Abstract
Neural crest cells appear early during embryogenesis and give rise to many structures in the mature adult. In particular, a specific population of neural crest cells migrates to and populates developing cranial tissues. The ensuing differentiation of these cells via individually complex and often intersecting signaling pathways is indispensible to growth and development of the craniofacial complex. Much research has been devoted to this area of development with particular emphasis on cell signaling events required for physiologic development. Understanding such mechanisms will allow researchers to investigate ways in which they can be exploited in order to treat a multitude of diseases affecting the craniofacial complex. Knowing how these multipotent cells are driven towards distinct fates could, in due course, allow patients to receive regenerative therapies for tissues lost to a variety of pathologies. In order to realize this goal, nucleotide sequencing advances allowing snapshots of entire genomes and exomes are being utilized to identify molecular entities associated with disease states. Once identified, these entities can be validated for biological significance with other methods. A crucial next step is the integration of knowledge gleaned from observations in disease states with normal physiology to generate an explanatory model for craniofacial development. This review seeks to provide a current view of the landscape on cell signaling and fate determination of the neural crest and to provide possible avenues of approach for future research.
Keywords: neural crest, development, BMP, Wnt, Hedgehog, craniofacial anomaly
Introduction
Neural crest cells are multi-potent cells that are transient during development. They emerge at the most dorsal aspect of the body around the time of neural tube closure and are thus named neural crest. Neural crest cells are formed on all axial levels, but those formed at craniofacial levels differentiate into several cell types contributing a large portion of adult craniofacial structures. Thus dysregulation during their development leads to congenital craniofacial disorders such as DiGeorge syndrome (also known as velocardiofacial syndrome and 22q11.2 deletion syndrome) and Treacher-Collins syndrome (mandibulofacial dysostosis) [1–3].
Signaling molecules such as growth factors play a critical role for cell fate determination, growth, differentiation, and survival. Genetic studies in both humans and model animals have revealed a number of growth factors and transcription factors regulated by growth factor signaling that are critical for development of cranial neural crest cells. In this review, we summarize recent progress on how growth factor signaling contributes to neural crest differentiation and to skull morphology. Due to limited space, we will not mention palatogenesis, another important craniofacial developmental process where neural crest cells play critical roles [4–6].
Origin of Neural Crest Cells
Neural crest progenitor cells are induced at regions of ectoderm between the neural plate and non-neural ectoderm. These cells undergo epithelial-mesenchymal transition to migrate ventrally to give rise to several different tissues including the peripheral nervous system. Unlike trunk neural crest cells that migrate to relatively deep levels of the body, the cranial neural crest cells migrate superficially. Neural crest cells emerging in the cranial region are distinct from those in trunk because they will give rise to osteoblasts and chondrocytes in addition to other cell types that trunk neural crest cells can differentiate into [7–10].
At the time of NC induction, growth factor signaling via bone morphogenetic proteins (BMPs), fibroblast growth factors (FGFs), and Wnt play pivotal roles [11–14]. Along with other signaling pathways such as Delta/Notch, retinoic acid, Hedgehog, and endothelin, the downstream targets of these pathways are transcription factors such as Msx1/2, Pax3/7, Zic1, Dlx3/5, Hairly2, Id3, and Ap2. These processes specify a border between neural plate and non-neural ectoderm. Soon after specification of neural crest progenitors, these signaling pathways induce a second set of transcription factors including Snail2, FoxD3, Sox9/10, Twist, cMyc and AP2. The combination of these transcription factors is believed to control EMT, migration, and differentiation of neural crest cells [15, 16].
Contribution of neural crest cells to skull bones
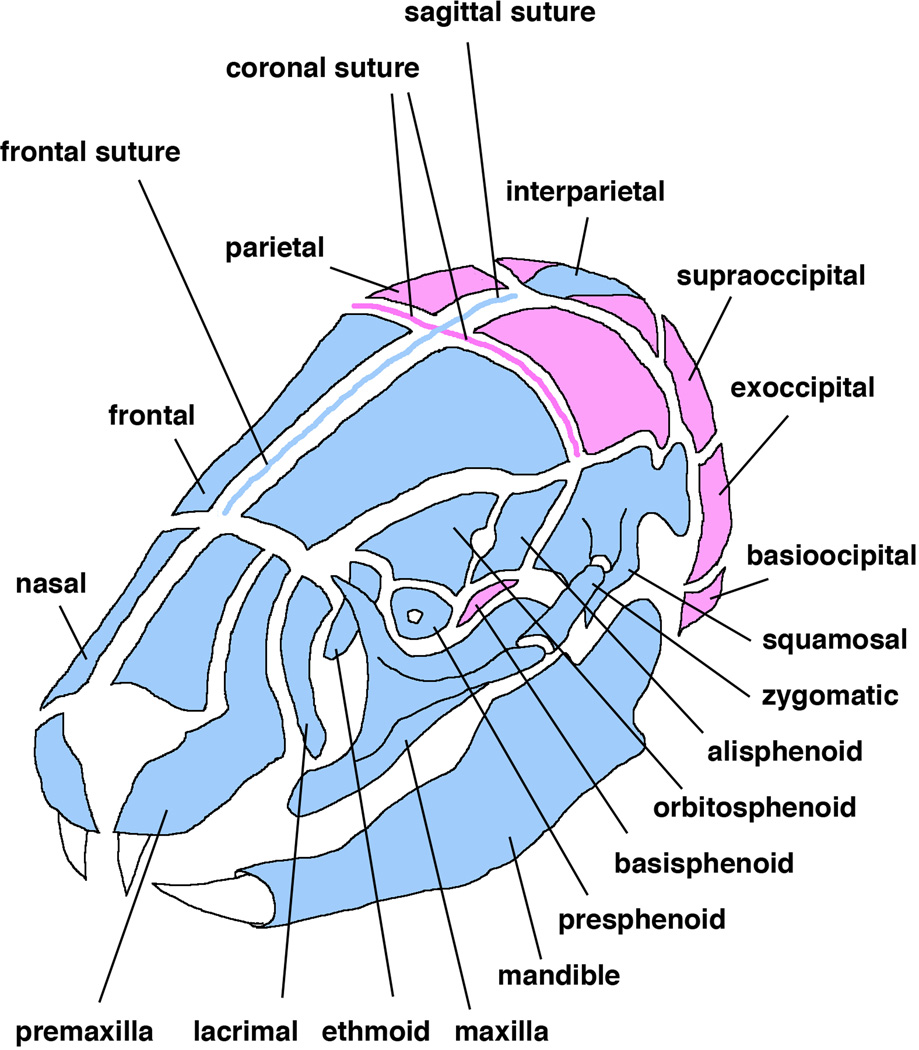
Craniofacial mesenchymal tissues have three origins: neural crest, paraxial mesoderm, and lateral mesoderm [17]. Cranial neural crest cells (CNCC) give rise to the majority of cranial bones and cartilage. The contribution of CNCC was initially investigated by performing chick-quail transplantation experiments [18, 19]. These assays revealed that the more anterior cranial bones are derived from neural crest whereas the posterior portion is from paraxial mesoderm [17]. The neural crest-mesoderm boundary lies within the frontal bone (Fig. 1). In the mouse, the Cre-lox system has allowed us to genetically label specific cell populations in order to trace their lineage. A handful of genes were found to be expressed in a neural crest-specific manner such as Wnt1 and protein zero (P0) and transgenic mouse lines that express Cre recombinase using these neural crest specific promoters have been generated [20–22]. These mice are used to label neural crest cell derived cells in combination with Cre-reporter mice. This can mark neural crest cells “permanently” even if they differentiate to other types of cells because the promoter used for the reporter is ubiquitously active. Using Wnt1-Cre transgenic mouse line, the fate of CNCC was extensively examined to reveal that, in the mouse, the neural crest-mesoderm boundary coincides with the coronal suture (Fig. 1) [17, 23–25]. These analyses also reveal that the interparietal bone is also derived from CNCC [24]. These results are not consistent with the findings from chick-quail transplantations. This may be due to differences in nomenclature of skull elements between avians and mammals or differences in experimental methods [17]. One likely explanation is that the position of the neural crest-mesoderm boundary remains constant relative to the brain and pharynx, that is, the avian frontal bone is more appropriately termed a frontoparietal bone [17].
Figure 1.
Origins of cranial bones and sutures. Blue indicates neural crest origin whereas pink indicates mesodermal origin. Not all bones comprising the cranial base are shown.
Skull osteogenesis and cranial sutures
Bones comprising the cranial vault, collectively referred to as the calvaria, are generated through intramembranous ossification. Some structures outside the calvaria, such as the body and ramus of the mandible, also undergo intramembranous ossification; however, most other cranial bones, such as bones in the cranial base, are formed by endochondral ossification. During endochondral ossification, mesenchymal cells differentiate to chondrocytes and form cartilage primordial whereas mesenchymal cells directly differentiate into osteoblast progenitors during intramembranous ossification. Osteoprogenitor cells further proliferate and differentiate to deposit fibrous matrix for bone formation.
