Abstract
Ubiquitin-dependent mechanisms have emerged as essential regulatory elements controlling cellular levels of Smads and TGFβ-dependent biological outputs such as epithelial-mesenchymal transition (EMT). Here, we identify a HECT E3 ubiquitin ligase known as WWP2 (Full-length WWP2-FL), together with two WWP2 isoforms (N-terminal, WWP2-N; C-terminal WWP2-C), as novel Smad binding partners. We show that WWP2-FL interacts exclusively with Smad2, Smad3 and Smad7 in the TGFβ pathway. Interestingly, the WWP2-N isoform interacts with Smad2 and Smad3, whereas WWP2-C interacts only with Smad7. In addition, WWP2-FL and WWP2-C have a preference for Smad7 based on protein turnover and ubiquitination studies. Unexpectedly, we also find that WWP2-N, which lacks the HECT ubiquitin ligase domain, can also interact with WWP2-FL in a TGFβ-regulated manner and activate endogenous WWP2 ubiquitin ligase activity causing degradation of unstimulated Smad2 and Smad3. Consistent with our protein interaction data, overexpression and knockdown approaches reveal that WWP2 isoforms differentially modulate TGFβ-dependent transcription and EMT. Finally, we show that selective disruption of WWP2 interactions with inhibitory Smad7 can stabilise Smad7 protein levels and prevent TGFβ-induced EMT. Collectively, our data suggest that WWP2-N can stimulate WWP2-FL leading to increased activity against unstimulated Smad2 and Smad3, and that Smad7 is a preferred substrate for WWP2-FL and WWP2-C following prolonged TGFβ stimulation. Significantly, this is the first report of an inter-dependent biological role for distinct HECT E3 ubiquitin ligase isoforms, and highlights an entirely novel regulatory paradigm that selectively limits the level of inhibitory and activating Smads.
Keywords: TGFβ, Smads, transcription, Ubiquitin ligase
INTRODUCTION
The TGFβ family controls a range of normal biological activities including cell growth, migration, differentiation and apoptosis (Heldin et al, 2009). Over-activation of TGFβ signalling is also implicated in many human diseases including fibrosis, heart disease and cancer. These pathologies are increasingly being linked to the TGFβ-dependent induction of cellular trans-differentiation and EMT (Xu et al., 2009b; Zavadil et al., 2005). TGFβs signal through Ser/Thr kinase receptors that phosphorylate TGFβ/activin/BMP pathway restricted R-Smads (Smads 1, 2, 3, 5, 8). Receptors for activin/TGFβ activate Smad2 and Smad3, and receptors for BMPs activate Smad1, Smad5 and Smad8. Down-regulation of TGFβ signalling is achieved, in part, by feedback mechanisms involving the inhibitory Smad6 and Smad7. Deciphering mechanisms that fine-tune and modulate Smads is critical for understanding TGFβ biological activity and complexity, and ubiquitin-dependent mechanisms have emerged as key regulatory elements in the control of cellular levels of both TGFβ receptors and Smads (Lönn et al., 2009).
Protein ubiquitination involves a cascade of events catalysed sequentially by E1 (ubiquitin-activating), E2 (ubiquitin-conjugating), and E3 (ubiquitin ligase) enzymes. The human genome contains two E1 ligases generated by alternate splicing of a single gene, approximately thirty E2 ligases, and over six hundred E3 ligases (Li et al., 2008). E3 ubiquitin ligases are key to target substrate specificity, and these can be segregated primarily into RING type and HECT domain ligases. Within the HECT-domain E3 ligases is a small sub-group known as the NEDD4 family. Smurf1 is a NEDD4 family E3 ligase, and this was the first E3 ligase to be implicated in the TGFβ signalling pathway. Smurf1 binds and regulates levels of un-stimulated BMP-regulated Smads 1 and 5 (Zhu et al., 1999). Smurf2 binds and ubiquitinylates primarily phospho-Smads 1 and 2 (Bonni et al., 2001), but is also recruited to the type 1 TGFβ receptor via Smad7 (Kavsak et al., 2000). The NEDD4 sub-group of HECT E3 ligases comprise three functional domains: an N-terminal Ca2+/phospholipid-binding C2 domain for membrane binding, a central region containing up to four WW (double-tryptophan) domains, and a C-terminal HECT domain for ubiquitin protein ligation (Bernassola et al, 2008). The WW domains are essential for target specificity and they interact with PPXY (also known as –PY−) motifs, phospho-Ser-Pro and Pro-Arg containing sequences in target substrates (Lu et al., 1999; Pirozzi et al., 1997). WWP2, which contains four WW domains, is the least well characterised of the HECT sub-family, although it has been implicated in regulating the turnover of the epithelial sodium channel (McDonald et al, 2002), the large subunit of RNA polymerase II (Li et al., 2007), divalent metal ion transporter DMT1 (Foot et al., 2008), and the stem-cell specific transcription factor Oct4 (Xu et al., 2004; Xu et al., 2009a).
Here we identify full-length WWP2 (WWP2-FL) and two novel WWP2 isoforms, WWP2-N and WWP2-C as Smad interacting proteins that selectively bind Smad2/3 and Smad7 and regulate TGFβ-induced biological responses. Mechanistically, we show that expression of WWP2-N can modulate TGFβ signalling activity by interacting with WWP2-FL and activating it’s intrinsic ubiquitin ligase activity. Collectively, our data indicate that the regulation of WWP2 isoform expression could provide a novel switch mechanism controlling cell context-dependent and disease-specific TGFβ signalling activity.
RESULTS
WWP2 interacts with Smad2, Smad3, and Smad7
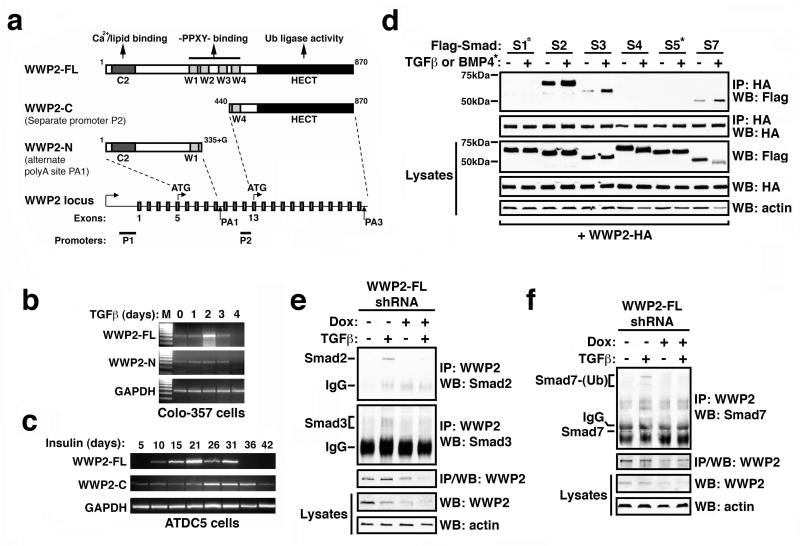
The WWP2 HECT E3 ubiquitin ligase emerged as a candidate Smad3 binding partner in a previous yeast two-hybrid screen of a mouse brain cDNA expression library using the MH1 and linker regions of Smad3 as bait (Wicks et al., 2005). Scrutiny of Mammalian Gene Collection (MGC) databases containing sequence-validated full-length protein-coding cDNA clones identified a unique and interesting feature of WWP2 in that there are three WWP2 isoforms arising from the same gene locus: full-length WWP2 (WWP2-FL, 870aa), an N-terminal isoform (WWP2-N, 336aa) presumably generated by failure to splice-out intron 9-10, and a C-terminal isoform (WWP2-C, 440aa) that is likely to be generated from a second promoter within intron 10-11 (Figure 1a). To independently confirm expression of these distinct and novel WWP2 isoforms we first used isoform-specific primers and RT-PCR to detect transcripts in Colo-357 pancreatic carcinoma cells that are know to be TGFβ responsive (Levy and Hill, 2005). Interestingly, expression of FL-WWP2 appears to be induced by TGFβ, peaking after 2 days stimulation and is absent after 4 days (Figure 1b). In contrast, WWP2-N expression does not change following TGFβ treatment, but does also disappear after 4 days (Figure 1b). Thus far, we have not been able to clearly detect the WWP2-C isoform in Colo-357 cells under the same experimental conditions, although we have seen robust expression in cartilage-derived ATDC5 cells that differentiate following prolonged exposure to insulin (Figure 1c). Therefore, these data demonstrate that all three WWP2 isoforms exist, and also suggest that they may be regulated independently in a cell-context-dependent manner.
