., 2011).
Our approach extends the Kriging framework for the integration of multiple omic data. Furthermore, this framework allows easy integration of prior information on the function of the variants by partitioning the genome and giving more weight to different subsets based on functional evidence. For example, known loci can be given more weight than the rest of the genome, or subsets of the genome with regulatory evidence of affecting gene expression (eQTLs) can be given larger weight. It differs from standard Kriging/BLUP in that it is not necessarily tied to an additive genetic/genomic model. Instead of using the maximum likelihood method to estimate the parameters we use a more pragmatic approach where we maximize the cross-validated prediction performance. In this sense, our approach is closely related to the semiparametric models using reproducing kernel Hilbert space (RKHS) regression proposed by Gianola et al. [2006] and de los Campos et al. [2010b] for WGP. To our knowledge, our method is the first to integrate multiple omics data using these semipara-metric methods based on similarity measures. Furthermore, our intuitive connection between Kriging and BLUP should allow investigators less familiar with the quantitative genetics field to appreciate the usefulness of the approach and encourage them to adopt these methods for their specific analyses.
In sum, we propose a novel systems approach to predict complex traits, which leverages and integrates similarity in genetic, transcriptomic, and/or other large scale omics data. Here, we describe our OmicKriging method, apply the method to several human complex traits (cellular growth, clinical statin response, and seven WTCCC diseases), and provide an R package for implementation.
Results
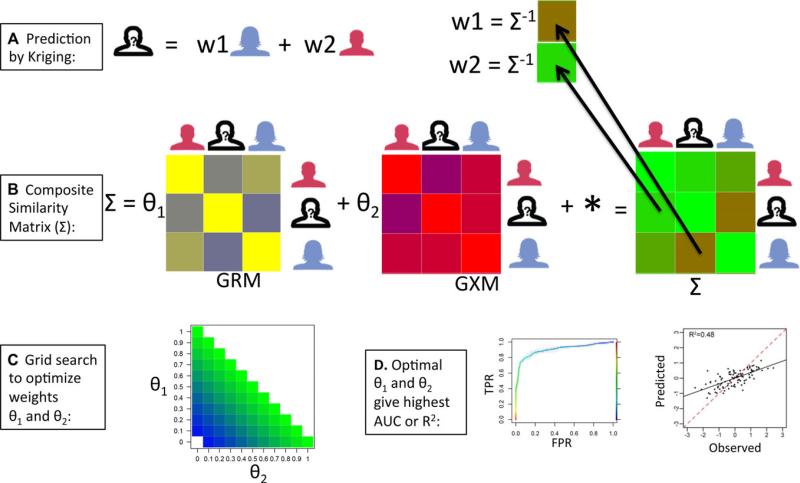
In implementing OmicKriging, the first step is to construct a similarity matrix or similarity matrices for use in the prediction. Such matrices may include a genetic relationship matrix (GRM) from a set of SNPs, a gene expression correlation matrix (GXM) from gene expression data, or any other correlation matrix derived from omics data. To predict the phenotype of an individual, the weighted average of the training set individuals’ phenotypes is calculated. In the case of SNP data, the weights are comprised of the GRM and pairwise genotype similarity of the unknown individual with the genotypes of those with observed phenotypes. Each similarity matrix can be tested individually or in a given weighted combination for phenotype predictive performance. When using a single omic component, we tested matrix weights between 0 and 1 (i.e. 0.1, 0.2, 0.3,..., 1 for the omic component and 1-weight for the environmental component) to find the matrix weight that produced optimal prediction. When two omic components (e.g. GRM and GXM) were combined, we performed a grid search to find the optimal prediction matrix weights θ1 and θ2, such that θ1 + θ2 ≤ 1 (1-θ1-θ2 for the environmental component). The optimal matrix weights for each omic component depend on the genetic architecture of the phenotype. A schematic of our procedure to find the optimal composite similarity matrix is shown in Figure 2.
Figure 2.
OmicKriging data integration and weighting. (A) The individual weights, depicted as w1 and w2, in the Kriging method are given by the product of the composite similarity matrix Σ and the correlation of omic data between the individual of unknown phenotype (?) and the individuals of known phenotype. (B) The composite similarity matrix Σ integrates different omic correlation matrices such as a genetic relationship matrix (GRM) derived from SNPs and a gene expression correlation matrix (GXM) derived from gene expression levels in this example. Σ also includes an environmental component, i.e. noise term (*). (C) In OmicKriging, we optimize the matrix weights, θ1 and θ2, by testing the θi values of the grid space depicted in color. (D) The optimal matrix weights θi give the highest values of AUC for binary traits and R2 for quantitative traits.
In applying OmicKriging, we used a 16-fold cross-validation approach with individuals assigned to the 16 subsets at random and repeating the procedure 500 time to assess the sampling variability of the prediction performance (see Methods). For quantitative traits, we computed the coefficient of determination R2 (equivalent to the square of the correlation) between the predicted and true values of the phenotype to assess prediction performance and for case/control traits, we computed the area under the receiver operating characteristic curve (AUC). Note that in the OmicKriging method, weighted averages are computed out-of-sample (only an individual's genotypes or gene expression levels are used to compute pairwise similarities) and should not be compared to reports where the R2 is computed using parameters estimated from the same data (in-sample).
Cellular Phenotype Applications
To assess the predictive performance of OmicKriging, we used the intrinsic growth rate (iGrowth) phenotype derived from multiple proliferation measurements in the commonly used HapMap lymphoblastoid cell lines (LCLs) (Im et al., 2012). This phenotype is relevant since it has been shown that genes associated with proliferation are strong prognostic factors in several types of cancers (Dai, 2005; Damasco et al., 2011; Rosenwald et al., 2003; Starmans et al., 2008) and such genes are differentially expressed in most cancer tissues (Rhodes et al., 2004; Ross et al., 2000; Whitfield et al., 2006). In addition, our unpublished data indicate that predicted drug-induced growth inhibition has predictive power on clinical response to drugs. iGrowth values from 99 LCLs from the HapMap CEU (Northern and Western European ancestry from Utah) and YRI (Yoruba from Ibadan, Nigeria) populations were used in the analysis. We tested common SNPs, gene (mRNA) expression levels, and microRNA expression levels for iGrowth predictive ability. The GRM was generated from 2.7 million common SNPs (minor allele frequency >0.05) from HapMap (Frazer et al., 2007) using GCTA (Yang et al., 2011) embedded in our OmicKriging R package. The GXM was generated from 13,080 transcript clusters from a previous genome-wide gene expression analysis (Zhang et al., 2008) as simply the correlation matrix of the expression data as described in the Methods. We also obtained expression measurements for 201 microRNAs (Gamazon et al., 2012) and generated a microRNA expression similarity matrix (MXM).
