Summary
Background
Many cellular processes operate in an “analog” regime in which the magnitude of the response is precisely tailored to the intensity of the stimulus. In order to maintain the coherence of such responses, the cell must provide for proportional expression of multiple target genes across a wide dynamic range of induction states. Our understanding of the strategies used to achieve graded gene regulation is limited.
Results
In this work, we document a relationship between stress responsive gene expression and the transcription factor Msn2 that is graded over a large range of Msn2 cocnentrations. We use computational modeling, in vivo, and in vitro analysis to dissect the roots of this relationship. Our studies reveal a simple and general strategy based on non-cooperative low-affinity interactions between Msn2 and its cognate binding sites, as well as competition over a large number of Msn2 binding sites in the genome relative to the number of Msn2 molecules.
Conclusions
In addition to enabling precise tuning of gene expression to the state of the environment, this strategy ensures co-linear activation of target genes, allowing for stoichiometric expression of large groups of genes without extensive promoter tuning. Furthermore, such a strategy enables precise modulation of the activity of any given promoter by addition of binding sites without altering the qualitative relationship between different genes in a regulon. This feature renders a given regulon highly ‘evolvable’.
Introduction
Cells respond to environmental cues and insults by adjusting gene expression to reorganize their proteome. Occasionally, such adjustments are drastic, as in the case of differentiation or entering a senescent state to survive or in anticipation of extreme stress. For model organisms such as S. cerevisiae, such emergency programs are well documented in response to changes in carbon sources, starvation, or large temperature shifts (1).
In addition to these rare and extreme cellular decisions, cells also must contend with a multitude of smaller, transient perturbations. These modest modulations require proportional adjustments to cellular metabolism and physiology, necessitating a graded regulatory system. Our understanding of such homeostatic responses does not match our extensive knowledge of cellular emergency responses, in part because experimental approaches have generally relied on large perturbations (2,3). As a result, the principles by which cellular responses can robustly achieve graded operation over a broad dynamic range remain largely obscure. Recent studies in the budding yeast pheromone pathway, the response to calcium stress, and a number of developmental systems have documented linear responses. In these contexts, mechanisms by which external signals are translated into linear patterns of gene expression occur at the signaling level and seem to involve abundant use of feedback loops (4,5, 6, 7).
To adapt to environmental perturbations, cells engage complex many-gene protective transcriptional programs. Although the specific implementations of these programs vary, the basic strategy is shared from yeast to mammals—stressful conditions impact the activity of a core set of kinases such as protein kinase A (PKA), TOR, or AMPK. These kinases modulate the activity of a range of transcription factors that, in turn, regulate the expression of cyto-protective genes. These networks of kinases, transcription factors, and their gene targets are crucial cellular homeostats that allow the organism to adapt to fluctuating environments. At least in one instance, NFkB signaling in mammalian cells, the regulatory structure of these stress responses, has been implicated in the graded responses that are observed in response to stimulus (8).
In the budding yeast S. cerevisiae, PKA and two if its target transcription factors, the paralogous proteins Msn2 and Msn4, play important roles in cellular homeostatic adaptation to carbon source fluctuations and other stresses (10). Activation of Msn2/4 turns on the transcription of at least 200 genes, including molecular chaperones, the trehalose and glycogen synthase machinery, oxidative stress response, mitochondrial components, and alternate glycolytic enzymes (11-13). Msn2 plays a dominant role under most conditions in the Environmental Stress Response (ESR), a general protective program triggered by many distinct stresses (1), raising the question about how a single factor can coordinate a large number of processes and whether target genes exhibit a spectrum of responses to the same Msn2 signal. Previous data documenting the behavior of the Msn2 transcriptional targets have generally been collected under strong stress conditions and show little obvious differentiation in timing of different target genes (1,14). Recently, however, more quantitative studies have uncovered differences in target gene response to the duration and frequency of Msn2 activity (13, 15).
Here, we use a combination of in vivo and in vitro measurements, guided by computational modeling to systematically dissect the response of target genes to Msn2 activity. We find that Msn2 exhibits non-cooperative activation of its targets, including those whose promoters contain a large number of Msn2 binding sites. This graded activation is the combined result of low-affinity interactions between Msn2 and its cognate binding site in gene promoters and competition over a large number of Msn2 binding sites in the genome relative to the number of Msn2 molecules. These effects result in a linear relationship between the concentration of active Msn2 protein and the expression of many of its target genes, leading to their co-linear and stoichiometric expression. Linearity is a robust feature of the system and extends over the full dynamic range of its operation, suggesting that Msn2 provides a proportional homeostatic response to stressful conditions. In addition to its properties as an ‘emergency’ stress response, these results position the ESR as a homeostatic system capable of providing a precisely balanced response across a range of environmental conditions. The strategy that positions Msn2 in this linear regime is general. It is implemented through simple constraints on binding affinity and copy number of the transcription factor as opposed to extensive tuning by feedback loops. As such, it may provide a fingerprint to identify these features in other cellular systems. Moreover, the Msn2 strategy constitutes a framework that may aid the design of synthetic homeostatic systems.
Results
A synthetic circuit allows for probing the relationship between Msn2 and its target genes
We sought to map the relationship between the responses of different target genes to the same Msn2 signal, by measuring expression of fluorescent proteins driven by prTPS1 and prPGM2, two stress responsive promoters that contain multiple consensus Msn2/4 binding sites also known as Stress Response Elements (STREs). To precisely control Msn2 activity, we developed synthetic tools that allowed us to tune the concentration of active Msn2 in the nucleus while avoiding the pleiotropic effects of Msn2 activation by stress. We used a constitutively active allele of MSN2, Msn2-5A, in which every PKA phosphosite is mutated to alanine (16). We controlled expression of this Msn2 mutant allele by placing it under a GAL1 promoter in a Δmsn2/4 strain expressing an estradiol-regulated Gal4 fusion protein (17). This construct allows for graded regulation of the abundance of Msn2-5A by addition of the small molecule estradiol. Since this estradiol-inducible synthetic circuit provides a general strategy for controlling the expression of any protein over a wide dynamic range, we further used it to drive the production of two negative regulators of the PKA pathway, the phosphodiesterase PDE2 which degrades cAMP and a dominant negative allele of RAS2 (S24N). These two constructs were integrated into wild type (MSN2/4) strains (Figure 1A), providing two additional means for tuning Msn2 activity.