Figure 1. Interaction of WWP2 with Smads.
(a) Schematic representation of the WWP2 gene locus and three WWP2 isoforms: full-length WWP2 (WWP2-FL, 870aa, Genbank accession no. NM_007014), an N-terminal isoform (WWP2-N, 336aa, Genbank accession no. NM_199423) presumably generated by failure to splice-out intron 9-10, and a C-terminal isoform (WWP2-C, 440aa, Genbank accession no. NM_199424) that is likely to be generated from a second internal promoter P2 within intron 10-11. (b) RNA isolated from Colo-357 cells stimulated with TGFβ for the indicated number of days, was isolated and reverse transcribed. 1.25 ng cDNA were utilised as template DNA in RT-PCR using primers specific for WWP2-FL, WWP2-N and GAPDH. Equal volumes of the PCR products were analysed by agarose gel electrophoresis. (c) RNA isolated from ATDC5 cells and stimulated with insulin for the indicated number of days, was isolated and reverse transcribed (as described in Atsumi et al, 1990). 1.25ng cDNA were utilised as template DNA in RT-PCR using primers specific for WWP2-FL, WWP2-C and GAPDH. Equal volumes of the PCR products were analysed by agarose gel electrophoresis. (d) HEK-293 cells were co-transfected with plasmids encoding Flag–tagged Smads 1-7 and HA-tagged WWP2. 48hr after transfection, the cells were treated with MG132 for 5hrs followed by human TGFβ (5ng/ml) or BMP-4 (10ng/ml) for a further 1hr. Cells were lysed and subjected to immunoprecipitation (IP) with an anti-HA antibody and then analysed by Western blotting (WB) with an anti-Flag antibody. Whole cell lysates were also analysed for positive expression of epitope-tagged proteins and β-actin as indicated (lower panels). (e) 107 confluent Colo-357 cells (WWP2-FL shRNA knockdown) were pre-treated with MG132 for 5hrs (+/− Doxycycline), and stimulated with 5ng/ml TGFβ for 1hr. Cleared lysates were immuno-precipitated with 3μg anti-WWP2 overnight, and analysed for the presence of Smad2 and Smad3. (f) As described in (e) above, except that cells were pre-treated for 18hrs with MG132 and stimulated for 48hr with 5ng/ml TGFβ. Anti-WWP2 immune-complexes were washed and analysed by Western blotting for Smad7. WWP2 and actin controls blots are also included.
To further characterise and confirm interactions between Smads and FL-WWP2 identified during the yeast two-hybrid screen, we then used co-expression of epitope-tagged proteins and co-immunoprecipitation experiments. Immunoprecipitation of WWP2 from cell lysates revealed strong binding of WWP2-FL to Smad2, Smad3 and Smad7, but not to BMP-regulated Smad1 and Smad5 (Figure 1d). Flag-WWP2-FL interactions with HA-Smad2 and HA-Smad3, and to some extent HA-Smad7, were also enhanced when cells were TGFβ stimulated (Figure 1d). Next, we examined whether WWP2 interacts with Smads endogenously at physiological levels of expression. In anti-WWP2 immunoprecipitates of lysates from Colo-357 cells, we observed both Smad2 and Smad3 co-precipitating with WWP2 in a TGFβ-dependent manner (Figure 1e). Specificity of these interactions was confirmed using stable and inducible shRNA-mediated knockdown of WWP2-FL in which Smad2 and Smad3 are both absent in WWP2 immunoprecipitated complexes (Figure 1e). Smad7 also bound to WWP2 endogenously although there are higher molecular weight Smad7 reactive bands following TGFβ stimulation that are not seen in WWP2-FL knockdown cells (Figure 1f). This may reflect low and undetectable levels of Smad7 due to enhanced Smad7 turnover following exposure to TGFβ, or that endogenous Smad7 bound to WWP2 is predominantly polyubiquitinated.
WWP2 isoforms bind differentially to R-Smads and I-Smads
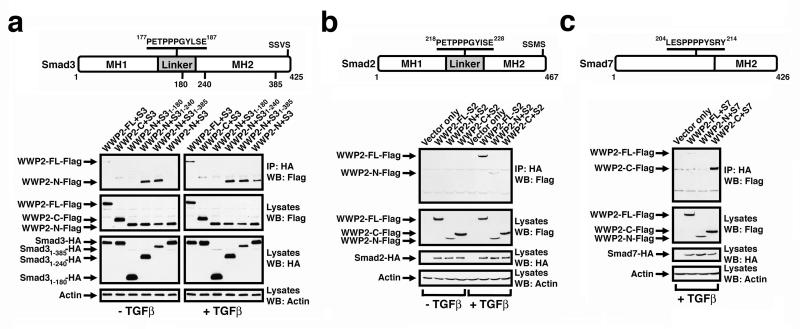
Since individual N- and C-terminal WWP2 isoforms contain separate and distinct WW domains, we next examined whether these domains conferred any differential binding specificity for individual Smad proteins. We co-expressed Flag-WWP2-FL, Flag-WW2-N or Flag-WWP2-C together with HA-Smad3, immunoprecipitated cell lysates with HA antibody and then probed Western blots with FLAG antibody. Interestingly, WWP2-N as well as WWP2-FL, but not WWP2-C, interact with Smad3 in a TGFβ-dependent manner (Figure 2a). Since WWP2-N contains the C2 domain and only a single WW domain (WW1), we surmise that WW1 in WWP2-N harbours the Smad3 interaction motif. We then used a series of Smad3 deletion constructs to map the region in Smad3 that binds the WW1 domain of WWP2-N. Truncated HA-Smad3 mutants comprising either residue 1-395, 1-240 or 1-180 were co-expressed in HEK-293 cells with Flag-WWP2-N, cell lysates immunoprecipitated with HA antibody and Western blots probed with Flag antibody. Smad31-395 and Smad31-240 bound efficiently to WWP2-N but not Smad31-180 (Figure 2a). The site in Smad3 that binds to WWP2-N is therefore within residues 180-240 which includes the WW-binding -PPGY- motif in the linker region. Interestingly, Smad3 C-terminal deletion proteins Smad31-395 and Smad31-240 interact independently of TGFβ stimulation. It is likely that this is due to removal of the receptor phosphorylation sites at the C-terminal -SSVS- motif relieving auto-inhibitory MH1/MH2 domain interactions that normally repress Smad3 function (Hata et al., 1997), and this supports our finding that the interactions between WWP2 isoforms and Smad3 is TGFβ-dependent. We next examined WWP2 isoform interactions with Smad2 and Smad7 using the same co-immunoprecipitation approach. Smad2 bound to WWP2-FL and WWP2-N, but not WWP2-C (Figure 2b). However, it is WWP2-C together with WWP2-FL, but not WWP2-N, which interact with Smad7 in a TGFβ-dependent manner (Figure 2c). Smad7 GST pull-down studies confirmed that WW4 domain in WWP2 interacts with the -PPXY- motif in Smad7 (Supplementary Figure S1). Therefore, it is the WW4 domain of WWP2-C that interacts with Smad7, and this difference in WW domain binding specificity may be due to the different composition of the PPXY motif sequence in Smad7 relative to Smad2 and Smad3.
Figure 2. Selective interaction of R-Smads and I-Smads with individual WWP2 isoforms.
HEK-293 cells were transfected with WWP2-Flag isoforms together with: (a) HA-tagged Smad3, or Smad3 deletion constructs, as indicated; (b) HA-tagged Smad2; (c) HA-tagged Smad7. In all cases, cells were pre-treated with MG132 for 5hrs, and then treated +/−5ng/ml TGFβ for 1hr and immunopreciptated with anti-HA. Samples were Western blotted against anti-Flag, anti-HA, or anti-actin as a loading control.