Both CNCC derived and paraxial mesoderm derived osteoprogenitor cells undergo intramembranous ossification to generate corresponding skull elements; however, they show some differences in osteogenic potential and skeletal regenerative capacity [26]. Osteoblasts from neural crest-derived bones such as the frontal bone feature a higher level of activation of FGF signaling pathways compared with osteoblasts from paraxial mesoderm-derived bones such as parietal bones [27, 28]. Osteoblasts from neural crest-derived bones such as frontal bones also show lower apoptotic response when stimulated by TGF-β signaling [29]. Interestingly, regenerative ability of skull defects in the frontal bone is higher than that in parietal bones [30]. Taken together, these results suggest that neural crest-derived bones are more proliferative and less apoptotic than paraxial-derived bones due to increases in FGF, BMP, and Wnt signaling pathways with a reduction in the TGF-β pathway [26].
Sutures are a fibrous connective tissue found between bones in the cranial vault and cranial base. Cells in the fibrous tissues differentiate from embryonic mesenchyme. Sutures are critical growth sites in the skull. Mesenchymal cells proliferate and differentiate into osteoblasts that deposit collagen fibers and minerals to the bony plates to increase their size. In addition to serving as growth sites in the skull through bone formation, sutures also function as joints to hold skull elements together. Genetic studies in mice demonstrate that nasal and metopic sutures, which connect nasal bones and frontal bones, are of neural crest origin [24]. Coronal sutures are of mesodermal origin and are formed between the neural crest derived frontal bones and the mesoderm derived parietal bones. The sagittal suture is formed between the two mesoderm-derived parietal bones and is of neural crest origin (Fig. 1). Sutures are critical for growth of the skull and fusion of sutures results in cessation of skull growth at the site of fusion. Premature fusion of sutures causes a pathological condition called craniosynostosis resulting in increased intracranial pressure and skull deformity [31, 32].
In humans, the metopic suture closes in the first two years after birth whereas the rest of the sutures may stay patent until early adulthood. The presence of cartilaginous tissues within the metopic suture has been confirmed in a five month human fetus [33]. A chondroid tissue, cartilaginous tissue mixed with bone matrix, is observed at the edges of the metopic suture both at birth as well as 17 months after birth. This tissue constitutes the first bridge between the two bones [33]. In the mouse, the corresponding suture to the metopic suture is the frontal suture and can be divided into two parts: the anterior frontal suture and the posterior frontal suture. The posterior frontal suture fuses in the first 45 days of life, whereas all other sutures, including the sagittal and coronal, remain patent in the mouse [34]. Like the metopic suture in human, cartilage formation is confirmed in the posterior frontal suture by expression of Sox9 [35]. These findings suggest that although skull bones are generated through intramembranous ossification, suture fusion undergoes either endochondral ossification or chondroid ossification.
Skull formation and Fibroblast Growth Factor (FGF) signaling
The FGF signaling pathway is active in facial epithelium as well as mesenchyme and functions to stimulate cell proliferation. FGF ligands, especially Fgf8, are expressed in developing craniofacial regions and their contribution to the growth and development of these areas is well documented [36–39]. Gain-of-function mutations in FGF signaling are known to cause some types of craniosynostosis [40, 41]. For example, two missense mutations (S252W and P253R) have been found in the IgII-LgIII linker region of FGFR2 and are associated with Apert syndrome [32, 40]. These mutations enhance binding of FGF ligands to FGFR2 and reduce specificity for ligand binding [42–44]. A closely related form of syndromic craniosynostosis is Crouzon Syndrome. In Crouzon syndrome several mutations in FGFR2 are also known to exist such as P263L in the IgII-LgIII linker region and C342Y in the IgIII loop [40, 41]. It is biochemically demonstrated these mutations make the FGFR2 signaling ligand-independent [45, 46].
It is known that subsequent to ligand binding FGF receptors activate mitogen-activated protein (MAP) kinase pathways. Phosphorylation levels of extracellular signal-regulated kinases 1 and 2 (ERK1/2), MAP kinases, are important to display FGF functions [47]. U0126, a small molecule inhibitor specific for MAP kinase kinase 1 and 2 (MEK1/2), can block phosphorylation and activation of ERK1/2. Heterozygous mice carrying Fgfr2S252W mutation develop an Apert-like syndrome including fusion of coronal suture and treatment of the mice with U0126 prevents the phenotype [48]. This in vivo evidence demonstrates activation of MEK-ERK pathway is a probable cause of the skull deformity found in patients and also offers a possibility to use small molecule inhibitors specific for FGF signaling in treating craniosynostosis induced by gain-of-function mutations in the FGF signaling pathway.
Another possible therapy would be to genetically suppress the impact of the mutations. Introduction of small hairpin RNA (shRNA) specific for the Fgfr2S252W mutation completely prevents Apert-like syndrome in heterozygous mice carrying Fgfr2S252W mutation [48]. It is important that only the transcript from the mutated allele is down regulated by the allele specific shRNA [48]. These results suggest that intervention by shRNA can suppress production of mutant proteins without affecting production of endogenous proteins, and thus this method may be more specific and therefore safer than administration of chemical inhibitors. In order to treat patients establishment of more efficient and site-directed delivery methods of shRNA are needed [49].
BMP signaling
Bone morphogenetic proteins (BMPs) were found by their propensity to induce ectopic bone formation [50, 51]. Currently, they are believed to be critical in regulating bone formation [52]. BMPs bind to membrane bound serine/threonine kinase receptors to activate signaling cascades. Msx2 is a transcription factor regulated by BMP-Smad signaling. A gain-of-function mutation in MSX2 can cause Boston-type craniosynostosis [53]. A second mutation in MSX2 has also been reported recently [54, 55]. Increased expression of Msx2 in mice enhances growth of parietal bones into the sagittal suture [56] reminiscent of premature fusion mechanisms found in human craniosynostosis. A BMP-responsive element is known to exist proximal to the promoter of Mxs2 [57]. Foxc1, a winged helix type transcription factor, directly interacts with this BMP responsive element to regulate expression of Msx2 [58]. Msx1 and Msx2 mutant mice show persistent calvarial foramina [59–61]. In the compound homozygous mutants for Msx1 and Msx2, the frontal and parietal bones are not formed [62]. These results suggest that both Msx1 and Msx2, as downstream targets of BMP signaling, play a critical role in the differentiation and proliferation of osteoblasts within the skull vault. Interestingly, when Msx1 and Msx2 are deleted in a neural crest-specific manner, “heterotopic” bones are formed within the frontal foramen [63]. Formation of heterotopic bones is associated with elevated BMP signaling. Taken together with cell labeling studies using vital dyes, these results suggest that MSX1 and MSX2 negatively regulate BMP signaling to help regulate cell fate determination in neural crest cells around day13.5 in utero [63], which is different from other stages and tissues.
BMP signaling is tightly regulated at several different levels. Extracellular proteins such as Noggin and Chordin bind to BMP ligands to prevent receptor binding [64]. Noggin is expressed only in patent sutures and ectopic expression of Noggin in the posterior frontal suture in mice prevents the fusion of the suture that normally occurs by postnatal day 45 [64]. Since Noggin expression in sutures is downregulated by FGF signaling, these findings suggest a possibility for the therapeutic use of Noggin to treat craniosynostosis caused by hyperactivation of FGF signaling [64]. Indeed, Noggin can suppress suture fusions experimentally induced by transplantation of osteoblasts expressing a mutant form of FGFR2 [65]. Noggin treatment also inhibits recurrence of suture fusion subsequent to suturectomy in a rabbit model of bilateral coronal synostosis [66]. Noggin treatment is not effective however in another rabbit model of delayed-onset craniosynostosis [67]. These results suggest divergence in the molecular causes of craniosynostosis.
Direct involvement of BMP signaling in pathogenesis of craniosynostosis has been recently demonstrated. A conditional transgenic mouse line that expresses a constitutively active form of BMP type 1A receptor (caBmpr1a) has been made [68] and is bred with a neural crest-specific Cre mouse line using protein zero (P0) promoter [22]. Enhanced BMP-Smad signaling in the neural crest cells of this model results in premature fusion of the anterior frontal suture leading to craniosynostosis [69]. It is interesting to contrast between this animal and caBmpr1a mice bred with osteoblast-specific Cre transgenic lines such as Col1-Cre and Osx-Cre where no craniosynostotic phenotypes are identified [69]. Within this model system, increased BMP signaling in osteoblasts and subsequent differentiation are not the direct causes of premature fusion of the suture. It is noteworthy that only a small increase in BMP signaling (50%) is enough to result in craniosynostosis in this animal model [69]. It is reasonable to speculate that large changes in expression of developmentally important genes such as growth factors may be deleterious for fetal survival. As detailed below, Genome-wide association studies (GWAS) indicate single-nucleotide polymorphisms (SNPs) associated with alterations in skull morphology are located in proximity to BMP related genes [70, 71]. Dysregulation of BMP signaling resulting in pathogenesis of craniosynostosis has not been convincingly demonstrated in man; however, upregulation of Msx2, a known downstream target gene of the BMP pathway, is a cause of Boston type craniosynostosis [53]. Furthermore, glypican-1 and glypican-3, which are known to negatively regulate BMP signaling, have been found in mesenchymal cells taken from sutures in craniosynostosis patients [72]. Therefore there is circumstantial evidence some human cases may be caused by upregulation of BMP signaling.