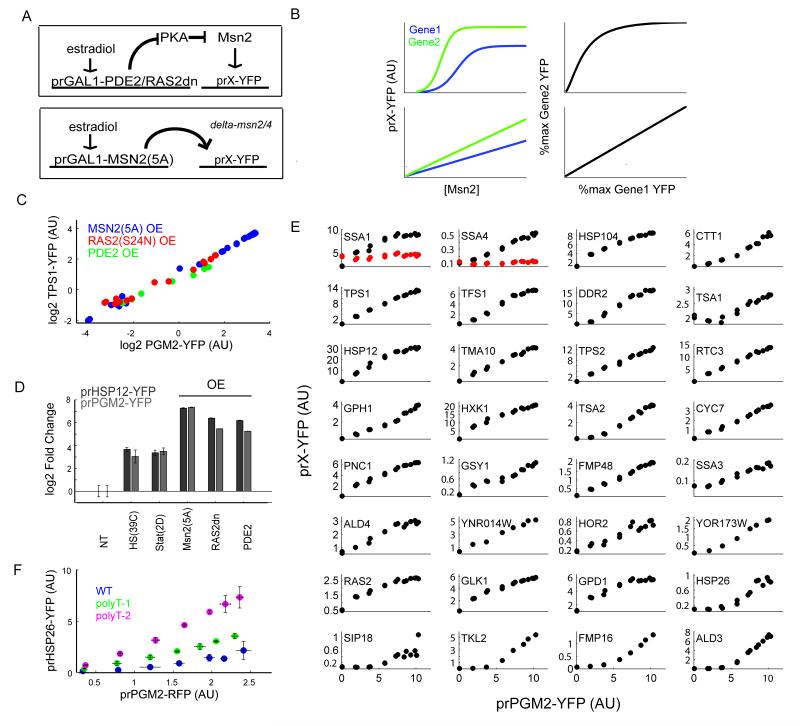
Figure 1. Msn2 Regulated Genes are Induced Co-Linearly.
(A) An estradiol-inducible synthetic circuit is used to overexpress constitutively active allele of MSN2 or of PKA inhibitors (PDE2 or RAS2dn). The resulting activation of Msn2 target gene can be measured by cloning the promoter in front of YFP (prX-YFP). (B) A simple model of gene expression reveals that if two genes (Gene 1 and Gene 2) have a sigmoidal dependence on the same transcription factor, then the ratio of their expression is not constant. The expression ratio of the two genes is constant either when the two genes have identical dependence on the transcription factor or if they are individually linearly dependent on the transcription factor. (C) The PGM2 and TPS1 promoters show co-linear activation over a wide range of expression levels upon overexpression of Msn2(5A) or PKA inhibitors using the estradiol inducible synthetic circuit. Each dot represents the average expression level from these promoters at a given concentration of the inducer estradiol. (D) The HSP12 and PGM2 promoters show similar susceptibility to Msn2(5A) overexpression but quantitatively differ in their response to PKA inhibition . (E) The expression level of different Msn2 target promoters plotted against the expression of prPGM2-YFP as the levels of Msn2(5A) are varied. The red dots in SSA1 and SSA4 show expression when promoters are mutated to remove Msn2 binding (STRE) elements. (F) The HSP26 promoter shows a sub-linear response to Msn2(5A) activation compared to PGM2. Insertion of one or two chromatin disrupting polyT elements into the HSP26 promoter converts HSP26-PGM2 to a monotonic linear relationship.
Using these tools, we were able to titrate the activity of Msn2 and measure the level of activity of fluorescent proteins driven by prTPS1 and prPGM2. Since these two promoters contain several STREs, we expected their expression to be a sigmoidal function of active Msn2, resulting in a nonlinear relationship between their respective expression profiles (Figure 1B). Contrary to this expectation, we found that the prTPS-prPGM2 relationship was linear over the whole range of Msn2 activity (Figure 1C). This result is particularly surprising given that both the direct titration of Msn2-5A and its activation in the Ras2(S24N) strain accessed the full dynamic range of the ESR system. In fact, estradiol mediated activation of Msn2 in these strains resulted in the induction of the two target promoters prPGM2 or prHSP12 to levels exceeding (~4 fold higher) those observed in heat shock or mid-stationary phase (Figure 1D).
Targets of Msn2 show co-linear activation
To test whether this linear relationship was a general feature of Msn2 transcriptional regulation of its targets, we measured the expression of a large number of other Msn2 target genes. We identified 40 such genes from microarray studies (1,13,14), and monitored their expression using a fluorescent reporter of their promoter activity in a strain harboring the estradiol responsive synthetic circuit driving either MSN2-5A, RAS2dn, or PDE2. Of these promoters, 32 showed measureable basal expression and greater than four-fold induction in one of the Msn2 perturbations, and lost at least 50% of this induction in a Δmsn2/4 background (Figure S1e). These results were reproducible and specific to PKA perturbations as overexpression of a second dominant negative allele of RAS2 (G22A) resulted in nearly identical induction as S24N (Figure S1a). Overexpression of a constitutively active allele of Msn4 (4A -- allele) also resulted in similar patterns to those generated by induction to Msn2(5A) (Figure S1a-d). We chose this set of 32 promoters for further characterization. For the bulk of these promoters, all perturbations gave similar results, therefore we carried out the rest of our analyses using Msn2(5A).
As with prTPS1, titration of Msn2-5A produced co-linear relationships between prPGM2 and most (26/32) of the 32 characterized promoters (Figure 1C, Figure S1a). Co-linear relationships were dependent on direct Msn2 binding, as removal of the Msn2 binding consensus STREs from the promoters of two such genes (SSA1 (2 sites) and SSA4 (3 sites)) ablated induction upon Msn2(5A) overexpression (Figure 1E, panels 1 and 2, mutants are in red).
In contrast to the 26 promoters that exhibited co-linearity, five promoters (last five panels of Figure 1E) showed a sub-linear behavior: expression from these promoters was insensitive to low amounts of Msn2(5A), while an increase of Msn2 beyond a certain threshold elicited a co-linear expression with that of prPGM2. Additionally, a couple of promoters (most notably GPD1) displayed a degree of saturating behavior, inducing linearly at low levels of Msn2(5A) but saturating before maximum induction from the synthetic circuit was reached. Of the five promoters that showed the sub-linear relationship with prPGM2, two (HSP26, SIP18) had been suggested in a previous study to be regulated by chromatin structure (15). To explore chromatin structure as the root of the sub-linear behavior, we sought to alter this structure by insertion of poly-T sequences, which have been shown to disrupt nucleosome positioning (17). We inserted either one or two poly-T sequences (12xdT) into the prHSP26 promoter and measured its co-expression with prPGM2. Consistent with chromatin playing a role in the threshold on activation of prHSP26, the insertion of these sequences increased the expression of prHSP26 and rendered it strongly co-linear with prPGM2 (Figure 1F). By contrast, insertion of poly-dT sequences into the promoters of three strongly co-linear genes (PGM2, TPS1, or PNC1) resulted in no substantive change in their expression (data not shown).
Taken together, these results indicate that Msn2 predominantly activates its downstream genes co-linearly. The exceptions to this linear regulation can be explained by other contributions such as chromatin architecture—as we observe for prHSP26—or possibly due to regulation by other transcription factors. Notably, these promoters show co-linear activation with other Msn2 targets once the threshold set by other regulatory factors is passed, and—at least in the case of HSP26—recover their co-linear activation for the full range once the chromatin thresholding influences are ablated. These results are consistent with earlier work on the kinetics of gene expression from Msn2 regulated genes that found most targets to be rapidly and coherently activated while a subset of genes showed more delayed kinetics (15).
Msn2 activates promoters in proportion to its concentration
Traditionally, the presence of multiple binding sites for transcriptional regulators in gene promoters has been thought to produce gene regulatory functions of increasing steepness (Hill coefficient) as a result of cooperative binding. Our results do not conform to this notion. Instead, the co-linearity in the expression of these promoters argues that gene expression is instead linearly related to the concentration of nuclear Msn2, even in cases where these promoters contain a large number of STREs.