WWP2 promotes Smad protein turnover and ubiquitination
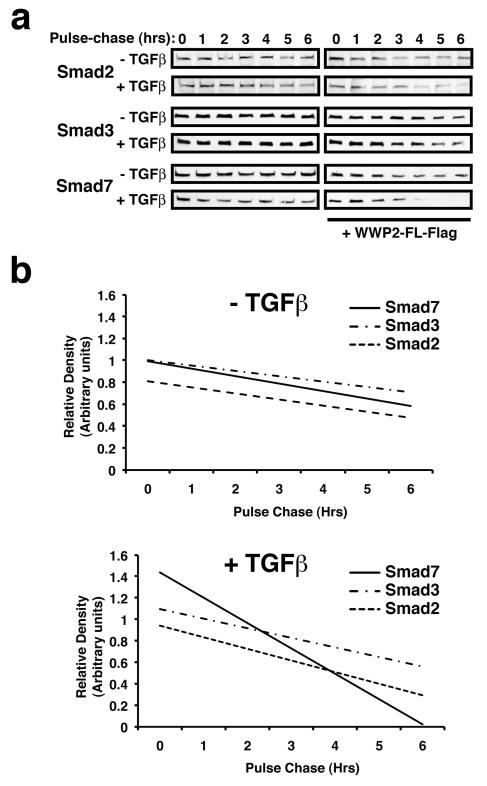
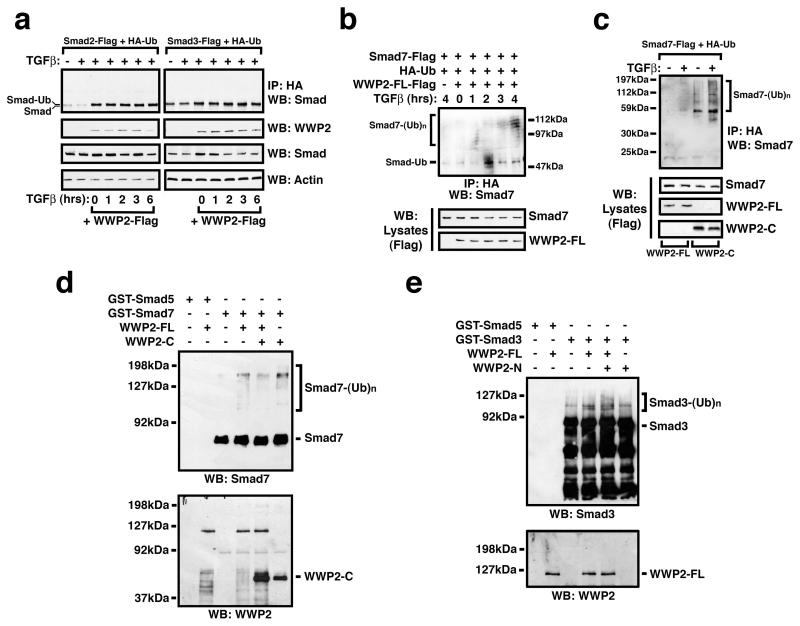
WWP2 is an E3 ubiquitin ligase that is able to ubiquitinate target substrates and can accelerate proteosomal-dependent protein turnover. To test whether Smad turnover rates are affected by WWP2, we performed cycloheximde chase experiments in HEK-293 cells expressing Flag-tagged Smads. Expression of WWP2 significantly enhanced turnover of Smad2, Smad3 and Smad7 proteins in a TGFβ-dependent manner (Figure 3a). The acceleration of Smad7 turnover was more pronounced than both Smad2 and Smad3, and following 6 hours cycloheximide chase Smad7 was undetectable (Figure 3b). We then tested whether increased levels of ubiquitination could explain the enhanced turnover of Smads in the presence of WWP2. HEK-293 cells were transfected with HA-ubiquitin, Flag-tagged WWP2 and Flag-tagged Smads 2,3, or 7, and cells were stimulated with TGFβ for up to 6 hours followed by anti-HA immunoprecipitation and anti-Smad immunoblotting. Co-expression with WWP2 followed by TGFβ stimulation clearly enhances the formation of Smad2- and Smad3-ubiquitinated complexes (Figure 4a). Interestingly, Smads 2 and 3 both appear to primarily undergo mono-ubiquitination within 6 hours of TGFβ stimulation, whereas we find typical high molecular weight laddering and smears with Smad7 within 4 hours suggesting increased polyubiquitination (Figure 4b). Although we have focussed on a timeframe for TGFβ stimulation that correlates with our protein turnover experiments, we do also find Smad2 and Smad3 polyubiquitination following prolonged stimulation (>16hours) with TGFβ (Supplementary Figure S2). Therefore, it is likely that mono-ubiquitinated Smad2 and Smad3 represent transient intermediates, and that more pronounced Smad7 polyubiquitination by WWP2 does indeed reflect preferred substrate specificity. We next examined whether Smad7 ubiquitination could also be induced by the WWP2-C isoform since WWP2-C interacts exclusively with Smad7 and also contains the E3 ubiquitin ligase catalytic domain. Interestingly, we found that WWP2-C can efficiently induce TGFβ-dependent Smad7 polyubiquitination, and this modification is more pronounced when compared alongside WWP2-FL in the same experiments (Figure 4c). To confirm our findings in transfected cells, we next used in vitro ubiquitination experiments with bacterially expressed WWP2 isoforms and GST-Smad fusion proteins to examine WWP2-FL ubiquitinylating activity against Smad substrates. Smad7 undergoes pronounced poly-ubiquitination in the presence of WWP2-FL and more so with WWP2-C (Figure 4d). Smad3-GST undergoes moderate ubiquitination in the presence of WWP2-FL and, interestingly, in these in vitro studies this is enhanced in the presence of the WWP2-N isoform that interacts selectively with R-Smads (Figure 4e).
Figure 3. WWP2 promotes Smad protein turnover.
(a) HEK293 cells transfected with 1μg Smad2, Smad3, or Smad7, were pre-treated with MG132 for 5 hrs. Cells washed in fresh growth medium and incubated in the absence of MG132 and in the presence of cycloheximide (20μg/ml) and TGFβ stimulation for the time-points indicated. Whole cell lysates were assessed for Smad expression levels. (b) Smad-specific Western blots were quantified by densitometric scanning and values were standardized for β-actin expression. Values were then standardized for WWP expression and Time=0 hr (which was set to 1), and graphs (−/+ TGFβ) were plotted using Excel and optimum best-fit trendlines determined.
Figure 4. WWP2 isoforms mediate Smad ubiquitination in vivo and in vitro.
HEK293 cells were transfected with tagged-Smad2 or Smad3 (a) or Smad7 (b), 100ng HA-Ubiquitin and 100ng WWP2-FL as indicated. Cells were pre-treated with MG132 for 5 hrs and then stimulated with TGFβ as indicated, followed by anti-HA immunoprecipitation and analysis of ubiquitination species by anti-Smad Western blot analysis. (c) HEK293 cells, transfected with 1μg Smad7-Flag, 100ng HA-Ubiquitin and either 100ng WWP2-FL or WWP2-C encoding plasmids (for 48 hours), were pre-treated with MG132 for 5hrs and then stimulated with TGFβ for 3 hours followed by anti-HA immunoprecipitation and analysis of Smad7-ubiquitination by Smad7 Western blotting. Included are whole cell lysate (WCL) control blots for WWP2 isoforms and Smad 7. In vitro ubiquitination assays were performed using GST-Smad7 (d) and GST-Smad3 (e) as substrates in the presence of purified WWP2 isoforms (as indicated). All reactions were performed identically at 30°C for 120 min, prior to being analysed by Western blotting using Smad specific antibodies. GST-Smad5 was used as a negative control.
