Abstract
Therapeutic monoclonal antibodies (mAb) constitute a group of highly effective agents for treating various refractory diseases. Nonetheless it is challenging to achieve selective and accurate quantification of mAb in pharmaceutical matrices, which is required by PK studies. Liquid chromatography/mass spectrometry under selected reaction monitoring mode (LC/SRM-MS) is emerging as an attractive alternative to immunoassays because of the high specificity and multiplexing capacity it provides, but may fall short in terms of sensitivity, reliability and quantitative accuracy. Moreover, the strategy for optimization of the MS conditions for many candidates of signature peptides (SP) and the selection of the optimal SP for quantification remains elusive. In this study, we employed a suite of technical advances to overcome these difficulties, which include i) a nano-LC/SRM-MS approach to achieve high analytical sensitivity, ii) a high-resolution nano-LC/LTQ/Orbitrap for confident identification of candidate peptides, iii) an on-the-fly orthogonal array optimization (OAO) method for the high-throughput, accurate and reproducible optimization for numerous candidate peptides in a single LC/MS run without using synthesized peptides, iv) a comprehensive evaluation of stability of candidates in matrix using the optimized SRM parameters, v) the use of two unique SP for quantification of one mAb to gauge possible degradation/modification in biological system and thus enhancing data reliability (e.g. rejection of data if the deviation between the two SP is greater than 25%) and vi) the utilization of purified target protein as the calibrator to eliminate the risk of severe negative biases that could occur when a synthesized peptide is used as calibrator. To show a proof of concept, this strategy is applied in the quantification of cT84.66, a chimeric, anti-CEA antibody, in preclinical mouse models. A low detection limit of the mAb down to 3.2 ng/mL was achieved, which is substantially more sensitive than established immunoassay methods for anti-CEA antibodies. The quantitative method showed good linearity (within the range of 12.9 ng/mL to 32.3 µg/mL in plasma), accuracy and precision. Additionally, the ultra-low sample consumption (2 µL plasma per preparation) permits the acquisition of an entire set of time course data from the same mouse, which represents a prominent advantage for PK study using small-animal models. The developed method enabled an accurate PK investigation of cT84.66 in mice following intravenous and subcutaneous administrations at relatively low doses over an extended period of time. The strategy employed in this study can be easily adapted to the sensitive and accurate analysis of other mAb and therapeutic proteins.
Keywords: monoclonal antibody, nano-flow LC, LC/MS, pharmacokinetics, quantification, anti-CEA antibody
1. Introduction
Monoclonal antibodies (mAb) constitute one of the most rapid-growing classes of cancer therapeutic agents. So far the FDA has approved 9 mAb drugs targeting different cancer antigens, while many more are currently under development [1–3]. To support pharmacokinetic (PK) studies in both preclinical and clinical development/evaluation of mAbs and to elucidate the underlying mechanism of drug action, an accurate, sensitive and selective quantification of these agents in pharmaceutical matrixes is essential [4,5]. Conventionally, enzyme-linked immunosorbent assay (ELISA) has been regarded as the method-of-choice for mAb quantification. However, developing high-quality ELISA method for each newly developed mAb can be both costly and time-consuming; furthermore, the assay accuracy, specificity and reproducibility of ELISA are often compromised by the cross reactivity with endogenous IgG and their fragments, and by the possible presence of anti-mAb antibody[6–8].
Recently, methods based on liquid chromatography/mass spectrometry (LC/MS) and selected-reaction-monitoring (SRM) have emerged as promising alternatives to immunoassays for the quantification of mAb in complex matrices[9–13]. The primary advantages of LC/SRM-MS-based methods include that the development of such methods is relatively more cost-efficient, and that these methods provide often high specificity and multiplexing capability[8,14–16]. Nevertheless, several technical difficulties have been preventing more prevalent use of this technique for quantification of therapeutic proteins. For an example, in order to achieve the desired selectivity, sensitivity and accuracy, it is critical to accurately identify the optimal proteolytic peptides for quantification (termed as the "signature peptide", SP) and to precisely optimize the optimal SRM conditions for potential SP candidates[14,15]. Unfortunately, an efficient and high-throughput strategy to achieve these goals remains elusive. A frequently-used approach for the selection of SP is in silico predication. While the algorithms have been advanced significantly over the last several years, in the absence of experimental data, it is highly challenging for these approaches to accurately predict many factors that are important for targeted quantification, such as the sensitivity achievable by different proteolytic peptides, the perplexing preferences by protease cleavage, peptide stability in the sample digest and chemical interferences in sample matrices [17]. Another prevalent approach of selecting SP is based on previous shotgun proteomic data, whereby the peptides that were reproducibly identified across biological replicates are considered as “the best representative” peptide and employed for targeted quantification. However, these peptides are not always favorable for SRM quantification probably due to the undersampling problem in data-dependent analysis [15,18,19]. This often leads to suboptimal choices [14,19].
Another challenge associated with method development is that it is not straightforward to optimize the SRM conditions (e.g. the optimal precursor/product ion transitions and the de-cluster/collision energy) for potential SP candidates unless the standard peptides are available. In order to enhance the likelihood of choosing a sensitive peptide as the SP, current approaches often synthesize multiple peptide candidates (i.e. > 5) for SRM optimization and evaluation, which could be costly and time-consuming [19,20].
Furthermore, existing LC/SRM-MS strategies for the quantification of therapeutic proteins may need to be improved to enhance quantitative reliability, accuracy and sensitivity. Firstly, most methods use a lone SP for the quantification of a protein, which may carry a significant risk of error where proteins are truncated biologically outside the SP domain [21,22] or certain residues within the SP domain are biologically modified[23]. Secondly, the majority of current approaches use synthesized peptides as calibrators, which may lead to severe negative biases, as we observed previously [24]. Finally, due to the high molecule weight of therapeutic proteins and their relatively low concentrations in pharmaceutical samples, the sensitivity achievable by a conventional LC/MS is often insufficient, especially for these studies employing low therapeutic doses [5].
The aim of this work was to address these difficulties by developing a strategy that enables sensitive, selective and reliable quantification of therapeutic mAb in pharmaceutical matrices. A highly sensitive nano-LC/Nanospray-MS method was employed to achieve a low lower limit of quantification (LLOQ). An on-the-fly orthogonal array optimization (OAO) method that we developed recently [24] was utilized for the high-throughput optimization of SRM conditions for each SP candidate. Without using synthesized peptides, the OAO approach is able to achieve batch-wise development of SRM methods, e.g. optimization for many SP candidate peptides in a complex digest mixture by a single LC/MS analysis. Additionally, the incorporation of orthogonal array design allowed the accurate determination as to how a combination of various key parameters affects the analytical sensitivity, and thus rapidly identified the optimum for each factor with a minimal number of trials. With the SRM method developed, the stability of SP candidates was readily evaluated in the target matrix. To enhance the quantitative reliability, two unique SPs that are highly sensitive and stable were selected for the quantification of one mAb. Pure mAb protein, rather than synthesized peptides, was used as the calibrator to prevent the quantitative biases that may otherwise occur when synthesized peptides are used for calibration curve.
We applied this strategy for sensitive, selective and accurate quantification of chimeric T84.66 (cT84.66) in mice plasma and conducted a preclinical PK study. The cT84.66 is a mouse/human chimeric mAb, which shows high specificity and affinity for tumor-associated carcinoembryonic antigen (CEA). Previous studies have shown that the CEA-specific mAbs can significantly suppress tumor growth and metastasis both alone and in combination with chemotherapy [25]. Additionally, these agents demonstrate higher clearance in tumor-bearing subjects than the healthy subjects, and thereby may serve as a sensitive probe for cancer detections (e.g. breast cancer, colorectal cancer etc) [4,26]. Clearly, the capacity of accurately quantifying anti-CEA mAbs in plasma not only enables the understanding of their PK characteristics, but also is of essential value to explore the therapeutic/diagnostic potentials of these agents.
