Abstract
Over 95% of all metazoan (animal) species comprise the “invertebrates,” but very few genomes from these organisms have been sequenced. We have, therefore, formed a “Global Invertebrate Genomics Alliance” (GIGA). Our intent is to build a collaborative network of diverse scientists to tackle major challenges (e.g., species selection, sample collection and storage, sequence assembly, annotation, analytical tools) associated with genome/transcriptome sequencing across a large taxonomic spectrum. We aim to promote standards that will facilitate comparative approaches to invertebrate genomics and collaborations across the international scientific community. Candidate study taxa include species from Porifera, Ctenophora, Cnidaria, Placozoa, Mollusca, Arthropoda, Echinodermata, Annelida, Bryozoa, and Platyhelminthes, among others. GIGA will target 7000 noninsect/nonnematode species, with an emphasis on marine taxa because of the unrivaled phyletic diversity in the oceans. Priorities for selecting invertebrates for sequencing will include, but are not restricted to, their phylogenetic placement; relevance to organismal, ecological, and conservation research; and their importance to fisheries and human health. We highlight benefits of sequencing both whole genomes (DNA) and transcriptomes and also suggest policies for genomic-level data access and sharing based on transparency and inclusiveness. The GIGA Web site (http://giga.nova.edu) has been launched to facilitate this collaborative venture.
Key words: biodiversity, comparative genomics, consortium, evolution, GIGA, invertebrates, metazoa
The last 600 million years of evolution have been marked by the diversification of animal life. Despite the range of body types and organisms observed among the Metazoa, spanning salps, sponges, shrimps, squids, and sea stars, all animals arose from a common ancestor. Metazoans share a number of features that distinguish them from other organisms: they are all mostly multicellular, heterotrophic, and chiefly motile eukaryotes with intercellular junctions and an extracellular matrix of collagen and glycoproteins. With the exception of species that also propagate asexually, some supposedly for a long time (Danchin et al. 2011), metazoans develop from embryos arising from a diploid zygote and passing through a blastula stage, which is followed by cell differentiation and morphogenesis (Slack et al. 1993; Valentine 2004; Nielsen 2012; Erwin and Valentine 2013). These processes are orchestrated by a conserved developmental toolkit, including a variety of transcription factors and signaling pathways, identified in most metazoan genomes studied so far (Srivastava et al. 2008, 2010).
Invertebrates, i.e. animals without backbones, encompass about 95% of metazoan diversity (Zhang 2011a). The concept of invertebrate was first proposed by Lamarck (1801) and is derived from our anthropocentric view of life—a biological equivalent of geocentrism that suggests that vertebrates hold a special status among metazoans. Although invertebrates clearly represent a paraphyletic assemblage, the term “invertebrate” persists, and the distinction between vertebrates and invertebrates is upheld in textbooks and university curricula. As a group of invertebrate zoologists, we have decided to maintain the distinction here for practical purposes.
Invertebrates play crucial roles in the functioning of most ecosystems, including many that affect people. Some are parasites and disease vectors that affect the health of humans, livestock, and plant crops. As invasive species, they can have devastating ecological and economic effects. But invertebrates also provide significant benefits, as many marine species are harvested or farmed for human consumption (Ponder and Lunney 1999). For the past four decades, marine invertebrates have been the focus of research that has led to the synthetic or recombinant production of drugs, molecular research tools (e.g., green fluorescent protein) (Chalfie et al. 1994), and biomedical research probes (Narahashi et al. 1994). Invertebrates also provide inspiration for a number of biomimetic materials, such as those modeled on spider silk, the hierarchical structure of glass sponge skeletons (Müller et al. 2013), and molluscan nacre, the composite lattice work comprising mother of pearl (Fratzl 2007). A better understanding of the genomes of these animals will enhance our ability to mitigate their negative impacts as parasites, disease vectors, and invasive species, and to sustainably manage them as providers of ecological services and economic benefits.
High throughput sequencing technologies provide us with an unprecedented opportunity to integrate traditional biological approaches with genomic data to describe new aspects of the functional and structural diversity of invertebrates. We aim to assemble a global consortium of scientists and institutions to evaluate the broad spectrum of invertebrate phylogenetic diversity suitable for whole-genome sequencing, to develop the standards and analytical tools necessary to maximize the utility of these genomes for comparative studies, and to sequence, assemble, and annotate whole genomes and/or transcriptomes of 7000 invertebrate species. As the first step toward this goal, we held an inaugural workshop in March 2013.
Although insects represent a lion’s share of animal species diversity, they are already the subject of targeted genome (Robinson et al. 2011) and transcriptome (http://www.1kite.org) initiatives, as are nematodes (Kumar et al. 2012). Conversely, noninsect/nonnematode invertebrates represent a vast phylogenetic and adaptive breadth and diversity in phenotypes (Figure 1; Edgecombe et al. 2011). Invertebrates span at least 30 very different body plans commonly referred to as “phyla.” Noninsect invertebrates inhabit marine, freshwater, and terrestrial realms. They are particularly diverse in the oceans, where all animal phyla originated and most continue to exist. The estimated number of marine animal species ranges from 275 000 to over 5 million (Appeltans et al. 2012; Collen et al. 2012; Scheffers et al. 2012), the vast majority of which are invertebrates.
Figure 1.
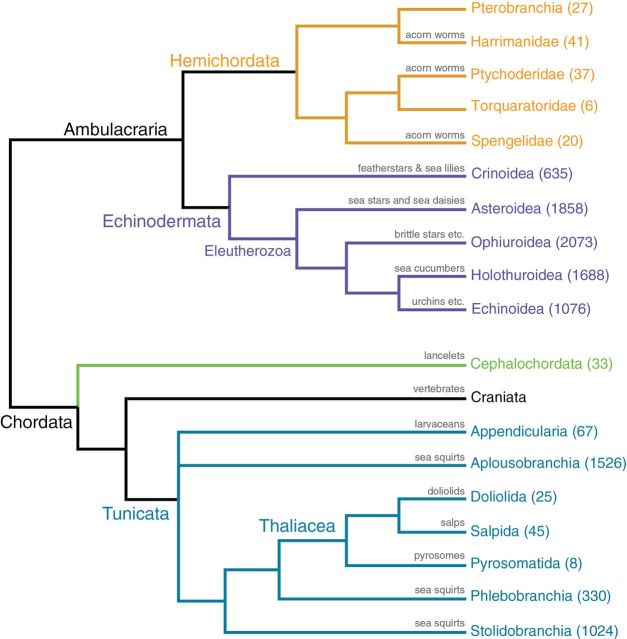
Metazoan relationships. This phylogeny of Metazoa is largely based on recent phylogenomic analyses by Dunn et al. (2008) and Hejnol et al. (2009). The basal nodes of the tree are shown as a polytomy based on varying results in recent analyses (Dunn et al. 2008; Hejnol et al. 2009; Philippe et al. 2009; Schierwater et al. 2009; Pick et al. 2010; Ryan et al. 2010; Nosenko et al. 2013). The position of Xenacoelomorpha is uncertain (dashed lines) and is based on recent studies; it could be the sister group to Bilateria (Hejnol et al. 2009), or as part of Deuterostomia (Philippe et al. 2011). There is no phylogenomic data for Mesozoa or Micrognathozoa, and their positions are based on a limited amount of direct sequencing data. Mesozoa (comprising Rhombozoa and Orthonectida) is shown as a single terminal but may not be a clade. The placement of Phoronida is based in the traditional placement of the group as close to Brachiopoda. The estimated numbers for extant described and valid species, indicated in parentheses after the terminal name, are from Zhang (2011a) and for marine groups, from queries of the World Register of Marine Species (June 2013), http://www.marinespecies.org/aphia.php?p=stats. Notes for terminals indicated by *; Mesozoa (comprising Rhombozoa and Orthonectida) is shown as a single terminal but may not be a clade. The species count for Rotifera includes Acanthocephala and for Cnidaria includes Myxozoa.
Invertebrates have long served as model organisms, providing insights into fundamental mechanisms of development, neurobiology, genetics, species diversification, and genome evolution. Two invertebrates—the fruit fly Drosophila melanogaster (Adams et al. 2000) and the nematode Caenorhabditis elegans ( C. elegans Sequencing Consortium 1998)—were the first animal species targeted for complete genome sequencing, setting the stage for other invertebrate-based studies such as i5K for insects (Robinson et al. 2011) and the 959 Nematode Genome program (Kumar et al. 2012), which target up to several thousand whole-genome sequencing projects.
Recent years have seen major advances in DNA sequencing technologies (Mardis 2011; Shendure and Lieberman Aiden 2012), bringing whole-genome sequencing capabilities beyond the sole province of well-funded laboratories and sequencing centers working on model organisms. Human-based studies such as the ENCODE (Encyclopedia of DNA Elements) (Ecker et al. 2012) and “Human Microbiome” projects (Turnbaugh et al. 2007) demonstrate the extraordinary power of genomic technologies to produce data resources that can promote hypothesis generation and more powerful analytical tools. However, the potential for greater insights stems from comparative research that can place genomic diversity into a phylogenetic context (e.g., Rubin et al. 2000). Technological advances and associated cost reductions now allow us to sequence whole genomes from a much wider spectrum of all organisms, broadening our capacity for comparative genomics.
The broad and basal phylogenetic placements of invertebrates also create opportunities to pose deeper, fundamental questions regarding classical versus mechanistic reductionist perspectives of biology and how genes actually shape each organism’s development and physiology (Woese 2004). Therefore, our group—the Global Invertebrate Genomics Alliance (GIGA)—represents a concerted effort toward sequencing invertebrate genomes/transcriptomes and developing informatics tools, resources, tissue repositories, and databases, which will be made publicly available.
The GIGA consortium herein reviews the phylogenetic status and adaptive and developmental features of potential species for whole-genome sequencing of invertebrate phyla. Careful taxon selection, the development of data standards, and facilitating comparative approaches will maximize the utility of genomes generated. Collecting whole-genome data is still a nontrivial task, both technologically and financially, and the computational constraints on assembling, annotating, and analyzing genomic data remain considerable. Therefore, GIGA also proposes a series of criteria (outlined below) for prioritizing invertebrate whole-genome sequencing projects, including standards for nominating species for whole-genome sequencing, specimen preparation, and processing, as well as general policies governing the distribution of genomic data as resources for the broader scientific community.
Scope and Goals
We propose to sequence, assemble, and annotate whole genomes and/or transcriptomes of 7000 invertebrate species, complementing ongoing efforts to sequence vertebrates, insects, and nematodes (Genome 10K Community of Scientists [G10KCOS], 2009; Robinson et al. 2011; Kumar et al. 2012) (Table 1). We have compiled a list of proposed species to sequence, which will be posted on the new web portal (http://giga.nova.edu, also available at http://giga-cos.org) created for distributing information about potential targets and projects. Selection and prioritization of taxa to be sequenced will occur through future discussion and coordination within GIGA. Thus, the target number is a somewhat arbitrary compromise between the desire to encompass as much phylogenetic, morphological, and ecological diversity as possible and the practical limitations of the initiative. Given the large population sizes of many invertebrate species, collection of a sufficient number of individuals may be relatively easy for the first set of targets. Collection of invertebrates usually involves fewer difficulties with regard to permits than collection of vertebrate tissues. However, some invertebrate taxa pose various logistic and technological challenges for whole-genome sequencing: many species live in relatively inaccessible habitats (e.g., as parasites, in the deep sea, or geographically remote) or are too small to yield sufficient amounts of DNA from single individuals. These challenges will be considered with other criteria as sequencing projects are developed and prioritized.
Table 1.