WWP2-N directly modulates WWP2-FL and induces breakdown of unstimulated R-Smads
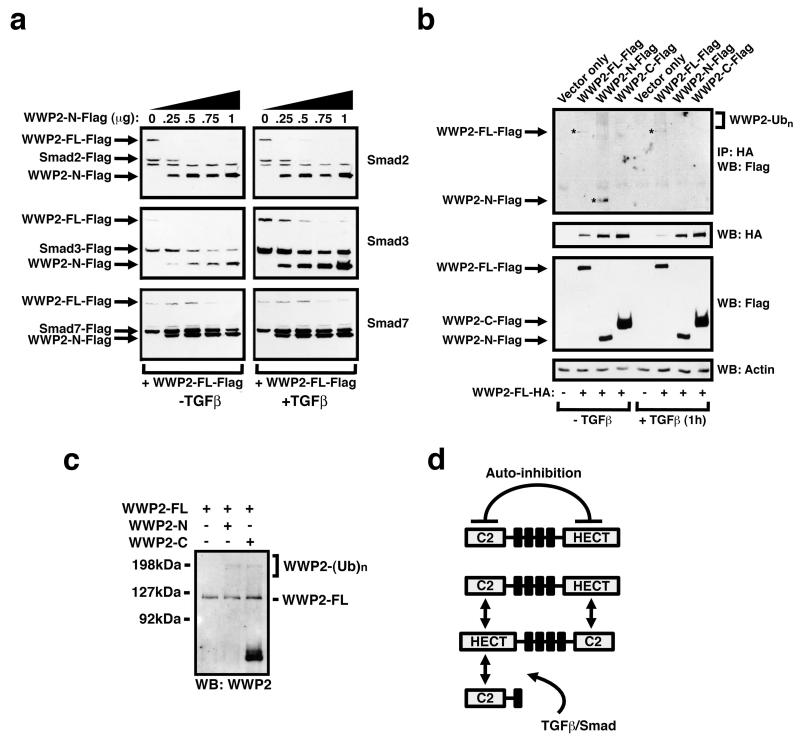
Since the WWP2-N isoform, which lacks the functional HECT E3 ligase domain, influenced the E3 ubiquitin ligase activity of WWP2-FL towards Smad3 in vitro, we next examined whether WWP2-N expression could influence Smad steady-state levels in the presence of WWP2-FL. Interestingly, dose-dependent expression of WWP2-N can potently reduce Smad2/3 steady-state levels in the presence of WWP2-FL, and independently of TGFβ stimulation (Figure 5a), and cycloheximide chase experiments confirm these findings (Supplementary Figure S3). In contrast, increased expression of WWP2-N has no effect on Smad7 levels (Figure 5a). Recent observations suggest that the C-terminal and N-terminal domains of related HECT ligases can form intramolecular auto-inhibitory links (Mund and Pelham, 2009; Wiesner et al., 2007). We therefore postulated that truncated WWP2 isoforms might associate with WWP2-FL, and could potentially interfere with intramolecular auto-inhibitory interactions. To test this possibility, we overexpressed Flag-tagged WWP2-FL with HA-tagged isoforms, and used co-immunoprecipitation to identify direct interactions. In the absence of TGFβ, WWP2-FL clearly associates with itself and with WWP2-N, and these interactions are disrupted following 1hour treatment of cells with TGFβ (Figure 5b). The appearance of high molecular weight species also suggests that WWP2-N might regulate WWP2-FL auto-ubiquitination (Figure 5b), a modification in WWP2 that is associated with ligase domain auto-activation (Mund and Pelham, 2009). To address whether the reduced levels of Smads in the presence of WWP2-N might reflect enhanced WWP2-FL auto-activation, we examined WWP2-FL auto-ubiquitination using in vitro ubiquitination experiments and find that WWP2-N, as well as WWP2-C, can promote WWP2-FL auto-ubiquitination (Figure 5c). We then assessed whether the reduced levels of Smads in the presence of WWP2-N might reflect enhanced WWP2-FL auto-activation in vivo, and expressed WWP2-FL together with WWP2-N in the presence of HA-tagged ubiquitin. Immunoprecipitation of HA-ubiquitin modified Smad2 and Smad3 demonstrates that expression of WWP2-N can increase the level of Smad ubiquination by endogenous WWP2 (Supplementary Figure S4). Enhanced WWP2-FL activity is concomitant with increased protein turnover seen in cycloheximide chase experiments (Supplementary Figure S5), and this is likely to reflect increased WWP2-N-mediated WWP2-FL auto-ubiquitination. Collectively, these data support the notion that WWP2-FL interacts with WWP2-N to induce WWP2-FL auto-activation, and that this complex is released following TGFβ stimulation (Figure 5d), presumably due to TGFβ-mediated R-Smad phosphorylation and preferential binding of phospho-R-Smads with WWP2.
Figure 5. WWP2-N inhibits TGFβ signalling by directly modulating endogenous WWP2-FL activity and enhancing Smad2/3 turnover.
(a) HEK-293 cells were co-transfected with 1μg of Smad expression plasmid and 0.25μg of WWP2-FL-Flag expression plasmid with the indicated increasing amounts of WWP2-N-Flag expression plasmid (DNA amounts were kept equivalent using empty vector pRK5). After 48 hrs, cells were harvested and analysed for Smad-Flag, FL-WWP2-Flag and WWP2-N-Flag expression by anti-Flag Western blotting. (b) HEK-293 cells were transfected with 1μg of WWP2-FL-HA expression construct (or empty vector control) together with 1μg of WWP2-FL-Flag, WWP2-N-Flag or WWP2-C-Flag. After 48hrs, cells were pre-treated with MG132 for 5hrs, and then stimulated (+/− 5ng/ml TGFβ for 1hr). Lysates were immunoprecipated with anti-HA antibody and Western blots probed with anti-Flag antibody. (c) In vitro ubiquitination assays were performed using bacterially purified GST-WWP2-FL in the presence of purified WWP2 isoforms. Reactions were performed identically at 30°C for 120 min, prior to being analysed by Western blotting using WWP2 specific antibodies. (d) Schematic representation indicating that WWP2-FL interacts with WWP2-N to relieve auto-inhibition in the absence of TGFβ, and that this complex is released following TGFβ stimulation.