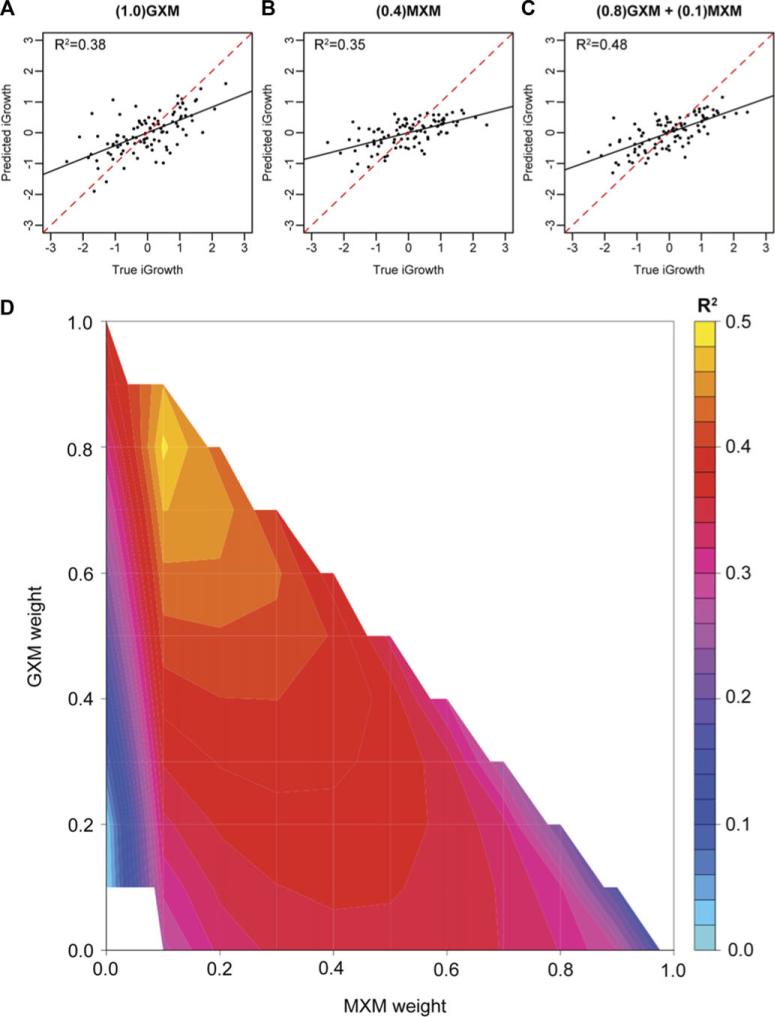
We first tested each single similarity matrix for predictive ability using OmicKriging. At every weight θGRM attempted (θGRM = 0.1, 0.2,..., 1 and θε = 1 – θGRM), the GRM did not show any predictive power (i.e. the correlation between the predicted and true iGrowth values did not differ from zero). However, for the GXM alone, the optimal prediction correlation was R2 = 0.38 [0.34, 0.43] when θGXM = 1 (Fig. 3A).
Figure 3.
iGrowth prediction using OmicKriging. Predicted versus true iGrowth (n = 99) using (A) the optimally weighted gene expression matrix (GXM) alone, (B) the optimally weighted microRNA expression matrix (MXM) alone, and (C) the optimally weighted combination of the two matrices from the grid search. The solid black lines represent the slopes of the regression between the predicted and true values. The red dashed lines are the identity lines representing perfect prediction (slope 1, intercept 0). (D) Results of the grid search that shows that the best iGrowth prediction correlation (R2 = 0.48 [0.45, 0.52]) was obtained with (MXM, GXM) matrix weights of (0.1, 0.8). The R2 values presented in the contour plot are the mean values from 500 random samplings of the data into 16 cross-validation folds.
The 95% confidence interval of each prediction [in brackets] was determined by 500 permutations of randomly partitioning the data into training and test sets as described in the Methods. In addition, for the MXM alone, the optimal prediction correlation was R2 = 0.35 [0.32, 0.38] when θMXM = 0.4 and θε = 0.6 (Fig. 3B). We then performed a grid search to determine if combining the GXM and MXM similarity matrices improved the iGrowth prediction. The optimal prediction from the grid search increased the correlation to R2 = 0.48 [0.45, 0.52], when θGXM = 0.8, θMXM = 0.1, and θε = 0.1 (Fig. 3C–D). The nonoverlapping confidence intervals indicate that combining genome-wide expression data improved the iGrowth predictive power of OmicKriging over using either the GXM or MXM alone.
We developed a baseline model similar to a polygenic score model, but with the top gene and microRNA expression associations rather than SNP associations, to compare to our OmicKriging results. Specifically, using the entire sample, we determined that 255 genes and 14 microRNAs associated with iGrowth by univariate linear regression after Bonferroni correction for multiple tests. Then, we performed 16-fold cross-validation by determining the top 255 genes and top 14 microRNAs by univariate linear regression in each training set and using the effect sizes to predict iGrowth in each test set. In addition to the top 255 genes and top 14 microRNAs from each training set, we also included the first 10 principal components derived from the genotype data in each multivariate prediction model. We repeated the cross-validation procedure 500 times to generate a confidence interval. The baseline model was unable to predict iGrowth, R2 = 0.0038 [–0.010, 0.064]. Therefore the maximum R2 = 0.48 obtained by OmicKriging represents a vast improvement in iGrowth prediction over the baseline model of top expression associations (Table 1).
Table 1.
iGrowth prediction is maximized by integration of expression data through OmicKriging
| Model: | OmicKriging GRM | OmicKriging GXM | OmicKriging MXM | OmicKriging GXM+MXM | Baseline |
|---|---|---|---|---|---|
| Mean R2 [95% CI] | 0.020 [–0.0083, 0.077] | 0.38 [0.34, 0.43] | 0.35 [0.32, 0.38] | 0.48 [0.45, 0.52] | 0.0038 [–0.010,0.064] |
| Matrix Weights | wgrm = 0.1, wE = 0.9 | wgxm = 1, wE = 0 | wmxm = 0.4, wE = 0.6 | wgxm = 0.8, wmxm = 0.1, wE = 0.1 | NA |
GRM, genetic relationship matrix; GXM, gene expression correlation matrix; MXM, microRNA expression matrix; Baseline, multivariate prediction model including top genes, top microRNAs, and first ten principal components.
Clinical Phenotype Applications
Clinical Statin Response
We also applied OmicKriging to the Cholesterol and Pharmacogenetics (CAP) Simvastatin Study, which contains both genome-wide genotype and expression data (Barber et al., 2010; Medina et al., 2012; Simon et al., 2006). DNA samples were either genotyped on the Illumina HumanHap 300K beadchip or the Illumina HumanHap 610K-Quad beadchip. A total of 562 individuals had their change in low-density lipoprotein cholesterol (dLDLC) after simvastatin treatment measured and also passed genotyping quality control. Additional SNP genotypes were imputed into these samples using data from the 1000 Genomes Project (Consortium, 2012). To assess potential predictive ability of the SNPs, we estimated the heritability of dLDLC captured by the 8.7 M imputed SNPs to be 0.69 (SE 0.61) using GCTA (Yang et al., 2011). Because of the large standard error obtained when all the SNPs were used, we sought to obtain a significant estimate of heritability by using a subset of SNPs known to be involved in statin response from a separate study. The Heart Protection Study identified 45 SNPs (Tables 2 and Supplementary Table S3 in Hopewell et al.) to be involved in LDLC response to simvastatin (Hopewell et al., 2013). We used SNPs within 50 kb of these 45, for a total of 10,925 SNPs, and obtained a heritability estimate for dLDLC of 0.16 (SE 0.079, P = 0.01). The estimated heritability is quite low but the fact that it is significantly different from zero indicated that significant predictive power might be attainable. When we applied OmicKriging to the CAP SNP data for dLDLC prediction, the optimal prediction correlation was R2 = 0.037 [0.026, 0.049] when θGRM = 0.8 using a single GRM calculated from the Hopewell 50 kb SNPs. Adding a second GRM of all imputed SNPs and performing a grid search did not improve the dLDLC prediction.
Table 2.