BMP signaling plays pivotal roles in many different aspects during embryogenesis [73]. Neural crest-specific disruption of Acvr1, one of the type 1 receptors for BMPs, results in multiple craniofacial defects including a hypomorphic mandible and lack of ossification in the squamous parts of frontal bones in addition to cleft palate [74]. Neural crest-specific disruption of another type 1 receptor, Bmpr1a, results in midgestation lethality due to malfunctions of cardiac neural crest [75, 76]. When this failure of cardiac function is compensated for by isoproterenol, a beta-adrenergic agonist, mutant embryos can survive until term, but the rescued embryos develop smaller heads and reduced projection of facial structures [77]. Increasing BMP signaling by knocking out its antagonist Noggin results in a microform of holoprosencephaly (HPE) [78] and compound mutations of Noggin and Chordin results in variable forms of HPE [79]. Since the specific overexpression of BMP signaling in neural crest cells does not exhibit HPE [69], candidate cell types sensitive for enhanced BMP signaling to cause HPE would differ from neural crest derived tissues. Since BMPs interact with nodal, another TGF-beta superfamily member important for development of the anterior primitive streak during gastrulation, it is possible that excess amount of BMP ligands due to the loss of their antagonists reduces Nodal signaling resulting in HPE [80, 81]. This implies that we may need to go back to an earlier stage of development in order to understand the molecular and cellular mechanisms of craniofacial development.
Wnt signaling
Wnt signaling is a critical player for generation and migration of neural crest cells [82, 83]. Wnt signaling is important for proliferation of the neural crest derived mesenchyme [82], and for the fusion of epithelium in facial prominences [84–86], thus deficiency of Wnt signaling frequently results in facial clefts. Beta-catenin is a central signaling component of the canonical Wnt signaling pathway and neural crest-specific disruption of b-catenin results in lack of skeletal structures derived from cranial neural crest [87]. On the other hand, activation of b-catenin causes morphological abnormalities in calvaria such as increased suture mesenchymal cells and expansion of immature, but not differentiated osteoblasts [88]. These results underscore the importance of Wnt canonical signaling for expansion of skeletal progenitor cells and subsequent differentiation towards the osteoblast lineage.
Axin2 acts as a negative regulator of the Wnt canonical pathway by promoting degradation of b-catenin. Axin2 is expressed in the osteogenic fronts and periosteum of developing cranial sutures and disruption of Axin2 results in premature fusion of the metopic suture in mice [89]. BMP signaling is upregulated in the Axin2 mutants and that may explain expansion of osteoprogenitors through a positive feedback mechanism [90]. The elevated BMP signaling alters subcellular localization of β-catenin towards a membrane fraction, which may play a critical role for cell-cell interaction during skull morphogenesis [90]. Expressions of signaling components in the FGF pathway are increased in Axin2 mutants [89]. However, compound mutant mice for Axin2 and Fgfr1 (Axin2−/−:Fgfr+/−) develop ectopic cartilage in the sagittal suture at postnatal day 7 that results in fusion of the sagittal suture at postnatal day 50 [91]. As mentioned earlier, the posterior frontal suture in the mouse fuses by postnatal day 45 through endochondral ossification [35]. In Axin2−/− mice cartilage is formed within the posterior frontal suture, but it soon goes away via apoptosis. This disappearance of cartilage may be a cause for delay of suture fusion [92]. It is known that both FGF and TGF-β signaling activity are high in the posterior frontal suture but not in the sagittal suture [26]. The signaling pattern in the compound mutant mice implies that both Wnt and FGF signaling work together to differentially regulate patency of sutures in a region-specific manner, possibly through regulating chondrogenesis.
Twist is a basic helix-loop-helix transcription factor with an indispensible role during neural crest differentiation [93]. Heterozygous null mutations of TWIST1 causes Saethre-Chotzen syndrome that features craniosynostosis [94, 95]. Heterozygous null mice for Twist1 show premature fusion of the coronal sutures that mimics the human condition [96, 97]. In the heterozygous mutants, an increase of Fgfr2 is observed suggesting a connection between TWIST and FGF signaling pathway in the etiology of craniosynostosis [98, 99]. Since Twist1 has been demonstrated as a down stream target of Wnt canonical signaling [100], it is possible to propose that Wnt canonical signaling positively regulates expression of Twist1 to suppress chondrogenesis in order to maintain patency of the sagittal suture and coronal sutures [101, 102].
Hedgehog signaling
The Hedgehog family has numerous roles during craniofacial development. Sonic hedgehog (Shh) is a critical factor for proliferation, survival and patterning of neural crest cells [103–105]. The amount of Hedgehog signaling is strongly associated with alterations in midline facial structures. For example, reductions in Hedgehog activity result in midline hypoplasia, and disruption of Shh in mice results in holoprosencephaly and cyclopia [104]. In humans, mutations in SHH are linked to holoprosencephaly [106–108]. On the other hand, increasing Hedgehog signaling leads to midline expansion including hypertelorism, and frontonasal dysplasia (FND) [109–112]. It has been found that augmenting the level of Hedgehog signaling past a certain level results in midline expansion and duplication of certain structures. The extreme end of the spectrum of midline expansion is total craniofacial duplication called diprosopus. Only 35 documented cases of diprosopus [113] combined with a lack of animal models has left the molecular mechanisms responsible for craniofacial duplication poorly understood.
Recent findings establish involvement of primary cilia in growth factor signaling and in particular for Hedgehog signaling. Primary cilia are microtubule-based organelles found in most types of cells in the body. There is mounting evidence that cilia are important regulators of signaling pathways during development [114]. It has been demonstrated that disruption of intraflagellar transport (IFT) genes important for formation of cilia and anterograde and retrograde movement within cilia compromises Shh signaling [115, 116]. Subsequently, it has been found that components of Shh signaling such as a Hh receptor Patched1 (Ptch1), a multi-pass membrane protein Smoothened (Smo), and transcription factors Gli1/2/3, are all localized in cilia [117, 118]. This has led to the current view of how the mammalian Hh pathway is regulated. In the absence of the Hs ligands, Ptch1 inhibits the accumulation of Smo to the cilia, then at the bottom of cilia, protein kinase A (PKA) along with kinesin Kif7 promotes proteolytic conversion of Gli3 to its repressor form. Concomitantly, suppressor of fused (Sufu) stabilizes Gli proteins to suppress transcriptional activity of Gli2. When Hh ligands bind to Ptch1, Ptch1 relieves inhibition of Smo and Smo accumulates in the cilia. This promotes the movement of Gli2/3, Sufu and Kif7 to the tip of cilium subsequently produce activated forms of Gli2/3 to initiate Hh dependent gene expressions [119].
Loss of one IFT, KIF3A, results in disruption of cilial function thus leading to reduction of Hedgehog signaling [115, 120, 121]. Interestingly, however, neural crestspecific disruption of Kif3a leads to a widened frontonasal prominence and cleft palate resembling FND [109]. These abnormalities are simultaneous with an increase in Shh signaling, likely due to the alteration of Gli3 processing leading to the reduction of a repressor from of Gli3 [122].
Ciliopathy
Ciliopathies are an inter-related cluster of genetic syndromes caused by dysfunctions in primary cilia [123]. As mentioned above, cilia play important roles during craniofacial development by serving as a hub for signaling regulation [123, 124]. Growth factor signaling pathways also play roles for cilial function, thus malfunction of the signaling components may potentially lead ciliopathy. FGF signaling is important for cilial length [125]. BMP signaling through ACVR1 is critical to form cilia at the node during gastrulation by negatively regulating cell cycle progression [126]. Planer Cell Polarity (PCP) signaling is a pathway critical for craniofacial development that utilizes some components of Wnt signaling pathway but is beta-catenin independent. Mice deficient for the PCP effector gene Fuzzy (Fuz) exhibit severe craniofacial defects along with reduction of Hedgehog and an increase in Wnt/beta-catenin signaling [127]. The mutants show a narrow palate and disrupted rugal organization due to excessive neural crest cells from the first branchial arch [128]. This phenotype resembles the high arched palate reported for human ciliopathy patients. Fuz is also critical to negatively regulate Fgf8 expression in neural crest derived tissues [128]. Interestingly, neural crest-specific disruption of Fuz does not result in high arched palate suggesting that negative regulation of FGF signaling by Fuz/cilia may be required before the induction of neural crest cells. These results suggest dynamic interaction among ciliogenesis, growth factor signaling, and tissue morphogenesis during normal craniofacial development.