WWP2 isoforms differentially modulate TGFβ-dependent transcription and EMT
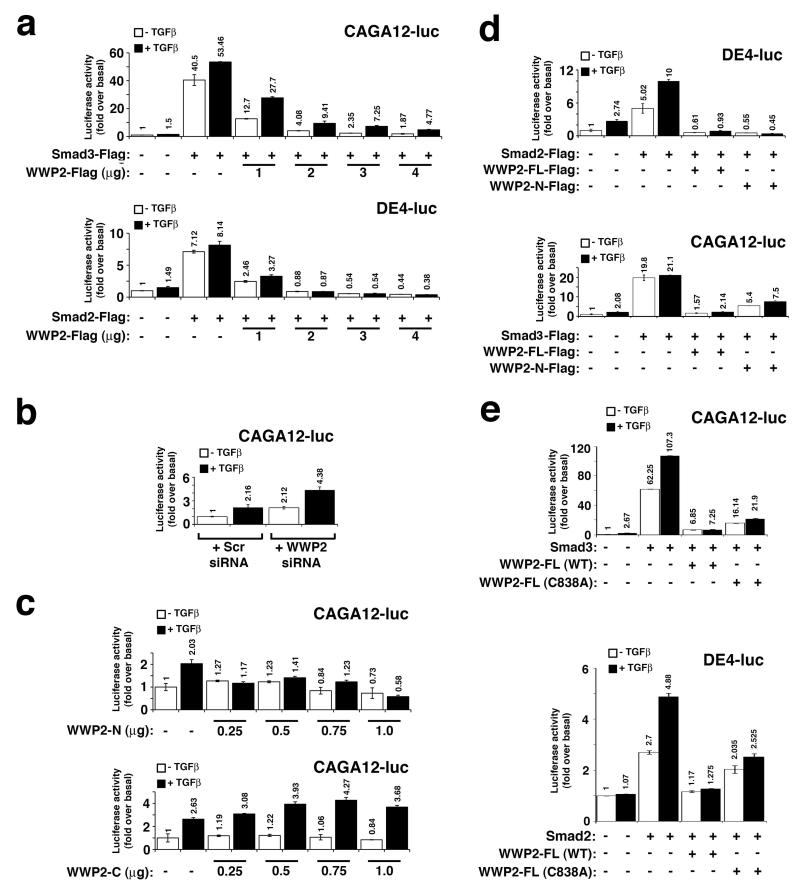
To characterise the effect of WWP2 isoforms on TGFβ-dependent biological responses, we first used CAGA12-luc and DE-luc luciferase reporter plasmids to measure effects on Smad3- and Smad2-dependent gene expression, respectively. In both instances, there is a significant reduction in TGFβ-dependent gene expression in cells overexpressing WWP2-FL in a dose-dependent manner (Figure 6a). Furthermore, ablation of endogenous WWP2 by transfection of cells with siRNA targeting WWP2 enhances TGFβ-dependent CAGA12-luc (Smad3) and DE4-luc (Smad2) activity, supporting the notion that WWP2 isoforms can modulate Smad transcriptional responses (Figure 6b). However, since the WWP2 siRNA could potentially affect endogenous levels of all three WWP2 isoforms, we cannot rule out differential effects on TGFβ-dependent gene expression by individual WWP isoforms. Therefore, we next expressed WWP2-C or WWP2-N isoforms and examined their effect on endogenous Smad3-dependent transcription using the CAGA12-luc reporter. Interestingly, over-expression of WWP2-N decreased CAGA12-luc activity following TGFβ stimulation for 6 hours in contrast to WWP2-C that showed an opposite response (Figure 6c). Moreover, expressed WWP2-N showed similar effects on Smad-driven DE4-luc and CAGA12-luc activity following Smad2 and Smad3 overexpression, respectively (Figure 6d). However, we cannot exclude effects of the truncated WWP2 isoforms acting independently of endogenous WWP2-FL, although this seems unlikely since in vitro ubiquitination erxperiments do indeed support the idea that WWP2-N functions by activating WWP2-FL (see Figure 4). To test whether the ubiquitin ligase activity of WWP2 is required for its effect on Smad-mediated transcription we next used a catalytically inactive Cys-838-Ala WWP2 mutant. We find partial reduction in the inhibitory effect on Smad2 and Smad3 gene reporter activity with this mutant compared to wild-type WWP2 (Figure 6e). This response may be due to activation of endogenous WWP2-FL wild-type enzyme by the overexpressed WWP2-mutant, in keeping with our observation that WWP2 dimerisation is associated with WWP2 auto-activation (see Figure 5).
Figure 6. Smad-dependent transcription is modulated by WWP2 isoforms.
(a) HEK293 cells transfected with 0.5μg WWP2-FL, were assayed for their ability to activate the Smad3 and Smad2-specific gene reporter constructs in the absence (white bars) and presence (black bars) of TGFβ stimulation (6hr). (b) HEK-293 cells were transfected with WWP-2 siRNA or scrambled siRNA and assayed for regulation of Smad3-mediated gene reporter regulation in the absence (white bars) and the presence (black bars) of TGFβ stimulation. HEK293 cells were transfected with CAGA12-luc or DE4-luc, and WWP2-N or WWP2-C as indicated (Panel c, endogenous Smads; Panel d, Smad overexpression), and treated with TGFβ for 6hrs. (e) HEK293 cells were transfected with Smad2-Flag, Smad3-Flag, CAGA12-luc or DE4-luc, and WWP2-FL or WWP2-FL-C838A as indicated. In all experiments, relative luciferase activity is expressed as fold over basal and data are representative of three independent experiments.
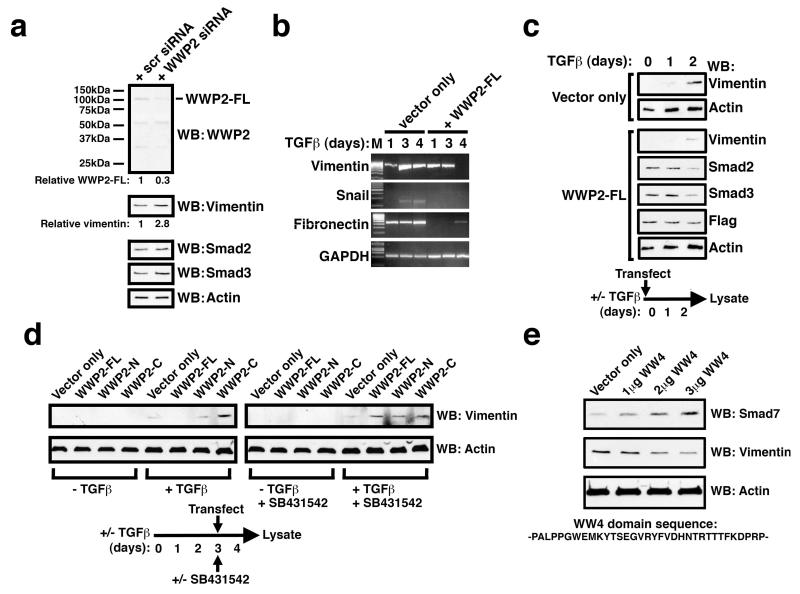
To examine further the biological role of WWP2 isoforms in the TGFβ pathway we next assessed the effects of WWP2 isoform expression on TGFβ-driven epithelial-to-mesenchymal transition (EMT). EMT represents an important differentiation programme that converts static epithelial cells into highly motile and invasive mesenchymal cells, a process that has been linked to cancer cell proliferation, dispersion and metastasis. We used the Colo-357 pancreatic carcinoma cell line that is known to undergo full EMT upon TGFβ stimulation (Ellenreider et al., 2001; Levy and Hill, 2005; see also Supplementary Figure 6). Firstly, we found that siRNA-mediated knockdown of WWP2 in Colo-357 cells treated with TGFβ enhances the expression of Vimentin by approximately 3-fold based on Western blotting of cell lysates and causes a moderate increase in Smad 2/3 levels (Figure 7a). We then analysed effects of WWP2-FL expression in Colo-357 cells on the TGFβ-dependent induction of EMT markers by RT-PCR. To allow for gene-specific temporal differences in expression patterns during EMT, cells were next pre-treated with TGFβ for 1, 3, or 4 days prior to transfection with WWP2-FL. The TGFβ-dependent induction of Vimentin, Snail and Fibronectin were significantly inhibited in the presence of WWP2-FL, although these effects were more pronounced following 4 days of pre-treatment with TGFβ (Figure 7b). This inhibition of TGFβ-induced Vimentin expression in the presence of expressed WWP2-FL was confirmed by Western blot analysis and correlates with a slight reduction in Smad 2/3 levels (Figure 7c). We then examined whether WWP2-FL expression 3 days after prior TGFβ stimulation could affect a TGFβ-mediated EMT program that is already underway, and at the same time also looked at the effect of WWP2-N and WWP2-C expression. Interestingly WWP2-FL, but not WWP2-N, can effectively block TGFβ-induced Vimentin expression whereas WWP2-C sustains moderately high Vimentin levels (Figure 7d). Interestingly, and in complete contrast to WWP2-FL expression, treating cells with a small molecule TGFβ receptor kinase inhibitor (SB431542) is not able to reverse a TGFβ-mediated EMT program that is already in progress (Figure 7d). Pre-treatment of cells with SB431542 also prevents any WWP2 isoform-mediated effects on Vimentin expression, presumably due to the TGFβ-dependent requirement for WWP2 isoform association with Smads.
Figure 7. WWP2 isoforms differentially affect TGFβ-induced EMT.