OmicKriging and other model performance for prediction of statin-induced change in LDL cholesterol levels
| Model: | OmicKriging Hopewell GRM (n = 562) | OmicKriging Hopewell GRM (n =461) | OmicKriging RHOA GXM (n = 461) | OmicKriging Hopewell GRM + RHOA GXM (n = 461) | Polyscore top1K (n = 562) | Baseline (n = 461) |
|---|---|---|---|---|---|---|
| Mean R2 [95% CI] | 0.037 [0.026, 0.049] | 0.024 [0.015, 0.035] | 0.0021 [–0.00086, 0.0057] | 0.025 [0.014, 0.036] | 0.016 [0.0048, 0.030] | –0.00014 [−0.0022, 0.0070] |
| Matrix Weights | wgrm = 0.8, wE = 0.2 | wgrm = 0.9, wE =0.1 | wgxm = 0.1, wE = 0.9 | wgrm = 0.6, wgxm = 0.3, wE = 0.1 | NA | NA |
Hopewell GRM, genetic relationship matrix of Hopewell et al. 50 kb SNPs; RHOA GXM, gene expression correlation matrix of RHOA regulatory pathway; Polyscore top l K, polygenic score model of top 1 K SNPs; Baseline, multivariate prediction model including top genes, the 45 Hopewell et al. SNPs, and first ten principal components.
To compare our OmicKriging method to existing WGP methods, we used polygenic score and attempted several sets of top SNPs for prediction of dLDLC using a 16-fold cross-validation approach. The best prediction had R2 = 0.016 [0.0048, 0.030] when the top 1,000 SNPs from each training set were used to predict the respective test set, which is less than half the prediction R2 obtained with OmicKriging (R2 = 0.037, Table 2).
Of the 562 individuals in the CAP study, 461 of them also had gene expression measurements from LCLs derived from their blood. Patient LCLs were exposed to simvastatin or vehicle control (baseline) for 24H and then gene expression was measured using the Illumina Ref8v3 beadchip (Medina et al., 2012). The differences between the treated and baseline expression values for each individual were used to generate a GXM. Including all 12,951 expressed genes in the GXM and performing a grid search with the Hopewell GRM did not improve the dLDLC prediction over the GRM alone. Like we did for the SNPs, we then chose to only include genes from candidate pathways known to be involved in lipid metabolism and inflammation in the GXM in an attempt to improve prediction. Combining a GXM of the 28 expressed genes from the PID RHOA REG PATHWAY from the Molecular Signatures Database (MSigDB) (Liberzon et al., 2011) with a GRM of the Hopewell SNPs produced an optimal R2 = 0.025 [0.014, 0.036] when θGXM = 0.3 and θGRM = 0.6. This prediction was not significantly different from using the Hopewell GRM alone with this reduced sample size (R2 = 0.024 [0.015, 0.035], when θGRM = 0.9). We also developed a baseline model using the 45 SNPs reported by Hopewell et al. combined with the first 10 principal components and the top 396 gene expression levels in a polygenic score prediction, but the R2 was negligible (R2 = –0.00014 [–0.0022, 0.0070]). Thus, combining the genotype and gene expression data did not improve the prediction (Table 2).
WTCCC Diseases
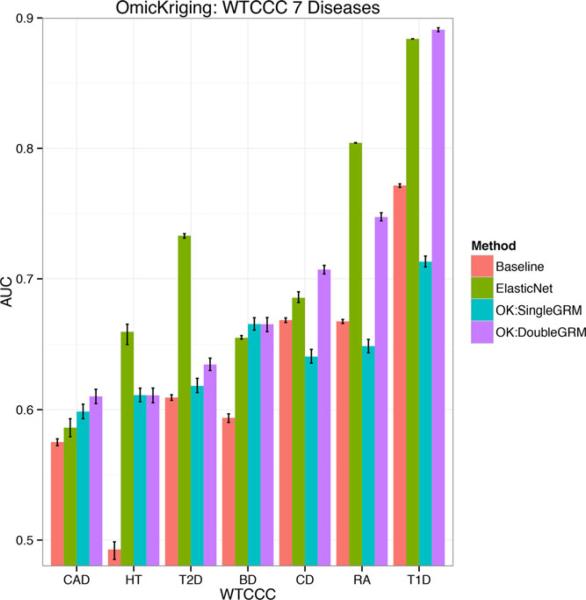
To test the predictive power of OmicKriging using larger clinical datasets, we turned to the seven diseases of the Well-come Trust Case Control Consortium (WTCCC) (Burton et al., 2007). For each disease, there are approximately 2000 cases and 3000 common controls. First, for each case/control dataset, all genotyped common SNPs (approximately 400,000) were used to generate a GRM. OmicKriging was run using these single GRMs (equivalent to G-BLUP) and case/control prediction was successful (AUC > 0.50 that would be considered random guess) for all seven diseases as has been shown previously (Zhou et al., 2013). In our analysis, mean areas under the ROC curve (AUCs) ranged from 0.598 [0.593, 0.604] when θGRM = 0.4 for coronary artery disease to 0.713 [0.709, 0.717] when θGRM = 0.4 for type 1 diabetes (Fig. 4, Table 3). The 95% confidence intervals [in brackets] were determined by randomly partitioning the data 500 times into 16 subsets and performing cross-validated prediction on every random partition to generate a distribution of 500 AUC values. While we did perform a grid search to determine the best AUC for each WTCCC disease, optimization typically resulted in minimal improvement. That is, the optimized θGRM did not improve the AUC greater than 0.02 over the default θGRM = 1.
Figure 4.
OmicKriging prediction performance for WTCCC disease risk prediction. Mean area under the ROC curve (AUC) for two implementations of OmicKriging for each disease from the WTCCC: a single common SNP genetic relationship matrix (OK:SingleGRM) and two optimallyweightedGRMsofcommonSNPsandknownloci(OK:DoubleGRM) for the predictions. The known loci were obtained from studies that did not include the WTCCC data to avoid overfitting. For comparison, we also show mean AUC results of the polygenic score method using genome-wide significant loci with 10 principal components (Baseline) and the lambda-optimized elastic-net penalized model (ElasticNet). Error bars represent the 95 % confidence intervals from multiple cross-validation runs (see Methods). BD, bipolar disorder; CAD, coronary artery disease; CD, Crohn's disease; HT, hypertension; RA, rheumatoid arthritis; T1D, type 1 diabetes; T2D, type 2 diabetes.
Table 3.