Perspective – new movements for more depth understanding
Genome-wide association studies (GWAS)
Due to ever increasing progress in DNA sequencing technologies, it has never been easier to get genome-wide sequence information for individuals and families [129]. This development opens a number of new avenues of exploration that were not possible even one decade ago. In the last a couple of years, GWAS in humans have revealed numbers of disease-associated mutations. For example, genome-wide information was collected from 201 case-parent trios and 13 nuclear families with non-syndromic sagittal craniosynostosis to identify several single-nucleotide polymorphisms (SNPs) [70]. Interestingly, the most significant SNP is located close to BMP2, possibly suggesting that this region may be an enhancer to increase BMP2 expression in the patient in a skull-specific manner [70]. Another significant group of SNPs are found within Bardet-Biedl Syndrome gene 9 (BBS9) of which mutations cause skeletal abnormalities, but not suture fusions [70]. BBS9 may be important for cilial functions; mutations found in BBS9 may alter cilial function and thus alter growth factor signaling to develop skull deformity. An exome sequencing approach was also utilized to reveal several additional mutations causative for skull deformity such as TCF12 for coronal craniosynostosis and ERF where all sutures are affected [130, 131].
Canine skull morphology is heterogeneous with variations such as brachycephaly (flattened head) and dolichocephaly (elongated head). GWAS has also been applied to compare different dog breeds and has led to identification of at least 5 loci responsible for cranioskeletal differences [71]. Two of the five loci are mapped to cGMP-dependent protein kinase 2 (PRKG2) and BMP3 with the SNP in BMP3 causes a missense mutation (F452L) [71]. Although no overt craniofacial abnormality is reported in Bmp3 mutant mice [132], knockdown of bmp3 in zebrafish by morpholino oligonucleotides results in severe deficiencies in jaw development [71]. These discrepancies may be explained by the different requirement of BMP signaling among different species or the F452L mutation alters signaling activity of BMP3 rather than diminishing it.
GWAS reveals an association of the SNP close to BMP2 with non-syndromic sagittal craniosynostosis [70]. Since the SNP locates outside of the coding exons, this region is expected to be a regulatory region for BMP2 expression. Mutations that lead to changes in amino acids may be too drastic to maintain viability of fetuses if the mutation happens to be in critical genes for development such as growth factor signaling. Based on these considerations, attempts to identify distant-acting enhancers that affect craniofacial morphology were made. P300 is an enhancer binding protein to increase gene expressions. Chromatin immunoprecipitation followed by sequencing with p300 is done using E11.5 mouse facial tissues and over 4,000 candidate enhancers were identified [133]. More than 200 candidates were examined for their enhancer activity in vivo and 121 of the sequences can lead to gene expression within craniofacial structure [133]. Targeted deletion of three candidates close to Msx1, Snai2 and Isl1, respectively, result in subtle but significant structural alterations in craniofacial morphology [133]. These results underscore the importance of small changes in gene expressions during embryogenesis for fine-tuning to develop normal craniofacial structures.
New tools and technologies
Many of the studies utilize Wnt1-Cre transgenic mice to investigate fate mapping of neural crest-derived tissues and functions of growth factor signaling in neural crest cells [20]. Recently, it has been shown that this transgenic line shows increased Wnt/beta-catenin signaling in the midbrain region associated with compromised development of the midbrain [21]. These results warrant revisiting results using Wnt1-Cre because newly generated Wnt1-Cre2 transgenic line shows the same cell type specificity without ectopic Wnt signaling [21]. P0-Cre transgenic mouse line is another frequently used mouse line that expresses Cre in a neural crest-specific manner [22]. Expression patterns of these lines (Wnt1-Cre and P0-Cre) are similar but not identical; expression domains of P0-Cre are found in epithelial layers of developing tooth germ and taste bud [134, 135]. Thus, comparisons of results among such Cre transgenic lines will be helpful.
Cranial neural crest culture
The ability to culture cells from the cranial neural crest and isolate stem cells from this population has been attempted for years. Once in hand, these cells would provide an invaluable tool for looking at the factors that are responsible for cell fate determination. Recently a major result has been reported in this area [136]. These researchers demonstrated the ability to isolate cranial neural crest stem cells and control their differentiation towards an osteogenic fate as well as others. This would have implications in dentistry with regards to bony defects. Currently, bone grafts are usually achieved with either lyophilized cadaver bone or xenografts [137–139] and often lead to repair in lieu of true regeneration. Ultimately, the ability to culture or induce a population of multi-potent progenitors from CNCC could lead to regenerative therapies for a multitude of pathologies arising in the craniofacial complex.
Acknowledgement
We thank Drs. Yoshihiro Komatsu and Sudha Rajderkar for critical reading and Yoshiko Mishina for her artwork. We are sorry for not including all critical the references due to the space limitation. Y.M. is supported by the National Institutes of Health (R01DE020843) and the Department of Defense (W81XWH-11-2-0073).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Achilleos A, Trainor PA. Neural crest stem cells: discovery, properties and potential for therapy. Cell Res. 2012;22:288–304. doi: 10.1038/cr.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trainor PA. Craniofacial birth defects: The role of neural crest cells in the etiology and pathogenesis of Treacher Collins syndrome and the potential for prevention. Am J Med Genet A. 2010;152A:2984–2994. doi: 10.1002/ajmg.a.33454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernandez L, Nevado J, Santos F, Heine-Suner D, Martinez-Glez V, Garcia-Minaur S, Palomo R, Delicado A, Pajares IL, Palomares M, Garcia-Guereta L, Valverde E, Hawkins F, Lapunzina P. A deletion and a duplication in distal 22q11.2 deletion syndrome region. Clinical implications and review. BMC Med Genet. 2009;10:48. doi: 10.1186/1471-2350-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bush JO, Jiang R. Palatogenesis: morphogenetic and molecular mechanisms of secondary palate development. Development. 2012;139:231–243. doi: 10.1242/dev.067082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu KJ. Future directions: molecular approaches provide insights into palatal clefting and repair. Front Oral Biol. 2012;16:147–154. doi: 10.1159/000337667. [DOI] [PubMed] [Google Scholar]
- 6.Levi B, Brugman S, Wong VW, Grova M, Longaker MT, Wan DC. Palatogenesis: engineering, pathways and pathologies. Organogenesis. 2011;7:242–254. doi: 10.4161/org.7.4.17926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhatt S, Diaz R, Trainor PA. Signals and switches in Mammalian neural crest cell differentiation. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a008326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuratani S. Cephalic neural crest cells and the evolution of craniofacial structures in vertebrates: morphological and embryological significance of the premandibular-mandibular boundary. Zoology (Jena) 2005;108:13–25. doi: 10.1016/j.zool.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Dupin E, Calloni GW, Le Douarin NM. The cephalic neural crest of amniote vertebrates is composed of a large majority of precursors endowed with neural, melanocytic, chondrogenic and osteogenic potentialities. Cell Cycle. 2010;9:238–249. doi: 10.4161/cc.9.2.10491. [DOI] [PubMed] [Google Scholar]
- 10.Le Douarin NM, Dieterlen-Lievre F. How studies on the avian embryo have opened new avenues in the understanding of development: a view about the neural and hematopoietic systems. Dev Growth Differ. 2013;55:1–14. doi: 10.1111/dgd.12015. [DOI] [PubMed] [Google Scholar]