(a) Colo-357 cells were transfected with WWP-2 siRNA or scrambled siRNA, treated with TGFβ for three days and cell lysates Western blotted against WWP2, anti-Vimentin, anti-Smad2, anti-Smad3 or anti-actin as indicated. The relative level of WWP2-FL and Vimentin under each experimental condition following WWP2 siRNA treatment was assessed by densitometric scanning. (b) Colo-357 cells were pre-treated with TGFβ for 1, 3, or 4 days as indicated and then transfected with 0.5μg WWP2-FL. RNA was isolated after a further 48hours, RT-PCR carried out using primers specific for Vimentin, Snail, fibronectin and GAPDH, and samples analysed by agarose gel electrophoresis. (c) Colo-357 cells transfected on Day 0 with pRK5 (vector only), WWP2-FL, or WWP2-N were treated under the following conditions: Unstimulated (−TGFβ) or TGFβ stimulated (+TGFβ). Cells were lysed on Day 4 and analysed for Vimentin, Smad2 and Smad3 expression using Western blotting analysis on the same gels (WB). The blots were also probed with β-actin to determine equal loading of lysate samples, and anti-Flag to detect isoform expression. (d) Colo-357 cells transfected on Day 3 with pRK5 (vector only), WWP2-FL, WWP2-N or WWP2-C were treated under the following conditions: Unstimulated (−TGFβ), TGFβ stimulated (+TGFβ) or TGFβ stimulated in the presence of a TGFβ receptor kinase inhibitor (+TGFβ+SB431542). Cells were lysed on Day 4 and analysed for Vimentin and β-actin expression using Western blotting analysis (WB). (e) Colo-357 cells were transfected with the indicated doses of expression plasmids encoding Flag-tagged WW4 domain, 24 hours after TGFβ stimulation. Cells were TGFβ stimulated for a further 48hrs post-transfection, harvested and whole cell lysates assessed for Smad7, vimentin and β-actin expression by Western blotting.
Since WWP2-C contains a single WW4 domain that mediates interactions between WWP2-C (or WWP2-FL) with inhibitory Smad7, we next examined whether TGFβ-induced EMT could be moderated by expression of an individual WW4 domain. We therefore expressed an isolated WWP2 WW4 domain tagged with Flag in a dose-dependent manner 24 hours after prior stimulation of Colo-357 cells with TGFβ, and 48 hours later examined expression levels of both Vimentin and Smad7. We find a clear dose-dependent increase in the levels of Smad7 in the presence of WW4 and a concomitant reduction in Vimentin (Figure 7e).
DISCUSSION
We have identified the WWP2 ubiquitin ligase as a novel Smad binding partner that selectively interacts with and ubiquitinates Smads 2, 3, and 7 in the TGFβ pathway. Furthermore, we have characterised two additional WWP2 isoforms generated from the WWP2 gene locus, WWP2-N and WWP2-C, and shown isoform-specific interactions with Smad2/3 (WWP2-N) and Smad7 (WWP2-C). Intriguingly, WWP2-N also interacts with WWP2-FL, and this complex is rapidly dissociated after TGFβ stimulation. Taken together, our data suggest that WWP2-N can stimulate WWP2-FL auto-activation and auto-ubiquitination in the absence of TGFβ leading to increased activity against target substrates Smad2 and Smad3 (but not Smad7), and that Smad7 is a preferred substrate for WWP2-FL and WWP2-C following TGFβ stimulation.
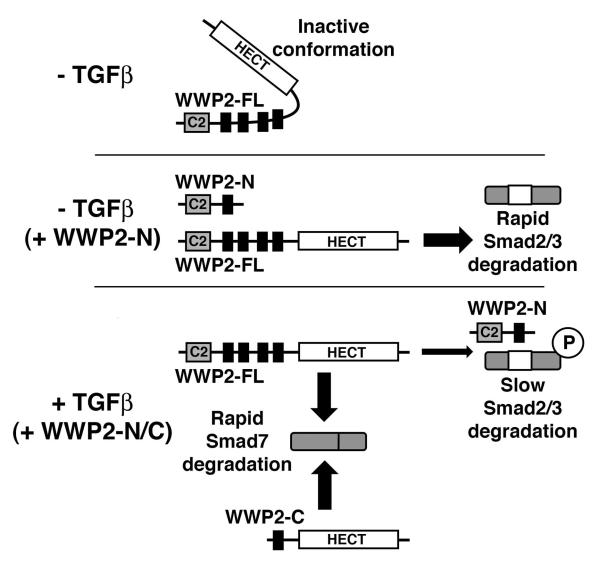
To our knowledge, this is the first report of an inter-dependent biological role for distinct E3 ligase isoforms, and demonstrates an entirely novel regulatory mechanism that selectively fine-tunes levels of both activating and inhibitory Smads. Based on our analysis, we propose a model in which expression of individual WWP2 isoforms could serve to drive tissue- and disease-specific changes in TGFβ signalling dynamics and biological responses such as EMT (Figure 8). Firstly, our data suggests that WWP2-N expression will deplete cellular levels of unstimulated Smads 2/3 presumably due to WWP2-N-dependent disruption of WWP2-FL auto-inhibitory interactions, and subsequent breakdown of these receptor-regulated Smads. Then, following TGFβ stimulation, WWP2-N is released from WWP2-FL to interact preferentially with Smads 2/3. Biologically, we predict that this will stabilise the early phase of pathway activation by WWP2-N competing for WWP2-FL:R-Smad interactions, in a process that would be further encouraged by the WWP2-FL-dependent breakdown of inhibitory Smad7 which we find to be a preferred substrate for WWP2. In the context of a prolonged TGFβ-induced EMT differentiation programme over several days, and the apparent disappearance of WWP2-N expression after 4 days of TGFβ exposure (see Figure 1b), the disruption of Smads2/3 interactions with WWP2-FL by WWP2-N would no longer occur, and this would then further stimulate Smad2/3 breakdown as the EMT differentiation programme comes into completion. Finally, expression of WWP2-C could be responsible for localised temporal activation of the TGFβ pathway due to efficient and selective breakdown of inhibitory Smad7.
Figure 8. A model of WWP2 isoform action on TGFβ signalling activity and EMT.
In the absence of TGFβ, WWP2-N expression causes rapid breakdown of unstimulated Smads 2/3 due to WWP2-N-dependent disruption of WWP2-FL auto-inhibitory interactions. Following TGFβ stimulation, WWP2-N is released from WWP2-FL to interact preferentially with Smads 2/3, initially slowing down R-Smad turnover. In the context of a prolonged TGFβ-induced EMT differentiation programme over several days, and the apparent disappearance of WWP2-N expression after 4 days of TGFβ exposure (see Fig 1b), the blockade of Smads2/3 interactions with WWP2-FL by WWP2-N would no longer occur, and this would then further stimulate Smad2/3 breakdown as the EMT differentiation programme comes to completion. Finally, expression of the WWP2-C isoform could be responsible for localised temporal activation of the TGFβ pathway due to efficient and selective breakdown of inhibitory Smad7.
The interconnections between Smad stability and individual WWP2 isoforms reported here could also provide a novel cross-talk mechanism linked to other signalling pathways that might control WWP2 isoform-specific expression. It is likely that WWP2-N is produced by an alternate splicing event that retains intron 9 in the mature transcript, enabling the use of an alternate polyadenylation site and premature stop codon. Regulation of alternate splicing/polyadenylation is a complex and poorly understood biological phenomenon that involves cell/tissue-specific expression of mRNA binding proteins and splicing factors (Lutz, 2008; Nilson and Graveley, 2010). These events are often controlled by extracellular stimuli and growth factor input, and abnormal splice factor activity is often the hallmark of disease states Danckwardt et al., 2008; Tazi et al., 2009). It is intriguing that the expression of WWP2-N disappears towards the final stages of TGFβ-induced EMT suggesting the existence of mechanisms that tightly control WWP2-N expression, at least during the progression and execution of this cellular differentiation programme. Although the generation of the WWP2-N transcript can be explained by alternative splicing events, the presence of a C-terminal isoform downstream of a TATA sequence and a putative start ATG codon suggests that like other proteins such as transcription factors p53/p73 (Murray-Zmijewski et al., 2006), WWP2-C isoform may arise due to the presence of intronic promoter activity. Other NEDD4 isoforms, variants of WWP1 and Nedd4-2 lacking the N-terminal regions have been described previously (Flasza et al., 2002; Raikwar et al., 2008), although these have not been ascribed biological functions. At present, the regulation of WWP2-C expression remains unclear and is the focus of on-going studies, although abundant expression has been detected in chondrocyte cell lines.
