Abstract
Recent genome-wide association studies (GWAS) have identified several common susceptibility loci associated with the risk of colorectal cancer (CRC). However, whether these loci affect clinical outcomes of CRC is not clear. In this study, we genotyped 26 single nucleotide polymorphisms (SNPs) in 10 GWAS-identified CRC susceptibility regions and evaluated their associations with survival and recurrence in 285 stage II and III patients receiving fluorouracil-based adjuvant chemotherapy. Only one SNP, rs10318 (15q13.3), was significantly associated with recurrence for patients with stage II disease. Three SNPs: rs10749971 (11q23.1), rs961253 (20p12.3) and rs355527 (20p12.3) in two regions were significantly associated with recurrence for patients with stage III disease. Five SNPs: rs961253 (20p12.3), rs355527 (20p12.3), rs4464148 (18q21.1), rs6983267 (8q24.21) and rs10505477 (8q24.21) in three regions were significantly associated with survival for patients with stage III disease. Cumulative effects of multiple unfavorable genotypes were observed for recurrence and survival in patients with stage III CRC. Our results suggest that cancer susceptibility loci may also affect the prognosis of CRC patients receiving fluorouracil-based adjuvant chemotherapy.
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer and the second leading cause of cancer death with an estimated 141 210 new cases and 49 380 deaths in 2011 in the USA (1). Traditionally, adjuvant chemotherapy after surgery is recommended for patients with high-risk stage II and stage III CRC. Patients with stage IV disease are usually offered palliative chemotherapy and supportive care. 5-Fluorouracil (5-FU) is the most frequently used chemotherapeutic agent for treating CRC. However, 5-FU alone will only result in an absolute benefit in overall survival of <10% following 6 months of adjuvant chemotherapy for both stage II and III patients (2,3). This small absolute benefit may be at the cost of treatment-related toxicities (4). Recent trials have failed to establish a new adjuvant treatment paradigm outside of 5-FU-based therapy (5). New predictive and prognostic biomarkers could facilitate tailoring of therapeutic strategies by maximizing therapeutic benefit and minimizing toxicities in CRC patients.
It is well documented that the development of CRC is a multi-step process involving accumulation of genetic and epigenetic alterations. Genetic variations in critical cellular pathways have been associated with the development and prognosis of CRC patients. Recent genome-wide association studies (GWAS) have identified single nucleotide polymorphisms (SNPs) in at least 10 genomic regions that were associated with CRC susceptibility, including 15q13.3 (GREM1), 20p12.3 (BMP2), 18q21.1 (SMAD7), 11q23.1 (LOC120376, FLJ45803, c11orf53, POU2AF1), 8q24.21 (POU5F1P1, DQ515897, MYC), 8q23.3 (c8orf53, EIF3H), 10p14 (FLJ3802842), 14q22.2 (BMP4), 16q22.1 (CDH1) and 19q13.11 (RHPN2) (6). Interestingly, GREM1, BMP2, BMP4, SMAD7 and RHPN2 are all components of transforming growth factor-β (TGF-β) pathway, suggesting that perturbation in TGF-β signaling pathway plays an important role in CRC susceptibility (7). TGF-β pathway is also involved in the prognosis of the CRC (8), and overexpression of TGF-β signaling genes has been known to play a vital role in the progression of CRC (9). However, whether these GWAS-identified cancer susceptibility SNPs affect CRC clinical outcomes remains unclear. In this study, we have included 510 patients with CRC to perform a comprehensive study to evaluate these CRC susceptibility SNPs with clinical outcome in stage II and III patients receiving 5-FU-based adjuvant chemotherapy.
Patients and methods
Patient population and clinical data collection
All patients were newly diagnosed (within 1 year of diagnosis before enrollment in this study) and histologically confirmed colorectal adenocarcinoma patients who were recruited from the University of Texas MD Anderson Cancer Center between January 1990 and June 2008. All participants signed an informed consent prior to participation in the study. There were no recruitment restrictions on age, gender, race, type of chemotherapy administered or cancer stage. Demographic and epidemiologic data were collected using a structured self-administered questionnaire. All study patients donated 10–20 ml blood sample for genomic DNA isolation and molecular analyses. Clinical and follow-up data were abstracted from medical records. The study end points were overall survival and recurrence. The study was approved by the Institutional Review Board of MD Anderson Cancer Center.