OmicKriging and other model performance for prediction of disease risk in the WTCCC
| Disease | Results | OmicKriging Single GRM | OmicKriging Double GRM | ElasticNet | BSLMM (SD)* | Baseline |
|---|---|---|---|---|---|---|
| BD | Mean AUC [95% CI] | 0.666 [0.661,0.670] | 0.665 [0.660, 0.670] | 0.655 [0.654,0.657] | 0.65 (0.02) | 0.594 [0.590,0.597] |
| Matrix Weights | wGRM = 0.3, wε = 0.7 | wGRM = 0.4, wGRMk = 0, wε = 0.6 | NA | NA | NA | |
| CAD | Mean AUC [95% CI] | 0.598 [0.593, 0.604] | 0.610 [0.605, 0.616] | 0.586 [0.579, 0.593] | 0.60 (0.03) | 0.575 [0.572, 0.578] |
| Matrix Weights | wGRM = 0.4, wε = 0.6 | wGRM = 0.6, wGRMk = 0.1, wε = 0.3 | NA | NA | NA | |
| CD | Mean AUC [95% CI] | 0.641 [0.636, 0.646] | 0.707 [0.704, 0.710] | 0.685 [0.682, 0.690] | 0.68 (0.02) | 0.669 [0.667, 0.670] |
| Matrix Weights | wGRM = 0.3, wε = 0.7 | wGRM = 0.4, wGRMk = 0.1, wε = 0.5 | NA | NA | NA | |
| HT | Mean AUC [95% CI] | 0.611 [0.606, 0.616] | 0.611 [0.605, 0.616] | 0.660 [0.650, 0.666] | 0.60 (0.02) | 0.493 [0.485, 0.499] |
| Matrix Weights | wGRM = 0.3, wε = 0.7 | wGRM = 0.3, wGRMk = 0, wε = 0.7 | NA | NA | NA | |
| RA | Mean AUC [95% CI] | 0.649 [0.644, 0.654] | 0.747 [0.744, 0.751] | 0.8042 [0.8041, 0.8044] | 0.72 (0.01) | 0.668 [0.666, 0.669] |
| Matrix Weights | wGRM = 0.2, wε = 0.8 | wGRM = 0.2, wGRMk = 0.5, wε = 0.3 | NA | NA | NA | |
| T1D | Mean AUC [95% CI] | 0.713 [0.709, 0.717] | 0.891 [0.889, 0.892] | 0.8839 [0.8838, 0.8840] | 0.88 (0.01) | 0.772 [0.770, 0.773] |
| Matrix Weights | wGRM = 0.4, wε = 0.6 | wGRM = 0.3, wGRMk = 0.4, wε = 0.3 | NA | NA | NA | |
| T2D | Mean AUC [95% CI] | 0.618 [0.613, 0.624] | 0.634 [0.630, 0.639] | 0.733 [0.731, 0.734] | 0.61 (0.03) | 0.609 [0.607, 0.611] |
| Matrix Weights | wGRM = 0.3, wε = 0.7 | wGRM = 0.7, wGRMk = 0.1, wε = 0.2 | NA | NA | NA |
Values reported in Zhou et al., ElasticNet, elastic-net penalized model; BSLMM, Bayesian sparse linear mixed model; SD, standard deviation; Baseline, polygenic score model of known GWAS SNPs and first ten principal components; AUC, area under the receiver operating characteristic curve; CI, confidence interval; GRM, genetic relationship matrix (all SNPs); GRMk, genetic relationship matrix of known GWAS SNPs; BD, bipolar disorder; CAD, coronary artery disease; CD, Crohn's disease; HT, hypertension; RA, rheumatoid arthritis; T1D, type 1 diabetes; T2D, type 2 diabetes.
In an attempt to further improve the prediction by integrating existing information on variants from previous studies, we generated a second GRM for each disease using the SNPs within 100 kb of known loci (identified outside of WTCCC studies for each disease) listed in the The National Human Genome Research Institute GWAS catalog (Hindorff et al., 2009) and the Database of Genotypes and Phenotypes (dbGAP) (Mailman et al., 2007). The optimal double GRM improved the predictive power of OmicKriging over using just the single common-SNP GRM alone slightly for coronary artery disease and type 2 diabetes and dramatically for Crohn's disease, rheumatoid arthritis, and type 1 diabetes (Fig. 4). The type 1 diabetes prediction showed the largest improvement when the second GRM was added: the AUC increased from 0.713 to 0.891 [0.889, 0.892] (Fig. 4, Table 3).
A comparison of different polygenic prediction approaches has been published by Abraham et al. [2013], where the authors report that the elastic-net approach slightly outperforms other methods. We compared OmicKriging to the elastic-net penalized model and to a baseline model that uses only genome-wide significant SNPs and the first ten principal components to calculate predicted phenotypes by the polygenic score method. Both OmicKriging models (single and double GRM) outperformed the elastic-net and baseline model for coronary artery disease and bipolar disorder (Fig. 4, Table 3). While both OmicKriging models outperformed the baseline model for hypertension and type 2 diabetes, elastic-net performed the best for these two diseases. The OmicKriging double GRM model greatly outperforms the baseline model for Crohn's disease, rheumatoid arthritis, and type 1 diabetes. The OmicKriging double GRM model also outperforms the elastic-net model for Crohn's disease and type 1 diabetes, while elastic-net was better for rheumatoid arthritis (Fig. 4, Table 3) .
Discussion
We propose a novel systems approach to predict complex traits, which leverages and integrates similarity in genetic, transcriptomic and/or other large-scale omics data. It can also integrate other sources of information such as prior evidence of association or function and even geographic proximity. We translate the genomic similarity into phenotypic similarity using a method called Kriging, commonly used in geostatistics and machine learning. Here, we construct the genomic similarity using a linear combination of omic similarity matrices (in addition to the environmental component), but more general similarity matrices can be used. Given the similarity matrix, the prediction is obtained by simply computing a weighted average of the phenotype of individuals in the training set. The individual weights are providedbytheKrigingmethod,whichcanbelooselyinterpreted as a converse of the linear regression method. Our method is a fast, simple, and flexible approach to polygenic, and more generally poly-omic, prediction. In addition, we provide an R package called OmicKriging and a tutorial explaining how to implement the Kriging algorithm at http://www.scandb.org/newinterface/tools/OmicKriging.html.
The key component of the method is the choice of the similarity matrix. If we assume certain conditions such as additive poly-omic models (as described in the Methods section) it is possible to estimate the matrix weights for each omic component as well as the environmental component using a (restricted) maximum likelihood approach. However, some of the modeling assumptions are quite stringent and we opted for a more pragmatic approach in which we use the matrix weights that maximize predictive performance. In this manuscript, we use a grid search approach to find reasonably close to optimal matrix weights for each similarity matrix component (GRM of known large effect SNPs, GRM of all SNPs, GXM, etc.).
Using mRNA (GXM) and microRNA (MXM) data in HapMap LCLs, we predict the cellular intrinsic growth phenotype with an out of sample R2 of 0.48 with just 99 samples. For comparison, previous studies using similar datasets had reported R2 values of the same magnitude but were computed in-sample, which is known to be highly upward-biased. The combined mRNA-microRNA prediction R2 was 0.10–0.13 higher than using either the GXM or MXM alone. Given that 30% of gene expression levels associate with the intrinsic growth phenotype (FDR < 0.1) (Im et al., 2012), it is perhaps not surprising that gene expression correlations are most useful in predicting this phenotype. However, using just the most significantly associated expression levels as was done in the baseline polyscore model, was not predictive. These results stress the importance of considering other potential biomarkers besides genotype, because the GRM was not helpful in predicting this particular phenotype at this sample size. LCLs from populations of both African and European descent were used in this analysis.
The issue of population stratification has slightly different implications in the prediction context than it does in the GWAS estimation context. If our goal is to achieve the best prediction possible, we are not concerned whether the predictive power comes from the genomic component or environmental and populations stratification components that are confounded with the genomic component. Naturally, we have to be aware of this limitation and not to expect similar levels of accuracy when predicting in different population mixtures.
Because the dataset contained genotype and gene expression data, we used the CAP simvastatin study to test the potential predictive ability of OmicKriging applied to a clinical phenotype with multiple types of omics-level data. While the out-of-sample prediction correlations were not large with OmicKriging, likely due to limited sample size, the best SNP-based prediction R2 was more than twice as high as that obtained from the polygenic score method (R2 = 0.037 vs. R2 = 0.016). While inclusion of expression data did not improve prediction over genetic variants alone, the CAP study examples demonstrate how OmicKriging can be applied to multiple types and subsets of omics data for phenotypes with such data available. This example demonstrates that polygenic prediction is possible when the SNPs used for prediction are selected based on top hits from prior studies even with limited sample size (i.e. less than the 1,000 individuals typically needed at minimum to estimate complex-trait heritability using whole-genome SNPs). In general, if prior information is not used to select SNPs for prediction, OmicK-riging will have similar sample size limitations as GCTA(Yang et al., 2011). The magnitude and standard error of the heritability estimate using either all SNPs or a selection of SNPs will anticipate the predictive power that can be achieved for a particular SNP set. This is, if the estimated heritability is low or the standard error is large, we do not expect significant predictive power of the phenotype by OmicKriging.