Elevated TGFβ signalling activity is associated with the pathobiology of many human diseases including fibrosis, heart disease and cancer metastasis. The main findings in this study highlight a key role for WWP2 in TGFβ-dependent EMT, and information reported regarding WWP2 isoform-specific interactions with activating Smads2/3 and inhibitory Smad7 could provide a novel approach to selectively moderate levels of R-Smads and I-Smads. Significantly, we have shown that over-expression of an isolated WWP2-WW4 domain can increase cellular levels of I-Smad7 and prevent EMT, presumably by disrupting interactions between Smad7 and both WWP2-FL and/or WWP2-C isoforms. Interestingly, elevating Smad7 levels is known to have therapeutic potential by inhibiting EMT (Javelaud et al., 2007; Valcourt et al., 2005), and supporting apoptotic events involving TNF, ATM and p53 in hyper-proliferating cells (Hong et al., 2007; Zhang et al., 2006). Therefore, similar benefits could be achieved in future strategies that specifically target Smad7 interactions with the WW4 domain in WWP2 and/or WWP2-C that would then stabilise inhibitory Smad7 levels.
In summary, this is the first characterisation of a regulatory mechanism involving individual E3 ubiquitin ligase isoforms that modulate TGFβ-induced EMT by selectively targeting activating and inhibitory Smads. In future studies, it will be important to address further the biological roles of individual isoforms using mis-expression approaches in stable cell lines. Additionally, it will be vital to establish normal and disease expression patterns of all three WWP2 isoforms, with particular emphasis on metastatic disease.
MATERIALS AND METHODS
Antibodies, DNA plasmids, reagents and cell lines
Rabbit anti-Smad3 (Chip grade, Abcam), Rabbit anti-Smad2 (Cell Signalling), Mouse anti-SMAD7 (New England Biolabs), high affinity Rat anti-HA (Roche), M2 Mouse anti-Flag (Sigma), mouse anti-human Vimentin (Sigma), mouse anti-β actin (Sigma) and goat anti-WWP2 (Santa Cruz Biotechnology) were used as recommended by the manufacturers. Scrambled (SCR) and siRNA for WWP-2 were obtained from Santa Cruz Biotechnology. Plasmids encoding the Flag-tagged derivatives of human WWP2, obtained from Fiona McDonald (University of Otago, NZ), were constructed by PCR cloning into pRK5 followed by the insertion of the Flag coding sequence at the 5′ terminal. Derivation of human HA-/Flag-tagged Smad2, Smad3 and Smad7 is described elsewhere (Wicks et al, 2005). Human HA-tagged ubiquitin (expressed in pCDNA3.1+) was a kind gift from Dirk Bohmann (University of Rochester, USA). MG132 was purchased from Sigma reconstituted in DMSO and used at 10μM, and SB431542 was purchased from Tocris Ltd and used at 10μM concentration. TGFβ and Bone Morphogenetic Protein-4 (BMP-4) were from R&D systems. For bacterial WWP2 expression plasmids, the WWP2 coding sequences were PCR amplified (see Supplementary Information) and cloned into pET28A (Novagen) or pGEX5X-1. Stable expression shRNA encoding plasmids specific for WWP2-FL were generated by annealing and cloning phosphorylated complementary oligonucleotides into Bgl II cleaved plasmid pTER (see supplementary information), and used to generate stable Colo-357-Tet repressor expressing cells (as described in Levy and Hill, 2005). Expression was induced by stimulation of cells with 2μM Doxycycline (Melford Laboratories) for 48 hours.
Semi-Quantitative RT-PCR
Total RNA was isolated from mammalian cells (Promega SV40 Total RNA Isolation) and 1μg converted to cDNA using random-hexamer primers and reverse transcriptase (Invitrogen). 1μl was removed and diluted into 40μl water, of which 1μl (1.25ng) was used as template in a 50μl reaction using specific primers (see Supplementary Information).
Immunoprecipitation Assays
Transfected cells treated with either 10μM MG132 for 5 hours in DMEM medium containing 2 % FCS and treated +/− 5ng/ml TGFβ for 1hr, washed in cold PBS, lysed in 1% v/v Igepal-630, 50mM Tris pH 8.0, 150mM NaCl, 10% v/v glycerol, 5mM EDTA, 1mM NaF, 1mM Na3VO4 and protease inhibitors (1% NP-40 LB), in the ratio 1 tablet (Roche): 30 ml 1% NP-40LB. Lysates were cleared by centrifugation and incubated with 0.5μg high affinity anti-HA antibody (Roche) and 20μl of protein-G agarose (Sigma) overnight at 4°C. Immune-complexes were harvested (2000 rpm; 30 sec), and repeatedly washed using 0.1% NP-40 LB. Immunoprecipitates were resuspended in 15μl Laemelli buffer (+10mM DTT), and samples analysed by Western blotting.
Cycloheximide Chase
HEK293 cells were transfected and after 48 hours were pre-treated with 10μM MG132 for 5 hours in DMEM (+ 2% serum). Cycloheximide (20μM) was added and cells incubated for 1 hr. The medium was removed and replaced with DMEM medium (+2% serum) and 20μg/ml cycloxeximide, and cells harvested at specified time-points. Cells were washed in ice cold PBS, lysed in 1% NP-40LB and analysed by Western blotting.
Bacterial WWP2 expression and in vitro ubiquitination assay
WWP2-pET28A protein expression was induced in BL-21 Lys-S (Novagen) cells and proteins purified using Ni-NTA agarose as recommended by the manufacturer (Invitrogen). Approximately 100ng of each purified protein was incorporated into the in vitro ubiquitination assay using 1μg of GST-Smad substrate and the quantities of His-Ub, rabbit E1, His-Ub-carrier protein 6 under conditions previously reported (Xu et al., 2004). Following incubation (30°C), each reaction divided into 2, and each half analysed by Western blotting for Smad and WWP2.
Luciferase Reporter Assays
The Smad3 reporter construct (pCAGAC12-luc) and the Smad2-specific reporter plasmid DE-luc were kindly provided by Caroline Hill (CRUK Laboratories, UK). For each plasmid transfection, 250ng pCAGAC12-luc and 15ng pRSV-β-galactosidase (pRSV-βgal) encoding plasmid were used in conjunction with Smad encoding plasmids as described previously (Wicks et al, 2005). Values were averaged, luciferase activity standardised for β-galactosidase activity and data expressed as relative fold changes in luciferase activity over basal activity. 250ng of DE-luc, 50ng pMIXER and 15ng pRSV-bgal were used in conjunction with other Smad expression plasmids for detection of Smad2-specific activity as described above.
Induction of EMT in Colo-357 cells
Colo-357 cells were seeded at 104 cells per 35 mm plastic well (Nunclon) and the next day left unstimulated, stimulated with 5ng/ml TGFβ, 5ng/ml TGFβ+SB431542, and everyday thereafter until day 6 (where the medium was changed every 2 days). Cells were then washed in PBS, lysed in 250μl 1% NP40-LB to which 125μl of 2× Lamelli buffer was added and samples boiled (10 min) and analysed by Western blotting.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Fiona McDonald, Caroline Hill, and Hans Clevers for generously providing cells/plasmids, and Ian Clark and Tracey Swingler for generously providing ATDC5 cDNA samples. This work was supported by the Association for International Research (AICR), the BigC Cancer Charity, the British Skin Foundation and the Dunhill Medical Trust.
Footnotes
CONFLICT OF INTEREST: The authors declare no conflict of interest.