2. Experimental
2.1 Materials and reagents
Sequencing-grade trypsin was from Promega (Madison WI). Protease, phosphatase and kinase inhibitor cocktail tablets were from Roche (Basel, Switzerland). Bicinchoninic acid (BCA) protein assay reagents were from Pierce (Rockford, IL, USA). HPLC grade methanol, acetonitrile, acetone, and water were from B&J (Muskegon, MI, USA). LC/MS grade formic acid was from Fluka (Buchs, Switzerland). Tris(2-carboxyethyl)phosphine (TCEP), Tris, iodoacetamide (IAA), and phosphate-buffered saline were obtained from Sigma-Aldrich (St. Louis, MO, USA). Isotope-labeled peptides, GPSVFPLAPSSK[15N,13C] and ASNLESGIPVR[15N,13C], were synthesized by New England Peptide (Gardner, MA, USA) and the actual content was accurately determined by quantitative amino acid analysis (AAA).
2.2 Production, purification and quality control of cT84.66 protein
Transfectomas secreting cT84.66 were developed and grown at 37°C in 1 L spinner flasks containing serum-free media (Hybridoma-SFM, Invitrogen, Grand Island, NY, USA)[27]. Following centrifugation and filtration to remove cells and cellular debris, cT84.66 was purified by protein-G affinity chromatography via Bio-Rad automated pressure system. The loading buffer was 20 mM Na2HPO4 (pH 7.0, Sigma Chemical), and the elution buffer was 100 mM glycine (pH 2.8, Sigma Chemical). Eluted antibody was collected in glass tubes containing 1 M Tris buffer to neutralize the solution and minimize antibody aggregation. The cT84.66 protein was then loaded on a dodecyl sulfate polyacrylamide (SDS-PAGE) electrophoresis to confirm that the preparation is free of impurity proteins, and the accurate purity was determined by quantitative amino acid analysis (AAA).
2.3. Nano-LC-LTQ/Orbitrap identification of SP candidates
The cT84.66 standard was spiked into blank mouse plasma at 10 µg/mL and digested (as described in 2.6). Peptide separation was performed on an Eksigent two-dimensional nano-LC system (Eksigent Technologies, Dublin, CA, USA). Solvents used were water/0.1% formic acid (mobile phase A) and 85% acetonitrile/0.1% formic acid (mobile phase B). Samples containing 6 µg of peptides were loaded onto a large-ID trap (300 µm ID × 5 mm, 3-µm C18) with 3% B at a flow rate of 10 µL/min, and the trap was washed for 3 min. A shallow gradient was used to back-flush the trapped samples onto the nano-LC column (75 µm ID × 25 cm, packed with Pepmap 3-µm C18 material): (i) a linear increase from 3% to 8% B over 12 min; (ii) an increase from 8 to 25% B over 53 min; (iii) an increase from 25 to 35% B over 20 min; (iv) an increase from 35 to 70% B over 20 min; (v) an increase from 70 to 97% B over 5 min; and (vi) isocratic flow at 97% B for 15 min. The flow rate was 250 nL/min. High resolution MS analysis was performed on an LTQ/Orbitrap-ETD hybrid mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA). MS/MS analysis was performed using a survey scan in FT mode (m/z 350 ~ m/z 1500) with a resolution of 60 000 and an ion accumulation target value of 5×106, followed by fragmentation of the 7 most intensive peaks in the linear ion trap. Identification of peptides was performed using BioWorks 3.3.1 embedded with Sequest (Thermo Fisher), searching against a FASTA database containing the sequences of target mAbs and murine proteins derived from the Swiss-Prot database. The precursor mass tolerance was 25 ppm and the mass tolerance for CID fragments was 1.0 mass unit. Cross correlation score (Xcorr) criteria were ≥4 for 4+ and higher charge states, ≥3 for 3+ ions, ≥2.2 for 2+ ions, and ≥1.9 for 1+ ions.
2.4 On-the-fly orthogonal array optimization
cT84.66 was spiked into pooled blank plasma at a concentration of 1.0 µg/mL, then processed and digested following an efficient acetone precipitation/on-pellet digestion procedure (described below). The digested sample was analyzed on a TSQ Quantum triple quadrupole mass spectrometer (Thermo Fisher Scientific, San Jose, CA) with the same nano-LC configuration and gradient condition used for peptide identification. Given the high reproducibility of the nano-LC separation, the SRM of peptide candidates were grouped according to retention time windows (segment), and one group was monitored in each window. A predefined L25 (35) orthogonal array design [28] was applied to evaluate the critical SRM parameters including product ion, collision energy (CE) and tube-lens voltage. The range of CE was estimated based on the empirical values we established through pilot analyses of 103 unique tryptic peptides [24]. The optimization was conducted by programming 25 independent SRM measurements with strategically varied parameters. For each SRM measurement, the dwell time was set as 25 ms. The peak area, peak height and S/N ratio were extracted from each SRM channel and exported to a statistic analysis module [24], which automatically calculate the effect curves for each parameter.
2.5 Peptide stability test
cT84.66 standard was spiked into pooled mouse plasma at 5 µg/mL each and digested as described below. Two aliquots of the mixtures were used for stability assessment. Peptide stabilities were evaluated by incubation in either (i) digestion buffer at 37°C for up to 18 h or (ii) in injection solution (i.e. digestion buffer acidified by 1% formic acid) at 4°C for 48h, mimicking the conditions to which the SP candidates are exposed during tryptic digestion or queuing in a cooled autosampler. For evaluation in digestion buffer, the solution was sampled at 0, 4, 12 and 18 h after the completion of digestion. At each time point, a 20 µL sample was taken, acidified by adding formic acid to a final concentration of 1% (v/v), and then analyzed immediately by nano-LC/MS using the optimized conditions obtained in the OAO procedure. For assessment in injection solution, the digestion mixture was acidified immediately upon completion of digestion, incubated in the refrigerated autosampler, and analyzed at 0, 4, 12, 24 and 48 h. Any candidate that degraded more than 20% within either evaluation period was disqualified.
2.6. Plasma sample preparation
An aliquot of 2 µL plasma was diluted 10-fold by PBS (100 mM, pH7.4) containing 0.5% SDS and a protease inhibitor cocktail. The diluted sample was reduced with 2 mM TCEP and then alkylated with 100 mM iodoacetamide at 37°C for 30 min in the darkness. The mixture was precipitated with 6 volumes of cold acetone in two steps and then incubated overnight at −20 °C. After centrifugation at 12,000 g for 20 min, the supernatant was removed carefully and the pellet was allowed to air dry, and a 70 µL of Tris buffer (50 mM, pH 8.2) containing the I.S. and trypsin was added to a final enzyme:substrate ratio of 1:30 (w/w). The solution was then incubated at 37°C and vortexed at 120 rpm for 6 h to dissolve the pellet. A second aliquot of trypsin was added at an enzyme:substrate ratio of 1:25 (w/w), and the mixture was incubated at 37 °C overnight to achieve complete digestion. Formic acid was added to a final concentration of 1% (v/v) to quench the digestion.