Published or in progress whole noninsect/nonnematode invertebrate genome projects
| Species | Major group | Subgroup | Size (MB) | URL | Citation |
|---|---|---|---|---|---|
| Helobdella robusta | Annelida | Clitellata | 215.44 | http://genome.jgi-psf.org/Helro1/Helro1.home.html | Simakov et al. (2013) |
| Capitella teleta | Annelida | Polychaeta | 276.69 | http://genome.jgi-psf.org/Capca1/Capca1.home.html | Simakov et al. (2013) |
| Ixodes scapularis | Arthropoda | Arachnida | 1,896.32 | http://www.ncbi.nlm.nih.gov/genome/?term=txid523 | Hill and Wikel (2005) |
| Rhipicephalus microplus | Arthropoda | Arachnida | 144.77 | http://www.ncbi.nlm.nih.gov/genome/?term=txid2797 | Guerrero et al. (2010) |
| Tetranychus urticae | Arthropoda | Arachnida | 89.6 | http://www.ncbi.nlm.nih.gov/genome/?term=txid2710 | Grbic et al. (2007), Grbić et al. (2011) |
| Strigamia maritima | Arthropoda | Myriapoda | 173.61 | http://www.ncbi.nlm.nih.gov/genome/?term=txid790 | Baylor College of Medicine (unpublished data) |
| Branchiostoma floridae | Chordata | Cephalochordata | 480.43 | http://genome.jgi-psf.org/Brafl1/Brafl1.home.html | Putnam et al. (2008) |
| Ciona intestinalis | Chordata | Tunicata | 115.24 | http://genome.jgi-psf.org/Cioin2/Cioin2.info.html | Dehal et al. (2002) |
| Ciona savignyi | Chordata | Tunicata | 401.48 | http://www.broadinstitute.org/annotation/ciona/ | Vinson et al. (2005) |
| Oikopleura dioica | Chordata | Tunicata | 66.54 | http://www.genoscope.cns.fr/externe/GenomeBrowser/Oikopleura | Seo et al. (2001) |
| Acropora digitifera | Cnidaria | Anthozoa | 364.99 | http://marinegenomics.oist.jp/genomes/viewer?project_id=3 | Shinzato et al. (2011) |
| Nematostella vectensis | Cnidaria | Anthozoa | 297.43 | http://genome.jgi-psf.org/Nemve1/Nemve1.home.html | Putnam et al. (2007) |
| Hydra magnipapillata | Cnidaria | Hydrozoa | 785.56 | http://www.ncbi.nlm.nih.gov/genome/?term=txid6085 | Chapman et al. (2010) |
| Daphnia pulex | Crustacea | Crustacea | 158.62 | http://wfleabase.org | Colbourne et al. (2011) |
| Mnemiopsis leidyi | Ctenophora | Lobata | 150.34 | http://research.nhgri.nih.gov/mnemiopsis/ | National Human Genome Research Institute (unpublished data) |
| Patiria miniata | Echinodermata | Asteroidea | 770.32 | http://www.ncbi.nlm.nih.gov/genome/?term=txid46514 | Baylor College of Medicine (unpublished data) |
| Lytechinus variegatus | Echinodermata | Echinoidea | 823.46 | http://www.ncbi.nlm.nih.gov/genome/?term=txid7654 | Baylor College of Medicine (unpublished data) |
| Strongylocentrotus purpuratus | Echinodermata | Echinoidea | 936.58 | http://www.spbase.org/SpBase/ | Sodergren et al. (2006) |
| Saccoglossus kowalevskii | Hemichordata | Enteropneusta | 642.31 | http://www.ncbi.nlm.nih.gov/genome/?term=txid10224 | |
| Crassostrea gigas | Mollusca | Bivalvia | 491.87 | http://www.ncbi.nlm.nih.gov/genome/?term=txid10758 | Zhang et al. (2012) |
| Pinctada fucata | Mollusca | Bivalvia | 1115 | http://marinegenomics.oist.jp/pinctada_fucata. | Takeuchi et al (2012) |
| Aplysia californica | Mollusca | Gastropoda | 737.86 | http://hgdownload.cse.ucsc.edu/downloads.html#seahare | Broad Institute (unpublished data) |
| Lottia gigantea | Mollusca | Mollusca | 298.9 | http://genome.jgi-psf.org/Lotgi1/Lotgi1.home.html | Simakov et al. (2013) |
| Trichoplax adhaerens | Placozoa | Trichoplax | 94.75 | http://www.ncbi.nlm.nih.gov/genome/?term=txid354 | Srivastava et al. (2008) |
| Schmidtea mediterranea | Platyhelminthes | Rhabditophora | 865.63 | http://www.ncbi.nlm.nih.gov/genome/?term=txid232 | Robb et al. (2008) |
| Clonorchis sinensis | Platyhelminthes | Trematoda | 547.11 | http://www.ncbi.nlm.nih.gov/genome/?term=txid2651 | Wang et al. (2011) |
| Schistosoma japonicum | Platyhelminthes | Trematoda | 369.09 | http://www.ncbi.nlm.nih.gov/genome/?term=txid237 | Zhou et al. (2009) |
| Schistosoma mansoni | Platyhelminthes | Trematoda | 329.51 | http://www.ncbi.nlm.nih.gov/genome/?term=txid236 | Berriman et al. (2009) |
| Amphimedon queenslandica | Porifera | Demospongiae | 144.88 | http://www.ncbi.nlm.nih.gov/genome/?term=txid400682 | Srivastava et al. (2010) |
| Adineta vaga | Rotifera | Bdelloidea | 218.1 | http://www.genoscope.cns.fr/adineta/ | Flot et al. (2013) |
Animal biodiversity is not constrained by political boundaries. Therefore, the geographic scope of the project in terms of participation, taxa collected, stored, and sequenced, data analysis and sharing, and derived benefits, requires global partnerships beyond the individuals and institutions represented at the inaugural workshop. Because international and interinstitutional cooperation is essential for long-term success, the new GIGA Web site will be used to foster cooperative research projects. For now, the GIGA Web site can serve as a community nexus to link projects and collaborators, but it could also eventually expand to host multiple shared data sets or interactive genome browsers. The broad scope of GIGA also necessitates growth in the genomics-enabled community overall. Sequencing and analyzing the large amount of resulting data pose significant bioinformatic and computational challenges and will require the identification and creation of shared bioinformatics infrastructure resources.
Why Focus on Invertebrates?
Phylogeny
Given the diversity of noninsect/nonnematode/nonvertebrate animals and the variety of invertebrate body plans, a first criterion for selecting taxa is phylogenetic. The taxa selected represent 36 metazoan phyla (Figure 1), acknowledging that conferring this taxonomic rank on a group can be dynamic and controversial. Whole or nearly whole-genome sequences (most genomes only asymptotically approach completeness but never fully reach it, Eddy 2013) have been published or submitted to pubic databases for one or a few representatives of 15 phyla (Porifera, Ctenophora, Cnidaria, Placozoa, Cephalochordata, Tunicata, Craniata, Echinodermata, Hemichordata, Nematoda, Arthropoda, Annelida, Mollusca, Platyhelminthes, and Rotifera), but there are currently no published genomes for the other 21 invertebrate phyla. We examined current phylogenetic hypotheses and selected key invertebrate species that span the phylogenetic diversity and morphological disparity on the animal tree of life (see Supplementary Material). New invertebrate genome data can reveal novel sequences with sufficient phylogenetic signal to resolve longstanding questions.
Developmental Biology
One of the most exciting approaches to understanding the origins and evolution of animal diversity is detailed comparative research on embryological development (i.e., evolutionary developmental biology, or evo-devo). Whole-genome and transcriptome information can facilitate the success of such research, just as Hox gene characterizations did decades before (Ikuta 2011). Gene content, synteny, gene copy number, and cis-regulatory information are some of the crucial data that may enable us to address the genotype–phenotype dilemma. Whole-genome sequencing provides these data in a comprehensive fashion. In the last decade, every invertebrate genome sequence paper has turned these select species into emerging and important developmental model systems (Dehal et al. 2002; Putnam et al. 2007, 2008; Srivastava et al. 2010). The additional evo-devo publications have generated major insights into the molecular basis of morphological diversity (Davidson and Erwin 2006; Hejnol 2010). Invertebrate taxa also include some of the longest living animals on the planet, such as the ocean Quahog clam Arctica islandica (maximum reported age 507 years), Lamellibrachia tube worms (~250 years) (Munro and Blier 2012), coralline demosponge Astrosclera willeyana (565 ± 70 years) (Wörheide 1998), gold coral Gerardia sp. (450–2742 years), black coral Leiopathes glaberrima (est. ~2377 years) (Roark et al. 2006), and the immortal Hydra (Boehm et al. 2012)
Speciation, Radiations, and Evolutionary Rates
Invertebrates have considerable potential to inform fundamental questions in other aspects of evolutionary biology, such as how new species form, why radiations occur, and the genetics of evolutionary stasis. Invertebrate sister species pairs now restricted to the Caribbean or eastern Pacific by the uplift of the Isthmus of Panama provide a superb model system for investigating rates and scales of speciation in the sea (Knowlton and Weigt 1998; Lessios 2008). Spectacular invertebrate radiations, such as the amphipod crustaceans of Lake Baikal (MacDonald et al. 2005), complement vertebrate lake radiations elsewhere. The sea star, Cryptasterina hystera, has clocked one of the fastest speciation rates (~6000 years) on record (Puritz et al. 2012). By contrast, invertebrates also include forms that appear to have remained at least superficially unchanged over long periods of geological time. Genomic analysis may reveal regulatory sequences involved in morphological stasis (e.g., horseshoe crabs, blue coral Heliopora, deep-sea lobster Neoglyphea inopinata, sclerosponges), as well as provide insight into rates of evolutionary change.
Ecology
Invertebrates are major components of marine, freshwater, and terrestrial ecosystems. Some invertebrates are ecosystem engineers, such as the scleractinian corals that construct reef habitats covering only 0.2% of all oceanic surface area yet supporting up to 25% of total marine biodiversity (Reaka-Kudla 1997). Invertebrates span all trophic levels, from primary consumers through widely utilized prey (e.g., planktonic copepods) to parasites and apex predators (e.g., Humboldt squid). In terms of biomass, some invertebrates such as krill (euphausiids) and copepods dominate pelagic food webs (Buitenhuis et al. 2006; Atkinson et al. 2009). Benthic and planktonic invertebrates play crucial roles in carbon and nutrient cycling in the ocean. In deeper waters without sunlight, water-column invertebrates cycle carbon through their trophic networks, and benthic animals (e.g., tubeworms, mussels, clams, gastropods) form partnerships with chemosynthetic bacteria to dominate and define ecosystems such as hydrothermal vent communities and methane seeps (German et al. 2011). Genomes of organisms that live in harsh environments (i.e., extremophiles) or in intimate symbiosis with other organisms (e.g., coral–algal symbioses, highly adapted parasites) have the potential to radically change our understanding of how these organisms survive (Russell et al. 2013; Flot et al. 2013).
Fisheries and Aquaculture
Invertebrates are becoming increasingly important sources of protein for human nutrition worldwide. Particularly with the collapse of a number of vertebrate fisheries, invertebrate fisheries and aquaculture dominated by mollusks and crustaceans provide food and employment for millions of people (Glaser and Diele 2004). Prominent examples are bivalves (oysters, clams, mussels), gastropods (abalone, queen conch), and cephalopods (squid, octopus, and cuttlefish) among the mollusks (Stoner 1997), and a variety of decapods (shrimps, crabs, lobsters, and crayfish) among crustaceans (Neiland et al. 2001). Gooseneck barnacles are among the most expensive crustacean seafood in Western Europe on a per-kilogram basis. Echinoderms (sea urchins and sea cucumbers) have important commercial fisheries, and species of Cnidaria, Annelida, Tunicata, Cephalochordata, and Sipuncula are all harvested for human consumption in some countries. Some invertebrates, such as krill, are harvested for human nutrition more indirectly, via animal feed or dietary supplements. Finally, not all fished species are food sources; pearl oysters, abalone, mussels, snails, black/red, and soft corals are harvested for their value in the jewelry or aquarium trades. A number of large mollusks are similarly fished to support the tourism market.