- 11.Stuhlmiller TJ, Garcia-Castro MI. Current perspectives of the signaling pathways directing neural crest induction. Cell Mol Life Sci. 69:3715–3737. doi: 10.1007/s00018-012-0991-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patthey C, Edlund T, Gunhaga L. Wnt-regulated temporal control of BMP exposure directs the choice between neural plate border and epidermal fate. Development. 2009;136:73–83. doi: 10.1242/dev.025890. [DOI] [PubMed] [Google Scholar]
- 13.Patthey C, Gunhaga L. Signaling pathways regulating ectodermal cell fate choices. Exp Cell Res. 2013 doi: 10.1016/j.yexcr.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 14.Meulemans D, Bronner-Fraser M. Gene-regulatory interactions in neural crest evolution and development. Dev Cell. 2004;7:291–299. doi: 10.1016/j.devcel.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 15.Simoes-Costa MS, McKeown SJ, Tan-Cabugao J, Sauka-Spengler T, Bronner ME. Dynamic and differential regulation of stem cell factor FoxD3 in the neural crest is Encrypted in the genome. PLoS Genet. 8:e1003142. doi: 10.1371/journal.pgen.1003142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Croze N, Maczkowiak F, Monsoro-Burq AH. Reiterative AP2a activity controls sequential steps in the neural crest gene regulatory network. Proc Natl Acad Sci U S A. 2011;108:155–160. doi: 10.1073/pnas.1010740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noden DM, Trainor PA. Relations and interactions between cranial mesoderm and neural crest populations. J Anat. 2005;207:575–601. doi: 10.1111/j.1469-7580.2005.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noden DM. The role of the neural crest in patterning of avian cranial skeletal, connective, and muscle tissues. Dev Biol. 1983;96:144–165. doi: 10.1016/0012-1606(83)90318-4. [DOI] [PubMed] [Google Scholar]
- 19.Evans DJ, Noden DM. Spatial relations between avian craniofacial neural crest and paraxial mesoderm cells. Dev Dyn. 2006;235:1310–1325. doi: 10.1002/dvdy.20663. [DOI] [PubMed] [Google Scholar]
- 20.Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- 21.Lewis AE, Vasudevan HN, O'Neill AK, Soriano P, Bush JO. The widely used Wnt1-Cre transgene causes developmental phenotypes by ectopic activation of Wnt signaling. Dev Biol. 2013;379:229–234. doi: 10.1016/j.ydbio.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamauchi Y, Abe K, Mantani A, Hitoshi Y, Suzuki M, Osuzu F, Kuratani S, Yamamura K. A novel transgenic technique that allows specific marking of the neural crest cell lineage in mice. Dev Biol. 1999;212:191–203. doi: 10.1006/dbio.1999.9323. [DOI] [PubMed] [Google Scholar]
- 23.Chai Y, Jiang X, Ito Y, Bringas P, Jr, Han J, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- 24.Jiang X, Iseki S, Maxson RE, Sucov HM, Morriss-Kay GM. Tissue origins and interactions in the mammalian skull vault. Dev Biol. 2002;241:106–116. doi: 10.1006/dbio.2001.0487. [DOI] [PubMed] [Google Scholar]
- 25.Chai Y, Maxson RE., Jr Recent advances in craniofacial morphogenesis. Dev Dyn. 2006;235:2353–2375. doi: 10.1002/dvdy.20833. [DOI] [PubMed] [Google Scholar]
- 26.Senarath-Yapa K, Li S, Meyer NP, Longaker MT, Quarto N. Integration of multiple signaling pathways determines differences in the osteogenic potential and tissue regeneration of neural crest-derived and mesoderm-derived calvarial bones. Int J Mol Sci. 2013;14:5978–5997. doi: 10.3390/ijms14035978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li S, Quarto N, Longaker MT. Activation of FGF signaling mediates proliferative and osteogenic differences between neural crest derived frontal and mesoderm parietal derived bone. PLoS One. 2010;5:e14033. doi: 10.1371/journal.pone.0014033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quarto N, Wan DC, Kwan MD, Panetta NJ, Li S, Longaker MT. Origin matters: differences in embryonic tissue origin and Wnt signaling determine the osteogenic potential and healing capacity of frontal and parietal calvarial bones. J Bone Miner Res. 2010;25:1680–1694. doi: 10.1359/jbmr.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li S, Meyer NP, Quarto N, Longaker MT. Integration of multiple signaling regulates through apoptosis the differential osteogenic potential of neural crest-derived and mesoderm-derived Osteoblasts. PLoS One. 2013;8:e58610. doi: 10.1371/journal.pone.0058610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Behr B, Panetta NJ, Longaker MT, Quarto N. Different endogenous threshold levels of Fibroblast Growth Factor-ligands determine the healing potential of frontal and parietal bones. Bone. 2010;47:281–294. doi: 10.1016/j.bone.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 31.Rice DP, Rice R, Thesleff I. Molecular mechanisms in calvarial bone and suture development, and their relation to craniosynostosis. Eur J Orthod. 2003;25:139–148. doi: 10.1093/ejo/25.2.139. [DOI] [PubMed] [Google Scholar]
- 32.Morriss-Kay GM, Wilkie AO. Growth of the normal skull vault and its alteration in craniosynostosis: insights from human genetics and experimental studies. J Anat. 2005;207:637–653. doi: 10.1111/j.1469-7580.2005.00475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manzanares MC, Goret-Nicaise M, Dhem A. Metopic sutural closure in the human skull. J Anat. 1988;161:203–215. [PMC free article] [PubMed] [Google Scholar]
- 34.Warren SM, Greenwald JA, Spector JA, Bouletreau P, Mehrara BJ, Longaker MT. New developments in cranial suture research. Plast Reconstr Surg. 2001;107:523–540. doi: 10.1097/00006534-200102000-00034. [DOI] [PubMed] [Google Scholar]
- 35.Sahar DE, Longaker MT, Quarto N. Sox9 neural crest determinant gene controls patterning and closure of the posterior frontal cranial suture. Dev Biol. 2005;280:344–361. doi: 10.1016/j.ydbio.2005.01.022. [DOI] [PubMed] [Google Scholar]
- 36.Creuzet S, Schuler B, Couly G, Le Douarin NM. Reciprocal relationships between Fgf8 and neural crest cells in facial and forebrain development. Proc Natl Acad Sci U S A. 2004;101:4843–4847. doi: 10.1073/pnas.0400869101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Macatee TL, Hammond BP, Arenkiel BR, Francis L, Frank DU, Moon AM. Ablation of specific expression domains reveals discrete functions of ectoderm- and endoderm-derived FGF8 during cardiovascular and pharyngeal development. Development. 2003;130:6361–6374. doi: 10.1242/dev.00850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trumpp A, Depew MJ, Rubenstein JL, Bishop JM, Martin GR. Cre-mediated gene inactivation demonstrates that FGF8 is required for cell survival and patterning of the first branchial arch. Genes Dev. 1999;13:3136–3148. doi: 10.1101/gad.13.23.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szabo-Rogers HL, Geetha-Loganathan P, Nimmagadda S, Fu KK, Richman JM. FGF signals from the nasal pit are necessary for normal facial morphogenesis. Dev Biol. 2008;318:289–302. doi: 10.1016/j.ydbio.2008.03.027. [DOI] [PubMed] [Google Scholar]
- 40.Passos-Bueno MR, Serti Eacute AE, Jehee FS, Fanganiello R, Yeh E. Genetics of craniosynostosis: genes, syndromes, mutations and genotype-phenotype correlations. Front Oral Biol. 2008;12:107–143. doi: 10.1159/000115035. [DOI] [PubMed] [Google Scholar]
- 41.Wilkie AO, Morriss-Kay GM. Genetics of craniofacial development and malformation. Nat Rev Genet. 2001;2:458–468. doi: 10.1038/35076601. [DOI] [PubMed] [Google Scholar]
- 42.Anderson J, Burns HD, Enriquez-Harris P, Wilkie AO, Heath JK. Apert syndrome mutations in fibroblast growth factor receptor 2 exhibit increased affinity for FGF ligand. Hum Mol Genet. 1998;7:1475–1483. doi: 10.1093/hmg/7.9.1475. [DOI] [PubMed] [Google Scholar]
- 43.Yu K, Herr AB, Waksman G, Ornitz DM. Loss of fibroblast growth factor receptor 2 ligand-binding specificity in Apert syndrome. Proc Natl Acad Sci U S A. 2000;97:14536–14541. doi: 10.1073/pnas.97.26.14536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ibrahimi OA, Zhang F, Eliseenkova AV, Itoh N, Linhardt RJ, Mohammadi M. Biochemical analysis of pathogenic ligand-dependent FGFR2 mutations suggests distinct pathophysiological mechanisms for craniofacial and limb abnormalities. Hum Mol Genet. 2004;13:2313–2324. doi: 10.1093/hmg/ddh235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neilson KM, Friesel RE. Constitutive activation of fibroblast growth factor receptor-2 by a point mutation associated with Crouzon syndrome. J Biol Chem. 1995;270:26037–26040. doi: 10.1074/jbc.270.44.26037. [DOI] [PubMed] [Google Scholar]