2.7 Quantification using nano-LC/SRM-MS
The chromatographic systems, column and trap used for quantification were the same as those employed for Orbitrap analysis and OAO. Nonetheless, the separation time was optimized to be much shorter to achieve a reasonable analytical throughput. The gradient consisted of a linear increase of mobile phase B from 3% to 10% in 4 min, followed by an increase to 25% B in 8 min, an increase to 60% B in 12 min, then another increase to 97% B within the next 4 min, and 97% B was held for the final 5 minutes of the gradient. The flow rate was 350 nL/min. The column temperature was maintained at 55°C. At the end of the run the trap column was back-flushed with 97%B and then equilibrated with 3% B at a flow rate of 10 µL/min. The analytical column was re-equilibrated at 3% B at 400 nL/min for 10 min. The nano-LC system was connected to a TSQ Quantum triple quadrupole mass spectrometer (same as in 2.4) equipped with a lab-made nano electrospray interface operated in the positive mode. The spray voltage was set at 2 kV and the capillary temperature was set at 320°C. Quantification was performed using selected reaction monitoring of the transitions m/z 593.8 → 846.4 for GPSVFPLAPSSK and m/z 571.8 → 757.5 for ASNLESGIPVR. The optimized collision energy and tube-lens voltage were set at 17 eV/85 V and 25 eV/85 V according to the OAO results (Fig 3). Additional transitions (e.g. m/z 593.8 → 699.4 and m/z 571.8 → 371.4) were monitored to confirm the accurate assignment of SP peak. IS peptides were detected using m/z 597.8 → 854.3 and m/z 576.8 → 767.5, respectively. Each transition was monitored with a 100 ms dwell time. Q1 and Q3 resolution were both set at 0.7 FWHM (full width at half maximum).
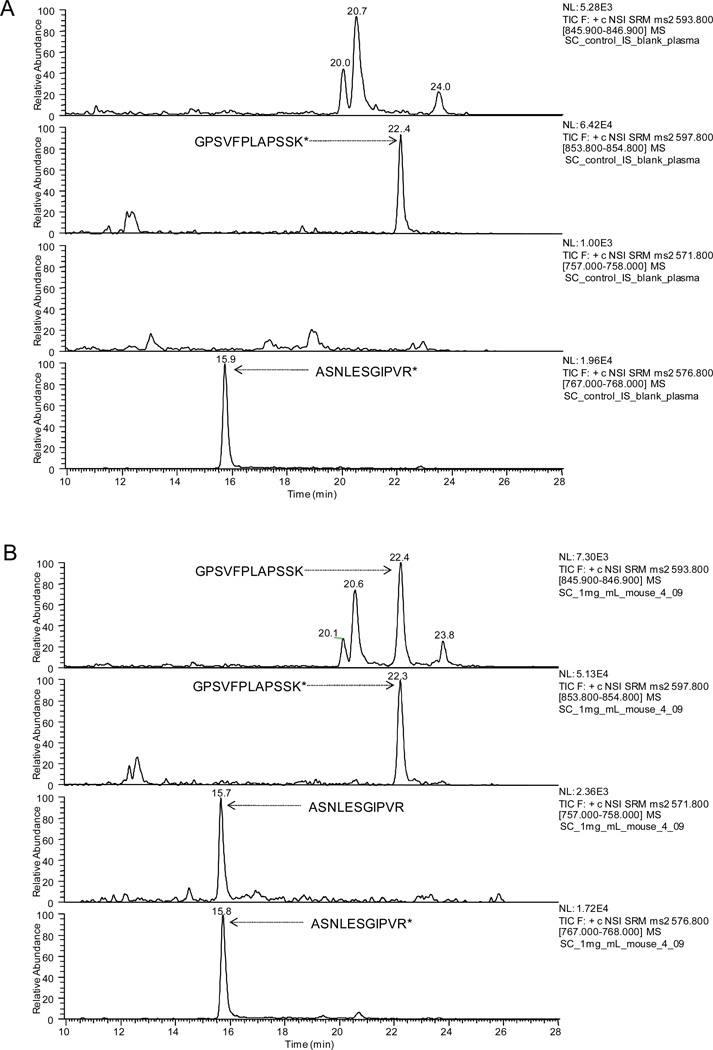
Figure 3.
Representative nanoLC/SRM-MS chromatograms of signature peptides for the quantification of cT84.66. (A) a blank plasma sample and (B) a sample procured at 288 h after S.C. administration of 1mg/kg cT84.66. Asterisk signifies isotope-coded Arg or Lys
2.8 Calibrations and method evaluation
Pooled plasma from saline-treated mice served as the blank matrix to prepare calibration curves. The concentrations used, based on purity determined by AAA method, were 12.9, 32.3, 64.5,129, 323, 645, 1290,3225, 6450, 12900 and 32250 ng/mL for cT84.66 in mouse plasma; all calibration samples were precipitated, digested and analyzed by nano-LC/SRM-MS. Isotope-labeled IS peptide was spiked into the samples prior to digestion at the concentration equivalent to 5 µg/mL protein. Calibration curves were constructed by plotting the peak area ratios of SP and I.S. versus the corresponding mAb concentrations. Linear regression with a 1/x2 weighting factor was employed. Method accuracy and precision were evaluated during three validation runs using quality control (QC) samples prepared by spiking mAb standard into blank matrix at three different levels (32.3, 645 and 29025 ng/mL).
2.9 Preclinical PK study in mouse model
Male athymic nude mice (20–25 g, 5–6 weeks old) were obtained from Harlan (Indianapolis, IN, USA). Protocols for animal use were approved by the Institutional Animal Care and Use Committee of the University at Buffalo. The target mAb cT84.66 was administered subcutaneously at 1 and 10 mg/kg (n=4) or intravenously via the penile vein at 1 mg/kg (n=4). Blood samples were collected from the retro-orbital plexus using calibrated capillary pipettes (Drummond Scientific Company, Cat # 2–000-020) pre-rinsed with EDTA at 1 h, 4 h, 8 h, 12h, 1d, 2d, 4d, 7d and 12 days. The blood was centrifuged at 13,000 rpm for 5 min to obtain plasma. Plasma samples were stored at −20°C until analyzed. Pharmacokinetic parameters were estimated by non-compartmental pharmacokinetic analysis using WinNonlin, version 5.0 (Pharsight Corporation, Palo Alto, CA, USA).
3. Results and Discussion
3.1 Development of the quantitative method
The procedure for method development consists of the following steps: i) confident identification of SP candidates by nano-LC/LTQ/Orbitrap, ii) a high-throughput, on-the-fly OAO optimization to obtain the optimal SRM conditions for each candidate; iii) evaluation of the sensitivity and stability of candidate peptides in plasma using the optimal SRM conditions, and the final selection of the two unique SP; iv) optimize the sample preparation and nano-LC/SRM-MS procedures for batch-wise quantification and v) method validation and evaluation. With the purpose of achieving high sensitivity and selectivity to the highest extent possible, each step was thoroughly optimized.
3.1.1 Confident identification of SP candidates by nano-LC/LTQ/Orbitrap
In order to select the optimal peptides as SP, it is important to obtain a comprehensive pool of candidates derived from the target mAb. Here an efficient precipitation/on-pellet-digestion method [29] was employed to digest the purified cT84.66. Peptides were resolved thoroughly with a shallow-gradient nano-LC, followed by sequencing with LTQ/Orbitrap MS. Under a set of stringent filtering criteria (cf. Experimental 2.3), 34 peptides derived from cT84.66 were identified with high confidence. Peptides that are not specific to the target mAbs (as revealed by the interrogation of the murine protein database) or bearing either miss-cleavage site(s) or known PTM positions (e.g. Glycosylation) were removed from the list. The peptides surviving this screening process were designated as the SP candidates that are subjected to OAO optimization followed by evaluations for sensitivity, stability and interference in the target matrices. The list of SP candidates is shown in Table 1. As the charge states of a peptide precursor markedly affect the sensitivity of tandem MS analysis [30], all identified charge states of a precursor were subjected to further evaluation. Because the following OAO procedure requires the inputs of retention times and abundant product ions for each SP candidate, this information was also recorded (Table 1).
Table 1.