Invasive and Pest Species
A number of invertebrates cause environmental problems around the world. Some are invasive species that have colonized and expanded their numbers into new habitats, often to the detriment of native species. Examples include the green crab (Carcinus maenas) and the zebra mussel (Dreissena polymorpha) in North America, the orange cup coral (Tubastraea coccinea) in the Gulf of Mexico and Caribbean, and the giant African land snail (Lissachatina fulica) in many tropical and subtropical areas. The zebra mussel causes millions of dollars of economic damage each year by clogging pipes carrying freshwater, and their destructive rapid growth and biofouling threaten native species of already endangered pearl mussels. Other invertebrate species are native but have recently become pests; in the ocean, this typically occurs because of increases in nutrient levels that support higher recruitment or growth rates of the pest species, or because overfishing of the predators of the pest species has allowed them to multiply uncontrolled (Galil 2012). Large aggregations of some medusa species negatively impact fisheries, tourism, and power plant cooling stations. On coral reefs, the most dramatic such pest is the crown-of-thorns starfish, Acanthaster planci, a major factor contributing to the decline of coral cover in the Indo-Pacific (De’ath et al. 2012).
Symbiosis
A wide range of invertebrates forms obligate associations with microorganisms to establish a holobiont (host + total symbiont community) that encompasses multiple symbiotic interactions. Symbiotic microbes carry out specific metabolic and physiological activities that are mutualistic, commensal, or neutral within their host. For example, the massive coral reef structures, critical to tropical marine ecosystems and aesthetically pleasing to tourists worldwide, could not develop without the photosynthetic activity of symbiotic algae living within coral tissues (Muscatine and Cernichiari 1969). As mentioned above, hydrothermal vent communities rely on chemosynthetic symbioses. Sacoglossan sea slugs consume algae as food, and also retain and integrate their chloroplasts into their tissues to continue photosynthesis for the host slug’s nutrition (Pierce and Curtis 2012). For humans, recent “Human Microbiome project” has significantly improved the scientific and public recognition of beneficial symbiont ecology and functions for host health and development (Human Microbiome Project 2012). Thus, the relatively small body sizes of many invertebrates coupled with their tendency to cultivate microbial symbioses increase their value as tractable models for exploring symbiosis studies in greater depth (Bosch and McFall-Ngai 2011). Genomic information on these symbiotic invertebrates are crucial if we are to understand host global regulatory networks that coordinate the expression of symbiotic factors involved in the establishment and communication between host and microbial symbionts. Furthermore, coupling metagenomic data of symbionts with corresponding host invertebrate genomes will enable a better understanding of symbiont–host interactions. The metagenomes of invertebrate symbioses with microbial consortia may also offer insights into understanding the production of metabolites with human health applications (see section “Relevance to Human Health” and Trindade-Silva et al. 2010).
Threatened, Endangered, and Remarkable Species
Relatively few invertebrates have been listed as extinct, but a number of them are threatened or endangered. Abalones (Haliotis spp.) were the first invertebrates listed under the Endangered Species Act, and all stony coral species are listed under Appendix II of the Convention on the Trade of Endangered Species (CITES). Other invertebrates fascinate the public by virtue of their size, such as the giant squid (Architeuthis dux), the giant clam (Tridacna gigas), and the nemertean bootlace worm Lineus longissimus, which may reach 54 m in length, perhaps the longest living organism (Ruppert et al. 2004). The decade-long Census of Marine Life showcased how small, beautiful, and unusual-looking invertebrates can engage the public (Knowlton 2010). Thus, genome sequences from these and other invertebrate species can be viewed as alternative or parallel measures to zoos and frozen tissue collections for the conservation of biological and genetic diversity (Ryder 2005).
Relevance to Human Health
Marine invertebrates are the source of tens of thousands of biochemical compounds with potential human health applications (Faulkner 2002; Blunt et al. 2013). In the last two decades alone (1990–2009), natural products research on Porifera (Demospongiae) and Cnidaria (Anthozoa) has resulted in the largest number of chemical “lead” molecular structures for discovery of novel pharmaceuticals (Leal et al. 2012). These include the sessile mangrove tunicate, Ecteinascidia turbinata, and the development of the anti-cancer drug, ET-743 (Yondelis®). Gastropod species of Conus produce more than 100 bioactive peptides in their potent venom, with little overlap of toxins among species (Olivera and Teichert 2007). The toxins exhibit a wide range of neurological effects, such as paralysis and analgesia, and one of the peptides is clinically available for treatment of chronic pain. With rare exceptions (Olivera and Teichert 2007), the targets and roles of these chemicals in the animals producing them are not known. Whole-genome sequences for invertebrate species that produce bioactive compounds will provide insights into their chemical roles in nature, their evolution, and the mechanisms that control their production. Sequences may also facilitate the discovery of other compounds or processes with human health or biotechnological applications (Russell et al. 2013). Rights to genomic data and source materials will be protected through GIGA’s compliance with accepted standards for recognition and protection of intellectual property rights and the Convention on Biological Diversity (CBD) and other regulations established by source countries (Kursar et al. 2007).
Policy
A main goal of GIGA is to build an international multidisciplinary community to pursue comparative invertebrate genomic studies. We seek to develop tools to enable and facilitate genomic research and encourage collaboration. We will develop standards that ensure data quality, comparability, and integration. By coordinating sample collecting and sequencing efforts among invertebrate biologists, we aim to avoid duplication of effort and leverage resources more efficiently. We envision a scientific commons where shared resources, data, data standards, and innovations move the generation and analysis of invertebrate genomic data to a level that likely could not be achieved with the traditional piecemeal single-investigator–driven approach.
GIGA embraces a transparent process of project coordination, collaboration, and data sharing that is designed to be fair to all involved parties. The ENCODE project may be emulated in this regard (Birney 2012). We are committed to the rapid release of genomic data, minimizing the period of knowledge latency prior to public release while protecting the rights of data product developers (Contreras 2010). The data accepted as part of GIGA resources will undergo quality control steps that will follow preestablished and evolving standards (see Standards section) prior to data release. Efforts such as those of Albertin et al. (2012) have addressed data sharing issues relevant to GIGA and other large-scale genomics consortia.
We also recognize the existence and formation of other recent genome science initiatives and coordination networks and will synchronize efforts with such groups through future projects. Because GIGA is an international consortium of scientists, agencies, and institutions, we will also abide by the rules of global funding agencies for data release (e.g., those proposed by the Global Research Council; http://www.globalresearchcouncil.org). We are aware that different nations have different constraints and regulations on the use of biological samples. Given the international nature of GIGA, we will work to ensure that national genomic legacies are protected and will consult with the pertinent governmental agencies in the countries from which samples originate. We will deposit sequence data in public databases (e.g., GenBank), as well as deposit DNA vouchers in publically accessible repositories (e.g. Global Genome Biodiversity Network, Smithsonian).
GIGA is an inclusive enterprise that invites all interested parties to join the effort of invertebrate genomics. We will attempt to capture the impact of the effort in the wider scientific and public arenas by following relevant publications and other products that result from GIGA initiatives.
Standards and Best Practices
GIGA has adopted a set of standards and best practices to help ensure that genomic resources, data, and associated metadata are acquired, documented, disseminated, and stored in ways that are directly comparable among projects and laboratories. These data should be easily and equitably shared among GIGA members and the broader scientific community, and GIGA will obey appropriate laws and regulations governing the protection of natural biodiversity. Briefly, all genome projects will report on a set of parameters that will allow assessment of genome assembly, annotation, and completeness (e.g., NG50, N50 of contigs and scaffolds, number of genes, assembled vs. estimated genome size) (Jeffery et al. 2013). Detailed descriptions of these standards and compliant protocols will be posted on the GIGA Web site. These will be revised periodically to facilitate the establishment and maintenance of current best practices common to many invertebrate genome and transcriptome sequencing projects and to help guide the researcher in selecting and assessing genomes for further analyses. The following recommendations summarize minimal project-wide standards designed to accommodate the large diversity of invertebrates, including extremely small and rare organisms, as well as those that live in close association with other organisms.
1. Permissions: GIGA participants must comply with treaties, laws, and regulations regarding acquisition of specimens or samples, publication of sequence data, and distribution or commercialization of data or materials derived from biological resources. Participants must acquire all necessary permits required for collection and transport of biological materials prior to the onset of the work. The CBD recognizes the sovereignty of each nation over its biological resources, and under the auspices of the CBD, many nations and jurisdictions rigorously regulate the use and distribution of bioIogical materials and data. GIGA participants must be aware of these regulations and respect the established rights of potential stakeholders, including nations, states, municipalities, commercial concerns, indigenous populations, and individual citizens, with respect to any materials being collected, to all derivatives and progeny of those materials, and to all intellectual property derived from them. GIGA participants must also familiarize themselves with the conservation status of organisms to be sampled and any special permits that may be required (e.g., CITES). Moreover, GIGA participants should collect in ways that minimize impacts to the sampled species and their associated environments.
2. Field collection and shipping: Methods for field collection and preservation of specimens and tissues should be compatible with recovery of high-quality (e.g., high molecular weight, minimally degraded) genomic DNA and RNA (Dawson et al. 1998; Riesgo et al. 2012; Wong et al. 2012). Many reagents commonly used for tissue and nucleic acid preservation (e.g., ethanol, dry ice) are regulated as hazardous and/or flammable materials. These reagents may be restricted from checked and carry-on luggage and may require special precautions for shipping or transport. GIGA participants should contact the appropriate airline carriers or shippers for information regarding safe and legal shipment of preserved biological materials. When possible, multiple samples will be collected so that extractions can be optimized and samples resequenced as technologies improve. Specimens of known origin (i.e., field-collected material) will be favored over specimens of unknown origin (e.g., material purchased from the aquarium trade). Collection data will include location (ideally, with GPS coordinates) and date, and also other data such as site photographs and environmental measurements (e.g., salinity) when relevant.
3. Selection and preparation of tissues: It is often advisable to avoid tissues that may contain high concentration of nucleases, foreign nucleic acids, large amounts of mucus, lipid, fat, wax, or glycogen or that are insoluble, chitinous, or mineralized. To obtain the highest quality material for sequencing or library construction, it may be preferable to extract nucleic acids from living or rapidly preserved tissue from freshly sacrificed animals, from gametes or embryos, or from cell lines cultivated from the target organism (Ryder 2005; Rinkevich 2011; Pomponi et al. 2013). When appropriate, select tissues or life history stages that will avoid contamination by symbionts, parasites, commensal organisms, gut contents, and incidentally associated biological and nonbiological material. Whenever possible, DNA or RNA will be sequenced from a single individual because many taxa display sufficient polymorphism among individuals to complicate assembly. Similarly, heterozygosity can also hinder assembly: inbreeding may be used to reduced heterozygosity (Zhang et al. 2012) or, when crossings are impossible (for instance in asexual species), haplotypes may have to be assembled separately (Flot et al. 2013).
4. Quantity and Quality: The quantity of DNA or RNA required for sequencing varies widely depending on the sequencing platform and library construction methods to be used and should be carefully considered. Recent consensus from the G10KCOS group of scientists suggests that at least 200 – 800 µg of high-quality genomic DNA is required to begin any project because of the requirement for large insert mate-pair libraries (Wong et al. 2012). However, these minimum quantities are expected to decline with improving technology. DNA quality can be assessed by size visualizatons and 260/280nm ratios. Quality of RNA will be checked using RNA integrity number (RIN > 7 is preferred); however, these values have been shown to appear degraded in arthropods due to artifacts during quantification (Schroeder et al. 2006; Winnebeck et al. 2010).
5. Taxonomic identity: The taxonomic identity of source organisms must be verified. Whenever possible, consensus should be sought from expert systematists, supportive literature, and sequence analysis of diagnostic genes (see next section).
6. Voucher specimens: As a prerequisite for inclusion as a GIGA sample, both morphological and nucleic acid voucher specimens must be preserved and deposited in public collections, and the associated accession numbers must be supplied to the GIGA database. Photographs should be taken of each specimen and cataloged along with other metadata. The GIGA Web site lists cooperating institutions willing to house voucher specimens for GIGA projects, such as the Smithsonian Institution or the Ocean Genome Legacy (http://www.oglf.org).
7. Documentation of projects, specimens, and samples: Unique alphanumeric identification numbers (GIGA accession numbers) will be assigned to each GIGA project and to each associated specimen or sample used as a source of genome or transcriptome material for analysis. A single database with a web interface will be established to accommodate metadata for all specimens and samples. Metadata recording will also aim to coordinate and comply with previously established standards in the community, such as those recommended by Genomics Standards Consortium (http://gensc.org/; Field et al. 2011).