- 46.Mangasarian K, Li Y, Mansukhani A, Basilico C. Mutation associated with Crouzon syndrome causes ligand-independent dimerization and activation of FGF receptor-2. J Cell Physiol. 1997;172:117–125. doi: 10.1002/(SICI)1097-4652(199707)172:1<117::AID-JCP13>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 47.Tsang M, Dawid IB. Promotion and attenuation of FGF signaling through the Ras-MAPK pathway. Sci STKE. 2004;2004:pe17. doi: 10.1126/stke.2282004pe17. [DOI] [PubMed] [Google Scholar]
- 48.Shukla V, Coumoul X, Wang RH, Kim HS, Deng CX. RNA interference and inhibition of MEK-ERK signaling prevent abnormal skeletal phenotypes in a mouse model of craniosynostosis. Nat Genet. 2007;39:1145–1150. doi: 10.1038/ng2096. [DOI] [PubMed] [Google Scholar]
- 49.Akita H, Harashima H. Advances in non-viral gene delivery: using multifunctional envelope-type nano-device. Expert Opin Drug Deliv. 2008;5:847–859. doi: 10.1517/17425247.5.8.847. [DOI] [PubMed] [Google Scholar]
- 50.Kamiya N, Mishina Y. New insights on the roles of BMP signaling in bone-A review of recent mouse genetic studies. Biofactors. 2011;37:75–82. doi: 10.1002/biof.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Urist MR. Bone: formation by autoinduction. Science. 1965;150:893–899. doi: 10.1126/science.150.3698.893. [DOI] [PubMed] [Google Scholar]
- 52.Bandyopadhyay A, Yadav PS, Prashar P. BMP signaling in development and diseases: a pharmacological perspective. Biochem Pharmacol. 2013;85:857–864. doi: 10.1016/j.bcp.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 53.Jabs EW, Muller U, Li X, Ma L, Luo W, Haworth IS, Klisak I, Sparkes R, Warman ML, Mulliken JB, et al. A mutation in the homeodomain of the human MSX2 gene in a family affected with autosomal dominant craniosynostosis. Cell. 1993;75:443–450. doi: 10.1016/0092-8674(93)90379-5. [DOI] [PubMed] [Google Scholar]
- 54.Florisson JM, Verkerk AJ, Huigh D, Hoogeboom AJ, Swagemakers S, Kremer A, Heijsman D, Lequin MH, Mathijssen IM, van der Spek PJ. Boston type craniosynostosis: Report of a second mutation in MSX2. Am J Med Genet A. 2013;161:2626–2633. doi: 10.1002/ajmg.a.36126. [DOI] [PubMed] [Google Scholar]
- 55.Janssen A, Hosen MJ, Jeannin P, Coucke PJ, De Paepe A, Vanakker OM. Second family with the boston-type craniosynostosis syndrome: Novel mutation and expansion of the clinical spectrum. Am J Med Genet A. 2013;161:2352–2357. doi: 10.1002/ajmg.a.36077. [DOI] [PubMed] [Google Scholar]
- 56.Liu YH, Tang Z, Kundu RK, Wu L, Luo W, Zhu D, Sangiorgi F, Snead ML, Maxson RE. Msx2 gene dosage influences the number of proliferative osteogenic cells in growth centers of the developing murine skull: a possible mechanism for MSX2-mediated craniosynostosis in humans. Dev Biol. 1999;205:260–274. doi: 10.1006/dbio.1998.9114. [DOI] [PubMed] [Google Scholar]
- 57.Brugger SM, Merrill AE, Torres-Vazquez J, Wu N, Ting MC, Cho JY, Dobias SL, Yi SE, Lyons K, Bell JR, Arora K, Warrior R, Maxson R. A phylogenetically conserved cis-regulatory module in the Msx2 promoter is sufficient for BMP-dependent transcription in murine and Drosophila embryos. Development. 2004;131:5153–5165. doi: 10.1242/dev.01390. [DOI] [PubMed] [Google Scholar]
- 58.Sun J, Ishii M, Ting MC, Maxson R. Foxc1 controls the growth of the murine frontal bone rudiment by direct regulation of a Bmp response threshold of Msx2. Development. 2013;140:1034–1044. doi: 10.1242/dev.085225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Satokata I, Maas R. Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat Genet. 1994;6:348–356. doi: 10.1038/ng0494-348. [DOI] [PubMed] [Google Scholar]
- 60.Satokata I, Ma L, Ohshima H, Bei M, Woo I, Nishizawa K, Maeda T, Takano Y, Uchiyama M, Heaney S, Peters H, Tang Z, Maxson R, Maas R. Msx2 deficiency in mice causes pleiotropic defects in bone growth and ectodermal organ formation. Nat Genet. 2000;24:391–395. doi: 10.1038/74231. [DOI] [PubMed] [Google Scholar]
- 61.Ishii M, Merrill AE, Chan YS, Gitelman I, Rice DP, Sucov HM, Maxson RE., Jr Msx2 and Twist cooperatively control the development of the neural crest-derived skeletogenic mesenchyme of the murine skull vault. Development. 2003;130:6131–6142. doi: 10.1242/dev.00793. [DOI] [PubMed] [Google Scholar]
- 62.Ishii M, Han J, Yen HY, Sucov HM, Chai Y, Maxson RE., Jr Combined deficiencies of Msx1 and Msx2 cause impaired patterning and survival of the cranial neural crest. Development. 2005;132:4937–4950. doi: 10.1242/dev.02072. [DOI] [PubMed] [Google Scholar]
- 63.Roybal PG, Wu NL, Sun J, Ting MC, Schafer CA, Maxson RE. Inactivation of Msx1 and Msx2 in neural crest reveals an unexpected role in suppressing heterotopic bone formation in the head. Dev Biol. 2010;343:28–39. doi: 10.1016/j.ydbio.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Warren SM, Brunet LJ, Harland RM, Economides AN, Longaker MT. The BMP antagonist noggin regulates cranial suture fusion. Nature. 2003;422:625–629. doi: 10.1038/nature01545. [DOI] [PubMed] [Google Scholar]
- 65.Shen K, Krakora SM, Cunningham M, Singh M, Wang X, Hu FZ, Post JC, Ehrlich GD. Medical treatment of craniosynostosis: recombinant Noggin inhibits coronal suture closure in the rat craniosynostosis model. Orthod Craniofac Res. 2009;12:254–262. doi: 10.1111/j.1601-6343.2009.01460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cooper GM, Curry C, Barbano TE, Burrows AM, Vecchione L, Caccamese JF, Norbutt CS, Costello BJ, Losee JE, Moursi AM, Huard J, Mooney MP. Noggin inhibits postoperative resynostosis in craniosynostotic rabbits. J Bone Miner Res. 2007;22:1046–1054. doi: 10.1359/jbmr.070410. [DOI] [PubMed] [Google Scholar]
- 67.Cray J, Jr, Burrows AM, Vecchione L, Caccamese JF, Jr, Losee JE, Moursi AM, Siegel MI, Cooper GM, Mooney MP. Blocking bone morphogenetic protein function using in vivo noggin therapy does not rescue premature suture fusion in rabbits with delayed-onset craniosynostosis. Plast Reconstr Surg. 2011;127:1163–1172. doi: 10.1097/PRS.0b013e318205f23b. [DOI] [PubMed] [Google Scholar]
- 68.Kamiya N, Ye L, Kobayashi T, Mochida Y, Yamauchi M, Kronenberg HM, Feng JQ, Mishina Y. BMP signaling negatively regulates bone mass through sclerostin by inhibiting the canonical Wnt pathway. Development. 2008;135:3801–3811. doi: 10.1242/dev.025825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Komatsu Y, Yu PB, Kamiya N, Pan H, Fukuda T, Scott GJ, Ray MK, Yamamura K, Mishina Y. Augmentation of Smad-dependent BMP signaling in neural crest cells causes craniosynostosis in mice. J Bone Miner Res. 2013;28:1422–1433. doi: 10.1002/jbmr.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Justice CM, Yagnik G, Kim Y, Peter I, Jabs EW, Erazo M, Ye X, Ainehsazan E, Shi L, Cunningham ML, Kimonis V, Roscioli T, Wall SA, Wilkie AO, Stoler J, Richtsmeier JT, Heuze Y, Sanchez-Lara PA, Buckley MF, Druschel CM, Mills JL, Caggana M, Romitti PA, Kay DM, Senders C, Taub PJ, Klein OD, Boggan J, Zwienenberg-Lee M, Naydenov C, Kim J, Wilson AF, Boyadjiev SA. A genome-wide association study identifies susceptibility loci for nonsyndromic sagittal craniosynostosis near BMP2 and within BBS9. Nat Genet. 2012;44:1360–1364. doi: 10.1038/ng.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schoenebeck JJ, Hutchinson SA, Byers A, Beale HC, Carrington B, Faden DL, Rimbault M, Decker B, Kidd JM, Sood R, Boyko AR, Fondon JW, 3rd, Wayne RK, Bustamante CD, Ciruna B, Ostrander EA. Variation of BMP3 contributes to dog breed skull diversity. PLoS Genet. 2012;8:e1002849. doi: 10.1371/journal.pgen.1002849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dwivedi PP, Grose RH, Filmus J, Hii CS, Xian CJ, Anderson PJ, Powell BC. Regulation of bone morphogenetic protein signalling and cranial osteogenesis by Gpc1 and Gpc3. Bone. 2013;55:367–376. doi: 10.1016/j.bone.2013.04.013. [DOI] [PubMed] [Google Scholar]
- 73.Kishigami S, Mishina Y. BMP signaling and early embryonic patterning. Cytokine Growth Factor Rev. 2005;16:265–278. doi: 10.1016/j.cytogfr.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 74.Dudas M, Sridurongrit S, Nagy A, Okazaki K, Kaartinen V. Craniofacial defects in mice lacking BMP type I receptor Alk2 in neural crest cells. Mech Dev. 2004;121:173–182. doi: 10.1016/j.mod.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 75.Nomura-Kitabayashi A, Phoon CK, Kishigami S, Rosenthal J, Yamauchi Y, Abe K, Yamamura K, Samtani R, Lo CW, Mishina Y. Outflow tract cushions perform a critical valve-like function in the early embryonic heart requiring BMPRIA-mediated signaling in cardiac neural crest. Am J Physiol Heart Circ Physiol. 2009;297:H1617–H1628. doi: 10.1152/ajpheart.00304.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stottmann RW, Choi M, Mishina Y, Meyers EN, Klingensmith J. BMP receptor IA is required in mammalian neural crest cells for development of the cardiac outflow tract and ventricular myocardium. Development. 2004;131:2205–2218. doi: 10.1242/dev.01086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Morikawa Y, Zehir A, Maska E, Deng C, Schneider MD, Mishina Y, Cserjesi P. BMP signaling regulates sympathetic nervous system development through Smad4-dependent and -independent pathways. Development. 2009;136:3575–3584. doi: 10.1242/dev.038133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lana-Elola E, Tylzanowski P, Takatalo M, Alakurtti K, Veistinen L, Mitsiadis TA, Graf D, Rice R, Luyten FP, Rice DP. Noggin null allele mice exhibit a microform of holoprosencephaly. Hum Mol Genet. 2011;20:4005–4015. doi: 10.1093/hmg/ddr329. [DOI] [PubMed] [Google Scholar]