SP candidates of cT84.66 identified by nano-LC/LTQ/Orbitrap and then filtered with a set of criteria
| Peptides | Domain | Charge state |
Precursor m/z |
Top 5 most intensive product ions | Stabilitya | RT (min) | |
|---|---|---|---|---|---|---|---|
| During digestion | In autosampler | ||||||
| FSGTGSR | Light Chain | 2+ | 356.175 | 347.1; 477.2; 267.2; 235.1; 207.2 | Good | Poor | 26.6 |
| IDPANGNSK | Heavy Chain | 1+ | 915.453 | 687.5; 670.4; 898.5; 405.2; 519.4 | Poor | Poor | 31.9 |
| 2+ | 458.230 | 687.5; 344.5; 229.2; 255.1; 519.4 | 31.9 | ||||
| VDNALQSGNSQESVTEQDSK | Light Chain | 2+ | 1068.488 | 707.4; 1495.6; 893.3; 1051.7; 1902.9 | Poor | Good | 35.1 |
| 3+ | 712.661 | 748.6; 698.0; 812.6; 641.3; 606.2 | 35.1 | ||||
| ALPAPIEK | Heavy Chain | 1+ | 838.503 | 654.3; 486.3; 563.3; 340.2; 508.3 | Good | Good | 40.2 |
| VYACEVTHQGLR | Light Chain | 2+ | 716.854 | 586.1; 810.5; 667.6; 1170.6; 610.5 | Good | Good | 44.9 |
| 3+ | 478.239 | 586.1; 235.2; 668.2; 550.5; 263.3 | 44.9 | ||||
| DTLMISR | Heavy Chain | 1+ | 835.434 | 720.4; 461.2; 619.4; 506.3; 574.2 | Good | Poor | 47.6 |
| 2+ | 418.220 | 409.4; 506.3; 619.4; 375.2; 262.1 | 47.6 | ||||
| DTYMHWVK | Heavy Chain | 1+ | 1079.498 | 933.4; 1061.4; 834.4; 964.6; 648.2 | Good | Poor | 50.7 |
| 2+ | 540.252 | 531.4; 432.3; 482.7; 700.3; 863.6 | 50.7 | ||||
| STSGGTAALGCLVK | Heavy Chain | 1+ | 1264.657 | 1321.7; 1303.6; 576.6; 1133.7; 760.5 | Good | Good | 52.8 |
| 2+ | 632.832 | 652.4; 576.3; 689.5; 760.5; 831.4 | 52.8 | ||||
| ASNLESGIPVR | Light Chain | 1+ | 1142.616 | 628.4; 1124.6; 371.4; 541.4; 870.6 | Good | Good | 54.5 |
| 2+ | 571.812 | 371.4; 757.5; 772.3; 628.6; 541.5 | 54.5 | ||||
| NQVSLTCLVK | Heavy Chain | 2+ | 552.808 | 916.6; 820.5; 342.1; 519.5; 572.1 | Good | Good | 61.2 |
| LSCTASGFNIK | Heavy Chain | 1+ | 1197.593 | 1179.7; 1067.5; 1051.5; 665.5; 578.4 | Good | Good | 63.7 |
| 2+ | 599.301 | 499.6; 665.5; 837.5; 736.5; 578.4 | 63.7 | ||||
| FNWYVDGVEVHNAK | Heavy Chain | 1+ | 1677.801 | 1678.7; 853.4; 1659.8; 1531.6; 1460.7 | Good | Good | 65.5 |
| 2+ | 839.405 | 968.5; 853.3; 1067.5; 1230.6; 697.4 | 65.5 | ||||
| 3+ | 559.939 | 709.1; 534.6; 615.8; 485.0; 262.1 | 65.5 | ||||
| GPSVFPLAPSSK | Heavy Chain | 1+ | 1186.647 | 699.4; 1168.6; 751.4; 769.4; 846.5 | Good | Good | 69.3 |
| 2+ | 593.827 | 699.5; 846.4; 769.4; 418.4; 470.2 | 69.3 | ||||
| TPEVTCVVVDVSHEDPEVK | Heavy Chain | 2+ | 1041.507 | 1667.8; 472.2; 1039.5; 1154.5; 1253.6 | Good | Poor | 76.1 |
| 3+ | 694.673 | 834.6; 472.2; 971.2; 680.3; 856.8 | 76.1 | ||||
| QRPEQGLEWIGR | Heavy Chain | 2+ | 734.886 | 726.4; 531.4; 921.5; 1184.6; 345.2 | Good | Good | 81.4 |
| 3+ | 490.260 | 480.2; 612.2; 562.6; 660.4; 830.5 | 81.4 | ||||
| GFYPSDIAVEWESNGQPENNYK | Heavy Chain | 2+ | 1272.569 | 764.4; 1150.6; 1465.6; 1780.7; 1764.8 | Good | Poor | 84.7 |
| 3+ | 848.715 | 798.2; 1089.3; 950.5; 764.4; 733.5 | 84.7 | ||||
| VVSVLTVLHQDWLNGK | Heavy Chain | 2+ | 904.507 | 997.5; 1110.5; 896.7; 617.2; 1192.6 | Good | Good | 87.3 |
| 3+ | 603.340 | 806.1; 805.4; 712.6; 656.1; 499.5 | 87.3 | ||||
| TVAAPSVFIFPPSDEQLK | Light Chain | 2+ | 973.517 | 913.4; 802.7; 1015.5; 1060.5; 1320.6 | Good | Poor | 88.2 |
| 3+ | 649.347 | 457.5; 913.4; 873.5; 517.3; 1060.7 | 88.2 | ||||
| AGESVDIFGVGFLHWYQQK | Light Chain | 3+ | 727.697 | 869.7; 962.8; 919.2; 812.3; 575.6 | Poor | Poor | 95.0 |
peptide stabilities were evaluated by incubation in either digestion buffer or injection buffer (c.f. Experimental 2.5); Any candidate that degraded more than 20% was considered as "poor" stability.
3.1.2. On-the-fly OAO optimization of candidate peptides
In order to achieve a sensitive and selective quantification with LC/SRM-MS, it is critical to employ the optimal SRM parameters (e.g. parent-to-product transitions, de-cluster energy and collision energy) for each analyte. However, though widely practiced, predicting the best SRM transitions based on the fragmentation data previously obtained by a discovery MS experiment may not be accurate[15,20]. Furthermore, it is often difficult to achieve an accurate in silico prediction of the optimal SRM conditions for a precursor ion, as these parameters are heavily dependent on numerous factors such as the chemical property and structure of the precursor[20]. By comparison, experimentally optimizing the SRM parameters using the target peptides provides highly accurate optimization [15,17]. So far such optimizations are often carried out using synthesized peptides, which may be cost-prohibitive since it is often necessary to synthesize multiple candidates (e.g. >5) for each protein, in order to be able to select an optimal SP for quantification [31–33].
One prominent advantage of SRM-based approach is its capacity of monitoring numerous SRM transitions in a single LC/MS run. However, this multiplexing capacity is often not fully exploited at the method development stage. Conventionally, the SRM conditions of each peptide are optimized in a one-factor-at-a-time manner, which may be time-consuming and error-prone because i) it is often necessary to optimize a number of SP candidates and their transitions in order to find an optimal SP [33], ii) the approach may not be able to accurately reveal the collective effects of different parameters, and (iii) replicate optimizations are often desirable.