8. Sequencing Standards: Standards for sequencing are platform and taxon specific, and sensitive to the requirements of individual sequencing facilities. For these reasons, best practices and standards will be established for individual applications. Coverage with high-quality raw sequence data is a minimal requirement to obtain reliable assemblies. An initial sequencing run and assembly will be used to estimate repeat structure and heterozygosity. These preliminary analyses will make it possible to evaluate the need for supplemental sequencing, with alternative technologies aimed at addressing specific challenges (e.g., mate-pair sequencing to resolve contig linkage). Moreover, all raw sequence reads generated as part of a GIGA project will be submitted to the NCBI Sequence Read Archive.
9. Sequence Assembly, Annotation, and Analyses: Because assemblies vary widely in quality and completeness, each assembly should be described using a minimum set of common metrics that may include: (1) N50 (or NG50) length of scaffolds and contigs (see explanation of N50 in Bradnam et al. 2013), (2) percent gaps, (3) percent detection of conserved eukaryotic genes (e.g., Core Eukaryotic Genes Mapping Approach (Parra et al. 2007), (4) statistical assessment of assembly (Howison et al, 2013), (5) alignment to any available syntenic or physical maps (Lewin et al. 2009), and (6) mapping statistics of any available transcript data (Ryan 2013).
10. The current paucity of whole invertebrate genome sequence projects can pose problems for gene calling, gene annotation, and identification of orthologous genes. In cases where the genome is difficult to assemble, we recommend that genome maps be developed for selected taxa via traditional methods or new methods (e.g., optical mapping of restriction sites) to aid and improve the quality of genome assembly (Lewin et al. 2009) and that GIGA genome projects be accompanied by transcriptome sequencing and analysis when possible. Such transcriptome data will assist open reading frame and gene annotation and are valuable in their own right.
Selection of Taxa
Our understanding of relationships among the major animal groups has improved in the last few decades by the use of ever more powerful genomic approaches (Figure 1). For example, we now recognize that arthropods and their relatives are more closely related to nematode worms than to the segmented annelid worms (Aguinaldo et al. 1997), a finding that contradicts views that dominated zoological literature for over a century. Subsequent studies have consistently recovered similar results (e.g., Dunn et al. 2008; Hejnol et al. 2009), although uncertainties of fundamental importance remain for understanding major evolutionary events. For example, relationships among some of the major lineages of the animal tree of life (Figures 1 and 2), Porifera (sponges), Cnidaria (corals, jellyfish, anemones, and their allies), Ctenophora (comb jellies), and Placozoa, are still not fully resolved (for recent reviews see Edgecombe 2011; Dohrmann and Wörheide 2013). Several recent studies have stirred debate regarding the relative positions of ctenophores, sponges, and the root of the Metazoa (Dunn et al. 2008; Hejnol et al. 2009; Philippe et al. 2009; Schierwater et al. 2009; Pick et al. 2010; Ryan et al. 2010; Nosenko et al. 2013). This controversy, compounded by the relatively slow evolutionary pace of mitochondrial genome evolution in several basal taxa (Huang et al. 2008), makes the genome sequencing of more sponges and ctenophores a high priority for resolving fundamental questions relating to the origins of animal life and the evolution of animal features such as the nervous and sensory systems, vision, and musculature. Whether some of these traits evolved once or multiple times can only be determined once nonbilaterian relationships have been resolved unequivocally.
Figure 2.
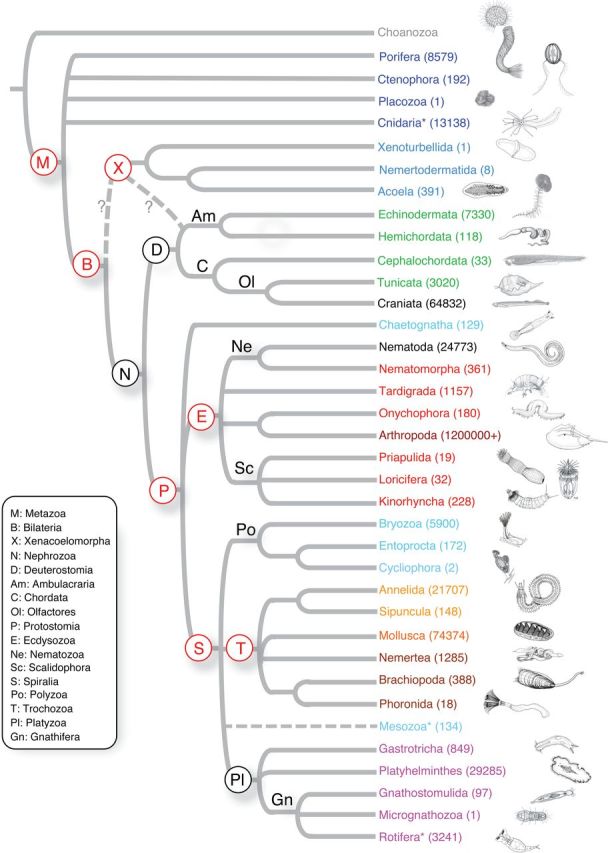
Consensus phylogeny of nonbilaterian animals. The Porifera tree and classification is based on Cárdenas et al. (2012), Nosenko et al. (2013), and Wörheide et al. (2012). The Cnidaria tree and classification is based on Collins (2009) and Evans et al. (2008, 2009). Following the terminal name, in parentheses, is the estimated number of extant described and valid species for that group based on the World Register of Marine Species as described in the Figure 1 legend, and Myxozoa estimates stem from Lom and Dyková (2006). When available, common names are written above the branches.
Many of the roughly 70 invertebrate species whose genomes have been sequenced belong to the Arthropoda or Nematoda, although the number of other invertebrate genomes continues to grow (e.g., Olson et al. 2012; Takeuchi et al. 2012; Zhang et al. 2012; Simakov et al. 2013; Tsai et al. 2013; Flot et al. 2013). We propose to focus on noninsect/nonnematode phyla, and specifically on an important group of currently neglected arthropods, the crustaceans. Below and in the Supplementary Material, we discuss relevant details of the phylogeny of major invertebrate taxa and their suitability for whole-genome sequencing.
Nonbilateria is a paraphyletic group of taxa that do not possess bilateral body symmetry and comprises the Porifera, Placozoa, Ctenophora, and Cnidaria. Porifera (sponges) are benthic aquatic (marine and freshwater) animals, with over 8500 described species distributed over four main extant lineages (Van Soest et al. 2012) (Figures 1 and 2). Their phylogeny at various levels, including their placement among the other nonbilaterian animals, is still intensively discussed (reviewed in Wörheide et al. 2012). Ctenophora (comb jellies) are exclusively marine organisms, ubiquitous in the global pelagic realm. Only 242 species have been formally described, and the group’s phylogenetic placement in the animal tree of life (Figures 1 and 2) is controversial (e.g., Dunn et al. 2008; Hejnol et al. 2009; Pick et al. 2010; Philippe et al. 2011; Nosenko et al. 2013; reviewed in Dohrmann and Wörheide 2013). Placozoa are small (up to few millimeters across) benthic marine animals that resemble a flat ciliated disc (Figure 1). Only one species has been formally described, but the phylum likely is more speciose (Voigt et al. 2004; Eitel and Schierwater 2010); their phylogenetic position among nonbilaterians is currently unresolved (e.g., Nosenko et al. 2013). Cnidaria occurs in all aquatic environments and includes more than 12 000 species, including the Portuguese man-o-war (Physalia), and reef-forming corals (Figure 2).
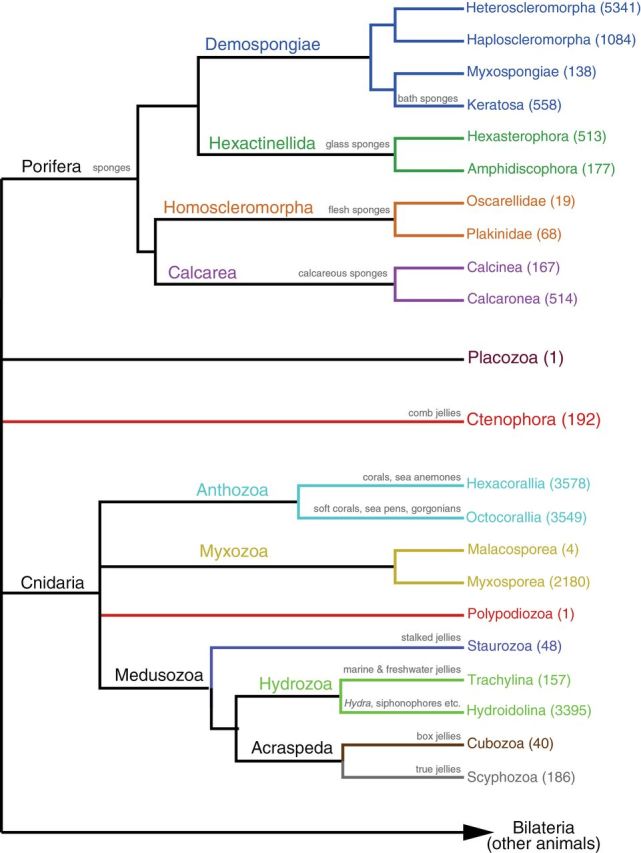
Ecdysozoa (molting animals) is a major protostome clade (Figure 1) proposed by Aguinaldo et al. (1997) that includes the large phyla Arthropoda (Figure 3) and Nematoda (both of tremendous ecological, economic, and biomedical importance) and their close relatives (Tardigrada [water bears, 1150 species], Nematomorpha [351 species], Onychophora [velvet worms; 182 species], Kinorhyncha [179 species], Loricifera [30 species], and Priapulida [19 species]). Ecdysozoans are characterized by their ability to molt the cuticle during their life cycle, and for having reduced epithelial ciliation, which requires locomotion via muscular action. They include segmented or unsegmented, acoelomate, pseudocoelomate, or coelomate animals; many have annulated cuticles and a mouth located at the end of a protrusible or telescopic proboscis, and some lack circular musculature (i.e., Nematoda, Nematomorpha, Tardigrada). Here we restrict the proposed sampling to the noninsect and nonnematode ecdysozoans. The clade includes the only animals (loriciferans) thought to complete their life cycles in anoxic environments (Danovaro et al. 2010). This group is also relevant for studies of extreme cell size reduction. Other than the mentioned arthropod and nematode genomes, no genome is available for any member of the Ecdysozoa.
Figure 3.
Consensus phylogeny of the major groups of Arthropoda, based on Regier et al. (2010) and von Reumont et al. (2012). Following the terminal name, in parentheses, is the estimated number of extant described and valid species for that group based on Zhang (2011b) and Ahyong et al. (2011). When available, common names are written above the branches.