SNP selection and genotyping
Twenty-six SNPs with minor allele frequency >0.05 in Caucasian population in 10 GWAS-identified CRC susceptibility regions were selected and genotyped with Taqman assay using the ABI 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA) (Supplementary Table 1 is available at Carcinogenesis Online) (6,10–16). The call rate was >95% for each SNP. Internal quality controls were used to ensure genotyping accuracy, and 5% of all samples were randomly selected and genotyped in duplicate with 100% concordance.
Statistical analysis
Stata software (version 10.2; StataCrop LP, College Station, TX) was used for the statistical analyses. The χ2 test or Fisher's exact test was applied to compare the distribution of selected demographic and clinical variables by recurrence and vital status. The Cox proportional hazard model was used to assess the effect of individual SNPs on overall survival and recurrence-free survival, defined as the time from the date of surgical resection to the date of death/recurrence or last follow-up. Hazard ratios (HRs) and 95% confidence intervals (CIs) were estimated by fitting the Cox model while adjusting for age, gender, race, tumor location and histology grade. Kaplan–Meier curves and log-rank tests were used to assess the differences in survival and recurrence-free survival times by different genotypes. For each SNP, three genetic models (dominant, recessive and additive) were used to assess their associations with end point events, and the model with the smallest P value was selected as the best-fitting model (17).
Results
Patient characteristics
Originally, 510 patients with CRC (271 of colon cancer and 239 of rectal cancer) were included in this analysis. All the participants were followed up until February 2010. Due to the limited sample size of patients with stage I and stage IV patients and the marked differences in outcome and end points by different stages and treatment, we focused on stage II and stage III patients receiving surgery and 5-FU-based adjuvant chemotherapy for further analyses. Patients receiving any 5-FU-containing combinatorial drug regimens were included. Table I shows the selected demographic and clinical characteristics of 285 patients who received surgery and 5-FU-based adjuvant chemotherapy. Among 115 patients with stage II disease, 27 had recurrence and 18 were dead; among 170 patients with stage III disease, 41 had recurrence and 26 were dead. The median follow-up time for stage II and III patients was 48.6 (range: 4.8–142.3) months.
Table I.
Demographic and clinical characteristics of stage II and III patients receiving 5-FU-based adjuvant chemotherapya
| Variables | Recurrence | No Recurrence | HRb (95% CI) | P value | Dead | Alive | HRb (95% CI) | P value |
| Age, mean (SD) | 58.12 (11.82) | 57.91 (13.78) | 1.00 (0.98–1.02) | 0.763 | 57.80 (12.27) | 59.52 (12.46) | 1.01 (0.99–1.03) | 0.477 |
| Gender, N (%) | ||||||||
| Male | 42 (23.46) | 137 (76.54) | 1 (reference) | 29 (16.20) | 150 (83.80) | 1 (reference) | ||
| Female | 26 (24.53) | 80 (75.47) | 1.02 (0.63–1.67) | 0.928 | 15 (14.15) | 91 (85.85) | 0.71 (0.38–1.33) | 0.282 |
| Race, N (%) | ||||||||
| Caucasian | 54 (22.50) | 186 (77.50) | 1 (reference) | 36 (15.00) | 204 (85.00) | 1 (reference) | ||
| African–American | 6 (27.27) | 16 (72.73) | 1.17 (0.51–2.73) | 0.709 | 4 (18.18) | 18 (81.82) | 1.15 (0.41–3.23) | 0.797 |