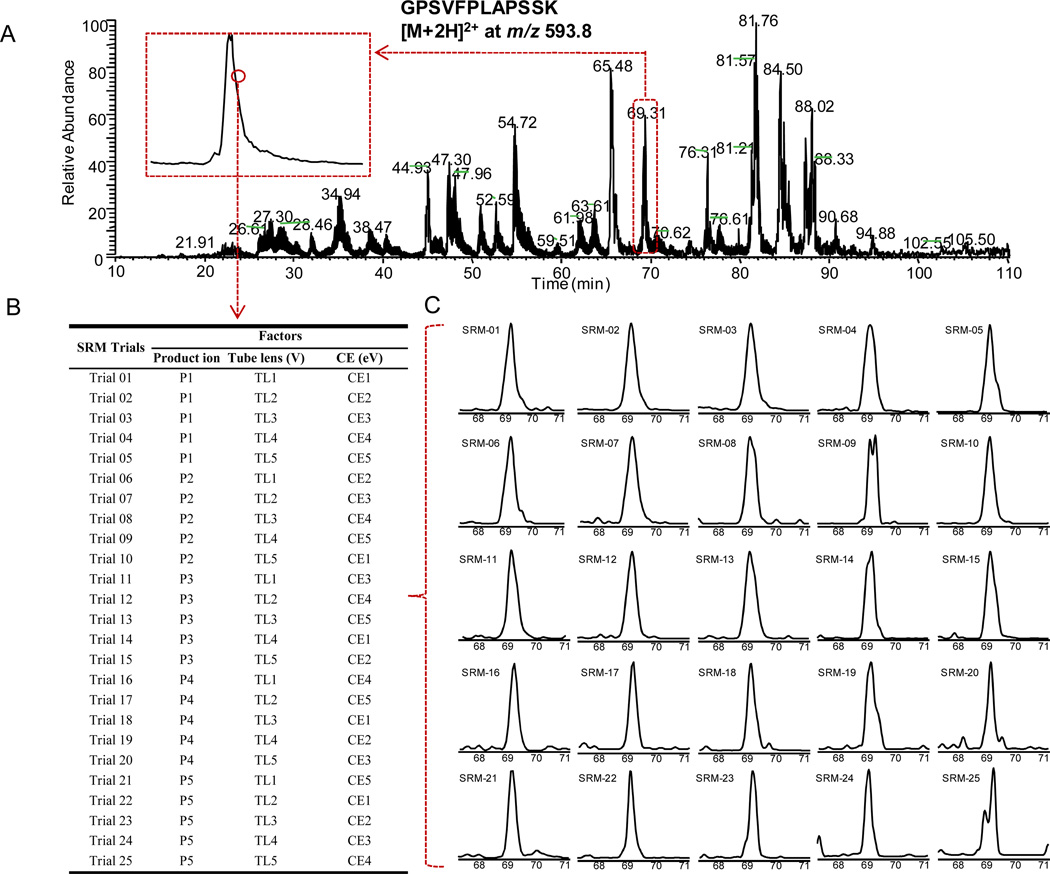
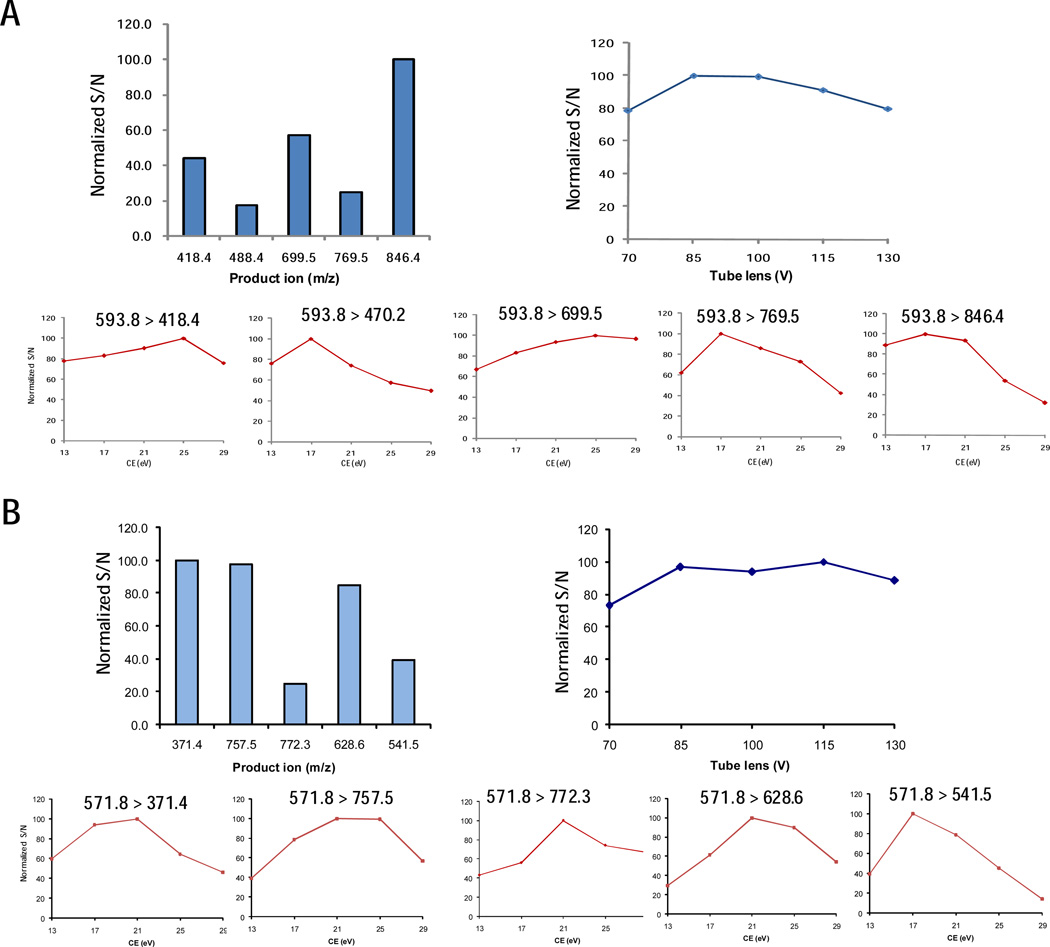
In this study, we implemented a high-throughput and accurate on-the-fly orthogonal array optimization (OAO) strategy that is capable of optimizing multiple key SRM parameters for many candidate peptides in a single LC/MS run, without using synthesized peptides. The rationale of OAO strategy was based on the fractional factorial design that aims to determine the combined effects of a large number of interactive factors with a minimal number of experiments [34]. As illustrated in Fig 1, the collective effects of the three key factors (fragment ions, collision energy and de-clustering potential) were systematically investigated for all 35 peptide precursors by performing OAO in a single 150-min LC/MS run. The candidate peptides were grouped into small retention time windows for OAO, and each peptide was optimized by performing a series of iterative SRM scan events (25 independent SRM trials with predefined combination of parameters, Fig 1B and 1C) across the retention time window. To accelerate the method development, an automated procedure was developed for the analysis of the large data sets acquired by OAO. The peak area, peak height and S/N ratio were obtained for each SRM trial and exported to a statistic analysis package, which computes the effect curves for each parameter in a batch-wise manner. Based on these results, the optimal SRM parameters for all candidate peptides were readily identified. The representative effect plots for SP candidates are shown in Fig 2.
Figure 1.
The illustration of the on-the-fly OAO strategy. (A) Total ion chromatogram (TIC) of all SRM trials for cT84.66 spiked-in mouse plasma digest separated by the nano-LC system. Time scheduled SRM was applied. In total, 875 SRM transitions corresponding to 19 peptide candidates for cT84.66 were monitored with a 150-min gradient. The full list of candidate peptides and the product ions used for SRM transitions is presented in Table 1. The insert depicts the zoomed-in profile of a typical peptide peak (doubly-charged GPSVFPLAPSSK). Each point on the profile represented one independent optimization cycle consisting of 25 trials. (B) The orthogonal array design (L25) for simultaneous optimization of key factors in SRM development. The numbers 1–5 in the table denote the five different levels for each factor. (C) A typical set of the chromatograms of 25 SRM trials conducted sequentially (took 50 ms each) acquired each data point. The XIC peak was used to evaluate the S/N for each trial.