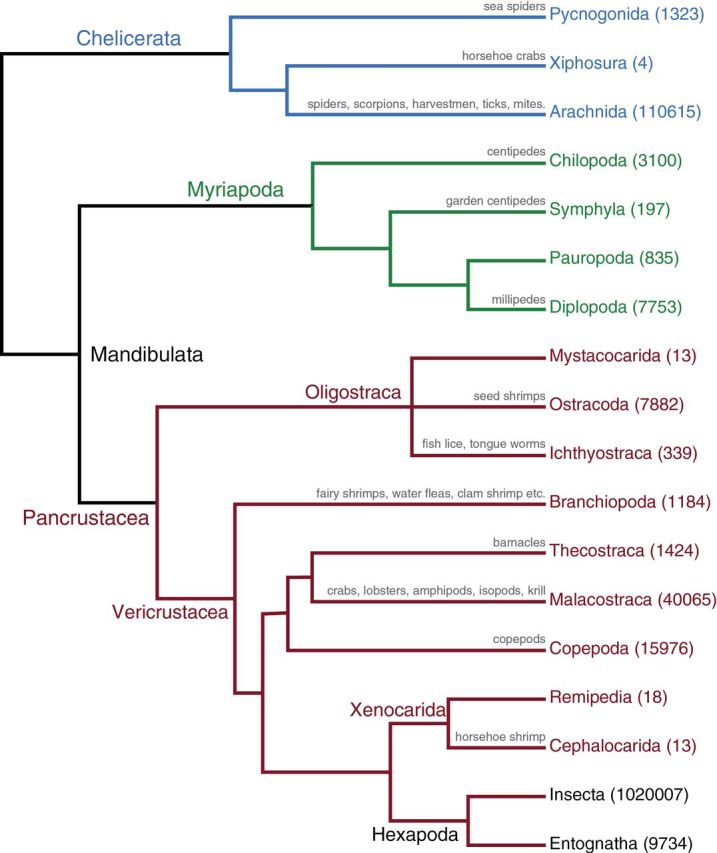
Spiralia encompasses Trochozoa (annelids, mollusks, brachiopods, and nemerteans), Platyzoa (flatworms, rotifers, and relatives), Polyzoa, and perhaps Mesozoa (Figure 1). Few of these taxa have been explored from a genomic perspective. Mollusca includes 117 358 extant described species, mostly marine (Figure 4). It currently has eight “classes”: Neomeniomorpha, Chaetodermomorpha, Polyplacophora, Monoplacophora, Scaphopoda, Gastropoda, Bivalvia, and Cephalopoda. Around 18 000 species of annelids are currently recognized (Rouse and Pleijel 2001), and most of them are polychaete worms. The group also contains earthworms and leeches, as well as the former phyla Echiura, Pogonophora, and Vestimentifera. Sipuncula (peanut worms) may be the sister group to annelids (Struck et al. 2011) (Figure 4). Nemertea (ribbon worms) can reach extraordinary lengths (tens of meters) and are almost all predators. Brachiopods were extremely abundant in the Paleozoic and are of prime importance as index fossils and for understanding the evolution of biomineralization (e.g., Brunton et al. 2001; Balthasar et al. 2011). Some spiralian phyla have unparalleled powers for budding (Entoprocta, Bryozoa, and Cycliophora), which could be useful for regeneration studies. In addition, groups like Cycliophora have complex life cycles in which individuals undergo metamorphosis, and larval stages are replaced by a distinct adult body plan (Funch and Kristensen 1995).
Figure 4.
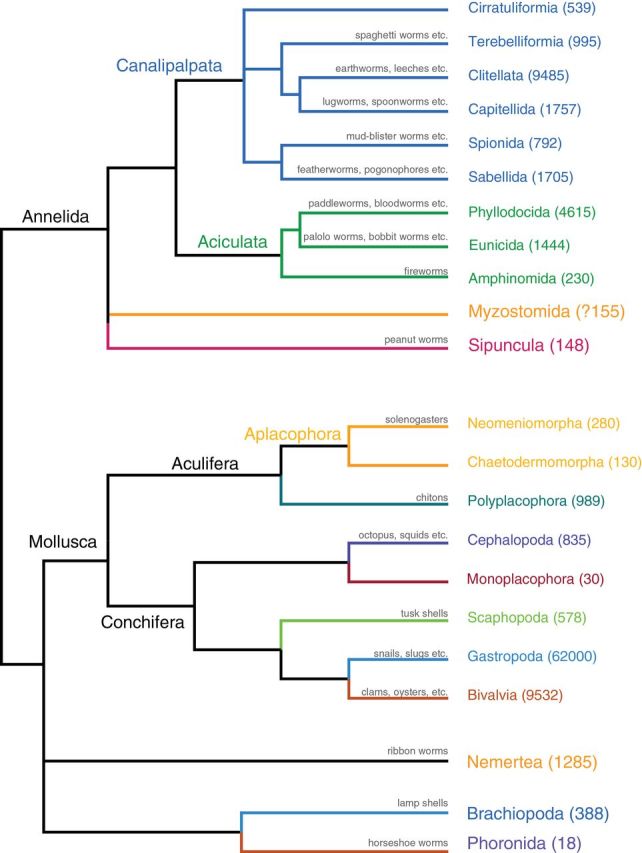
Combined phylogeny of Mollusca and Annelida. The Mollusca tree is drawn from Smith et al. (2011), while the consensus phylogeny of Annelida and Sipuncula is based on Rouse and Pleijel (2001) and Struck et al. (2011). As in Figure 1, following the terminal name, in parentheses, is the estimated number of extant described and valid species for that group based on the World Register of Marine Species. Additional references for groups with nonmarine species are Bivalvia, which also includes many freshwater species (Graf 2013); Gastropoda, which includes numerous freshwater and terrestrial species (Cameron 2013); and Clitellata, which includes many freshwater and terrestrial species (Sket and Trontelj 2007; Wetzel and Reynolds 2011). The species count for the taxon Capitellida includes taxa such as Echiura and polychaetes formerly included in Scolecida, a taxon now shown to be polyphyletic (Struck et al. 2011). When available, common names are written above the branches.
Deuterostomia is one of the major clades of bilaterian animals and includes the vertebrates (Figures 1 and 5). Among its nonvertebrate members, Deuterostomia comprises a wide diversity of forms ranging from microscopic to huge, familiar to bizarre, gelatinous to armor-plated, and worm-like to plant-like. Apart from Craniata (including Vertebrata), the group includes four major clades: Echinodermata, Hemichordata, Tunicata, and Cephalochordata, all exclusively marine. Echinodermata and Hemichordata form a clade known as Ambulacraria (Figure 5), characterized by similar dipleurula larval morphology and supported in most phylogenetic analyses. Tunicata are the closest relatives to Craniata and along with Cephalochordata form Chordata (Figures 1 and 5). It also has been suggested that Xenacoelomorpha are the sister group to Ambulacraria rather than being placed in a more basal position as seen in Figure 1.
Figure 5.
Phylogeny of Deuterostomia. Note that Xenacoelomorpha is excluded from this figure, but has been proposed to be the sister group to Ambulacraria (see Figure 1). The tree for Hemichordata is from Worsaae et al. (2012), for Echinodermata from Janies et al. (2011), for Chordata from Delsuc et al. (2006), and for Tunicata from Swalla et al. (2000). As in Figure 1, the estimated numbers for extant described and valid species are indicated in parentheses after the terminal name and are from the World Register of Marine Species. When available, common names are written above the branches.
Conclusions
The answers to fundamental questions about the origins of animal life and the evolution of their diverse phenotypes may be held in the genomes of distinct invertebrate phyla. Seldom-studied taxa may be key to discovering how neurons, muscles, vision, hormones, immunity, limb regeneration, camouflage, bioluminescence, cooperation, organismal longevity, and physiological complexities evolved (Dehal et al. 2002; Hofmann et al. 2005; Putnam et al. 2007; 2008; Srivastava et al. 2010). Definitive answers to these questions have eluded biologists for centuries, but technological breakthroughs have now made it feasible to perform genome-scale studies of nonmodel organisms.
Genomic research in the 21st century touches many areas of biological enquiry. The biggest challenge facing the scientific community has shifted from the acquisition of sequence data to the analysis of the large data sets generated. GIGA will thus attempt to provide genomics support allowing the invertebrate biology community to overcome these challenges while applying the latest advances from the community of computational biologists.
A collective effort to sequence thousands of invertebrate genomes will only be feasible with participation and commitment from the scientific community. The large breadth of invertebrate diversity will require taxon-specific expertise and integration of traditional biology with molecular advances, both in data generation and analysis. The GIGA team has already expanded beyond initial participants in the first GIGA planning workshop, where the cooperative spirit and ability to work in concert to establish a common platform for data sharing and analysis were demonstrated. The long-range impact of GIGA will go beyond understanding phylogenetic relationships among invertebrate taxa, leading to new avenues of research in comparative developmental biology, environmental genomics, biodiversity, and climate change research, as is routinely done in many recent ecological studies focusing on gene expression data under stress conditions (DeSalvo et al. 2008; Bellantuono et al. 2012; Förster et al. 2012; Moya et al. 2012; Barshis et al. 2013; Pérez-Porro et al. 2013; Vidal-Dupiol et al. 2013).
Future efforts will concentrate on expanding the GIGA community, refining its goals, developing hypotheses, and establishing genomics research and educational resources. GIGA welcomes all members of the scientific community who wish to contribute to comparative approaches for understanding the genomic diversity of the large number of still underexplored animal phyla. We also invite new members with expertise in rare taxa and with the collection experience to ensure proper identification and preservation standards for successful genome sequencing and interpretations. The link to join can be found at http://giga.nova.edu. This is an opportunity for those who have devoted their lives to the study of particular taxa to merge their deeper insights of the evolutionary history of their organisms into a genomics context. A second, larger GIGA workshop is currently planned for late 2014. Funding the sequencing of thousands of invertebrate genomes will require creative and collaborative approaches that go beyond the traditional funding mechanisms of individual principal investigator grants (Oleksyk et al. 2012). While the GIGA team has started exploring possible funding opportunities, we invite feedback and participation from the scientific community for joint fund-seeking strategies and ideas.
Supplementary Material
Supplementary material can be found at http://www.jhered.oxfordjournals.org/.
Funding
American Genetic Association with a Special Event Award that provided the primary funding for the maiden GIGA workshop; Theodosius Dobzhansky Center for Genome Bioinformatics (Russia Ministry of Science Mega grant 11.G34.31.0068 to S.J. O’Brien, Principal Investigator); Life Technologies and BioNanoGenomics; National Science Foundation’s “Assembling the Tree of Life” (DEB awards 0732903, 0829763, 0829783, 0829791, 0829986).
Supplementary Material
Acknowledgments
GIGA also greatly appreciates the help of NSU staff, including the Office of Information Technology for Web site support, and several NSU students, Brittnee Barris, Andrea Bernhard, Alexandra Campbell, Andia Chaves Fonnegra, Kirk Kilfoyle, Jonathan Lanzas, Rebecca Mulheron, Bryce Parrish, Renee Potens, Kristian Taylor, and Christine Testermann, for assisting with GIGA workshop I logistics. For helpful feedback and comments on early drafts, the GIGA Community of Scientists (COS) thank Drs. Harris Lewin, Carmen Ablan, C. Titus Brown, Rob Steele, and Scott Nichols, and the Genome10K COS, including Warren Johnson. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
Appendix
Coauthor Committee chairs (alphabetical): Heather Bracken-Grissom (Florida International University, Miami, FL); Allen G. Collins (National Museum of Natural History, Smithsonian Institution, Washington, DC); Timothy Collins (Florida International University, Miami, FL); Keith Crandall (George Washington University, Washington, DC); Daniel Distel (Ocean Genome Legacy, Ipswich, MA); Casey Dunn (Brown University, Providence, RI); Gonzalo Giribet (Museum of Comparative Zoology, Harvard University, Cambridge, MA); Steven Haddock (Monterey Bay Aquarium Research Institute, Moss Landing, CA); Nancy Knowlton (National Museum of Natural History, Smithsonian Institution, Washington, DC); Mark Martindale (Whitney Laboratory, University of Florida, St Augustine, FL); Mónica Medina (Pennsylvania State University, State College, PA); Charles Messing (Oceanographic Center, Nova Southeastern University, Dania Beach, FL); Stephen J. O’Brien (Oceanographic Center, Nova Southeastern University, Dania Beach, FL and the Dobzhansky Center for Genome Bioinformatics); Gustav Paulay (Florida Museum of Natural History, University of Florida, Gainesville, FL); Nicolas Putnam (Rice University, Houston, TX); Timothy Ravasi (King Abdullah University of Science and Technology, Saudi Arabia); Greg W. Rouse (Scripps Institution of Oceanography, University of California, San Diego, CA); Joseph F. Ryan (Sars International Centre for Marine Molecular Biology, Bergen, Norway); Anja Schulze (Texas A&M University at Galveston, TX); Gert Wörheide (Ludwig-Maximilians-Universität München, München, Germany).