| Others | 8 (34.78) | 15 (65.22) | 1.58 (0.75–3.32) | 0.227 | 4 (17.39) | 19 (82.61) | 1.13 (0.40–3.17) | 0.821 |
| Tumor location, N (%) | ||||||||
| Distal | 12 (21.43) | 44 (78.57) | 1 (reference) | 8 (14.29) | 48 (85.71) | 1 (reference) | ||
| Proximal | 12 (20.69) | 46 (79.31) | 0.96 (0.43–2.14) | 0.923 | 8 (13.79) | 50 (86.21) | 0.91 (0.34–2.43) | 0.852 |
| Rectal | 41 (24.70) | 125 (75.30) | 1.10 (0.58–2.08) | 0.782 | 25 (15.06) | 141 (84.94) | 0.96 (0.43–2.13) | 0.913 |
| Stage, N (%) | ||||||||
| Stage II | 27 (23.48) | 88 (76.52) | 1 (reference) | 18 (15.65) | 97 (84.35) | 1 (reference) | ||
| Stage III | 41 (24.12) | 129 (75.88) | 1.13 (0.69–1.83) | 0.626 | 26 (15.29) | 144 (84.71) | 1.11 (0.61–2.03) | 0.728 |
| Histology grade, N (%) | ||||||||
| Well-differentiated | 3 (42.86) | 4 (57.14) | 1 (reference) | 2 (28.57) | 5 (71.43) | 1 (reference) | ||
| Moderate-differentiated | 55 (23.40) | 180 (76.60) | 0.40 (0.12–1.28) | 0.122 | 34 (14.47) | 201 (85.53) | 0.63 (0.15–2.69) | 0.534 |
| Poorly-differentiated | 8 (22.86) | 27 (77.14) | 0.39 (0.10–1.48) | 0.167 | 6 (17.14) | 29 (82.86) | 0.79 (0.16–4.02) | 0.778 |
The numbers in some strata may not add up to the total patient number due to missing values.
HR unadjusted.
Single SNPs associated with clinical outcomes in patients with CRC stage II and stage III
The association of each SNP with recurrence (recurrence included both local recurrence and distant metastasis) and survival was analyzed in stage II and III patients separately (Table II and Table III).
Table II.
Significant SNPs associated with recurrence in CRC stage II and III patients receiving 5-FU-based adjuvant chemotherapy
| SNP | Region (gene) | No recurrence/recurrence | HRb (95% CI) | P value | Log-rank P |
| Stage II | |||||
| rs10318 | 15q13.3 (GREM1) | ||||
| GG | 71/15 | 1 (Reference) | |||
| GA + AA | 17/12 | 2.98 (1.27–6.99) | 0.012 | 0.047 | |
| Stage III | |||||
| rs10749971 | 11q23.1 (LOC120376, FLJ45803, c11orf53, POU2AF1) | ||||
| AA | 47/19 | 1 (Reference) | |||
| AG | 56/21 | 0.73 (0.36–1.48) | 0.382 | ||
| GG | 25/1 | 0.06 (0.01–0.55) | 0.012 | 0.038 | |
| Trend | 0.46 (0.27–0.8) | 0.006 | |||
| rs961253b | 20p12.3 (BMP2) | ||||
| CC | 50/20 | 1 (Reference) | |||
| CA + AA | 79/21 | 0.46 (0.22–0.96) | 0.038 | 0.201 | |
| rs355527b | 20p12.3 (BMP2) | ||||
| GG | 57/21 | 1 (Reference) | |||
| GA + AA | 71/20 | 0.48 (0.23–0.99) | 0.048 | 0.360 |
HR adjusted by age, gender, race, tumor location and histology grade.
Linkage SNPs.
Table III.
Significant SNPs associated with overall survival in stage II and III CRC patients receiving 5-FU-based adjuvant chemotherapy
| SNP | Region (gene) | Alive/Dead | HRa (95% CI) | P value | Log-rank P |
| Stage III | |||||
| rs961253b | 20p12.3 (BMP2) | ||||
| CC | 55/15 | 1 (Reference) | |||
| CA + AA | 89/11 | 0.24 (0.09–0.68) | 0.007 | 0.054 | |
| rs355527b | 20p12.3 (BMP2) | ||||
| GG | 63/15 | 1 (Reference) | |||
| GA + AA | 80/11 | 0.29 (0.1–0.81) | 0.019 | 0.179 | |
| rs4464148 | 18q21.1 (SMAD7) | ||||
| AA + AG | 133/21 | 1 (Reference) | |||
| GG | 10/5 | 4.34 (1.46–12.89) | 0.008 | 0.020 | |
| rs6983267b | 8q24.21 (POU5F1P1, DQ515897, MYC) | ||||
| CC | 53/3 | 1 (Reference) | |||
| CA + AA | 91/23 | 4.2 (1.13–15.64) | 0.032 | 0.007 | |
| rs10505477b | 8q24.21 (POU5F1P1, DQ515897, MYC) | ||||
| AA | 51/3 | 1 (Reference) | |||
| AG + GG | 93/23 | 4.2 (1.13–15.64) | 0.032 | 0.010 |
HR adjusted by age, gender, race, tumor location and histology grade.