The relationship between Msn2 and a promoter it regulates, such as prPGM2, depends on at least two distinct kinetic steps: the translocation of Msn2 into the nucleus and binding of nuclear Msn2 to prPGM2, resulting in the production of RFP. The most parsimonious model accounting for these two steps represents the translocation of Msn2 in and out of the nucleus as first order processes proceeding at a rate kin and kout respectively, and the rate of production of proteins (RFP in this example) as a simple linear function of nuclear Msn2,[Msn2n]. In addition, in this model, Msn2 is assumed to be produced in the cytoplasm at a constant rate and degraded both in the cytoplasm and nucleus with first order kinetics. The degradation rate in the nucleus is denoted γn and the total amount of Msn2 in nucleus and cytoplasm is denoted by [Msn2total] (Supplementary materials). This model predicts that at steady-state, the nuclear concentration of Msn2 is given by:
As a result, the amount of fluorescence produced by the PGM2 promoter is proportional to [Msn2total] (Figure 2A):
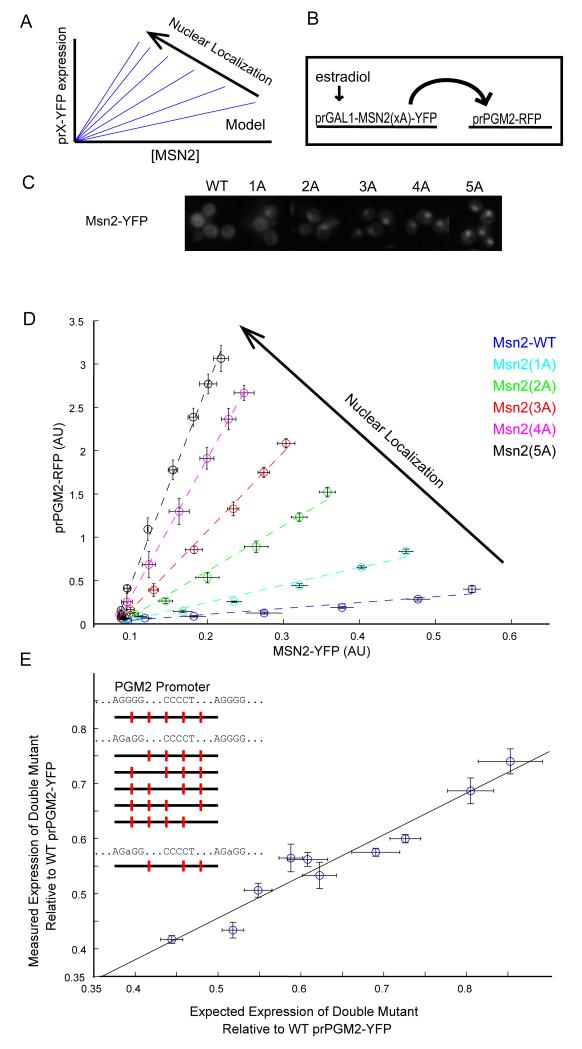
With such simplifying assumptions, the equation above predicts that if the nuclear residence of Msn2 were to be increased by either increasing kin or decreasing kout, then the relationship between prPGM2 expression and total Msn2 would still be linear, but with an increased slope (Figure 2A). This prediction can be validated by taking advantage of different Msn2 alleles with an increasing number of PKA phosphosite substitutions (Msn2-Xa, where X=0,1,2,3,4,5). These alleles span a range of nuclear localizations, from limited (Msn2-0A) to constitutive nuclear localization (Msn2-5A) (Figure 2C, S2A). We simultaneously monitored in the same cell the expression of Msn2(Xa)-YFP whose concentration can be titrated using the estradiol synthetic circuit and prPGM2-RFP expression (Figure 2B). Induction by estradiol resulted in increasing concentrations of Msn2, and revealed that a strong linear relationship indeed exists between prPGM2 expression and total Msn2 concentration irrespective of the Msn2 allele used (Figure 2D). The slope of the line relating prPGM2-RFP expression and any one Msn2 allele was an increasing function of the allele’s nuclear localization. These patterns were not unique to prPGM2, as the same linear and ordered relationship also existed between the different Msn2 alleles and prTPS1 (Figure S2). This uniformity of behavior across the spectrum of Msn2 alleles demonstrate that neither the protein’s nuclear import nor export dynamics contribute to the linear activation behavior.
Figure2. Gene expression is linear as a function of Msn2.
(A) A model that assumes a linear relationship between Msn2 level and gene expression of its target promoters predicts a continued linear relationship with a higher slope upon overexpression of increasingly active Msn2 alleles. (B) An estradiol-inducible synthetic circuit drives the expression of Msn2 alleles (Msn2(xA)-YFP) that have different nuclear localization levels. The activity of a downstream PGM2-RFP promoter is monitored. (C) Cells expressing Msn2(xA)-YFP are imaged after 2hrs of induction with estradiol. Msn2 alleles with multiple phosphosite substitutions show increased nuclear localization of Msn2. (D) prPGM2-RFP fluorescence as a function of Msn2-YFP for the different Msn2 alleles. As predicted by the model, the linear relationship persists but with a different slope for different alleles. (E) Measured expression level from a double STRE mutant PGM2 promoter plotted against the expression level predicted from a multiplicative model that assumes independent binding to every STRE. Mutations were made to each of the five consensus Msn2 binding sites. Expression levels measured for these single mutants were used to predict the expression level of the double mutants. All ten possible double mutants were then constructed and their expression measured. The best fit line of a linear model is also plotted. All expression levels are normalized to WT pr-PGM2 expression and error bars represent standard error (n=3).
Further, we note that total Msn2 levels consistently decreased in abundance for the same estradiol induction in the more active alleles, confirming previous observations of an increased rate of degradation of Msn2 in the nucleus (19,20).
Binding sites in target promoters show no interactions
The proportional activation of Msn2 to its target genes over the whole dynamic range of the system suggests that Msn2 should bind to different STREs in the same promoter independently in the regime we are investigating, which also spans the physiological regime of the system. To directly test this notion, we focused on the Pgm2 promoter, which has five consensus STREs in the 500bp preceding the start codon. If the binding of Msn2 to any one STRE is independent from its binding to other STREs present in the same promoter, prPGM2 expression in any two single STRE mutants should be predictive of expression of the double mutant. To test this notion, we mutated each individual STRE in series—we chose to use a minimal single base pair substitution (AGGGG->AGaGG) that ablates in vivo activity of the binding site—creating five single mutant alleles. We then constructed all ten possible double mutant alleles and measured the expression of each of the wild type and 15 mutant prPGM2 promoters. The expression of all double mutant promoters was approximated as a product of the activity of the constituent single mutants (Figure 2E). Therefore, no pair of STREs depended on each other for activity.
A simple binding model predicts the regimes in which a linear relationship between gene expression and TF concentration exists
Next, we sought to probe the root of the linear relationship between gene expression and Msn2 concentration. Using mass action kinetics equations, the fraction of bound Msn2 to STRE can be expressed by the Michaelis-Menten relationship (Supplementary material):
Here, Msn2f is the concentration of free nuclear Msn2 and is related to total nuclear Msn2 by the equation:
In the limit that Kd ⟫ [Msn2n] or [Msn2n] ⟪ [STREtotal], simple calculations demonstrate that
In this case (See supplementary materials), gene expression is a linear function of [Msn2n]. Furthermore, since [Msn2n] ∝ [Msn2total], the experimentally measured linear relationship between gene expression and [Msn2total] is recapitulated at steady-state if either Kd is large, Msn2 is small, or both.