Additional authors: Maja Adamska (Sars International Centre for Marine Molecular Biology, Bergen, Norway); Xavier Bailly (Roscoff Marine Lab, France); Jesse Breinholt (Florida Museum of Natural History, University of Florida, Gainesville, FL); William E. Browne (University of Miami, Miami, FL); M. Christina Diaz (Oceanographic Center, Nova Southeastern University, Dania Beach, FL); Nathaniel Evans (Florida Museum of Natural History, University of Florida, Gainesville, FL); Jean-François Flot (Max Planck Institute for Dynamics and Self-Organization, Göttingen, Germany); Nicole Fogarty (Oceanographic Center, Nova Southeastern University, Dania Beach, FL); Matthew Johnston (Oceanographic Center, Nova Southeastern University, Dania Beach, FL); Bishoy Kamel (University of California, Merced, CA); Akito Y. Kawahara (Florida Museum of Natural History, University of Florida, Gainesville, FL); Tammy Laberge (Rosenstiel School of Marine and Atmosphere Sciences, Miami, FL); Dennis Lavrov (Iowa State University, Ames, IA); François Michonneau (Florida Museum of Natural History, University of Florida, Gainesville, FL); Leonid L. Moroz (Neuroscience, University of Florida, Gainesville and St. Augustine); Todd Oakley (University of California Santa Barbara, Santa Barbara, CA); Karen Osborne (National Museum of Natural History, Smithsonian Institution, Washington, DC); Shirley A. Pomponi (Harbor Branch Oceanographic Institute at Florida Atlantic University, Ft Pierce, FL); Adelaide Rhodes (Harte Research Institute Gulf of Mexico Studies, Texas A&M University, Corpus Christi, TX); Mauricio Rodriguez-Lanetty (Florida International University, Miami, FL); Scott R. Santos (Auburn University, Auburn, AL); Nori Satoh (Okinawa Institute of Science and Technology Graduate University, Okinawa, Japan); Robert W. Thacker (University of Alabama at Birmingham, Birmingham, AL); Yves Van de Peer (Ghent University, Ghent, Belgium); Christian R. Voolstra (King Abdullah University of Science and Technology, Saudi Arabia); David Mark Welch (Marine Biological Laboratory, Woods Hole, MA); Judith Winston (Virginia Museum of Natural History, Martinsville, VA); Xin Zhou (Beijing Genomics Institute, Shenzhen, China).
References
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. 2000. The genome sequence of Drosophila melanogaster . Science. 287:2185–2195 [DOI] [PubMed] [Google Scholar]
- Aguinaldo AM, Turbeville JM, Linford LS, Rivera MC, Garey JR, Raff RA, Lake JA. 1997. Evidence for a clade of nematodes, arthropods and other moulting animals. Nature. 387:489–493 [DOI] [PubMed] [Google Scholar]
- Ahyong ST, Lowry JK, Alonso M, Bamber RN, Boxshall GA, Castro P, Gerken S, Karaman GS, Goy JW, Jones DS, et al. 2011. Subphylum Crustacea Brünnich, 1772. In: Zhang ZQ, editor. Animal biodiversity: an outline of higher-level classification and survey of taxonomic richness. Vol. 3148 Zootaxa; p. 165–191 [Google Scholar]
- Albertin CB, Bonnaud L, Brown CT, Crookes-Goodson WJ, da Fonseca RR, Di Cristo C, Dilkes BP, Edsinger-Gonzales E, Freeman RM, Jr, Hanlon RT, et al. 2012. Cephalopod genomics: a plan of strategies and organization. Stand Genomic Sci. 7:175–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appeltans W, Ahyong ST, Anderson G, Angel MV, Artois T, Bailly N, Bamber R, Barber A, Bartsch I, Berta A, et al. 2012. The magnitude of global marine species diversity. Curr Biol. 22:2189–2202 [DOI] [PubMed] [Google Scholar]
- Atkinson A, Siegel V, Pakhomov EA, Jessopp MJ, Loeb V. 2009. A re-appraisal of the total biomass and annual production of Antarctic krill. Deep-Sea Res Pt I. 56:727–740 [Google Scholar]
- Balthasar U, Cusack M, Faryma L, Chung P, Holmer LE, Jin J, Percival IG, Popov LE. 2011. Relic aragonite from Ordovician-Silurian brachiopods: implications for the evolution of calcification. Geology. 39: 967–970 [Google Scholar]
- Barshis DJ, Ladner JT, Oliver TA, Seneca FO, Traylor-Knowles N, Palumbi SR. 2013. Genomic basis for coral resilience to climate change. Proc Natl Acad Sci USA. 110:1387–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellantuono AJ, Granados-Cifuentes C, Miller DJ, Hoegh-Guldberg O, Rodriguez-Lanetty M. 2012. Coral thermal tolerance: tuning gene expression to resist thermal stress. PLoS One. 7:e50685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriman M, Haas BJ, LoVerde PT, Wilson RA, Dillon GP, Cerqueira GC, Mashiyama ST, Al-Lazikani B, Andrade LF, Ashton PD, et al. 2009. The genome of the blood fluke Schistosoma mansoni . Nature. 460:352–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E. 2012. The making of ENCODE: lessons for big-data projects. Nature. 489:49–51 [DOI] [PubMed] [Google Scholar]
- Blunt JW, Copp BR, Keyzers RA, Munro MH, Prinsep MR. 2013. Marine natural products. Nat Prod Rep. 30:237–323 [DOI] [PubMed] [Google Scholar]
- Boehm AM, Khalturin K, Anton-Erxleben F, Hemmrich G, Klostermeier UC, Lopez-Quintero JA, Oberg HH, Puchert M, Rosenstiel P, Wittlieb J, et al. 2012. FoxO is a critical regulator of stem cell maintenance in immortal Hydra. Proc Natl Acad Sci USA. 109:19697–19702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch TC, McFall-Ngai MJ. 2011. Metaorganisms as the new frontier. Zoology (Jena). 114:185–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradnam KR, Fass JN, Alexandrov A, Baranay P, Bechner M, Birol I, Boisvert S, Chapman JA, Chapuis G, Chikhi R, et al. 2013. Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species. Gigascience. 2:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunton CHC, Cocks RM, Long SL, editors. 2001. Brachiopods past and present. Vol. 63 London: Taylor & Francis, The Systematics Association; ISBN 0748 409211 p. 13, 441 [Google Scholar]
- Buitenhuis E, Le Quere C, Aumont O, Beaugrand G, Bunker A, Hirst A, Ikeda T, O’Brien T, Piontkovski S, Straile D. 2006. Biogeochemical fluxes through mesozooplankton. Global Biogeochem Cy. 20:GB2003. 10.1029/2005GB002511 [Google Scholar]
- Cameron RAD. 2013. The diversity of land molluscs–questions unanswered and questions unasked*. Am Malacol Bull. 31:169–180 [Google Scholar]
- Cárdenas P, Pérez T, Boury-Esnault N. 2012. Sponge systematics facing new challenges. Adv Mar Biol. 61:79–209 [DOI] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 282:2012–2018 [DOI] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. 1994. Green fluorescent protein as a marker for gene expression. Science. 263:802–805 [DOI] [PubMed] [Google Scholar]
- Chapman JA, Kirkness EF, Simakov O, Hampson SE, Mitros T, Weinmaier T, Rattei T, Balasubramanian PG, Borman J, Busam D, et al. 2010. The dynamic genome of Hydra. Nature. 464:592–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbourne JK, Pfrender ME, Gilbert D, Thomas WK, Tucker A, Oakley TH, Tokishita S, Aerts A, Arnold GJ, Basu MK, et al. 2011. The ecoresponsive genome of Daphnia pulex . Science. 331:555–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collen B, Böhm M, Kemp R, Baillie JEM. 2012. Spineless: status and trends of the world’s invertebrates. London: Zoological Society of London [Google Scholar]
- Collins AG. 2009. Recent insights into cnidarian phylogeny. 38:139–149 [Google Scholar]
- Contreras JL. 2010. Information access. Prepublication data release, latency, and genome commons. Science. 329:393–394 [DOI] [PubMed] [Google Scholar]
- Danchin EGJ, Flot J-F, Perfus-Barbeoch L, Van Doninck K. 2011. Genomic perspectives on the long-term absence of sexual reproduction in animals. In: Pontarotti P, editor. Evolutionary biology—concepts, biodiversity, macroevolution and genome evolution. Berlin, Heidelberg: Springer, p. 223–242 [Google Scholar]
- Danovaro R, Dell’Anno A, Pusceddu A, Gambi C, Heiner I, Kristensen RM. 2010. The first metazoa living in permanently anoxic conditions. BMC Biol. 8:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH, Erwin DH. 2006. Gene regulatory networks and the evolution of animal body plans. Science. 311:796–800 [DOI] [PubMed] [Google Scholar]
- Dawson MN, Raskoff KA, Jacobs DK. 1998. Field preservation of marine invertebrate tissue for DNA analyses. Mol Mar Biol Biotechnol. 7:145–152 [PubMed] [Google Scholar]
- De’ath G, Fabricius KE, Sweatman H, Puotinen M. 2012. The 27-year decline of coral cover on the Great Barrier Reef and its causes. Proc Natl Acad Sci USA. 109:17995–17999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, Davidson B, Di Gregorio A, Gelpke M, Goodstein DM, et al. 2002. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 298:2157–2167 [DOI] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Chourrout D, Philippe H. 2006. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 439:965–968 [DOI] [PubMed] [Google Scholar]
- DeSalvo MK, Voolstra CR, Sunagawa S, Schwarz JA, Stillman JH, Coffroth MA, Szmant AM, Medina M. 2008. Differential gene expression during thermal stress and bleaching in the Caribbean coral Montastraea faveolata . Mol Ecol. 17:3952–3971 [DOI] [PubMed] [Google Scholar]
- Dohrmann M, Wörheide G. 2013. Novel scenarios of early animal evolution–is it time to rewrite textbooks? Integr Comp Biol. 53:503–511 [DOI] [PubMed] [Google Scholar]
- Dunn CW, Hejnol A, Matus DQ, Pang K, Browne WE, Smith SA, Seaver E, Rouse GW, Obst M, Edgecombe GD, et al. 2008. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 452:745–749 [DOI] [PubMed] [Google Scholar]
- Ecker JR, Bickmore WA, Barroso I, Pritchard JK, Gilad Y, Segal E. 2012. Genomics: ENCODE explained. Nature. 489:52–55 [DOI] [PubMed] [Google Scholar]
- Eddy SR. 2013. The ENCODE project: missteps overshadowing a success. Curr Biol. 23:R259–R261 [DOI] [PubMed] [Google Scholar]
- Edgecombe GD, Giribet G, Dunn CW, Hejnol A, Kristensen RM, Neves RC, Rouse GW, Worsaae K, Sørensen MV. 2011. Higher-level metazoan relationships: recent progress and remaining questions. Org Divers Evol. 11:151–172 [Google Scholar]
- Eitel M, Schierwater B. 2010. The phylogeography of the Placozoa suggests a taxon-rich phylum in tropical and subtropical waters. Mol Ecol. 19:2315–2327 [DOI] [PubMed] [Google Scholar]
- Erwin DH, Valentine JW. 2013. The Cambrian Explosion: The Construction of Animal Biodiversity. Greenwood Village (CO): Roberts and Company Publishers [Google Scholar]
- Evans NM, Lindner A, Raikova EV, Collins AG, Cartwright P. 2008. Phylogenetic placement of the enigmatic parasite, Polypodium hydriforme, within the phylum Cnidaria. BMC Evol Biol. 8:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans NM, Lindner A, Raikova EV, Collins AG, Cartwright P. 2009. Correction: phylogenetic placement of the enigmatic parasite, Polypodium hydriforme, within the phylum Cnidaria. BMC Evol Biol. 9:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner DJ. 2002. Marine natural products. Nat Prod Rep. 19:1–48 [DOI] [PubMed] [Google Scholar]
- Field D, Amaral-Zettler L, Cochrane G, Cole JR, Dawyndt P, Garrity GM, Gilbert J, Glöckner FO, Hirschman L, Karsch-Mizrachi I, et al. 2011. The Genomic Standards Consortium. PLoS Biol. 9:e1001088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flot JF, Hespeels B, Li X, Noel B, Arkhipova I, Danchin EG, Hejnol A, Henrissat B, Koszul R, Aury JM, et al. 2013. Genomic evidence for ameiotic evolution in the bdelloid rotifer Adineta vaga. Nature. 500:453–457 [DOI] [PubMed] [Google Scholar]
- Förster F, Beisser D, Grohme MA, Liang C, Mali B, Siegl AM, Engelmann JC, Shkumatov AV, Schokraie E, Müller T, et al. 2012. Transcriptome analysis in tardigrade species reveals specific molecular pathways for stress adaptations. Bioinform Biol Insights. 6:69–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratzl P. 2007. Biomimetic materials research: what can we really learn from nature’s structural materials? J R Soc Interface. 4:637–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funch P, Kristensen RM. 1995. Cycliophora is a new phylum with affinities to Entoprocta and Ectoprocta. Nature. 378:711–714 [Google Scholar]
- G10KCOS. 2009. Genome 10K: a proposal to obtain whole-genome sequence for 10,000 vertebrate species. J Hered. 100:659–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galil BS. 2012. Truth and consequences: the bioinvasion of the Mediterranean Sea. Integr Zool. 7:299–311 [DOI] [PubMed] [Google Scholar]