We also successfully predicted seven clinical disease risk phenotypes with OmicKriging. For WTCCC disease prediction, we show that our OmicKriging method yields performance similar to or better than polygenic score and similar to elastic-net. In addition, OmicKriging when restricted to genotypic data performs just as well as the computationally more intensive BSLMM method (Zhou et al., 2013) (Table 3). Our goal is not to present just another polygenic prediction method but a simple and flexible method that can integrate across multiple heterogeneous sources of data, which spans prior information on GWAS results, other omic data or even geographic similarity.
The average OmicKriging double GRM run time on a Xeon E5345 processor is 14 min, whereas the average GEMMA software (Zhou and Stephens, 2012) run time for BSLMM is 28 hr on the Xeon L5420 processor, which is a slighly newer, but comparable processor (Zhou et al., 2013). Most of the BSLMM run time is used for the Markov chain Monte Carlo (MCMC) iterations, whereas in OmicKriging, we specify the sparse effects (known GWAS loci) before the run. For the known autoimmune diseases (Crohn's disease, rheumatoid arthritis, type 1 diabetes), adding a second GRM of known loci increased the prediction AUC values over a single common SNP GRM alone to AUC values slightly higher (not significant) than those obtained by BSLMM (Table 3). (Zhou et al., 2013). These autoimmune diseases are known to have multiple associated loci of relatively strong effects, so this prior knowledge was used to improve prediction performance (Visscher et al., 2012). Unless replicated in an independent study, the results from the WTCCC data were not used to select top SNPs to avoid overfitting the data.
In the double GRM OmicKriging model we have used all SNPs to build the first similarity matrix and a subset of SNPs with prior evidence of association to build the second similarity matrix. The SNPs that were previously implicated are used in both similarity matrices. The prediction is not affected by this choice. The main reason we use this approach is to make the underlying model directly comparable to the Bayesian Sparse Linear Mixed Model, BSLMM (Zhou et al., 2013), where the effect sizes of all SNPs can be represented as a mixture of two distributions: one with small effect sizes for all SNPs and one with large effect sizes for a subset of SNPs. This way the variance explained by the second GRM is directly comparable to the PGE value (the proportion of variance explained by the sparse terms) in BSLMM. The total variance explained by the known variants will be the sum of the PGE and the proportion of the variance explained by the small effect sizes of the known variants from the first GRM.
While we recognize that assuming that a binary trait is continuous is not statistically optimal, we do so here in our initial modeling for computational reasons, as have others (Lee et al., 2011; Zhou et al., 2013). Linear probability models for binary outcomes are considered to be adequate approximations when the proportion of cases (and controls) exceed 25% given the approximate linearity of the logit or probit functions near the origin (Zhou et al., 2013). It has been reported that the gain in statistical efficiency is hard to realize because of the added computational burden and consequent loss in numerical accuracy (Visscher et al., 1996; Zhou et al., 2013). Unlike us, Vazquez et al. used a probit link model for skin cancer incidence, but needed to restrict their analysis to only 41 K SNPs due to computer memory limitations (Vazquez et al., 2012). In their dataset, the linear probability model would have been less appropriate since the proportion of cases was between 11 and 24%.
We use an additive poly-omic model to motivate the main functional form of the overall similarity matrix but find the matrix weights for each omic component (and the environmental component) by maximizing the cross-validated prediction performance. Thus our approach is closely related to the semi-parametric models using RKHS regression proposed by Gianola et al. [2006] and the kernel averaging approach proposed by de los Campos et al. [2010b] for WGP. We use an additive poly-omic model to motivate the structure of the composite similarity matrix, but have ignored the cross-correlation terms between different omic components by assuming independence of the effects from different sources. Approaches to include the cross-correlation term merit further investigation. It may be possible to use eQTL information to restrict the number of nonzero correlations, but even then further assumptions must be made to be able to estimate the needed parameters given available data and computational limitations.
Methods
OmicKriging Approach
We propose to use an extension of the Kriging framework to integrate different omic data as well as prior information on function such as existing GWAS studies, eQTL information (genetic markers associated with gene expression levels), regulatory evidence such as provided by ENCODE studies, etc. We build the similarity matrix as a linear combination of the similarity matrices from each omic component where the coefficients or weights for each component are chosen so that prediction performance is maximized. Given a similarity matrix, the usual Kriging formulas are used to compute the predicted values. Prediction is performed by randomly partitioning the samples into 16 subsets and using each subset as the testing set and the remaining 15 sets as the training set. This is repeated 500 times to assess the sampling variability. The correlation squared between the true and predicted values are used as performance measures. For binary/disease traits we use the area under the receiving operating curve. In this work, we assume a linear probability model for the disease status following Lee et al. [2011] and Zhou et al. [2013].
Omic Similarity Matrix
For each omic dataset used for prediction we compute the corresponding similarity matrix. For convenience, we will denote the similarity matrix constructed from genetic data as GRM, the one constructed by mRNA expression profile data as GXM, and the ones constructed with microRNA expression data as MXM. We assume that the environmental component is independent across individuals and is represented by the identity matrix, . In the current implementation of the OmicKriging R package, we are computing the similarity matrix for genetic data by invoking the GCTA (Yang et al., 2011) software, whereas for other omic data we compute the similarity matrix directly in R. More specifically, the ij component of the GRM is computed as
and the ij component of the other omics data is computed as
where i and j denote individuals, is the number of reference alleles of individual i at marker l, pl is the reference allele frequency of marker l, is the level of omic marker l (l is a dummy index and there is no one to one correspondence between genetic and omic markers indices), M is the number of genetic markers, L is the number of genes or omic markers, , and .
By using the correlation without prior centering and standardizing the gene expression and other continuous omic traits, we are effectively giving more weight to the traits that have larger variance. The effects of different choices of the similarity matrix on the prediction accuracy will be investigated in future work.
Optimal Similarity Matrix Under an Additive Model
A key component of the success of OmicKriging is understanding which proximity measures translate best into phenotypic similarity. The optimal similarity matrix depends on the underlying genetic and epigenetic architecture of the complex trait.
We will use an additive poly-omic model to motivate our choice of the similarity matrix. The phenotype for individual i, Yi (a scalar) is represented as
| (1) |
where
a is a constant,
is the additive genetic component (assumed to be known), is the effect size of the standardized genotype , and M is the total number of genetic markers,
(assumed to be known), is the effect size of the standardized gene expression and L is the total number genes,
is the additive (other) omic component (assumed to be known), is the effect size of the standardized omic level , and L’ is the total number of omic markers,
and εi is a noise term (iid, independent and identically distributed).
For notational convenience let us define Xi without a superscript to denote all three omic data such that if l ≤ M, if M < l ≤ M + L and if M + L < l ≤ M + L + L’ and similarly for coefficients β's such that if l ≤ M, if M < l ≤ M + L and if M + L < l ≤ M + L + L’.