Linkage SNPs.
Among 26 SNPs examined, only one SNP, rs10318 (15q13.3, GREM1), was associated with recurrence in stage II patients treated with 5-FU-based adjuvant chemotherapy: the variant genotypes were significantly associated with an increased risk (HR = 2.98, 95% CI 1.27–6.99; P = 0.012) compared with the homozygous common genotype. Patients carrying the variant allele of this SNP had significantly shorter recurrence-free survival time (log-rank test P = 0.047). Three SNPs: rs10749971 (11q23.1, POU2AF1) and two linked SNPs (r 2 = 0.80) on 20p12.3 (rs961253 and rs355527, BMP2) were associated with recurrence in patients with stage III CRC receiving 5-FU-based adjuvant chemotherapy (Table II). rs10749971 was associated with a reduced risk of recurrence (HR = 0.46 in an additive model, 95% CI, 0.27–0.8; P for trend = 0.006).
For survival analyses, no SNP was significant among 115 patients with stage II disease. Among 170 patients with stage III disease, the variant allele containing genotypes of two linked SNPs on 20p12.3 (BMP2), rs961253 and rs355527, was associated with a significantly reduced risk of death with HRs of 0.24 (95% CI, 0.09–0.68; P = 0.007) and 0.29 (95% CI, 0.1–0.81; P = 0.019), respectively, consistent with the protective effect of these two variants on recurrence in stage II patients. On the other hand, the wild-type-containing genotypes of rs4464148 (SMAD7) was associated with an increased risk of death in patients with stage III disease, with HR of 4.34 (95% CI, 1.46–12.98; P = 0.008), compared with the variant containing genotype. The variant containing genotypes of two linked SNPs (r 2 = 0.94) on 8q24, rs6983267 and rs10505477, were associated with increased risks of death, with HRs of 4.2 (95% CI, 1.13–15.64; P = 0.032) and 4.2 (95% CI, 1.13–15.64; P = 0.032), respectively, compared with the homozygous wild-type genotypes.
Cumulative effects of unfavorable genotypes associated with the clinical outcomes in CRC stage III patients
Given that two SNPs on 20p12.3, rs961253 and rs355527, were in strong linkage disequilibrium (r 2 = 0.80) and the two SNPs on 8q24.21, rs6983267 and rs10505477, were also in strong linkage disequilibrium (r 2 = 0.94), we therefore only included rs961253 and rs6983267 in this cumulative analysis. Cumulative effects of unfavorable genotypes were evaluated by rs10749971 (11q23.1) and rs961253 (20p12.3, BMP2) in the recurrence of stage III disease by three SNPs, rs961253 (20p12.3, BMP2), rs4464148 (18q21.1, SMAD7) and rs6983267 (8q24.21) in the survival of stage III disease. There was a significant trend of increasing recurrence risk (P for trend = 0.004) and death risk (P for trend = 0.0006) with increasing number of putative high-risk alleles (Table IV). Kaplan–Meier analysis showed that there was a significant trend for shorter median survival time with increasing number of risk alleles for the survival in patients with stage III CRC receiving 5-FU-based adjuvant chemotherapy (log-rank test P = 0.0013; Figure 1A). The cumulative effect did not reach statistical significance for recurrence (log-rank test P = 0.279; Figure 1B).
Table IV.