- 79.Klingensmith J, Matsui M, Yang YP, Anderson RM. Roles of bone morphogenetic protein signaling and its antagonism in holoprosencephaly. Am J Med Genet C Semin Med Genet. 2010;154C:43–51. doi: 10.1002/ajmg.c.30256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang YP, Anderson RM, Klingensmith J. BMP antagonism protects Nodal signaling in the gastrula to promote the tissue interactions underlying mammalian forebrain and craniofacial patterning. Hum Mol Genet. 2010;19:3030–3042. doi: 10.1093/hmg/ddq208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang YP, Klingensmith J. Roles of organizer factors and BMP antagonism in mammalian forebrain establishment. Dev Biol. 2006;296:458–475. doi: 10.1016/j.ydbio.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 82.Brugmann SA, Goodnough LH, Gregorieff A, Leucht P, ten Berge D, Fuerer C, Clevers H, Nusse R, Helms JA. Wnt signaling mediates regional specification in the vertebrate face. Development. 2007;134:3283–3295. doi: 10.1242/dev.005132. [DOI] [PubMed] [Google Scholar]
- 83.Basch ML, Bronner-Fraser M. Neural crest inducing signals. Adv Exp Med Biol. 2006;589:24–31. doi: 10.1007/978-0-387-46954-6_2. [DOI] [PubMed] [Google Scholar]
- 84.Song L, Li Y, Wang K, Wang YZ, Molotkov A, Gao L, Zhao T, Yamagami T, Wang Y, Gan Q, Pleasure DE, Zhou CJ. Lrp6-mediated canonical Wnt signaling is required for lip formation and fusion. Development. 2009;136:3161–3171. doi: 10.1242/dev.037440. [DOI] [PubMed] [Google Scholar]
- 85.Lan Y, Ryan RC, Zhang Z, Bullard SA, Bush JO, Maltby KM, Lidral AC, Jiang R. Expression of Wnt9b and activation of canonical Wnt signaling during midfacial morphogenesis in mice. Dev Dyn. 2006;235:1448–1454. doi: 10.1002/dvdy.20723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Geetha-Loganathan P, Nimmagadda S, Antoni L, Fu K, Whiting CJ, Francis-West P, Richman JM. Expression of WNT signalling pathway genes during chicken craniofacial development. Dev Dyn. 2009;238:1150–1165. doi: 10.1002/dvdy.21934. [DOI] [PubMed] [Google Scholar]
- 87.Brault V, Moore R, Kutsch S, Ishibashi M, Rowitch DH, McMahon AP, Sommer L, Boussadia O, Kemler R. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 2001;128:1253–1264. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- 88.Mirando AJ, Maruyama T, Fu J, Yu HM, Hsu W. beta-catenin/cyclin D1 mediated development of suture mesenchyme in calvarial morphogenesis. BMC Dev Biol. 2010;10:116. doi: 10.1186/1471-213X-10-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yu HM, Jerchow B, Sheu TJ, Liu B, Costantini F, Puzas JE, Birchmeier W, Hsu W. The role of Axin2 in calvarial morphogenesis and craniosynostosis. Development. 2005;132:1995–2005. doi: 10.1242/dev.01786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu B, Yu HM, Hsu W. Craniosynostosis caused by Axin2 deficiency is mediated through distinct functions of beta-catenin in proliferation and differentiation. Dev Biol. 2007;301:298–308. doi: 10.1016/j.physletb.2003.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Maruyama T, Mirando AJ, Deng CX, Hsu W. The balance of WNT and FGF signaling influences mesenchymal stem cell fate during skeletal development. Sci Signal. 2010;3:ra40. doi: 10.1126/scisignal.2000727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Behr B, Longaker MT, Quarto N. Absence of endochondral ossification and craniosynostosis in posterior frontal cranial sutures of Axin2(−/−) mice. PLoS One. 2013;8:e70240. doi: 10.1371/journal.pone.0070240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen ZF, Behringer RR. twist is required in head mesenchyme for cranial neural tube morphogenesis. Genes Dev. 1995;9:686–699. doi: 10.1101/gad.9.6.686. [DOI] [PubMed] [Google Scholar]
- 94.el Ghouzzi V, Le Merrer M, Perrin-Schmitt F, Lajeunie E, Benit P, Renier D, Bourgeois P, Bolcato-Bellemin AL, Munnich A, Bonaventure J. Mutations of the TWIST gene in the Saethre-Chotzen syndrome. Nat Genet. 1997;15:42–46. doi: 10.1038/ng0197-42. [DOI] [PubMed] [Google Scholar]
- 95.Howard TD, Paznekas WA, Green ED, Chiang LC, Ma N, Ortiz de Luna RI, Garcia Delgado C, Gonzalez-Ramos M, Kline AD, Jabs EW. Mutations in TWIST, a basic helix-loop-helix transcription factor, in Saethre-Chotzen syndrome. Nat Genet. 1997;15:36–41. doi: 10.1038/ng0197-36. [DOI] [PubMed] [Google Scholar]
- 96.Bourgeois P, Bolcato-Bellemin AL, Danse JM, Bloch-Zupan A, Yoshiba K, Stoetzel C, Perrin-Schmitt F. The variable expressivity and incomplete penetrance of the twist-null heterozygous mouse phenotype resemble those of human Saethre-Chotzen syndrome. Hum Mol Genet. 1998;7:945–957. doi: 10.1093/hmg/7.6.945. [DOI] [PubMed] [Google Scholar]
- 97.Carver EA, Oram KF, Gridley T. Craniosynostosis in Twist heterozygous mice: a model for Saethre-Chotzen syndrome. Anat Rec. 2002;268:90–92. doi: 10.1002/ar.10124. [DOI] [PubMed] [Google Scholar]
- 98.Connerney J, Andreeva V, Leshem Y, Mercado MA, Dowell K, Yang X, Lindner V, Friesel RE, Spicer DB. Twist1 homodimers enhance FGF responsiveness of the cranial sutures and promote suture closure. Dev Biol. 2008;318:323–334. doi: 10.1016/j.ydbio.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rice DP, Aberg T, Chan Y, Tang Z, Kettunen PJ, Pakarinen L, Maxson RE, Thesleff I. Integration of FGF and TWIST in calvarial bone and suture development. Development. 2000;127:1845–1855. doi: 10.1242/dev.127.9.1845. [DOI] [PubMed] [Google Scholar]
- 100.Howe LR, Watanabe O, Leonard J, Brown AM. Twist is up-regulated in response to Wnt1 and inhibits mouse mammary cell differentiation. Cancer Res. 2003;63:1906–1913. [PubMed] [Google Scholar]
- 101.Behr B, Longaker MT, Quarto N. Differential activation of canonical Wnt signaling determines cranial sutures fate: a novel mechanism for sagittal suture craniosynostosis. Dev Biol. 2010;344:922–940. doi: 10.1016/j.ydbio.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 102.Behr B, Longaker MT, Quarto N. Craniosynostosis of coronal suture in twist1 mice occurs through endochondral ossification recapitulating the physiological closure of posterior frontal suture. Front Physiol. 2011;2:37. doi: 10.3389/fphys.2011.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ahlgren SC, Bronner-Fraser M. Inhibition of sonic hedgehog signaling in vivo results in craniofacial neural crest cell death. Curr Biol. 1999;9:1304–1314. doi: 10.1016/s0960-9822(00)80052-4. [DOI] [PubMed] [Google Scholar]
- 104.Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, Beachy PA. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- 105.Jeong J, Mao J, Tenzen T, Kottmann AH, McMahon AP. Hedgehog signaling in the neural crest cells regulates the patterning and growth of facial primordia. Genes Dev. 2004;18:937–951. doi: 10.1101/gad.1190304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Belloni E, Muenke M, Roessler E, Traverso G, Siegel-Bartelt J, Frumkin A, Mitchell HF, Donis-Keller H, Helms C, Hing AV, Heng HH, Koop B, Martindale D, Rommens JM, Tsui LC, Scherer SW. Identification of Sonic hedgehog as a candidate gene responsible for holoprosencephaly. Nat Genet. 1996;14:353–356. doi: 10.1038/ng1196-353. [DOI] [PubMed] [Google Scholar]
- 107.Roessler E, Belloni E, Gaudenz K, Jay P, Berta P, Scherer SW, Tsui LC, Muenke M. Mutations in the human Sonic Hedgehog gene cause holoprosencephaly. Nat Genet. 1996;14:357–360. doi: 10.1038/ng1196-357. [DOI] [PubMed] [Google Scholar]