- German CR, Ramirez-Llodra E, Baker MC, Tyler PA; ChEss Scientific Steering Committee. 2011. Deep-water chemosynthetic ecosystem research during the census of marine life decade and beyond: a proposed deep-ocean road map. PLoS One. 6:e23259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser M, Diele K. 2004. Asymmetric outcomes: assessing central aspects of the biological, economic and social sustainability of a mangrove crab fishery, Ucides cordatus (Ocypodidae), in North Brazil. Ecol Econ. 49:361–373 [Google Scholar]
- Graf DL. 2013. Patterns of freshwater bivalve global diversity and the state of phylogenetic studies on the Unionoida, Sphaeriidae, and Cyrenidae. Am Malacol Bull. 31:135–153 [Google Scholar]
- Grbic M, Khila A, Lee KZ, Bjelica A, Grbic V, Whistlecraft J, Verdon L, Navajas M, Nagy L. 2007. Mity model: Tetranychus urticae, a candidate for chelicerate model organism. Bioessays. 29:489–496 [DOI] [PubMed] [Google Scholar]
- Grbić M, Van Leeuwen T, Clark RM, Rombauts S, Rouzé P, Grbić V, Osborne EJ, Dermauw W, Ngoc PC, Ortego F, et al. 2011. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature. 479:487–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero FD, Moolhuijzen P, Peterson DG, Bidwell S, Caler E, Bellgard M, Nene VM, Djikeng A. 2010. Reassociation kinetics-based approach for partial genome sequencing of the cattle tick, Rhipicephalus (Boophilus) microplus. BMC Genomics. 11:374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hejnol A. 2010. A twist in time–the evolution of spiral cleavage in the light of animal phylogeny. Integr Comp Biol. 50:695–706 [DOI] [PubMed] [Google Scholar]
- Hejnol A, Obst M, Stamatakis A, Ott M, Rouse GW, Edgecombe GD, Martinez P, Baguñà J, Bailly X, Jondelius U, et al. 2009. Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc Biol Sci. 276:4261–4270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CA, Wikel SK. 2005. The Ixodes scapularis Genome Project: an opportunity for advancing tick research. Trends Parasitol. 21:151–153 [DOI] [PubMed] [Google Scholar]
- Hofmann GE, Burnaford JL, Fielman KT. 2005. Genomics-fueled approaches to current challenges in marine ecology. Trends Ecol Evol. 20:305–311 [DOI] [PubMed] [Google Scholar]
- Howison M, Dunn CW, Zapata F. 2013. Toward a statistically explicit understanding of de novo sequence assembly. Bioinformatics. 10.1093/bioinformatics/btt525 [DOI] [PubMed] [Google Scholar]
- Huang D, Meier R, Todd PA, Chou LM. 2008. Slow mitochondrial COI sequence evolution at the base of the metazoan tree and its implications for DNA barcoding. J Mol Evol. 66:167–174 [DOI] [PubMed] [Google Scholar]
- Human Microbiome Project. 2012. Structure, function and diversity of the healthy human microbiome. Nature. 486:207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikuta T. 2011. Evolution of invertebrate deuterostomes and Hox/ParaHox genes. Genomics Proteomics Bioinformatics. 9:77–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janies DA, Voight JR, Daly M. 2011. Echinoderm phylogeny including Xyloplax, a progenetic asteroid. Syst Biol. 60:420–438 [DOI] [PubMed] [Google Scholar]
- Jeffery NW, Jardine CB, Gregory TR. 2013. A first exploration of genome size diversity in sponges. Genome. 56:451–456 [DOI] [PubMed] [Google Scholar]
- Knowlton N. 2010. Citizens of the sea: wondrous creatures from the census of marine life. Washington (DC): National Geographic [Google Scholar]
- Knowlton N, Weigt LA. 1998. New dates and new rates for divergence across the Isthmus of Panama. Proc R Soc B. 265:2257–2263 [Google Scholar]
- Kumar S, Schiffer PH, Blaxter M. 2012. 959 Nematode Genomes: a semantic wiki for coordinating sequencing projects. Nucleic Acids Res. 40:D1295–D1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kursar TA, Caballero-George CC, Capson TL, Cubilla-Rios L, Gerwick WH, Heller MV, Ibañez A, Linington RG, McPhail KL, Ortega-Barría E, et al. 2007. Linking bioprospecting with sustainable development and conservation: the Panama case. Biodivers Conserv. 16:2789–2800 [Google Scholar]
- Lamarck JBd. 1801. Systeme des Animaux sans vertebres, ou tableau général des classes, des ordres et des genres de ces animaux. Paris: Chez l’Auteur, au Muséum d’Histoire Naturelle [Google Scholar]
- Leal MC, Puga J, Serôdio J, Gomes NC, Calado R. 2012. Trends in the discovery of new marine natural products from invertebrates over the last two decades–where and what are we bioprospecting? PLoS One. 7:e30580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessios HA. 2008. The Great American schism: divergence of marine organisms after the rise of the Central American Isthmus. Annu Rev Ecol Evol Syst. 39:63–91 [Google Scholar]
- Lewin HA, Larkin DM, Pontius J, O’Brien SJ. 2009. Every genome sequence needs a good map. Genome Res. 19:1925–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lom J, Dyková I. 2006. Myxozoan genera: definition and notes on taxonomy, life-cycle terminology and pathogenic species. Folia Parasitol (Praha). 53:1–36 [PubMed] [Google Scholar]
- MacDonald KS, 3rd, Yampolsky L, Duffy JE. 2005. Molecular and morphological evolution of the amphipod radiation of Lake Baikal. Mol Phylogenet Evol. 35:323–343 [DOI] [PubMed] [Google Scholar]
- Mardis ER. 2011. A decade’s perspective on DNA sequencing technology. Nature. 470:198–203 [DOI] [PubMed] [Google Scholar]
- Moya A, Huisman L, Ball EE, Hayward DC, Grasso LC, Chua CM, Woo HN, Gattuso JP, Forêt S, Miller DJ. 2012. Whole transcriptome analysis of the coral Acropora millepora reveals complex responses to CO2-driven acidification during the initiation of calcification. Mol Ecol. 21:2440–2454 [DOI] [PubMed] [Google Scholar]
- Müller WE, Schröder HC, Burghard Z, Pisignano D, Wang X. 2013. Silicateins–a novel paradigm in bioinorganic chemistry: enzymatic synthesis of inorganic polymeric silica. Chemistry. 19:5790–5804 [DOI] [PubMed] [Google Scholar]
- Munro D, Blier PU. 2012. The extreme longevity of Arctica islandica is associated with increased peroxidation resistance in mitochondrial membranes. Aging Cell. 11:845–855 [DOI] [PubMed] [Google Scholar]
- Muscatine L, Cernichiari E. 1969. Assimilation of photosynthetic products of zooxanthellae by a reef coral. Biol Bull. 137:506–523 [DOI] [PubMed] [Google Scholar]
- Narahashi T, Roy ML, Ginsburg KS. 1994. Recent advances in the study of mechanism of action of marine neurotoxins. Neurotoxicology. 15:545–554 [PubMed] [Google Scholar]
- Nielsen C. 2012. Animal evolution: interrelationships of the living phyla. 3rd edn Oxford: Oxford University Press [Google Scholar]
- Neiland AE, Soley N, Varley JB, Whitmarsh DJ. 2001. Shrimp aquaculture: economic perspectives for policy development. Marine Policy. 25:265–279. 10.1016/S0308-597X(01)00017-3 [Google Scholar]
- Nosenko T, Schreiber F, Adamska M, Adamski M, Eitel M, Hammel J, Maldonado M, Müller WE, Nickel M, Schierwater B, et al. 2013. Deep metazoan phylogeny: when different genes tell different stories. Mol Phylogenet Evol. 67:223–233 [DOI] [PubMed] [Google Scholar]
- Oleksyk TK, Pombert JF, Siu D, Mazo-Vargas A, Ramos B, Guiblet W, Afanador Y, Ruiz-Rodriguez CT, Nickerson ML, Logue DM, et al. 2012. A locally funded Puerto Rican parrot (Amazona vittata) genome sequencing project increases avian data and advances young researcher education. Gigascience. 1:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivera BM, Teichert RW. 2007. Diversity of the neurotoxic Conus peptides: a model for concerted pharmacological discovery. Mol Interv. 7:251–260 [DOI] [PubMed] [Google Scholar]
- Olson PD, Zarowiecki M, Kiss F, Brehm K. 2012. Cestode genomics—progress and prospects for advancing basic and applied aspects of flatworm biology. Parasite Immunol. 34:130–150 [DOI] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Korf I. 2007. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 23:1061–1067 [DOI] [PubMed] [Google Scholar]
- Pérez-Porro AR, Navarro-Gómez D, Uriz MJ, Giribet G. 2013. A NGS approach to the encrusting Mediterranean sponge Crella elegans (Porifera, Demospongiae, Poecilosclerida): transcriptome sequencing, characterization and overview of the gene expression along three life cycle stages. Mol Ecol Resour. 13:494–509 [DOI] [PubMed] [Google Scholar]
- Philippe H, Brinkmann H, Copley RR, Moroz LL, Nakano H, Poustka AJ, Wallberg A, Peterson KJ, Telford MJ. 2011. Acoelomorph flatworms are deuterostomes related to Xenoturbella. Nature. 470:255–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H, Derelle R, Lopez P, Pick K, Borchiellini C, Boury-Esnault N, Vacelet J, Renard E, Houliston E, Quéinnec E, et al. 2009. Phylogenomics revives traditional views on deep animal relationships. Curr Biol. 19:706–712 [DOI] [PubMed] [Google Scholar]
- Pick KS, Philippe H, Schreiber F, Erpenbeck D, Jackson DJ, Wrede P, Wiens M, Alié A, Morgenstern B, Manuel M, et al. 2010. Improved phylogenomic taxon sampling noticeably affects nonbilaterian relationships. Mol Biol Evol. 27:1983–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce SK, Curtis NE. 2012. Cell biology of the chloroplast symbiosis in sacoglossan sea slugs. Int Rev Cell Mol Biol. 293:123–148 [DOI] [PubMed] [Google Scholar]
- Pomponi SA, Jevitt A, Patel J, Diaz MC. 2013. Sponge hybridomas: applications and implications. Integr Comp Biol. 53:524–530 [DOI] [PubMed] [Google Scholar]
- Ponder WF, Lunney D, editors. 1999. The other 99%: the conservation and biodiversity of invertebrates. Mosman (Australia): Royal Zoological Society of New South Wales [Google Scholar]
- Puritz JB, Keever CC, Addison JA, Byrne M, Hart MW, Grosberg RK, Toonen RJ. 2012. Extraordinarily rapid life-history divergence between Cryptasterina sea star species. Proc Biol Sci. 279:3914–3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu JK, et al. 2008. The amphioxus genome and the evolution of the chordate karyotype. Nature. 453:1064–1071 [DOI] [PubMed] [Google Scholar]
- Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV, et al. 2007. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 317:86–94 [DOI] [PubMed] [Google Scholar]
- Reaka-Kudla ML. 1997. The global biodiversity of coral reefs: A comparison with rain forests. In: Reaka-Kudla ML, Wilson DE, Wilson EO, Keener CS, editors. Biodiversity II: understanding and protecting our biological resources. Washington (DC): Joseph Henry Press [Google Scholar]
- Regier JC, Shultz JW, Zwick A, Hussey A, Ball B, Wetzer R, Martin JW, Cunningham CW. 2010. Arthropod relationships revealed by phylogenomic analysis of nuclear protein-coding sequences. Nature. 463:1079–1083 [DOI] [PubMed] [Google Scholar]
- Riesgo A, Pérez-Porro AR, Carmona S, Leys SP, Giribet G. 2012. Optimization of preservation and storage time of sponge tissues to obtain quality mRNA for next-generation sequencing. Mol Ecol Resour. 12:312–322 [DOI] [PubMed] [Google Scholar]
- Rinkevich B. 2011. Cell cultures from marine invertebrates: new insights for capturing endless stemness. Mar Biotechnol (NY). 13:345–354 [DOI] [PubMed] [Google Scholar]
- Roark EB, Guilderson TP, Dunbar RB, Ingram BL. 2006. Radiocarbon-based ages and growth rates of Hawaiian deep-sea corals. Marine Ecol Prog Series. 327:1–14 [Google Scholar]
- Robb SM, Ross E, Sánchez Alvarado A. 2008. SmedGD: the Schmidtea mediterranea genome database. Nucleic Acids Res. 36:D599–D606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson GE, Hackett KJ, Purcell-Miramontes M, Brown SJ, Evans JD, Goldsmith MR, Lawson D, Okamuro J, Robertson HM, Schneider DJ. 2011. Creating a buzz about insect genomes. Science. 331:1386. [DOI] [PubMed] [Google Scholar]
- Rouse GW, Pleijel F. 2001. Polychaetes. Oxford; New York: Oxford University Press [Google Scholar]
- Rubin GM, Yandell MD, Wortman JR, Gabor Miklos GL, Nelson CR, Hariharan IK, Fortini ME, Li PW, Apweiler R, Fleischmann W, et al. 2000. Comparative genomics of the eukaryotes. Science. 287:2204–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppert EE, Fox RS, Barnes RD. 2004. Invertebrate zoology, a functional evolutionary approach. Belmont (CA): Thomson, Brooks/Cole [Google Scholar]
- Russell CW, Bouvaine S, Newell PD, Douglas AE. 2013. Shared metabolic pathways in a coevolved insect-bacterial symbiosis. Appl Environ Microbiol. 79:6117–6123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan JF. 2013. Baa. pl: a tool to evaluate de novo genome assemblies with RNA transcripts. arXiv 1309:2087 [q-bio].