We assume a random effects model for the β's. For convenience we also assume that the X's have been centered and standardized. If we further assume that the betas are independent, i.e. that then the covariance matrix of the n-vector Y will have components
| (2) |
where δij is the kronecker delta (1 if i = j and 0 otherwise) and θG , θT, and θO are non negative. If all modeling assumptions were met and we assumed normality of the environmental term, this covariance matrix should be used as the similarity matrix to compute the best linear unbiased prediction (BLUP). However, these assumptions are quite strong and do not account for correlations of between marker effects, gene– gene interactions, gene–environment interactions, etc. Thus, we adopt a pragmatic approach in which we use a combination of the covariance matrices for each omic component but allow the weights θ to vary and pick the combination that provides the best predictive performance.
Independence assumption for all betas is clearly too restrictive. The effect size of genetic marker , , that influences gene expression level is likely to be correlated with . For unconstrained values of the cov(βk,βl) the covariance matrix has the form
In case prior expression quantitative trait loci (eQTL, genetic markers that have an effect on gene expression traits) information is available, it may be possible to restrict the nonzero cross-correlation terms to known eQTL pairs . Additional restrictions in the values of the cov(βl, βk) must be imposed to be able to characterize them given existing data and care must be taken to preserve positive definiteness of Σ (all eigenvalues must be >0). This is a complex topic that merits further research. In this paper, to keep computations within reach, we ignore the cross-correlation terms and find the coefficient thetas that maximize prediction performance.
Composite Similarity Matrix
Based on the form of the optimal similarity matrix under an additive poly-omic model, we propose to use a composite similarity matrix that integrates different omic components to be used for Kriging that is a linear combination of each component similarity matrix (Ss)
where the weights θ's will be determined as the ones providing optimal prediction. All coefficients θ are constrained to be nonnegative. The environmental component is known as the nugget term in geostatistical applications.
Kriging Formula
Within the Kriging framework, the predicted phenotype of a test individual is computed as the weighted average of the phenotype of the individuals in the training set.
| (3) |
where the weights ωi are a function of all n(n + 1)/2 pairs of similarity measures. In the simplest case where no covariates are needed, the weights prescribed by the Kriging method are given by
| (4) |
where ρ is the similarity vector between the test individual and the training individuals and Σ is the similarity matrix of the individuals in the training set (Cressie, 1993). Covariates are easily included in the method by using the so called universal Kriging approach (Cressie, 1993). Assuming there are p covariates (if only the intercept is considered, p = 1), let z be the p by 1 vector with covariates 1 to p corresponding to the test individual and be the n by p matrix with the p covariates for the n individuals in the training set. The weights become
| (5) |
where .
Prediction Performance
We measure prediction performance for binary traits with the area under the receiving operator characteristic curve (AUC). For quantitative traits, we use R2, the correlation coefficient between true and predicted values squared.
Grid Search
When using a single similarity matrix, we compared the prediction performance measures for similarity matrix weights, θ1 = 0, 0.1, 0.2,... 1, and environmental component weights, θ1 = 1 – θ1. For two similarity matrices (e.g. GRM and GXM) we allow matrix weights θ1 = 0, 0.1, 0.2,... 1 and θ2 = 0, 0.1, 0.2,... 1, with the constraint that θ1 + θ2 ≤ 1. When two similarity matrices are used, the environmental component weights are θε = 1 – θ1 – θ2.
Sampling Variability
To assess the uncertainty of the R2 or AUC estimates due to sampling variability, we randomly partitioned each dataset 500 times (except elastic-net) into 16 subsets and performed cross-validation on every random partition. That is, within each cross-validation fold, 1/16 of the data was used as the test set and 15/16 of the data was used as the training set. This random sampling and cross-validation generated a distribution of 500 R2 or AUC values for each prediction method and trait from which 95% confidence intervals were calculated (as the 0.025 and 0.975 percentiles of the distribution of R2 or AUC values). All analyses were performed using the R statistical language and environment (Team, 2005).
Clinical Statin Response Analysis
Genotype Imputation
Self-reporting Caucasian individuals from the Cholesterol and Pharmacogenetics Study (Simon et al., 2006) were geno-typed on the Illumina HumanHap 300 K beadchip (n = 305) or the Illumina HumanHap 610K-Quad beadchip (n = 282). Prior to imputation, we performed standard GWAS quality control, removing poorly called SNPs and SNPs in Hardy– Weinberg disequilibrium (HWD, P < 0.001). We also removed related individuals and outliers for heterozygosity or principal components. This left 562 individuals, who also had baseline and postsimvastatin treatment LDLC measurements, for imputation. Prior to imputation we prephased the genotype data using SHAPEIT (Delaneau et al., 2011) using the recommended settings. Then we used IMPUTE2 (Howie et al., 2009) to impute genotypes from the 1000 Genomes Project (Consortium, 2012) using the default settings for prephased data. A total of 8.7 M SNPs with IMPUTE2-info scores >0.3 and minor allele frequency (MAF) > 0.001 and genotypes with probabilites > 0.9 were used in the heritability estimation and prediction analyses.
Phenotype
The change in low-density lipoprotein cholesterol (dLDLC) phenotype was calculated by subtracting the log-transformed mean (over the two baseline measurements) of the baseline LDLC plasma levels from the log-transformed mean (over the two visits posttreatment) of the LDLC plasma levels collected while patients were on simvastatin (Simon et al., 2006).
Expression Pathway Analysis
To test whether the gene expression of pathways potentially related to simvastatin-induced LDLC response could predict dLDLC, we chose a few canonical pathways from the MSigDB (Liberzon et al., 2011) to test in our OmicK-riging model as proof-of-concept for future more comprehensive pathway analyses. The pathways tested for prediction ability include BIOCARTA INFLAM PATHWAY, PID RHOA PATHWAY, PID RHOA REG PATHWAY, and REACTOME CHOLESTEROL BIOSYNTHESIS. While RHOA has been previously implicated in lipid metabolism and thus makes a plausible candidate pathway, we focus on it here over many other potential lipid metabolism pathways, because recent functional work in the CAP LCLs have revealed specific effects of RHOA (ras homolog family member A) in modulating the cholesterol-lowering effects of statin (Medina et al., 2012).
WTCCC Disease Analysis
We performed standard quality control for all WTCCC data sets. All WTCCC data sets were merged into a single bed file where we removed all individuals recommended by the WTCCC. This left 2,937 common controls, 1,868 bipolar disorder cases, 1,926 cardiovascular disease cases, 1,748 Crohn's disease cases, 1,952 hypertension cases, 1,860 rheumatoid arthritis cases, 1,963 type 1 diabetes cases, and 1,924 type 2 diabetes cases. We removed SNPs in HWD P < 0.0005 and MAF < 0.01. This resulted in approximately 388 K SNPs after pruning. In addition, we computed the GRM for all WTCCC and identified a pair of cases with unusually high relatedness that were not included in the WTCCC removal list. The duplicate individual was removed.
OmicKriging Models
We selected SNPs from dbGAP and NHGRI to be used in the double GRM model (Hindorff et al., 2009; Mailman et al., 2007). We pruned all SNPs from studies that contained WTCCC datasets. We fit OmicKriging models with single (all SNPs) and double GRMs (all SNPs and known GWAS SNPs) in 16-fold cross-validation. We chose 16-fold because OmicKriging can be multithreaded and for these analyses, we used dual Xeon E5620 processors with 16 logical cores.
We performed a grid search (as described previously) to identify the optimal weights for single and double GRM models (Table 3). Prediction performance of all OmicKriging and baseline models was measured by area under the receiver operating characteristic (ROC) curve (AUC) with the package ROCR in R (Sing et al., 2005). We used the R package ggplot2 (Wickham, 2009) to generate Figure 4.