Figure 2.
The representative effect curves for key parameters obtained by the on-the-fly OAO of candidate peptides. The curves (A) of candidate peptide GPSVFPLAPSSK and (B) of candidate peptide ASNLESGIPVR are shown. Both were selected as the SP after further evaluation.
Compared to the one-factor-at-a-time approaches conventionally employed for SRM method development, the OAO strategy holds several distinct advantages. First, it provides both high throughput and multiplex capacity. By dividing candidates into successive retention time windows, at least 50 precursors can be evaluated with ease by a single nano-LC/MS analysis, allowing an array-style optimization for multiple peptides; second, for each OAO optimization (comprised of 25 SRM trials), the duty cycle is quite short (< 1s), which enables the completion of at least 20 replicates of OAO optimization for a precursor across the elution time window of a typical tryptic peptide, contributing to a highly reproducible optimization in only a single run. third, this approach evaluates the collective effects of multiple factors, rather than one factor at a time, and thereby providing more comprehensive and accurate optimization; finally, the optimization was carried out in the plasma, rendering the optimized conditions directly applicable for the analysis of plasma samples.
To validate the accuracy of optimization by OAO, we compared the optimal parameters of selected peptides obtained by OAO against these by optimization via direct infusion of synthesized peptides. While the OAO method is much more cost-effective, and provides far superior throughput, the optimization results by the OAO and direct infusion agreed very well.
Under the optimized conditions, it was discovered that the achievable sensitivity for cT84.66 depended heavily upon the peptide chosen, which varied in a 146-fold range in plasma. In addition to SRM settings, many factors may also exert perplexing affects on the sensitivity achievable for each peptide, such as the efficiencies of enzyme digestion, chromatographic separation, and interference from the sample matrix. This observation underscores the importance of experimentally evaluating the sensitivity for SP candidates in the target matrix.
3.1.3. Evaluation of the stability of SP candidates and selection of two SP
Whereas the risks of degradation and modification of SP in complex matrix are not readily predictable from the sequence of a peptide, they may severely compromise the accuracy, sensitivity and reproducibility of protein quantification [21], which necessitates experimental examination of the stability of SP candidates in the digested target matrices prior to selection of SP. In this study, two sets of spiked plasma were subjected to the precipitation/on-pellet-digestion [29], and then used for stability evaluation. Upon the completion of the digestion, the tryptic peptide mixtures were further incubated in either (i) digestion buffer at 37°C for up to 18 h or (ii) in the injection solution (i.e. the digested buffer acidified with 1% formic acid) at 4°C for up to 48h, respectively mimicking the environments to which the tissue digests are exposed during tryptic digestion or queuing in a cooled autosampler. Subsequent LC/MS measurements revealed that 3 were instable in plasma digest after incubation at 37°C for 18h. Additionally, 8 peptides fell out of the acceptable range (signal decrease < 20%) after sitting under autosampler conditions for 48h (Table 1).
Further investigation revealed that the use of these unstable peptides for quantification introduced substantial quantitative biases, which cannot be compensated for by the addition of isotopic-labeled peptides prior to digestion (data not shown), likely due to the delayed decay of the newly-released peptides from the protein compared to the spiked IS [21,24]. While the study of degradation mechanisms of unstable peptides is beyond the scope of this work, the results suggested the considerable prevalence of unstable tryptic peptides from an mAb. Furthermore, among the unstable peptides, some exhibited high LC/MS response (e.g. TVAAPSVFIFPPSDEQLK), which highlights the risk of selecting SP merely based on the sensitivity achieved.
Based on the sensitivity and stability of the candidates, the peptide GPSVFPLAPSSK and ASNLESGIPVR were selected as the two SP for the quantification of cT84.66 in mouse plasma.
3.1.4. Sample preparation and Nano-LC/SRM-MS strategy for batch-wise quantification
Owing to the use of a highly sensitive nano-LC/SRM-MS approach (discussed below) and the selection/optimization of the most sensitive peptides by OAO, enrichments on either protein level (e.g. affinity enrichment using Protein A beads) or peptide level (e.g. SISCAPA [35,36]) were unnecessary for the proposed pre-clinical PK study. For the preparation of the plasma sample, we employed and optimized a straightforward and reproducible precipitation/on-pellet digestion procedure [29] to efficiently remove non-protein interference (e.g. lipids, endogenous small molecules, salt etc) from plasma while maintaining a high recovery of tryptic peptides. Details are specified in the Experimental section. The key parameters such as the enzyme-to-substrate ratios and the incubation durations for each of the two digestion steps were extensively optimized to achieve the highest yields of the two SP to the extent possible. As demonstrated by the validation data (Table 2), the optimized preparation approach helped to achieve an accurate and reproducible quantification.
Table 2.
Precision and accuracy of the nano-LC/SRM-MS method to determine cT84.66 in mouse plasma
| Nominal concentration (ng/ml) |
Calculated concentration (ng/ml) (mean ± SD) |
Intra-day RSD (%) |
Inter-day RSD (%) |
Relative error (%) |
|---|---|---|---|---|
| 32.3 | 30.3 ± 4.1 | 11.2 | 10.3 | −5.9 |
| 645 | 686 ± 52 | 7.9 | 13.7 | 6.4 |
| 29025 | 27516 ± 2537 | 7.3 | 10.1 | −5.2 |
In order to achieve the sensitivity necessary to quantify the anticipated low mAb levels in plasma for an extended period (288h) after a single treatment, a sensitive nano-LC/nano-spray configuration was employed. A low-flow chromatography, when coupled to electrospray ionization MS, can drastically increase the analytical sensitivity for analysis in complex mixtures, as we demonstrated previously [24,37–41]. The unique designs of this setup help to alleviate problems often associated with low-flow LC systems and thus enable a robust quantification. One of the unique features is the use of a large-inner-diameter (300 µm ID) trap. Because of the special flow path configuration and the utilization of retrograde trapping/analysis flow directions, as described in our previous reports [29,42], this setup does not result in band broadening of the peptides. The large-ID trap provided a larger loading capacity and therefore significantly improved quantitative sensitivity and dynamic range over a regular trap (e.g. 75 µm ID). Another prominent benefit associated with the use a large-ID trap is that it provides exceptional reproducibility for nano-LC separation by dampening the pump noise that is prevalent in commercially available nano-LC systems. Moreover, an elevated separation temperature (55°C) was employed to improve the peak resolution and separation reproducibility, and to reduce the column back-pressure [43].
In order to achieve a reasonable throughput for batch-wise quantification, the gradient conditions were optimized to reduce the cycle time from that used for the LTQ/Orbitrap identification and OAO procedures, while maintaining sufficient selectivity of the SP peaks. In this process, the peaks of SP in plasma was unambiguously assigned by both comparing the elution profile with that of isotope-coded IS peptide and examining the ratio of quantification and confirmation transitions (variation within ±25% compared to reference standards). Eventually, a throughput of less than 45 min per sample (e.g. > 30 samples per day) was achieved. The optimized conditions are specified in the Experimental section. Typical chromatograms for the analysis of plasma samples are shown in Fig 3.
3.2. Selectivity and sensitivity of the nano-LC/SRM-MS approach
In order to assess the method selectivity, blank plasma samples from control group (saline treated, n=3) were experimentally processed and analyzed. No interference at the retention times of the two SP was observed (Fig 3A). This high degree of selectivity is attributable to the efficient sample cleanup, sufficient chromatographic separation, and the accurate selection of specific SP based on the OAO performed in plasma matrix.