- Ryan JF, Pang K, Mullikin JC, Martindale MQ, Baxevanis AD; NISC Comparative Sequencing Program. 2010. The homeodomain complement of the ctenophore Mnemiopsis leidyi suggests that Ctenophora and Porifera diverged prior to the ParaHoxozoa. Evodevo. 1:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder OA. 2005. Conservation genomics: applying whole genome studies to species conservation efforts. Cytogenet Genome Res. 108:6–15 [DOI] [PubMed] [Google Scholar]
- Scheffers BR, Joppa LN, Pimm SL, Laurance WF. 2012. What we know and don’t know about Earth’s missing biodiversity. Trends Ecol Evol. 27:501–510 [DOI] [PubMed] [Google Scholar]
- Schierwater B, Eitel M, Jakob W, Osigus HJ, Hadrys H, Dellaporta SL, Kolokotronis SO, Desalle R. 2009. Concatenated analysis sheds light on early metazoan evolution and fuels a modern “urmetazoon” hypothesis. PLoS Biol. 7:e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, Lightfoot S, Menzel W, Granzow M, Ragg T. 2006. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HC, Kube M, Edvardsen RB, Jensen MF, Beck A, Spriet E, Gorsky G, Thompson EM, Lehrach H, Reinhardt R, et al. 2001. Miniature genome in the marine chordate Oikopleura dioica . Science. 294:2506. [DOI] [PubMed] [Google Scholar]
- Shendure J, Lieberman Aiden E. 2012. The expanding scope of DNA sequencing. Nat Biotechnol. 30:1084–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzato C, Shoguchi E, Kawashima T, Hamada M, Hisata K, Tanaka M, Fujie M, Fujiwara M, Koyanagi R, Ikuta T, et al. 2011. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature. 476:320–323 [DOI] [PubMed] [Google Scholar]
- Simakov O, Marletaz F, Cho SJ, Edsinger-Gonzales E, Havlak P, Hellsten U, Kuo DH, Larsson T, Lv J, Arendt D, et al. 2013. Insights into bilaterian evolution from three spiralian genomes. Nature. 493:526–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sket B, Trontelj P. 2007. Global diversity of leeches (Hirudinea) in freshwater. Hydrobiologia. 595:129–137 [Google Scholar]
- Slack JM, Holland PW, Graham CF. 1993. The zootype and the phylotypic stage. Nature. 361:490–492 [DOI] [PubMed] [Google Scholar]
- Smith SA, Wilson NG, Goetz FE, Feehery C, Andrade SC, Rouse GW, Giribet G, Dunn CW. 2011. Resolving the evolutionary relationships of molluscs with phylogenomic tools. Nature. 480:364–367 [DOI] [PubMed] [Google Scholar]
- Sodergren E, Weinstock GM, Davidson EH, Cameron RA, Gibbs RA, Angerer RC, Angerer LM, Arnone MI, Burgess DR, Burke RD, et al. 2006. The genome of the sea urchin Strongylocentrotus purpuratus . Science. 314:941–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, Kuo A, Mitros T, Salamov A, Carpenter ML, et al. 2008. The Trichoplax genome and the nature of placozoans. Nature. 454:955–960 [DOI] [PubMed] [Google Scholar]
- Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier ME, Mitros T, Richards GS, Conaco C, Dacre M, Hellsten U, et al. 2010. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 466:720–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoner A. 1997. The status of queen conch, Strombus gigas, research in the Caribbean. Marine Fisheries Rev. 59:14–23 [Google Scholar]
- Struck TH, Paul C, Hill N, Hartmann S, Hösel C, Kube M, Lieb B, Meyer A, Tiedemann R, Purschke G, et al. 2011. Phylogenomic analyses unravel annelid evolution. Nature. 471:95–98 [DOI] [PubMed] [Google Scholar]
- Swalla BJ, Cameron CB, Corley LS, Garey JR. 2000. Urochordates are monophyletic within the deuterostomes. Syst Biol. 49:52–64 [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Kawashima T, Koyanagi R, Gyoja F, Tanaka M, Ikuta T, Shoguchi E, Fujiwara M, Shinzato C, Hisata K, et al. 2012. Draft genome of the pearl oyster Pinctada fucata: a platform for understanding bivalve biology. DNA Res. 19:117–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trindade-Silva AE, Lim-Fong GE, Sharp KH, Haygood MG. 2010. Bryostatins: biological context and biotechnological prospects. Curr Opin Biotechnol. 21:834–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai IJ, Zarowiecki M, Holroyd N, Garciarrubio A, Sanchez-Flores A, Brooks KL, Tracey A, Bobes RJ, Fragoso G, Sciutto E, et al. ; Taenia solium Genome Consortium. 2013. The genomes of four tapeworm species reveal adaptations to parasitism. Nature. 496:57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. 2007. The human microbiome project. Nature. 449:804–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine JW. 2004. On the origin of phyla. Chicago: The University of Chicago Press [Google Scholar]
- Van Soest RW, Boury-Esnault N, Vacelet J, Dohrmann M, Erpenbeck D, De Voogd NJ, Santodomingo N, Vanhoorne B, Kelly M, Hooper JN. 2012. Global diversity of sponges (Porifera). PLoS One. 7:e35105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal-Dupiol J, Zoccola D, Tambutté E, Grunau C, Cosseau C, Smith KM, Freitag M, Dheilly NM, Allemand D, Tambutté S. 2013. Genes related to ion-transport and energy production are upregulated in response to CO2-driven pH decrease in corals: new insights from transcriptome analysis. PLoS One. 8:e58652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson JP, Jaffe DB, O’Neill K, Karlsson EK, Stange-Thomann N, Anderson S, Mesirov JP, Satoh N, Satou Y, Nusbaum C, et al. 2005. Assembly of polymorphic genomes: algorithms and application to Ciona savignyi . Genome Res. 15:1127–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt O, Collins AG, Pearse VB, Pearse JS, Ender A, Hadrys H, Schierwater B. 2004. Placozoa—no longer a phylum of one. Curr Biol. 14:R944–R945 [DOI] [PubMed] [Google Scholar]
- von Reumont BM, Jenner RA, Wills MA, Dell’ampio E, Pass G, Ebersberger I, Meyer B, Koenemann S, Iliffe TM, Stamatakis A, et al. 2012. Pancrustacean phylogeny in the light of new phylogenomic data: support for Remipedia as the possible sister group of Hexapoda. Mol Biol Evol. 29:1031–1045 [DOI] [PubMed] [Google Scholar]
- Wang XY, Chen WJ, Huang Y, Sun JF, Men JT, Liu HL, Luo F, Guo L, Lv XL, Deng CH, et al. 2011. The draft genome of the carcinogenic human liver fluke Clonorchis sinensis . Genome Biol. 12:R107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel MJ, Reynolds JW. 2011. Nomenclatura Oligochaetologica [Internet]. Available from: http://www.inhs.uiuc.edu/~mjwetzel/Nomen.Oligo.html
- Winnebeck EC, Millar CD, Warman GR. 2010. Why does insect RNA look degraded? J Insect Sci. 10:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR. 2004. A new biology for a new century. Microbiol Mol Biol Rev. 68:173–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong PB, Wiley EO, Johnson WE, Ryder OA, O’Brien SJ, Haussler D, Koepfli KP, Houck ML, Perelman P, Mastromonaco G, et al. ; G10KCOS. 2012. Tissue sampling methods and standards for vertebrate genomics. Gigascience. 1:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wörheide G. 1998. The reef cave dwelling ultraconservative coralline demosponge Astrosclera willeyana Lister 1900 from the Indo-Pacific–Micromorphology, Ultrastructure, Biocalcification, Isotope Record, Taxonomy, Biogeography, Phylogeny. Facies. 38:1–88 [Google Scholar]
- Wörheide G, Dohrmann M, Erpenbeck D, Larroux C, Maldonado M, Voigt O, Borchiellini C, Lavrov DV. 2012. Deep phylogeny and evolution of sponges (phylum Porifera). Adv Mar Biol. 61:1–78 [DOI] [PubMed] [Google Scholar]
- Worsaae K, Sterrer W, Kaul-Strehlow S, Hay-Schmidt A, Giribet G. 2012. An anatomical description of a miniaturized acorn worm (hemichordata, enteropneusta) with asexual reproduction by paratomy. PLoS One. 7:e48529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Fang X, Guo X, Li L, Luo R, Xu F, Yang P, Zhang L, Wang X, Qi H, et al. 2012. The oyster genome reveals stress adaptation and complexity of shell formation. Nature. 490:49–54 [DOI] [PubMed] [Google Scholar]
- Zhang Z-Q. 2011a. Animal biodiversity: An outline of higher-level classification and survey of taxonomic richness. Zootaxa. 3148:237. [DOI] [PubMed] [Google Scholar]
- Zhang Z-Q. 2011b. Phylum Arthropoda von Siebold, 1848. In: Zhang ZQ, editor. Animal biodiversity: an introduction to higher-level classification and taxonomic richness. Zootaxa. 4138:99–103 [Google Scholar]
- Zhou Y, Zheng HJ, Chen YY, Zhang L, Wang K, Guo J, Huang Z, Zhang B, Huang W, Jin K, et al. 2009. The Schistosoma japonicum genome reveals features of host-parasite interplay. Nature. 460:345–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.