Polygenic Score Models
We applied the polygenic score method by fitting the first 10 principal components and p genome-wide-significant loci jointly in 16-fold cross-validation (baseline model). Specifically, we fit Y~PC1 + · · · + PC10 + SNP1 + + SNPp in each training set (15/16th of the dataset). With· · · the remaining test set (1/16th of the dataset), the m =p+10 estimated regression coefficients are multiplied by an nxm matrix of n individuals and m principal components/genotype dosages (Zil) and each individual's predicted phenotype (polygenic score) is the sum of the respective individual's products: .
Elastic-Net Models
We applied the elastic-net regularized regression method implemented by the glmnet package in R (Friedman et al., 2010) to the WTCCC data. In the WTCCC data, glmnet traverses a lambda penalty path for 100 iterations. We performed three replications of 16-fold cross-validation to estimate mean AUC and calculate 95% confidence intervals for prediction performance. Lambda penalty was chosen to be the value that maximized the AUC estimate for each WTCCC disease. The elastic-net mixing parameter alpha was set to 0.5.
Acknowledgments
We thank Peter McCullagh for useful comments on the manuscript. The Cholesterol and Pharmacogenetics Study data used for the analyses described in this manuscript were obtained through an agreement with the investigators and the Pharmacogenomics Research Network (PGRN) Statistical Analysis Resource Workshop and from the database of Genotype and Phenotype (dbGaP) found at http://www.ncbi.nlm.nih.gov/gap through db-GaP accession number phs000481.v1.p1. This work was supported in part by U19 HL069757-11: Pharmacogenomics and Risk of Cardiovascular Disease (PARC). We acknowledge the PARC investigators and research team, supported by NHLBI, for collection of data from the Cholesterol and Pharmacogenetics clinical trial. This study makes use of data generated by the Wellcome Trust Case-Control Consortium. A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. Funding for the project was provided by the Wellcome Trust under award 076113 and 085475. This study was in part supported by the Pharmacogenetics of Anticancer Agents Research (PAAR) Group (NIH/NIGMS grant UO1GM61393), the Genotype-Tissue Expression project (GTeX) (R01 MH101820 and R01 MH090937), the University of Chicago DRTC (Diabetes Research and Training Center; P60 DK20595; P30 DK020595), the University of Chicago Cancer Center Support Grant (NCI P30 CA014599-36), the PGRN Statistical Analysis Resource (U19 HL065962), and the Conte Center grant P50MH094267. HEW was supported in part by the National Research Service Award F32CA165823. HKI was supported in part by Award Number K12CA139160 from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
References
- Abraham G, Kowalczyk A, Zobel J, Inouye M. SparSNP: fast and memory-efficient analysis of all SNPs for phenotype prediction. BMC Bioinformatics. 2012;13:88. doi: 10.1186/1471-2105-13-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham G, Kowalczyk A, Zobel J, Inouye M. Performance and robustness of penalized and unpenalized methods for genetic prediction of complex human disease. Genet Epidemiol. 2013;37(2):184–195. doi: 10.1002/gepi.21698. [DOI] [PubMed] [Google Scholar]
- Barber MJ, Mangravite LM, Hyde CL, Chasman DI, Smith JD, McCarty CA, Li X, Wilke RA, Rieder MJ, Williams PT. Genome-wide association of lipid-lowering response to statins in combined study populations. PLoS One. 2010;5(3):e9763. doi: 10.1371/journal.pone.0009763. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton PR, Clayton DG, Cardon LR, Craddock N, Deloukas P, Duncanson A, Kwiatkowski DP, McCarthy MI, Ouwehand WH, Samani NJ. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium TGP. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cressie NA. Statistics for Spatial Data, revised edition , volume 928. Wiley; New York: 1993. [Google Scholar]
- Dai H. A cell proliferation signature is a marker of extremely poor outcome in a subpopulation of breast cancer patients. Cancer Res. 2005;65(10):4059–4066. doi: 10.1158/0008-5472.CAN-04-3953. [DOI] [PubMed] [Google Scholar]
- Damasco C, Lembo A, Somma MP, Gatti M, Di Cunto F, Provero P. A signature inferred from Drosophila mitotic genes predicts survival of breast cancer patients. PLoS One. 2011;6(2):e14737. doi: 10.1371/journal.pone.0014737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Campos G, Gianola D, Allison D. Predicting genetic predisposition in humans: the promise of whole-genome markers. Nat Rev Genet. 2010a;11(12):880–886. doi: 10.1038/nrg2898. [DOI] [PubMed] [Google Scholar]
- de los Campos G, Gianola D, Rosa GJ. Reproducing kernel hilbert spaces regression: a general framework for genetic evaluation. J Anim Sci. 2009;87(6):1883–1887. doi: 10.2527/jas.2008-1259. [DOI] [PubMed] [Google Scholar]
- de los Campos G, Gianola D, Rosa GJM, Weigel KA, Crossa J. Semi-parametric genomic-enabled prediction of genetic values using reproducing kernel Hilbert spaces methods. Genet Res Camb. 2010b;92(4):295–308. doi: 10.1017/S0016672310000285. [DOI] [PubMed] [Google Scholar]
- Delaneau O, Marchini J, Zagury J-F. A linear complexity phasing method for thousands of genomes. Nat Methods. 2011;9(2):179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- Evans DM, Visscher PM, Wray NR. Harnessing the information contained within genome-wide association studies to improve individual prediction of complex disease risk. Hum Mol Genet. 2009;18(18):3525–3531. doi: 10.1093/hmg/ddp295. [DOI] [PubMed] [Google Scholar]
- Fisher RA. The correlation between relatives on the supposition of mendelian inheritance. Trans R Soc Edinburgh. 1918;52(02):399–433. [Google Scholar]
- Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–861. doi: 10.1038/nature06258. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Software. 2010;33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
- Gamazon ER, Ziliak D, Im HK, LaCroix B, Park DS, Cox NJ, Huang RS. Genetic architecture of microRNA expression: implications for the transcriptome and complex traits. Am J Hum Genet. 2012;90(6):1046–63. doi: 10.1016/j.ajhg.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianola D. Genomic-assisted prediction of genetic value with semiparametric procedures. Genetics. 2006;173(3):1761–1776. doi: 10.1534/genetics.105.049510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberger AS. Best linear unbiased prediction in the generalized linear regression model. J Am Stat Assoc. 1962;57(298):369–375. [Google Scholar]
- Habier D, Fernando R, Dekkers J. The impact of genetic relationship information on genome-assisted breeding values. Genetics. 2007;177(4):2389–2397. doi: 10.1534/genetics.107.081190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harville DA. Discussion on interpolation and estimation. In: David HA, David HT, editors. Statistics: An Appraisal. Iowa State Univ Press; Ames: 1984. pp. 281–286. [Google Scholar]
- Henderson CR. Estimation of genetic parameters. Ann Math Stat. 1950;21:309–310. [Google Scholar]
- Henderson CR. Best linear unbiased estimation and prediction under a selection model. Biometrics. 1975;31:423–447. [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci. 2009;106(23):9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoerl AE, Kennard RW. Ridge regression: applications to nonorthogonal problems. Technometrics. 1970;12(1):69–82. [Google Scholar]