Cumulative effect of unfavorable genotypes in stage III patients receiving 5-FU-based adjuvant chemotherapya
| No. of unfavorable genotypes | No. event/Event | HRb (95% CI) | P value | Log-rank P |
| Recurrence | ||||
| Low risk (0) | 50/12 | 1 (reference) | ||
| Medium risk (1) | 59/19 | 2.33 (0.97–5.62) | 0.059 | |
| High risk (2) | 19/10 | 4.67 (1.60–13.64) | 0.005 | 0.237 |
| Trend | 0.004 | |||
| Survival | ||||
| Low risk (0) | 26/1 | 1 (reference) | ||
| Medium risk (1) | 81/10 | 1.69 (0.19–14.97) | 0.640 | |
| High risk (2–3) | 36/15 | 9.84 (1.23–78.65) | 0.031 | 0.0013 |
| Trend | 0.0006 |
Unfavorable genotypes: rs10749971 (AA) and rs961253 (CC) for recurrence; rs961253 (CC), rs4464148 (GG) and rs6983267 (CA + AA) for survival.
HR adjusted by age, gender, race, tumor location and histology grade.
Fig. 1.
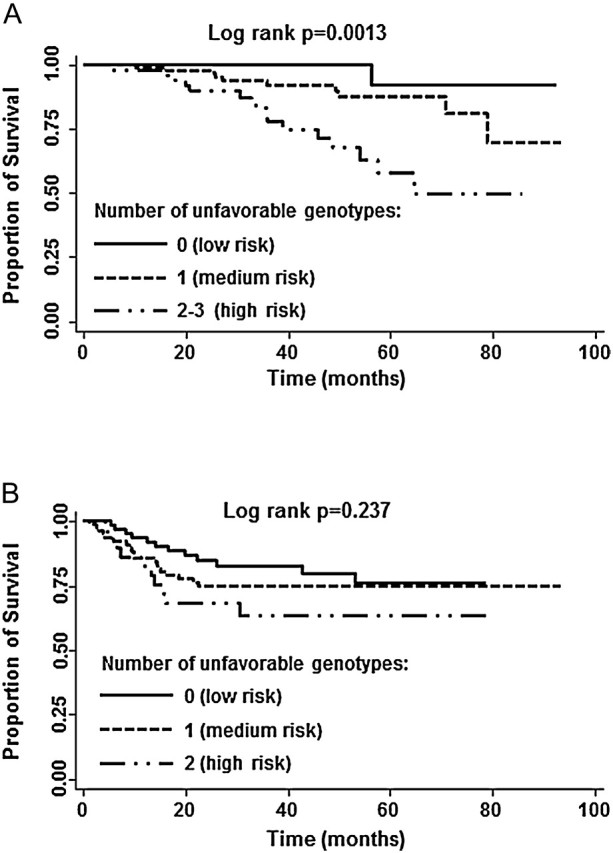
Kaplan–Meier curves of stage III CRC patients who received 5-FU-based adjuvant chemotherapy stratified by the number of unfavorable genotypes. (A) Overall survival; (B) recurrence-free survival.
Discussion
In this study, we found that GWAS-identified CRC susceptibility SNPs may individually or jointly affect clinical outcomes for CRC patients. To the best of our knowledge, this is the first study to report the association of CRC susceptibility loci with clinical outcomes.
Though advances in the treatment of metastatic CRC have evolved in the last few years, few developments have occurred in the adjuvant treatment of stage II and III patients. 5-FU-based chemotherapy is routinely used in stage III disease, but recurrence rates remained high (18). Risk stratification based on germline genotype status would provide patients’ genetic background information and help for individualized chemotherapy. In the present study, the genotypes of SNPs: rs10318 (GREM1), rs6983267 (POU5F1P1, DQ515897, MYC) and rs4464148 (SMAD7), identified subgroups of patients with poor clinical outcome in patients with stage II and stage III CRC treated with 5-FU-based adjuvant chemotherapy. Recent studies showed that SNP rs6983267 is a functional SNP on 8q24, and the variant genotype of rs6983267 confers increased CRC risk through the mechanism of Wnt signaling by disrupting an enhancer element and interacting with the promoter of MYC oncogene (19–21). MYC is an important oncogene and overexpressed in numerous tumors, including colorectal tumors (22). In line with the etiologic studies of CRC, we found that the variant allele of rs6983267 was associated with the increased risk of death in stage III CRC patients.