The number of Msn2 molecules in the cell has been estimated to be 125 (21). Even overexpressed from a strong GAL1 promoter, we estimate that there are no more than 2000-3000 Msn2 molecules in the nucleus (see supplementary materials). For a nuclear volume of 4-10pl, [Msn2total], the concentration of total Msn2 is ~0.1uM to low micromolar. Therefore, a linear relationship can arise between [Msn2total] and the bound STRE fraction if either Kd or [STREtotal] were large compared to this [Msn2total].
Msn2 binds STREs with low affinity
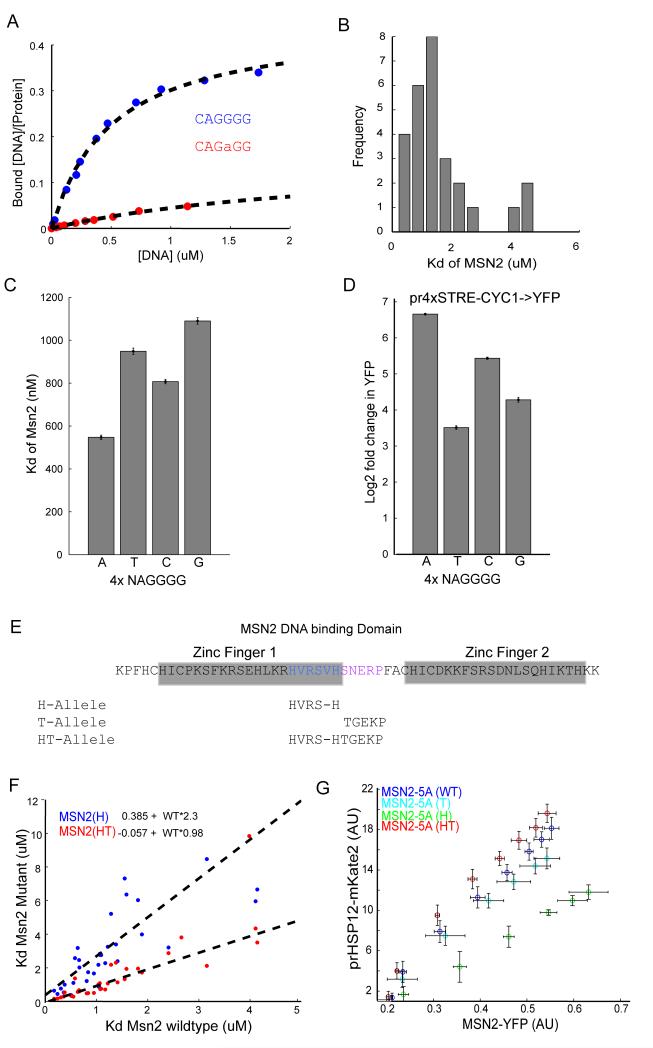
To measure directly the binding affinity of Msn2 to STREs and extract the Kd value, we took advantage of the MITOMI 2.0 system, an in vitro microfluidic technique that determines the absolute binding affinity of a given transcription factor to its cognate binding sites (22, 23). The MITOMI 2.0 procedure works as follows: Individual cells of the microfluidic device are programmed with fluorescently labeled double-stranded oligonucleotides, arrayed in a series of dilutions. Using a separate fluorophore, labeled His-tagged protein (Msn2) is introduced into the device, solubilizing the deposited DNA sequences. Within each cell, anti-His antibodies, deposited below a “button” valve, recruit Msn2 protein, allowing the Msn2-DNA binding interactions to be mechanically trapped by the activation of the button valve. Untrapped material is then washed away, leaving trapped complexes at their equilibrium concentrations. By imaging, the ratio of bound protein and trapped DNA may be determined beneath the button. The binding occupancy curves and the free DNA concentration are then fit and Kd of each interaction measured (Figure 3A). Using this approach, we measured the in vitro affinity of Msn2 for 27 oligonucleotide sequences each consisting of 40bp centered at a single genomic consensus STRE binding motif (‘AGGGG’). In the event that two STREs fell into this range we mutated one of the sequences to a non-consensus site. These sequences were taken from endogenous promoters of genes that are strongly responsive to Msn2 activity (PGM2, HSP12, TPS1, HSP26, RTC3, or CYC7, supplementary table 4, Figure S3). Measured Kd values ranged from 0.2-4μM (Figure 3B), corresponding to a relatively low affinity of Msn2 to STREs.
Figure3. Msn2 affinity measurements of STREs by MITOMI show low affinity.
(A) MITOMI measurements of Msn2 affinity for a cognate or mismatched motif. The plots show the bound fraction of DNA as a function of total DNA in the microfluidic chamber. These data can be used to extract an absolute binding constant Kd (B) Histogram of Kd for a range of binding motifs from promoters that are in vivo targets of Msn2. (C) MITOMI measurements of Kd show that Msn2 exhibits different binding preferences as the first nucleotide in the binding motif (NAGGGG) changes. Error bars show 95% CI of fit to data pooled from at least three replicate experiments. (D) in vivo measurements of expression from a crippled CYC1 promoter with four copies of the NAGGGG sequence (pr4XSTRE-CYC1). Data are reported as fold change of fluorescence upon maximal Msn2(5A) overexpression using the estradiol inducible circuit. Errorbars represent standard error (n=3). (E) Design of Msn2 alleles with higher and lower binding affinity. The regions we targeted for mutation a highlighted in blue (histidine spacing) and pink (linker). (F) Kd values for the H and HT Msn2 alleles plotted against the Kd of the WT allele for the same motifs. The mutants show the same binding preference as the WT but different binding affinities. Lines of best fit are for each mutant are plotted and the equations given. (G) Expression of prHSP12 for the different Msn2 alleles as the levels of Msn2-YFP (X-axis) is varied using the estradiol inducible synthetic circuit. The H and T alleles induce less prHSP12 expression while the HT allele induces slightly increased expression. Errorbars depict standard error (n=3).
To further explore the contribution of flanking nucleotides, we selected a single binding site from the PGM2 promoter and measured Msn2 binding affinity for different bases in the first position of the ‘NAGGGG’ sequence (Figure 3C). Consistent with previous data, Msn2 had a significantly lower Kd for the ‘AAGGGG’ motif than for motifs that have C/G/T in the first position (Figure 3C; Fordyce et al., 2010). To validate these measurements, we determined expression upon Msn2 overexpression of a promoter containing four copies of each of these motifs fused to a crippled CYC1-YFP promoter. In agreement with the in vitro data, the “AAGGGG” motif, which has the lowest Kd, showed substantially higher activity, followed by “CAGGGG”, “GAGGGG” and “TAGGGG” (Figure 3D). Examining the yeast genome, we find that almost two-fifths (38%) of NAGGGG motifs have ‘A’ in the first position and that this fraction increases slightly as one examines only promoters with four or more binding sites (Figure S3A).