The limit of detection (LOD), defined as the concentration in plasma that produce an S/N of approximately 3, was determined as 3.2 ng/mL (protein concentration, same below). The lower limit of quantification (LLOQ) was established as 12.9 ng/mL by using the digests derived from only 0.2 µL of plasma per injection (each preparation used 2 µL of plasma, cf. Experimental). The method developed here is considerably more sensitive compared with published immunoassays for anti-CEA antibodies (e.g. ELISA [44,45] and RIA [46]). Therefore, it permits an accurate PK assessment of cT84.66 for an extended period of time at relatively low dosages (e.g. 1.0 mg/kg, discussed below).
3.3. Linearity, accuracy and precision
To avoid quantitative bias that would have been introduced when synthesized peptides were used as the calibrator, as we observed in the preliminary studies in this work as well as in a previous project [24], purified cT84.66 protein with purities accurately measured by quantitative amino acid analysis (AAA), were used to prepare calibration curve in blank mouse plasma. For both of the SP, good linearity was observed over a range of three orders of magnitude, with regression coefficients (r2) ranged from 0.9819~0.9946. The wide dynamic range and good linearity are attributable to the quantitative sample preparation procedure, the specific and sensitive nano-LC/SRM-MS analysis and the use of isotope-labeled internal standards.
Precision and accuracy were evaluated by analyzing quality control (QC) samples at three concentration levels in the target matrix. The results are summarized in Table 2. The intra-assay and inter-assay precisions were within 11.2% and 13.7%, respectively. The accuracy, as estimated by the deviation between nominal and calculated concentrations of QC samples, ranged from −5.9 to 6.4%. As to the validation of LLOQ (12.9 ng/mL in plasma), the variation and accuracy were respectively 15.5% and 11.9%, as measured with 6 replicates.
3.4. Sensitive quantification of cT84.66 in mice after I.V. or S.C. injections
A comprehensive study of the PK of tumor-specific mAbs is critical to evaluate their therapeutic effects and to explore the diagnostic potentials. Nonetheless, due to the insufficient sensitivity and/or selectivity of the previously developed immune-based methods [44–46], it was difficult to characterize the PK of anti-CEA mAb where the systemic levels are low, e.g. when a low-dose regimen is used or a study over an extended period of time post treatment is required.
In this study, using the highly sensitive and selective nano-LC/SRM-MS strategy developed in this study, we were able to reliably investigate the PK of cT84.66 in mouse plasma over a 288h period after low-dose treatments. Using athymic Foxnu mice, three treatment groups were studied: the single I.V. at 1 mg/kg and the single S.C. treatments respectively at 1 and 10 mg/kg. The very low plasma sample consumption (2 µL per preparation and 0.2 µL per injection) posed a tremendous advantage in PK study based on mouse models, because it permits the collection of the entire serial of plasma samples from the same animal, and thus not only avoids the use of a much larger number of animals but also helps to diminish inter-individual variability.
The nano-LC/SRM-MS method is sufficiently sensitive to quantify the cT84.66 in all animals at each point, as expressed by the fact that the S/N are greater than 40 in all samples. The two SP were monitored and quantified in parallel for each sample and the mean of the two quantitative values was reported as the measured concentration. In any of the samples, the calculated deviation between the concentrations obtained independently by the two SP was lower than 25%, indicating the two SP selected here afforded highly reliable quantitative results.
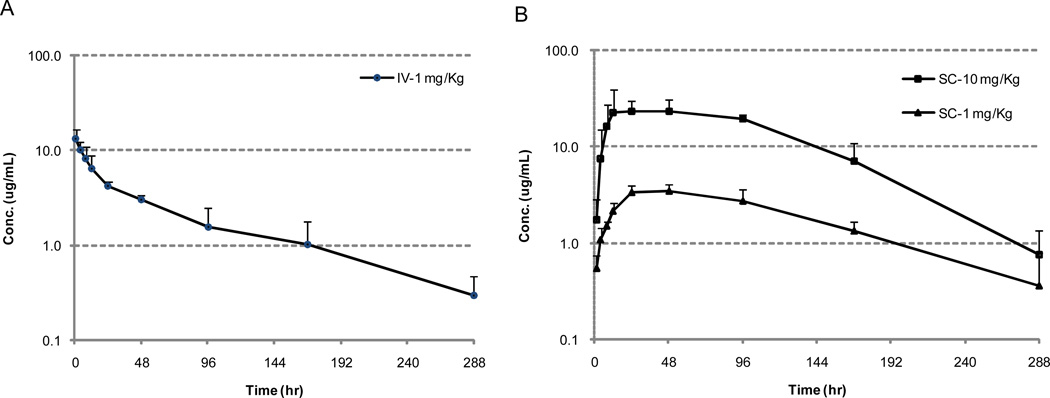
The profile of mean plasma concentration of cT84.66 vs. time after 1 mg/kg I.V. is presented in Fig. 4A, while the profiles after dosing 1 mg/kg and 10 mg/kg via S.C. are shown in Fig 4B. Based on the results, a non-compartmental analysis was conducted to estimate PK parameters (Table 3).
Figure 4.
Mean-plasma-concentration (logarithmic scale) vs. time (decimal scale) plots of cT84.66 mAb in athymic Foxnu mouse (A) after a single I.V. administration of 1 mg/kg cT84.66; (B) after a single S.C. administration of 1mg/kg and 10 mg/kg cT84.66, respectively (n = 4).
Table 3.
Pharmacokinetic parameter of cT84.66 following IV or SC administration to mice calculated by noncompartmental analysis (n = 4)
| Parameters | IV 1 mg/kg | SC 1 mg/kg | SC 10 mg/kg |
|---|---|---|---|
| tmax (h) | 1.0 | 36 | 36 |
| Cmax (µg/mL) | 13.2 ± 3.7 | 3.8 ± 0.6 | 29.9 ± 10.4 |
| AUC0-t (µg·h/mL) | 540.2 ± 175.4 | 530.4 ± 99.5 | 3401.3 ± 513.9 |
| AUC0-∝ (µg·h/mL) | 575.0 ± 197.0 | 592.4 ± 161.4 | 3456.1 ± 549.4 |
| t1/2 (h) | 73.0 ± 19.7 | 77.4 ± 34.6 | 45.4 ± 7.1 |
| CL (mL/h/kg) | 2.0 ± 0.7 | 1.9 ± 0.4 | 3.0 ± 0.5 |
In this particular mouse model, cT84.66 showed a ploy-exponential elimination following I.V. administration at 1 mg/kg dose. The terminal half-life (t1/2) was approximate 3 days. After S.C. administration at the same dose, cT84.66 demonstrated a slower but near-complete absorption with the bioavailability higher than 98% and a peak-concentration (Cmax) of 36 h. The t1/2 and clearance rate of this group were both consistent with those of the I.V. group. Interestingly, whereas Cmax increased proportionally with the dosage from the 1 mg/kg vs. 10 mg/kg S.C. group, mean AUC0~t did not increased proportionally (from 530.4 µg·h/mL to 3401.3 µg·h/mL). As both the extent (dose normalized Cmax) and the rate (tmax) of absorption remains similar between the two S. C. groups, the less-than-proportional increase of AUC may be primarily attributed to the elimination processes, as cT84.66 showed increased clearance and reduced t1/2 at 10 mg/kg. Many factors such as FcRn saturation may contribute to this phenomenal [4,26]. Mechanistic investigation of determinants of cT84.66 elimination kinetics, including evaluation of FcRn and off-target binding, are currently underway.