- 108.Solomon BD, Bear KA, Wyllie A, Keaton AA, Dubourg C, David V, Mercier S, Odent S, Hehr U, Paulussen A, Clegg NJ, Delgado MR, Bale SJ, Lacbawan F, Ardinger HH, Aylsworth AS, Bhengu NL, Braddock S, Brookhyser K, Burton B, Gaspar H, Grix A, Horovitz D, Kanetzke E, Kayserili H, Lev D, Nikkel SM, Norton M, Roberts R, Saal H, Schaefer GB, Schneider A, Smith EK, Sowry E, Spence MA, Shalev SA, Steiner CE, Thompson EM, Winder TL, Balog JZ, Hadley DW, Zhou N, Pineda-Alvarez DE, Roessler E, Muenke M. Genotypic and phenotypic analysis of 396 individuals with mutations in Sonic Hedgehog. J Med Genet. 2012;49:473–479. doi: 10.1136/jmedgenet-2012-101008. [DOI] [PubMed] [Google Scholar]
- 109.Brugmann SA, Allen NC, James AW, Mekonnen Z, Madan E, Helms JA. A primary cilia-dependent etiology for midline facial disorders. Hum Mol Genet. 2010;19:1577–1592. doi: 10.1093/hmg/ddq030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Balk K, Biesecker LG. The clinical atlas of Greig cephalopolysyndactyly syndrome. Am J Med Genet A. 2008;146A:548–557. doi: 10.1002/ajmg.a.32167. [DOI] [PubMed] [Google Scholar]
- 111.Hu D, Helms JA. The role of sonic hedgehog in normal and abnormal craniofacial morphogenesis. Development. 1999;126:4873–4884. doi: 10.1242/dev.126.21.4873. [DOI] [PubMed] [Google Scholar]
- 112.Costa MA, Borzabadi-Farahani A, Lara-Sanchez PA, Schweitzer D, Jacobson L, Clarke N, Hammoudeh J, Urata MM, Magee WP, 3rd, et al. Partial craniofacial duplication: A review of the literature and case report. J Craniomaxillofac Surg. 2013 doi: 10.1016/j.jcms.2013.05.016. [DOI] [PubMed] [Google Scholar]
- 113.Suryawanshi P, Deshpande M, Verma N, Mahendrakar V, Mahendrakar S. Craniofacial duplication: a case report. J Clin Diagn Res. 2013;7:2025–2026. doi: 10.7860/JCDR/2013/5658.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Eggenschwiler JT, Anderson KV. Cilia and developmental signaling. Annu Rev Cell Dev Biol. 2007;23:345–373. doi: 10.1146/annurev.cellbio.23.090506.123249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, Anderson KV. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- 116.May SR, Ashique AM, Karlen M, Wang B, Shen Y, Zarbalis K, Reiter J, Ericson J, Peterson AS. Loss of the retrograde motor for IFT disrupts localization of Smo to cilia and prevents the expression of both activator and repressor functions of Gli. Dev Biol. 2005;287:378–389. doi: 10.1016/j.ydbio.2005.08.050. [DOI] [PubMed] [Google Scholar]
- 117.Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DY, Reiter JF. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 118.Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nozawa YI, Lin C, Chuang PT. Hedgehog signaling from the primary cilium to the nucleus: an emerging picture of ciliary localization, trafficking and transduction. Curr Opin Genet Dev. 2013;23:429–437. doi: 10.1016/j.gde.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kondo S, Sato-Yoshitake R, Noda Y, Aizawa H, Nakata T, Matsuura Y, Hirokawa N. KIF3A is a new microtubule-based anterograde motor in the nerve axon. J Cell Biol. 1994;125:1095–1107. doi: 10.1083/jcb.125.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Spassky N, Han YG, Aguilar A, Strehl L, Besse L, Laclef C, Ros MR, Garcia-Verdugo JM, Alvarez-Buylla A. Primary cilia are required for cerebellar development and Shh-dependent expansion of progenitor pool. Dev Biol. 2008;317:246–259. doi: 10.1016/j.ydbio.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kolpakova-Hart E, Jinnin M, Hou B, Fukai N, Olsen BR. Kinesin-2 controls development and patterning of the vertebrate skeleton by Hedgehog- and Gli3-dependent mechanisms. Dev Biol. 2007;309:273–284. doi: 10.1016/j.ydbio.2007.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zaghloul NA, Brugmann SA. The emerging face of primary cilia. Genesis. 2011;49:231–246. doi: 10.1002/dvg.20728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Brugmann SA, Cordero DR, Helms JA. Craniofacial ciliopathies: A new classification for craniofacial disorders. Am J Med Genet A. 2010;152A:2995–3006. doi: 10.1002/ajmg.a.33727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Neugebauer JM, Amack JD, Peterson AG, Bisgrove BW, Yost HJ. FGF signalling during embryo development regulates cilia length in diverse epithelia. Nature. 2009;458:651–654. doi: 10.1038/nature07753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Komatsu Y, Kaartinen V, Mishina Y. Cell cycle arrest in node cells governs ciliogenesis at the node to break left-right symmetry. Development. 2011;138:3915–3920. doi: 10.1242/dev.068833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang Z, Wlodarczyk BJ, Niederreither K, Venugopalan S, Florez S, Finnell RH, Amendt BA. Fuz regulates craniofacial development through tissue specific responses to signaling factors. PLoS One. 2011;6:e24608. doi: 10.1371/journal.pone.0024608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tabler JM, Barrell WB, Szabo-Rogers HL, Healy C, Yeung Y, Perdiguero EG, Schulz C, Yannakoudakis BZ, Mesbahi A, Wlodarczyk B, Geissmann F, Finnell RH, Wallingford JB, Liu KJ. Fuz mutant mice reveal shared mechanisms between ciliopathies and FGF-related syndromes. Dev Cell. 2013;25:623–635. doi: 10.1016/j.devcel.2013.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Clarke AJ, Cooper DN, Krawczak M, Tyler-Smith C, Wallace HM, Wilkie AO, Raymond FL, Chadwick R, Craddock N, John R, Gallacher J, Chiano M. 'Sifting the significance from the data' -the impact of high-throughput genomic technologies on human genetics and health care. Hum Genomics. 2012;6:11. doi: 10.1186/1479-7364-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Twigg SR, Vorgia E, McGowan SJ, Peraki I, Fenwick AL, Sharma VP, Allegra M, Zaragkoulias A, Sadighi Akha E, Knight SJ, Lord H, Lester T, Izatt L, Lampe AK, Mohammed SN, Stewart FJ, Verloes A, Wilson LC, Healy C, Sharpe PT, Hammond P, Hughes J, Taylor S, Johnson D, Wall SA, Mavrothalassitis G, Wilkie AO. Reduced dosage of ERF causes complex craniosynostosis in humans and mice and links ERK1/2 signaling to regulation of osteogenesis. Nat Genet. 2013;45:308–313. doi: 10.1038/ng.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sharma VP, Fenwick AL, Brockop MS, McGowan SJ, Goos JA, Hoogeboom AJ, Brady AF, Jeelani NO, Lynch SA, Mulliken JB, Murray DJ, Phipps JM, Sweeney E, Tomkins SE, Wilson LC, Bennett S, Cornall RJ, Broxholme J, Kanapin A, Johnson D, Wall SA, van der Spek PJ, Mathijssen IM, Maxson RE, Twigg SR, Wilkie AO. Mutations in TCF12, encoding a basic helix-loop-helix partner of TWIST1, are a frequent cause of coronal craniosynostosis. Nat Genet. 2013;45:304–307. doi: 10.1038/ng.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Daluiski A, Engstrand T, Bahamonde ME, Gamer LW, Agius E, Stevenson SL, Cox K, Rosen V, Lyons KM. Bone morphogenetic protein-3 is a negative regulator of bone density. Nat Genet. 2001;27:84–88. doi: 10.1038/83810. [DOI] [PubMed] [Google Scholar]
- 133.Attanasio C, Nord AS, Zhu Y, Blow MJ, Li Z, Liberton DK, Morrison H, Plajzer-Frick I, Holt A, Hosseini R, Phouanenavong S, Akiyama JA, Shoukry M, Afzal V, Rubin EM, FitzPatrick DR, Ren B, Hallgrimsson B, Pennacchio LA, Visel A. Fine tuning of craniofacial morphology by distant-acting enhancers. Science. 2013;342:1241006. doi: 10.1126/science.1241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang SK, Komatsu Y, Mishina Y. Potential contribution of neural crest cells to dental enamel formation. Biochem Biophys Res Commun. 2011;415:114–119. doi: 10.1016/j.bbrc.2011.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Liu HX, Komatsu Y, Mishina Y, Mistretta CM. Neural crest contribution to lingual mesenchyme, epithelium and developing taste papillae and taste buds. Dev Biol. 2012;368:294–303. doi: 10.1016/j.ydbio.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ishii M, Arias AC, Liu L, Chen YB, Bronner ME, Maxson RE. A stable cranial neural crest cell line from mouse. Stem Cells Dev. 2012;21:3069–3080. doi: 10.1089/scd.2012.0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhang X, Xie C, Lin AS, Ito H, Awad H, Lieberman JR, Rubery PT, Schwarz EM, O'Keefe RJ, Guldberg RE. Periosteal progenitor cell fate in segmental cortical bone graft transplantations: implications for functional tissue engineering. J Bone Miner Res. 2005;20:2124–2137. doi: 10.1359/JBMR.050806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Colnot C. Skeletal cell fate decisions within periosteum and bone marrow during bone regeneration. J Bone Miner Res. 2009;24:274–282. doi: 10.1359/jbmr.081003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chappuis V, Gamer L, Cox K, Lowery JW, Bosshardt DD, Rosen V. Periosteal BMP2 activity drives bone graft healing. Bone. 2011;51:800–809. doi: 10.1016/j.bone.2012.07.017. [DOI] [PubMed] [Google Scholar]