Low affinity binding is not caused by non-canonical Zinc-Finger linker arrangements
The Msn2 DNA binding domain consists of two tandem zinc fingers connected by a short linker. For many zinc finger transcription factors, this linker region is strongly conserved and has been shown to exert an influence on DNA binding affinity (24). The linker region of Msn2 is divergent from consensus sequences, having both increased spacing between invariant histidine residues immediately before the linker and also a strongly diverged sequence within the linker itself (Figure 3E; Figure S3). To investigate whether the Msn2 divergent linker might explain the protein’s relatively low binding affinity to DNA, we engineered Msn2 alleles with linkers conforming to the consensus sequences. Previous work that has explored the function of the linker residues suggests that perturbations to the linker should not significantly affect the protein’s DNA binding specificity, but can impact its binding affinity (24-27). Drawing on this extensive literature, we constructed three MSN2 mutants with altered linker arrangements. One construct (H allele) converted the HX4H spacing to a more conventional HX3H spacing by removing the final valine residue (V669), the second construct (T allele) converted the linker sequence to the consensus (S671T, N672G,R674K), and the final construct combined these two mutations (HT allele).
Based on previous data, we suspected that the H and T alleles would by themselves reduce binding affinity, but that in combination, they would move the protein towards the consensus and therefore increase DNA binding affinity without altering specificity (Figure 3E, see supplement for details). Affinity measurements by MITOMI 2.0 show that, as expected, the H allele reduces affinity by ~2-fold. Somewhat surprisingly, the HT allele showed relatively small binding site-dependent change in affinity but on average appeared to show similar affinity to wild type. Further, as one would predict given that the linker residues are not expected to contact DNA, these alleles appear not to have altered preference for DNA binding (Figure 3F, Figure S3). Consistent with our predictions and binding affinities, we found that the H allele had a strongly decreased and the T allele had a slightly decreased ability to activate the prHSP12 promoter in vivo (Figure 3G). The HT allele has a slightly increased ability to activate prHSP12, despite our inability to detect a significant change of affinity in vitro. Importantly, overexpression of these alleles preserved the linear inductions of prHSP12 (Figure 3G), although as expected the HT and H allele showed increased and reduced activity, respectively. These data suggest that perturbations to the Msn2 linker arrangement that change the binding affinity (at least within the affinity regimes that are experimentally accessible) are not sufficient to break a robust linear relationship between Msn2 and its target genes.
Competitive interactions of Msn2 with binding sites may contribute to linearity
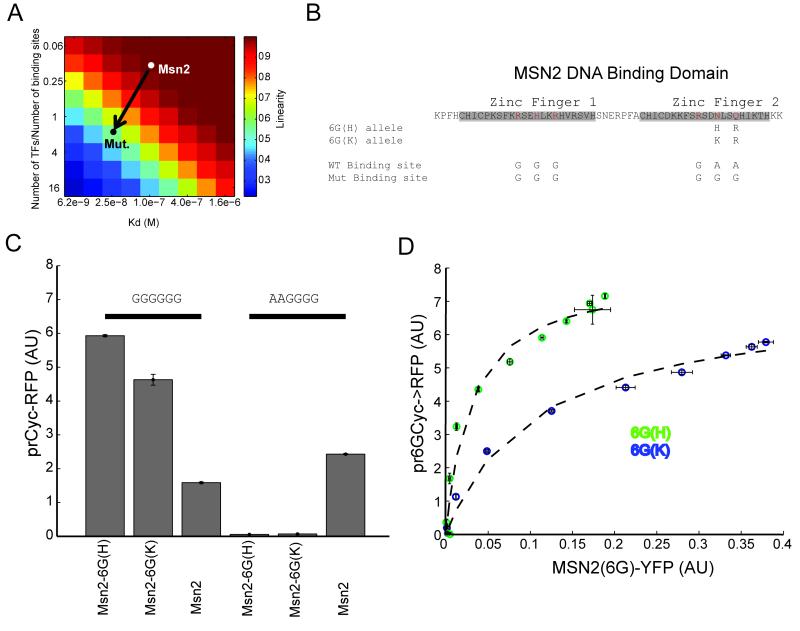
As discussed above, in addition to low binding affinity, linearity in the relationship between Msn2-dependent gene expression and the total available Msn2 can result from a low Msn2 to STRE ratio, resulting in competition between STREs for binding to Msn2 (see supplementary materials for details, Figure 4A). There are 8450 consensus STREs (AGGGG) in the genome, roughly half of which (4122) are present in promoter regions (defined here as DNA sequences 700bp upstream of a start codon). Consistent with these sequence based estimates, recent in vivo measurements of Msn2 binding identified 1290 loci—many of which contain multiple STRE motifs—that associate with Msn2 during stress conditions (13).
Figure4. Saturated binding of Msn2 to DNA can be induced by altering its DNA binding site preference and affinity.
(A)A mass-action model shows that linearity in the relationship between gene expression and transcription factor concentration can be achieved by either low affinity binding or plentiful binding sites. Msn2 appears to be centered in this regime in respect to both criteria. (B)Two point mutations in the second zinc finger of Msn2 convert its specificity from AAGGGG to GGGGGG. (C) Expression from a CYC1 promoter containing three consecutive GGGGGG or AAGGGG motifs. The Msn2 mutant alleles (Msn2-6G(H) or Msn2-6G(K)) specifically activate the promoter containing the three GGGGGG motifs, but not the wild type (AAGGGG) Msn2 binding motifs. Msn2-6G(H) activates the promoter more strongly than Msn2-6G(K) (D) Expression from pr6GCYC1-RFP as a function of Msn2-6G(H)) (green) and Msn2-6G(K) (blue). The expression shows saturating activation of the transcriptional reporter, with the Msn2-6G(K) allele showing ~2 fold reduced affinity. Dashed lines represent fits to the [RFP]/([RFP]+kd). Titration of the corresponding Msn2 allele is achieved using the estradiol inducible circuit.
Testing the prediction that an excess of binding sites contributes to the observed linear relationship would require reducing the number of Msn2 binding sites in the genome, which is not experimentally tractable. An alternative experimental approach would be to engineer the Msn2 protein to recognize DNA sequences that are less abundant in the genome.
To define such a low occurrence sequence motif, we scanned a database of the S. cerevisiae promoter sequences for the number of occurrences of GNNGNN, a sequence motif known to be bound by canonical zinc fingers. Interestingly, the least common motif ‘GTCGGG’ (416 occurrences) matches the consensus of the proteosomal regulator Rpn4 and the fifth least common motif ‘GCGGGG’ matches the Mig1 consensus (12,28). These results hint that random transcription factor binding might be deleterious, perhaps causing selection pressure to eliminate non-specific sites. Of the least common such sequences, the 9th on this list (‘GGGGGG’, 540 occurrences) is not target of any of the known transcription factors in yeast. To switch Msn2 recognition from AAGGGG to GGGGGG, we made two mutations to its second zinc finger (Q693R and N690H--the Msn2-6G(H) allele) or (Q693R and N690K--the Msn2-6G(K) allele). These mutations are predicted to alter Msn2 specificity from AAG to GGG, with high affinity for Msn2-6G(H) and low affinity for Msn2-6G(K) (Figure 4B; 29).