- Hopewell JC, Parish S, Offer A, Link E, Clarke R, Lathrop M, Armitage J, Collins R. Impact of common genetic variation on response to simvastatin therapy among 18,705 participants in the heart protection study. Eur Heart J. 2013;34(13):982–992. doi: 10.1093/eurheartj/ehs344. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im HK, Gamazon ER, Stark AL, Huang RS, Cox NJ, Dolan ME. Mixed effects modeling of proliferation rates in cell-based models: consequence for pharmacogenomics and cancer. PLoS Genet. 2012;8(2):e1002525. doi: 10.1371/journal.pgen.1002525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im HK, Stein ML, Zhu Z. Semiparametric estimation of spectral density with irregular observations. J Am Stat Assoc. 2007;102(478):726–735. [Google Scholar]
- Janss L, de los Campos G, Sheehan N, Sorensen D. Inferences from genomic models in stratified populations. Genetics. 2012;192(2):693–704. doi: 10.1534/genetics.112.141143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet. 2011;88(3):294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Ritland K. Estimation of pairwise relatedness with molecular markers. Genetics. 1999;152(4):1753–1766. doi: 10.1093/genetics/152.4.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, Hao L, Kiang A, Paschall J, Phan L. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39(10):1181–1186. doi: 10.1038/ng1007-1181. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makowsky R, Pajewski N, Klimentidis Y, Vazquez A, Duarte C, Allison D, de los Campos G. Beyond missing heritability: prediction of complex traits. PLoS Genet. 2011;7(4):e1002051. doi: 10.1371/journal.pgen.1002051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina MW, Theusch E, Naidoo D, Bauzon F, Stevens K, Mangravite LM, Kuang Y-L, Krauss RM. RHOA is a modulator of the cholesterol-lowering effects of statin. PLoS Genet. 2012;8(11):e1003058. doi: 10.1371/journal.pgen.1003058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen T, Hayes B, Goddard M. Prediction of total genetic value using genome-wide dense marker maps. Genetics. 2001;157(4):1819–1829. doi: 10.1093/genetics/157.4.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre J, Johnson T, Bryc K, Kutalik Z, Boyko A, Auton A, Indap A, King K, Bergmann S, Nelson M. Genes mirror geography within europe. Nature. 2008;456(7218):98–101. doi: 10.1038/nature07331. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober U, Ayroles J, Stone E, Richards S, Zhu D, Gibbs R, Stricker C, Gianola D, Schlather M, Mackay T. Using whole-genome sequence data to predict quantitative trait phenotypes in Drosophila melanogaster. PLoS Genet. 2012;8(5):e1002685. doi: 10.1371/journal.pgen.1002685. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober U, Erbe M, Long N, Porcu E, Schlather M, Simianer H. Predicting genetic values: a kernel-based best linear unbiased prediction with genomic data. Genetics. 2011;188(3):695–708. doi: 10.1534/genetics.111.128694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park T, Casella G. The Bayesian Lasso. J Am Stat Assoc. 2008;103(482):681–686. [Google Scholar]
- Paulino Pérez, Gustavo de los Campos, José Crossa, Daniel Gianola. Genomic-enabled prediction based on molecular markers and pedigree using the Bayesian linear regression package in R. Plant Genome J. 2010;3(2):106. doi: 10.3835/plantgenome2010.04.0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Wray N, Stone J, Visscher P, O'Donovan M, Sullivan P, Sklar P, Ruderfer D, McQuillin A, Morris D. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460(7256):748–752. doi: 10.1038/nature08185. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan A. Large-scale meta-analysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc Natl Acad Sci USA. 2004;101(25):9309. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson GK. That BLUP is a good thing: the estimation of random effects. Stat Sci. 1991;6(1):15–32. [Google Scholar]
- Rosenwald A, Wright G, Wiestner A, Chan WC, Connors JM, Campo E, Gascoyne RD, Grogan TM, Muller-Hermelink H, Smeland EB. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell. 2003;3(2):185–197. doi: 10.1016/s1535-6108(03)00028-x. others. [DOI] [PubMed] [Google Scholar]
- Ross D, Scherf U, Eisen M, Perou C, Rees C, Spellman P, Iyer V, Jeffrey S, Van de Rijn M, Waltham M. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24(3):227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- Simon J, Lin F, Hulley S, Blanche P, Waters D, Shiboski S, Rotter J, Nickerson D, Yang H, Saad M. Phenotypic predictors of response to simvastatin therapy among African-Americans and Caucasians: the cholesterol and pharmacogenetics (CAP) study. Am J Cardiol. 2006;97(6):843. doi: 10.1016/j.amjcard.2005.09.134. others. [DOI] [PubMed] [Google Scholar]
- Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21(20):3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- Starmans MHW, Krishnapuram B, Steck H, Horlings H, Nuyten D SA, Vijver MJVD, Seigneuric R, Buffa FM, Harris AL, Wouters BG, Lambin P. Robust prognostic value of a knowledge-based proliferation signature across large patient microarray studies spanning different cancer types. Br J Cancer. 2008;99(11):1884. doi: 10.1038/sj.bjc.6604746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein ML. Interpolation of Spatial Data: Some Theory for Kriging. Springer Verlag; New York: 1999. [Google Scholar]
- Team RD. R: a language and environment for statistical computing. Technical report, ISBN 3-900051-07-0. R Foundation for Statistical Computing; Vienna, Austria: 2005. 2013. url: http://www.R-project.org. [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the Lasso. J R Stat Soc B. 1996;58(1):267–288. [Google Scholar]
- VanRaden P. Efficient methods to compute genomic predictions. J Dairy Sci. 2008;91(11):4414–4423. doi: 10.3168/jds.2007-0980. [DOI] [PubMed] [Google Scholar]
- Vazquez AI, de los Campos G, Klimentidis YC, Rosa GJM, Gianola D, Yi N, Allison DB. A comprehensive genetic approach for improving prediction of skin cancer risk in humans. Genetics. 2012;192(4):1493–1502. doi: 10.1534/genetics.112.141705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012;90(1):7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Haley CS, Knott SA. Mapping QTLs for binary traits in backcross and F2 populations. Genet Res. 1996;68(01):55–55. [Google Scholar]
- Whitfield M, George L, Grant G, Perou C. Common markers of proliferation. Nat RevCancer. 2006;6(2):99–106. doi: 10.1038/nrc1802. [DOI] [PubMed] [Google Scholar]
- Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer Publishing Company; New York: 2009. Incorporated. [Google Scholar]
- Wright S. Systems of mating. parts I-V. Genetics. 1921;6(2):111–178. doi: 10.1093/genetics/6.2.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy B, Gordon S, Henders A, Nyholt D, Madden P, Heath A, Martin N, Montgomery G. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42(7):565–569. doi: 10.1038/ng.608. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee S, Goddard M, Visscher P. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Duan S, Kistner EO, Bleibel WK, Huang RS, Clark TA, Chen TX, Schweitzer AC, Blume JE, Cox NJ. Evaluation of genetic variation contributing to differences in gene expression between populations. Am J Hum Genet. 2008;82(3):631–640. doi: 10.1016/j.ajhg.2007.12.015. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Liu J, Ding X, Bijma P, de Koning D-J, Zhang Q. Best linear unbiased prediction of genomic breeding values using a trait-specific marker-derived relationship matrix. PLoS One. 2010;5(9):e12648. doi: 10.1371/journal.pone.0012648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Carbonetto P, Stephens M. Polygenic modeling with Bayesian sparse linear mixed models. PLoS Genet. 2013;9(2):e1003264. doi: 10.1371/journal.pgen.1003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 2012;44(7):821–824. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H, Hastie T. Regularization and variable selection via the elastic net. J R Stat Soc B. 2005;67(2):301–320. [Google Scholar]