REFERENCES
- Atsumi T, Miwa Y, Kimata K, Ikawa Y. A chondrogenic cell line derived from a differentiating culture of AT805 teratocarcinoma cells. Cell Differ Dev. 1990;30:109–116. doi: 10.1016/0922-3371(90)90079-c. [DOI] [PubMed] [Google Scholar]
- Bernassola F, Karin M, Ciechanover A, Melino G. The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell. 2008;14:10–21. doi: 10.1016/j.ccr.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Bonni S, Wang HR, Causing CG, Kavsak P, Stroschein SL, Luo K, et al. TGF-beta induces assembly of a Smad2-Smurf2 ubiquitin ligase complex that targets SnoN for degradation. Nat Cell Biol. 2001;3:587–595. doi: 10.1038/35078562. [DOI] [PubMed] [Google Scholar]
- Danckwardt S, Hentze MW, Kulozik AE. 3′ end mRNA processing: molecular mechanisms and implications for health and disease. EMBO Journal. 2008;27:482–498. doi: 10.1038/sj.emboj.7601932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenrieder V, Hendler SF, Boeck W, Seufferlein WT, Menke A, Ruhland C, et al. Transforming growth factor beta1 treatment leads to an epithelial-mesenchymal transdifferentiation of pancreatic cancer cells requiring extracellular signal-regulated kinase 2 activation. Cancer Res. 2001;61:4222–4228. [PubMed] [Google Scholar]
- Flasza M, Gorman P, Roylance R, Canfield AE, Baron M. Alternative splicing determines the domain structure of WWP1, a Nedd4 family protein. Biochem Biophys Res Commun. 2002;290:431–437. doi: 10.1006/bbrc.2001.6206. [DOI] [PubMed] [Google Scholar]
- Foot NJ, Dalton HE, Shearwin-Whyatt LM, Dorstyn L, Tan SS, Yang B, et al. Regulation of the divalent metal ion transporter DMT1 and iron homeostasis by a ubiquitin-dependent mechanism involving Ndfips and WWP2. Blood. 2008;112:4268–4275. doi: 10.1182/blood-2008-04-150953. [DOI] [PubMed] [Google Scholar]
- Kavsak P, Rasmussen RK, Causing CG, Bonni S, Zhu H, Thomsen GH, et al. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol Cell. 2000;6:1365–1375. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- Hata A, Lo RS, Wotton D, Lagna G, Massagué J. Mutations increasing autoinhibition inactivate tumour suppressors Smad2 and Smad4. Nature. 1997;388:82–87. doi: 10.1038/40424. [DOI] [PubMed] [Google Scholar]
- Heldin CH, Landström M, Moustakas A. Mechanism of TGF-beta signaling to growth arrest, apoptosis, and epithelial-mesenchymal transition. Curr Opin Cell Biol. 2009;21:166–176. doi: 10.1016/j.ceb.2009.01.021. M. [DOI] [PubMed] [Google Scholar]
- Hong S, Lee C, Kim SJ. Smad7 sensitizes tumor necrosis factor induced apoptosis through the inhibition of anti-apoptotic gene expression by suppressing activation of the nuclear factor-kappaB pathway. Cancer Res. 2007;67:9577–9583. doi: 10.1158/0008-5472.CAN-07-1179. [DOI] [PubMed] [Google Scholar]
- Javelaud D, Mohammad KS, McKenna CR, Fournier P, Luciani F, Niewolna M, et al. Stable expression of Smad7 in human melanoma cells impairs bone metastasis. Cancer Research. 2007;67:2317–2324. doi: 10.1158/0008-5472.CAN-06-3950. [DOI] [PubMed] [Google Scholar]
- Levy L, Hill CS. Smad4 dependency defines two classes of transforming growth factor-β target genes and distinguishes TGF-β-induced epithelial-mesenchymal transition from its antiproliferative and migratory responses. Mol Cell Biol. 2005;25:8108–8125. doi: 10.1128/MCB.25.18.8108-8125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Bengtson MH, Ulbrich A, Matsuda A, Reddy VA, Orth A, et al. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS ONE. 2008;3(1):e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhang Z, Wang B, Zhang J, Zhao Y, Jin Y. Wwp2-mediated ubiquitination of the RNA polymerase II large subunit in mouse embryonic pluripotent stem cells. Mol Cell Biol. 2007;27:5296–5305. doi: 10.1128/MCB.01667-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lönn P, Morén A, Raja E, Dahl M, Moustakas A. Regulating the stability of TGFbeta receptors and Smads. Cell Res. 2009;19:21–35. doi: 10.1038/cr.2008.308. [DOI] [PubMed] [Google Scholar]
- Lu PJ, Zhou XZ, Shen M, Lu KP. Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science. 1999;283:1325–1328. doi: 10.1126/science.283.5406.1325. [DOI] [PubMed] [Google Scholar]
- Lutz CS. Alternative polyadenylation: A twist on mRNA 3′ end formation. ACS Chemical Biology. 2008;3:609–617. doi: 10.1021/cb800138w. [DOI] [PubMed] [Google Scholar]
- McDonald FJ, Western AH, McNeil JD, Thomas BC, Olson DR, Snyder PM. Ubiquitin-protein ligase WWP2 binds to and downregulates the epithelial Na(+) channel. Am J Physiol Renal Physiol. 2002;283:F431–6. doi: 10.1152/ajprenal.00080.2002. [DOI] [PubMed] [Google Scholar]
- Mund T, Pelham HR. Control of the activity of WW-HECT domain E3 ubiquitin ligases by NDFIP proteins. EMBO Rep. 2009;10:501–507. doi: 10.1038/embor.2009.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray-Zmijewski F, Lane DP, Bourdon JC. p53/p63/p73 isoforms: an orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ. 2006;13:962–972. doi: 10.1038/sj.cdd.4401914. [DOI] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirozzi G, McConnell SJ, Uveges AJ, Carter JM, Sparks AB, Kay BK, et al. Identification of novel human WW domain-containing proteins by cloning of ligand targets. J Biol Chem. 1997;272:14611–14616. doi: 10.1074/jbc.272.23.14611. [DOI] [PubMed] [Google Scholar]
- Raikwar NS, Thomas CP. Nedd4-2 isoforms ubiquitinate individual epithelial sodium channel subunits and reduce surface expression and function of the epithelial sodium channel. Am J Physiol Renal Physiol. 2008;294:1157–1165. doi: 10.1152/ajprenal.00339.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazi J, Bakkour N, Stamm S. Alternative splicing and disease. Biochimica et Biophysica Acta. 2009;1792:14–26. doi: 10.1016/j.bbadis.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valcourt U, Kowanetz M, Niimi H, Heldin CH, Moustakas A. TGF-beta and the Smad signalling pathway support transcriptomic reprogramming during epithelial-mesenchymal cell transition. Mol Biol Cell. 2005;16:1987–2002. doi: 10.1091/mbc.E04-08-0658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicks S, Haros K, Maillard M, Song L, Cohen RE, ten Dijke P, Chantry A. The deubiquitinating enzyme UCH37 interacts with Smads and regulates TGFβ signalling. Oncogene. 2005;24:8080–8084. doi: 10.1038/sj.onc.1208944. [DOI] [PubMed] [Google Scholar]
- Wiesner S, Ogunjimi AA, Wang HR, Rotin D, Sicheri F, Wrana JL, et al. Autoinhibition of the HECT-type ubiquitin ligase Smurf2 through its C2 domain. Cell. 2007;130:651–662. doi: 10.1016/j.cell.2007.06.050. [DOI] [PubMed] [Google Scholar]
- Xu H, Wang W, Li C, Yu H, Yang A, Wang B, et al. WWP2 promotes degradation of transcription factor OCT4 in human embryonic stem cells. Cell Res. 2009a;19:561–573. doi: 10.1038/cr.2009.31. [DOI] [PubMed] [Google Scholar]
- Xu HM, Liao B, Zhang QJ, Wang BB, Li H, Zhong XM, et al. Wwp2, an E3 ubiquitin ligase that targets transcription factor Oct-4 for ubiquitination. J Biol Chem. 2004;279:23495–23503. doi: 10.1074/jbc.M400516200. [DOI] [PubMed] [Google Scholar]
- Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009b;19:156–172. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavadil J, Böttinger EP. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24:5764–5774. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- Zhang S, Ekman M, Thakur N, Bu S, Davoodpour P, Grimsby S, et al. TGFbeta-induced activation of ATM and p53 mediates apoptosis in a Smad7-dependent manner. Cell Cycle. 2006;5:2787–2795. doi: 10.4161/cc.5.23.3523. [DOI] [PubMed] [Google Scholar]
- Zhu H, Kavsak P, Abdollah S, Wrana JL, Thomsen GH. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature. 1999;400:687–693. doi: 10.1038/23293. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.