The Msn2-6G(H) and Msn2-6G(K) alleles, tested in a msn2/4Δ strain, specifically bound and activated a CYC1 promoter whose UAS was modified to contain three 6xG sequences (pr6GCYC-YFP). At the same time, these alleles did not induce any activity in a CYC1 promoter containing the consensus WT STRE (AGGGG, Figure 4C). However, the wild type Msn2 allele was able to bind and activate the 6xG promoter, albeit to a lesser degree than our engineered alleles, further emphasizing Msn2’s binding promiscuity. These data suggest that the mutant alleles can be used to investigate whether the Msn2 binding linearity can be compromised by a combination of increased binding affinity and decreased number of competing genomic binding sites (Figure 4A, arrow).
To do so, we titrated Msn2-6G(H) and Msn2-6G(K) using the estradiol synthetic circuit and measured the expression of a pr6GCYC-RFP. In contrast to WT Msn2, these alleles produced a nonlinear and saturating expression profile (Figure 4D). This relationship was well fit by a simple Michaelis-Menten binding model (R2>0.95). Additionally, the Msn2-6G(K) allele showed weaker binding with less maximum expression and a two-fold lower apparent Kd for the promoter (Figure 4C). Our alterations of the Msn2 DNA binding domain did not affect its cooperativity since the MSN2-6G allele induced graded and proportional increase in gene expression from a synthetic promoter containing 1, 2, or 3 binding sites (Figure S4). Furthermore, a transcription factor chimera that substitutes the Msn2 DNA binding domain with that of the Gal4 transcription factor while preserving the Msn2 activation domain shows strong saturating interactions with the GAL1 promoter (Figure S4). Since Gal4 is known to bind strongly to less than 20 loci in the genome (30), these data further corroborate low affinity competitive interactions as the root of the linear relationship between Msn2 and its target genes.
Discussion
In many contexts, cells respond to stimuli with decisive commitment to a phenotypic state. It is usually assumed that genes that drive this transition exist in just two alternative functional states, active and inactive, and that the switch between these two states occurs decisively in a narrow regime of transcription factor concentration. In addition to making decisive choices, cells and organisms also need to continuously adjust to the demands of their environment. Systems that are responsible for homeostasis or graded developmental processes may need to operate in an “analog” regime where a response is tailored to the exact intensity of the stimulus in order to prevent deleterious over or under-reactions (4,6,31,32). In fact, many nutrient and stress responsive systems are tightly embedded in negative feedback loops that ensure regulated and limited expression (5, 9, 33-35). For example, feedback loops present in the budding yeast pheromone and calcium sensing pathways have been shown to play a role in maintaining the graded relationship between stimulus and response in these systems (5, 9).
In this work, we document a relationship between stress responsive gene expression and the transcription factor Msn2 that generates graded behavior over a large range of Msn2 concentrations. We demonstrate that a large number of ESR genes show a linear expression response as a function of Msn2. Using synthetic biology tools to precisely set the concentrations of the active transcription factor Msn2, we were able to probe gene activation quantitatively and measure dose responses over the whole physiological regime of the ESR system while avoiding pleitropic effects of its stress-mediated induction. Our investigations, coupled with computational modeling and in vitro analysis, revealed a robust and simple strategy for the linear relationship between Msn2 and its targets. Specifically, we established that the interaction between Msn2 and its cognate binding motif is positioned in its linear regime through a combination of weak non-cooperative binding and a limited number of Msn2 molecules relative to binding sites in the genome. Supporting this model, we demonstrated that modifications of Msn2 that increase its binding affinity and change its binding preference to an infrequently occurring motif in the genome abrogate this linearity and induce dose-response saturation. The conversion to a rapidly saturating factor could be achieved with two amino acid substitutions in the second zinc finger of Msn2, suggesting that the graded nature of Msn2 binding to its promoters is an organizational feature of the system, not a consequence of its biophysical constraints.
In addition to enabling precise tuning of gene expression to the state of the environment, there are at least two additional benefits for maintaining such a linear response in the ESR system. First, linearity as a function of Msn2 implies co-linear activation of target genes, allowing for stoichiometric expression of large groups of genes without extensive promoter tuning. Secondly, low affinity linear interactions allow for precise tuning of promoter activity by modification of binding site affinity or addition of new binding sites. These features render the Msn2 regulon easily ‘evolvable’ as a change in a given promoter, for example addition by a binding site, can modify the slope of the linear relationship that relates Msn2 and the gene output. However, such a change does not affect the coherence of the regulon. Consistent with this notion, stress responsive networks in fungi show rapid rewiring across evolutionary timescales (36-39).
While our studies were focused on the steady-state input output relationship between Msn2 and its target genes, the linearity we uncovered has important implications for the dynamic operation of the system. This aspect needs to be considered in the context of the PKA regulatory system, which produces dynamic changes in Msn2 activity in response to stress. Low affinity interactions of Msn2 with DNA induce rapid (sub-second) binding and un-binding of Msn2 to its response elements, allowing for rapid dynamic control of gene expression if the rate limiting step for promoter activation is transcription factor binding. Further dynamic control can be achieved by layering additional rate limiting steps such as chromatin remodeling. Indeed, prHSP26 that showed a threshold-linear relationship with Msn2 has also been previously shown to exhibit dynamics that are different than other Msn2-responsive genes (at the minute timescales) in response to Msn2 activity (15). More broadly, such a strategy might be useful for inducing temporal coherence for a group of genes while still allowing for select genes to be ‘late responders’. It will be interesting to determine whether tightly regulated temporal processes, such as the cell cycle (38), employ similar mechanisms to implement temporally regimented gene induction profiles.
In summary, we have shown that the ESR in the yeast S. cerevisiae constitutes a system where gene activation is linearly commensurate with the cell’s perception of environmental stress as encoded by the level of active Msn2. This linearity, which endows the system with many kinetic and dynamic properties, is implemented through a simple and robust strategy relying on competitive and weak binding of the transcription factor. As a result, we anticipate this strategy to be a recurring feature of many systems where homeostatic regulation is important.
Methods
Yeast Strains
All yeast strains used for these experiments are derived from W303A-1 in which the ade2 marker was reverted to ADE2+ to reduce the autofluorescence. Promoter constructs were integrated at the TRP1 locus of a HIS3+ MATa strain. Overexpression constructs were integrated into the TRP1 locus of a LEU2+ MATalpha strain which contained the estradiol inducible construct. These strains were then mated and diploids selected in SD-leu/his media. All strains were constructed using standard yeast protocols and LioAc/PEG transformation. For a complete list of strains and plasmids see supplementary tables 1 and 2.
For all strains expressing a single florophore YFP(Venus) was used due to its superior maturation rate and brightness, for two color experiments RFP (mKate2) was used for the promoter construct and the fast maturing YFP for the transcription factor.
Growth and fluorescence measurements by flow cytometry
For all measurements, cells were grown to saturation in 96-shallow well plates (Costar) and then diluted into fresh media, grown at 30C on orbital shakers (Elim) for 12hrs to an OD of ~0.5. Cells were subsequently diluted and estradiol added as necessary and grown for 6hrs to an OD of ~0.05 before measurement. Expression of the estradiol regulated system was activated by addition of 0-200nm estradiol (stock of 1.6mM in 90/10 mixture of EtOH and DMSO), typically applied in log1.6 titration series. Expression of the tetracyclin regulated system was activated by addition of 0-100ng/ml anhydrotetracycline (Sigma, stock of 200ug/ml in 90/10 mixture of EtOH and DMSO), typically applied in log2 titration series.