GREM1, BMP2 and SMAD7 are all signaling molecules in the TGF-β pathway, which plays a central but paradoxical role in the predisposition and progression of CRC (8). TGF-β suppresses the early event in CRC by inhibiting normal colonic epithelial cell proliferation and inducing apoptosis (8,23). However, many CRCs escape the tumor suppressor effects of TGF-β and are resistant to TGF-β-induced growth inhibition (24). During the late stage of CRC, TGF-β serves as a tumor promoter for the survival, invasion and metastasis of CRC cells, thereby acting as an oncogene (8). Boulay et al. (25) found that deletion of SMAD7 caused carcinoma cells more sensitive to cell growth apoptotic effect of TGF-β, whereas rescue of SMAD7 function resulted in TGF-β resistance. Moreover, SMAD7 deletion associated with a favorable outcome for CRC patients, whereas SMAD7 duplication had a hazardous effect on survival of these patients (25). In our study population, we found the homozygous variant genotype of SNP rs4464148, which located in SMAD7, was associated with a significant better survival in CRC stage III patients, as compared with the homozygous wild-type and heterozygous genotypes of rs4464148.
GREM1 encodes a secreted bone morphogenetic protein (BMP) antagonist (26,27). GREM1 plays an important role in CRC tumorigenesis through the TGF-β/BMP pathway (15). SNP rs10318 is located in the 3′-untranslated region of GREM1. Our study reported for the first time that GREM1 SNP associated with the recurrence in patients with stage II CRC. BMP2 was involved in colon cancer aggressiveness through PI3 kinase/Akt pathway (28). SNPs rs961253 and rs355527 are located in the chromosome 20p12.3, ∼500 kb proximal to BMP2. These two SNPs were associated with both recurrence and survival in stage III patients. The functional significance of these SNPs and biological mechanisms underlying their associations with CRC risk as well as outcome warrant further study.
Given the complex process of CRC tumorigenesis, it is unlikely that any single gene or SNP would have a dramatic effect on CRC clinical outcomes. Our data support the cumulative effects of two or more SNPs in modulating clinical outcomes of CRC that would otherwise be undetectable or have minimal effect in the analysis of individual SNPs.
In conclusion, this is the first study investigating the association of GWAS-identified CRC susceptibility SNPs with clinical outcomes of patients with early-stage colorectal adenocarcinoma. We have also performed cumulative analysis to establish a gene-dosage effect. Future independent validations in larger populations are necessary before these findings can be translated into clinics.
Supplementary material
Supplementary Table 1 can be found at http://carcin.oxfordjournals.org/.
Funding
This work was supported by a Multidisciplinary Research Program grant on colorectal cancer from the University of Texas MD Anderson Cancer Center. The study sponsors were not involved in the study design, in the collection, analysis and interpretation of data or in the writing of the manuscript.
Supplementary Material
Acknowledgments
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- BMP
bone morphogenetic protein
- CI
confidence interval
- CRC
colorectal cancer
- GWAS
genome-wide association studies
- HR
hazard ratio
- SNP
single nucleotide polymorphism
- TGF-β
transforming growth factor-β
- 5-FU
5-fluorouracil
References
- 1.Siegel R, et al. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J. Clin. 2011;61:212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.Andre T, et al. Oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment for colon cancer. N. Engl. J. Med. 2004;350:2343–2351. doi: 10.1056/NEJMoa032709. [DOI] [PubMed] [Google Scholar]
- 3.Andre T, et al. Improved overall survival with oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment in stage II or III colon cancer in the MOSAIC trial. J. Clin. Oncol. 2009;27:3109–3116. doi: 10.1200/JCO.2008.20.6771. [DOI] [PubMed] [Google Scholar]
- 4.Pohl A, et al. Pharmacogenomics and -genetics in colorectal cancer. Adv. Drug Deliv. Rev. 2009;61:375–380. doi: 10.1016/j.addr.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Segura C, et al. [Adjuvant treatment of colorectal cancer] Bull Cancer. 2006;93:683–690. [PubMed] [Google Scholar]
- 6.Houlston RS, et al. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat. Genet. 2008;40:1426–1435. doi: 10.1038/ng.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tenesa A, et al. New insights into the aetiology of colorectal cancer from genome-wide association studies. Nat. Rev. Genet. 2009;10:353–358. doi: 10.1038/nrg2574. [DOI] [PubMed] [Google Scholar]
- 8.Xu Y, et al. TGF-beta signaling alterations and susceptibility to colorectal cancer. Hum. Mol. Genet. 2007;16(Spec no 1):R14–R20. doi: 10.1093/hmg/ddl486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim CJ, et al. Overexpression of serine-threonine receptor kinase-associated protein in colorectal cancers. Pathol. Int. 2007;57:178–182. doi: 10.1111/j.1440-1827.2007.02078.x. [DOI] [PubMed] [Google Scholar]
- 10.Broderick P, et al. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat. Genet. 2007;39:1315–1317. doi: 10.1038/ng.2007.18. [DOI] [PubMed] [Google Scholar]
- 11.Haiman CA, et al. A common genetic risk factor for colorectal and prostate cancer. Nat. Genet. 2007;39:954–956. doi: 10.1038/ng2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomlinson I, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat. Genet. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 13.Tomlinson IP, et al. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat. Genet. 2008;40:623–630. doi: 10.1038/ng.111. [DOI] [PubMed] [Google Scholar]
- 14.Zanke BW, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat. Genet. 2007;39:989–994. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 15.Jaeger E, et al. Common genetic variants at the CRAC1 (HMPS) locus on chromosome 15q13.3 influence colorectal cancer risk. Nat. Genet. 2008;40:26–28. doi: 10.1038/ng.2007.41. [DOI] [PubMed] [Google Scholar]
- 16.Tenesa A, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat. Genet. 2008;40:631–637. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng SL, et al. Cumulative association of five genetic variants with prostate cancer. N. Engl. J. Med. 2008;358:910–919. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 18.Mano MS, et al. Colon cancer: update on adjuvant therapy. Clin. Colorectal Cancer. 2008;7:178–183. doi: 10.3816/CCC.2008.n.023. [DOI] [PubMed] [Google Scholar]
- 19.Pomerantz MM, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat. Genet. 2009;41:882–884. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tuupanen S, et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat. Genet. 2009;41:885–890. doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]
- 21.Harismendy O, et al. Elucidating the role of 8q24 in colorectal cancer. Nat. Genet. 2009;41:868–869. doi: 10.1038/ng0809-868. [DOI] [PubMed] [Google Scholar]
- 22.Hutter CM, et al. Characterization of the association between 8q24 and colon cancer: gene-environment exploration and meta-analysis. BMC Cancer. 2010;10:670. doi: 10.1186/1471-2407-10-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Engle SJ, et al. Transforming growth factor beta1 suppresses nonmetastatic colon cancer at an early stage of tumorigenesis. Cancer Res. 1999;59:3379–3386. [PubMed] [Google Scholar]
- 24.Hoosein NM, et al. Differential sensitivity of subclasses of human colon carcinoma cell lines to the growth inhibitory effects of transforming growth factor-beta 1. Exp. Cell Res. 1989;181:442–453. doi: 10.1016/0014-4827(89)90101-8. [DOI] [PubMed] [Google Scholar]
- 25.Boulay JL, et al. SMAD7 is a prognostic marker in patients with colorectal cancer. Int. J. Cancer. 2003;104:446–449. doi: 10.1002/ijc.10908. [DOI] [PubMed] [Google Scholar]
- 26.Bellam N, et al. Tgf-beta signaling alterations and colon cancer. Cancer Treat. Res. 2010;155:85–103. doi: 10.1007/978-1-4419-6033-7_5. [DOI] [PubMed] [Google Scholar]
- 27.Kupfer SS, et al. Genetic heterogeneity in colorectal cancer associations between African and European americans. Gastroenterology. 2010;139:1677–1685. doi: 10.1053/j.gastro.2010.07.038. , e1–e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang MH, et al. Inhibition of PI3 kinase/Akt pathway is required for BMP2-induced EMT and invasion. Oncol. Rep. 2009;22:525–534. doi: 10.3892/or_00000467. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