All cytometry measurements were made on a Becton Dickinson LSRII flow cytometer, along with an autosampler device (HTS) to collect data over a sampling time of 6-12 seconds, typically corresponding to 2000-10000 cells. GFP and YFP were excited at 488nm, and fluorescence was collected through a HQ530/30 bandpass filters (Chroma), mCherry and mKate2 were excited at 561 nm and fluorescence collected through 610/20 bandpass filter (Chroma).
Microscopy and image analysis
Cells expressing Msn2-YFP or related constructs were plated in SD complete media onto ConcanavalinA coated 96 well glass bottom plates, allowed to settle and then washed twice with fresh media. Samples were imaged on a Nikon Ti inverted scope with arc-lamp illumination using RFP(560/40nm excitation, 630/75nm emission, Chroma) and YFP (510/10nm excitation, 542/27nm emission, Semrock) filters. Images were processed and analyzed with ImageJ and custom built Matlab scripts. Nuclear enrichment was computed by dividing the average intensity of the brightest 10pixels in the cell by the median intensity of the cell.
Flow cytometry Data analysis
All data was analyzed with custom Matlab software. Raw cytometry data were filtered to remove errors due to uneven sampling and cell size corrected for as previously described (Stewart-Ornstein et al., 2012).
Sequence Analysis
The latest S288C genome was downloaded from SGD (yeastgenome.org) and the DNA sequence was processed and examined with custom matlab scripts. Promoters were operationally defined as 700bp upstream of the start (ATG) condon of the gene.
MITOMI devices and experiments
MITOMI devices were made as described in Maerkl and Quake, 2007 and Fordyce et al., 2010. Devices were based on the designs from Maerkl and Quake, 2007. The two layers of the device were made from RTV615 PDMS casts from the silicon molds. The two-layer device was aligned and bonded to a glass substrate with a contact-printed array of the DNA library (ssDNA template strands were ordered from IDT Coralville, Iowa, and Alexa-647 end-labeled second strands were synthesized with Klenow exo- enzyme). Finished devices were run as described previously (Maerkl and Quake 2007, Fordyce et al Nat Biotech 2010 and PNAS 2012). His-tagged Msn2 protein was synthesized in wheat germ extract (Promega), and fluorescently labeled by incorporation of Bodipy-lysine. After running the devices the fluorescence intensities were scanned using an arrayWoRx scanner. Fluorescence data for bound DNA and protein and free DNA in the DNA chamber were extracted from the scanned images with Genepix 6.1 . A dilution series of the labeled primer flowed onto the DNA chambers was used as a standard curve to calibrate the relationship of Alexa-fluor signal to free DNA concentration in the DNA storage chamber on the devices. Binding curves were fit to a hyperbolic saturation curve with global nonlinear regression in Graphpad Prizm 4.00, to derive Kd values.
Supplementary Material
Highlights.
→The Transcription Factor Msn2 induces the expression of targets proportional to its abundance
→Graded activation of Msn2 targets relies on low affinity binding and low molecular numbers
→Proportional activation of Msn2 targets allow for stoichiometric activation of its diverse targets
Acknowledgements
We would like to thank Charles Biddle-Snead, Raj Bhatnagar, David Pincus, Onn Brandmann, and the Weissman and El-Samad labs for scientific discussion and comments on the manuscript. This work was funded by the NIGMS system biology center (P50 GM081879) and the David and Lucille Packard Foundation (H.E.S.), the Howard Hughes Medical Institute (J.S.W.), and an NSERC post-graduate scholarship (J.S.O). J.S.O and H.E.S designed the majority of the experiments with input from J.S.W., which J.S.O carried out and analyzed. J.S.O and C.N. designed and C.N. preformed the MITOMI experiments with supervision from J.D. The manuscript was prepared by J.S.O with input from H.E.S., C.N., J.S.W., and J.D.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic Expression Programs in the Response of Yeast Cells to Environmental Changes. Molecular Biology of the Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tirosh I, Weinberger A, Carmi M, Barkai N. A genetic signature of interspecies variations in gene expression. Nat Genet. 2006;38(7):830–4. doi: 10.1038/ng1819. [DOI] [PubMed] [Google Scholar]
- 3.Sangurdekar DP, Srienc F, Khodursky AB. A classification based framework for quantitative description of large-scale microarray data. Genome Biol. 2006;7(4) doi: 10.1186/gb-2006-7-4-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colman-Lerner A, Gordon A, Serra E, Chin T, Resnekov O, Endy D, Pesce CG, Brent R. Regulated cell-to-cell variation in a cell-fate decision system. Nature. 2005;437(7059):699–706. doi: 10.1038/nature03998. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi S, Pryciak PM. Membrane localization of scaffold proteins promotes graded signaling in the yeast MAP kinase cascade. Curr Biol. 2008;18(16):1184–91. doi: 10.1016/j.cub.2008.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rogers KW, Schier AF. Morphogen gradients: from generation to interpretation. Annu Rev Cell Dev Biol. 2011;27:377–407. doi: 10.1146/annurev-cellbio-092910-154148. [DOI] [PubMed] [Google Scholar]
- 7.Giorgetti L, Siggers T, Tiana G, Caprara G, Notarbartolo S, Corona T, Pasparakis M, Milani P, Bulyk ML, Natoli G. Noncooperative interactions between transcription factors and clustered DNA binding sites enable graded transcriptional responses to environmental inputs. Mol Cell. 2010;37(3):418–28. doi: 10.1016/j.molcel.2010.01.016. [DOI] [PubMed] [Google Scholar]
- 8.Cai L, Dalal CK, Elowitz MB. Frequency-modulated nuclear localization bursts coordinate gene regulation. Nature. 2008;455(7212):485–90. doi: 10.1038/nature07292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu RC, Pesce CG, Colman-Lerner A, Lok L, Pincus D, Serra E, Holl M, Benjamin K, Gordon A, Brent R. Negative feedback that improves information transmission in yeast signalling. Nature. 2008;456(7223):755–61. doi: 10.1038/nature07513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Broach JR. Nutritional control of growth and development in yeast. Genetics. 2012;192(1):73–105. doi: 10.1534/genetics.111.135731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boy-Marcotte E, Garmendia C, Garreau H, Lallet S, Mallet L, Jacquet M. The transcriptional activation region of Msn2p, in Saccharomyces cerevisiae, is regulated by stress but is insensitive to the cAMP signalling pathway. Mol Genet Genomics. 2006;275(3):277–87. doi: 10.1007/s00438-005-0017-4. [DOI] [PubMed] [Google Scholar]
- 12.Zhu C, Byers KJ, McCord RP, Shi Z, Berger MF, Newburger DE, Saulrieta K, Smith Z, Shah MV, Radhakrishnan M, Philippakis AA, Hu Y, De Masi F, Pacek M, Rolfs A, Murthy T, Labaer J, Bulyk ML. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 2009;19(4):556–66. doi: 10.1101/gr.090233.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huebert DJ, Kuan PF, Keles S, Gasch AP. Dynamic changes in nucleosome occupancy are not predictive of gene expression dynamics but are linked to transcription and chromatin regulators. Mol Cell Biol. 2012;32(9):1645–53. doi: 10.1128/MCB.06170-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capaldi AP, Kaplan T, Liu Y, Habib N, Regev A, Friedman N, O’Shea EK. Structure and function of a transcriptional network activated by the MAPK Hog1. Nat Genet. 2008;40(11):1300–6. doi: 10.1038/ng.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hao N, O’Shea EK. Signal-dependent dynamics of transcription factor translocation controls gene expression. Nat Struct Mol Biol. 2011;19(1):31–9. doi: 10.1038/nsmb.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Görner W, Durchschlag E, Martinez-Pastor MT, Estruch F, Ammerer G, Hamilton B, Ruis H, Schüller C. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 1998;12:586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stewart-Ornstein J, Weissman JS, El-Samad H. Cellular noise regulons underlie fluctuations in Saccharomyces cerevisiae. Mol Cell. 2012;45(4):483–93. doi: 10.1016/j.molcel.2011.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raveh-Sadka T, Levo M, Shabi U, Shany B, Keren L, Lotan-Pompan M, Zeevi D, Sharon E, Weinberger A, Segal E. Manipulating nucleosome disfavoring sequences allows fine-tune regulation of gene expression in yeast. Nature Genetics. 2012;44(7):743–50. doi: 10.1038/ng.2305. [DOI] [PubMed] [Google Scholar]
- 19.Chi Y, Huddleston MJ, Zhang X, Young RA, Annan RS, Carr SA, Deshaies RJ. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 2001;15(9):1078–92. doi: 10.1101/gad.867501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Durchschlag E, Reiter W, Ammerer G, Schüller C. Nuclear localization destabilizes the stress-regulated transcription factor Msn2. J Biol Chem. 2004;279(53):55425–32. doi: 10.1074/jbc.M407264200. [DOI] [PubMed] [Google Scholar]
- 21.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425(6959):737–41. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 22.Fordyce PM, Gerber D, Tran D, Zheng J, Li H, DeRisi JL, Quake SR. De novo identification and biophysical characterization of transcription-factor binding sites with microfluidic affinity analysis. Nat Biotechnol. 2010;28(9):970–5. doi: 10.1038/nbt.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maerkl SJ, Quake SR. A systems approach to measuring the binding energy landscapes of transcription factors. Science. 2007;315(5809):233–7. doi: 10.1126/science.1131007. [DOI] [PubMed] [Google Scholar]
- 24.Wuttke DS, Foster MP, Case DA, Gottesfeld JM, Wright PE. Solution structure of the first three zinc fingers of TFIIIA bound to the cognate DNA sequence: determinants of affinity and sequence specificity. J Mol Biol. 1997;273(1):183–206. doi: 10.1006/jmbi.1997.1291. [DOI] [PubMed] [Google Scholar]
- 25.Bernstein BE, Hoffman RC, Horvath S, Herriott JR, Klevit RE. Structure of a histidine-X4-histidine zinc finger domain: insights into ADR1-UAS1 protein-DNA recognition. Biochemistry. 1994;33(15):4460–70. doi: 10.1021/bi00181a005. [DOI] [PubMed] [Google Scholar]
- 26.Laity JH, Dyson HJ, Wright PE. DNA-induced alpha-helix capping in conserved linker sequences is a determinant of binding affinity in Cys(2)-His(2) zinc fingers. J Mol Biol. 2000;295(4):719–27. doi: 10.1006/jmbi.1999.3406. [DOI] [PubMed] [Google Scholar]
- 27.Kochoyan M, Keutmann HT, Weiss MA. Alternating zinc fingers in the human male-associated protein ZFY: HX3H and HX4H motifs encode a local structural switch. Biochemistry. 1991;30(39):9396–402. doi: 10.1021/bi00103a002. [DOI] [PubMed] [Google Scholar]
- 28.Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, Jennings EG, Zeitlinger J, Pokholok DK, Kellis M, Rolfe PA, Takusagawa KS, Lander ES, Gifford DK, Fraenkel E, Young RA. transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Segal DJ, Dreier B, Beerli RR, Barbas CF., III Toward controlling gene expression at will, selection and design of zinc finger domains recognizing each of the 5′-GNN-3′ DNA target sequences. Proc. Natl Acad. Sci. USA. 1999;96:2758–2763. doi: 10.1073/pnas.96.6.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhee HS, Pugh BF. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell. 2011;147(6):1408–19. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spitz F, Furlong EEM. Transcription factors: from enhancer binding to developmental control. Nature Reviews Genetics. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 32.Gregor T, Wieschaus EF, McGregor AP, Bialek W, Tank DW. Stability and nuclear dynamics of the bicoid morphogen gradient. Cell. 2007;130(1):141–152. doi: 10.1016/j.cell.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sadeh A, Movshovich N, Volokh M, Gheber L, Aharoni A. Fine-tuning of the Msn2/4-mediated yeast stress responses as revealed by systematic deletion of Msn2/4 partners. Mol Biol Cell. 2012;22(17):3127–38. doi: 10.1091/mbc.E10-12-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, Xu H, Ha SW, Ju D, Xie Y. Proteasomal degradation of Rpn4 in Saccharomyces cerevisiae is critical for cell viability under stressed conditions. Genetics. 2010;184(2):335–42. doi: 10.1534/genetics.109.112227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lahav G, Rosenfeld N, Sigal A, Geva-Zatorsky N, Levine AJ, Elowitz MB, Alon U. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat Genet. 2004 Feb;36(2):147–50. doi: 10.1038/ng1293. 2004. [DOI] [PubMed] [Google Scholar]
- 36.Wapinski I, Pfeffer A, Friedman N, Regev A. Natural history and evolutionary principles of gene duplication in fungi. Nature. 2007;449(7158):54–61. doi: 10.1038/nature06107. [DOI] [PubMed] [Google Scholar]
- 37.Tirosh I, Wong KH, Barkai N, Struhl K. Extensive divergence of yeast stress responses through transitions between induced and constitutive activation. Proc Natl Acad Sci U S A. 2011;108(40):16693–8. doi: 10.1073/pnas.1113718108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, Wapinski I, Roy S, Lin MF, Heiman DI, Young SK, Furuya K, Guo Y, Pidoux A, Chen HM, Robbertse B, Goldberg JM, Aoki K, Bayne EH, Berlin AM, Desjardins CA, Dobbs E, Dukaj L, Fan L, FitzGerald MG, French C, Gujja S, Hansen K, Keifenheim D, Levin JZ, Mosher RA, Müller CA, Pfiffner J, Priest M, Russ C, Smialowska A, Swoboda P, Sykes SM, Vaughn M, Vengrova S, Yoder R, Zeng Q, Allshire R, Baulcombe D, Birren BW, Brown W, Ekwall K, Kellis M, Leatherwood J, Levin H, Margalit H, Martienssen R, Nieduszynski CA, Spatafora JW, Friedman N, Dalgaard JZ, Baumann P, Niki H, Regev A, Nusbaum C. Comparative functional genomics of the fission yeasts. Science. 2008;332(6032):930–6. doi: 10.1126/science.1203357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eser U, Falleur-Fettig M, Johnson A, Skotheim JM. Commitment to a cellular transition precedes genome-wide transcriptional change. Mol Cell. 2011;43(4):515–27. doi: 10.1016/j.molcel.2011.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.