4. Conclusions
While the study of mAb PK necessitates a sensitive, selective and accurate quantification in complex matrices, the strategy for developing such a method using LC/SRM-MS remains elusive due to a number of technical difficulties (cf. Introduction). In this study, we employed a suite of technical advances to address the challenges. Firstly, an optimized nano-LC approach, which features relatively large loading capacity, high reproducibility and sensitivity, was employed for the analysis of mAb. Secondly, a high-throughput OAO procedure has been implemented for rapid and accurate peptide selection and SRM optimization. The procedure takes full advantage of the multiplex capability of SRM for optimization and employs an orthogonal array design to systematically evaluate the collective effects of several important parameters without using synthesized peptides. As demonstrated in this work, the OAO is capable of providing an accurate and reproducible optimization for numerous candidates in a single nano-LC/SRM-MS run. Additional benefits of the OAO procedure include that the experimental design and data processing can be readily automatized, and that the optimums are directly applicable to the biological analysis because the optimization is conducted in the target matrix. Thirdly, using the OAO-optimized parameters, the stability of SP candidates were evaluated in the plasma digest prior to SP selection. Whereas using unstable SP may introduce severe quantitative bias and variation, the issue of SP instability is often overlooked. In this study, we discovered that a substantial number of candidate peptides were not stable in the digestion or analysis conditions, including some with high responses, which demonstrates the risk of selecting SP merely based on the sensitivity without assessing stability.
Finally, in terms of the design of quantitative approaches, two unique SP from different domains of the mAb were used to quantify cT84.66, and the purified protein (as opposed to synthesized peptides) was employed for the calibrators. The dual-SP strategy enhances data reliability by gauging possible degradation and modification of the target protein, e.g. if deviation between the results by the two SP is greater than 25%, the data will be rejected. Using the purified protein for calibration would eliminate the severe quantitative bias that is likely to occur when synthesized peptides alone are used for calibrators.
The procedures for sample preparation and nano-LC/SRM-MS analysis were extensively optimized and the quantitative method was evaluated and validated. A high degree of sensitivity and selectivity were achieved and the method showed excellent linearity, accuracy and precision.
To show a proof of concept, the developed strategy was applied in the PK study of cT84.66 for a 288h-period after treatments with relatively low doses. The analytical method provides substantially higher sensitivity over the immune-based methods reported previously, and enabled the accurate measurements of cT84.66 in all of the samples with high S/N. Furthermore, the ultra-low sample size (2 µL plasma) permits the acquisition of an entire set of time course data from the same mouse, which represents a prominent advantage for PK study using small animals. Some interesting PK characteristics of I.V and S.C. administrations at various doses were observed, which are of high values for the subsequent modeling.
Based on the PK data (Fig 4) and the LLOQ achieved in this study, we speculate that the method is well capable to conduct a PK study at a much lower dose (e.g. 0.1 mg/kg) and/or over a substantial longer period. Therefore, this study demonstrates the feasibility of studying mAb PK at ultra-low doses using LC/MS-based methods without employing immuno-enrichment prior to analysis. Overall, the streamlined procedure developed here is generally adaptable to high-throughput development of sensitive and accurate methods for the quantification of mAb and other therapeutic proteins in various pharmaceutical and clinical matrices.
Supplementary Material
Highlights.
OAO procedure allowed for rational selection of optimal signature peptide and high-throughput method development;
The use of nano-LC/MS enabled highly sensitive quantification of mAb in plasma;
The use of two unique SP and protein calibrator enhanced quantitative accuracy;
The extremely-low sample consumption is desirable for small-animal models;
The method provided superior selectivity and sensitivity over immunoassays.
Acknowledgements
This work was mainly supported by an industrial consortium grant sponsored by the Center of Protein Therapeutics, SUNY-Buffalo to JQ and JPB, and also was supported principally by NIH grants CA118213 to JPB and DA027528 and U54HD071594 to JQ, a CTSA and an AHA award 12SDG9450036 to JQ. This work was also supported in part by the Pilot Studies program of the University at Buffalo Clinical and Translational Research Center and the Buffalo Translational Consortium.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mellman I, Coukos G, Dranoff G. Nature. 2011;480:480. doi: 10.1038/nature10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reichert JM. MAbs. 2012;4:1. doi: 10.4161/mabs.4.1.18719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nelson AL, Dhimolea E, Reichert JM. Nat Rev Drug Discov. 2010;9:767. doi: 10.1038/nrd3229. [DOI] [PubMed] [Google Scholar]
- 4.Wang W, Wang EQ, Balthasar JP. Clin Pharmacol Ther. 2008;84:548. doi: 10.1038/clpt.2008.170. [DOI] [PubMed] [Google Scholar]
- 5.Damen CW, Schellens JH, Beijnen JH. Hum Antibodies. 2009;18:47. doi: 10.3233/HAB-2009-0206. [DOI] [PubMed] [Google Scholar]
- 6.Hoofnagle AN, Wener MH. J Immunol Methods. 2009;347:3. doi: 10.1016/j.jim.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ezan E, Dubois M, Becher F. Analyst. 2009;134:825. doi: 10.1039/b819706g. [DOI] [PubMed] [Google Scholar]
- 8.Ezan E, Bitsch F. Bioanalysis. 2009;1:1375. doi: 10.4155/bio.09.121. [DOI] [PubMed] [Google Scholar]
- 9.Hagman C, Ricke D, Ewert S, Bek S, Falchetto R, Bitsch F. Anal Chem. 2008;80:1290. doi: 10.1021/ac702115b. [DOI] [PubMed] [Google Scholar]
- 10.Lesur A, Varesio E, Hopfgartner G. Anal Chem. 2010;82:5227. doi: 10.1021/ac100602d. [DOI] [PubMed] [Google Scholar]
- 11.Dubois M, Fenaille F, Clement G, Lechmann M, Tabet JC, Ezan E, Becher F. Anal Chem. 2008;80:1737. doi: 10.1021/ac7021234. [DOI] [PubMed] [Google Scholar]
- 12.Heudi O, Barteau S, Zimmer D, Schmidt J, Bill K, Lehmann N, Bauer C, Kretz O. Anal Chem. 2008;80:4200. doi: 10.1021/ac800205s. [DOI] [PubMed] [Google Scholar]
- 13.Ji C, Sadagopan N, Zhang Y, Lepsy C. Anal Chem. 2009;81:9321. doi: 10.1021/ac901800f. [DOI] [PubMed] [Google Scholar]
- 14.Gallien S, Duriez E, Domon B. J Mass Spectrom. 2011;46:298. doi: 10.1002/jms.1895. [DOI] [PubMed] [Google Scholar]
- 15.Pan S, Aebersold R, Chen R, Rush J, Goodlett DR, McIntosh MW, Zhang J, Brentnall TA. J Proteome Res. 2009;8:787. doi: 10.1021/pr800538n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prely LM, Paal K, Hermans J, van der Heide S, van Oosterhout AJ, Bischoff R. J Chromatogr A. 2012 doi: 10.1016/j.chroma.2012.02.076. in press. [DOI] [PubMed] [Google Scholar]
- 17.Stergachis AB, MacLean B, Lee K, Stamatoyannopoulos JA, MacCoss MJ. Nat Methods. 2011;8:1041. doi: 10.1038/nmeth.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mallick P, Schirle M, Chen SS, Flory MR, Lee H, Martin D, Ranish J, Raught B, Schmitt R, Werner T, Kuster B, Aebersold R. Nat Biotechnol. 2007;25:125. doi: 10.1038/nbt1275. [DOI] [PubMed] [Google Scholar]
- 19.Picotti P, Rinner O, Stallmach R, Dautel F, Farrah T, Domon B, Wenschuh H, Aebersold R. Nat Methods. 2010;7:43. doi: 10.1038/nmeth.1408. [DOI] [PubMed] [Google Scholar]
- 20.Maclean B, Tomazela DM, Abbatiello SE, Zhang S, Whiteaker JR, Paulovich AG, Carr SA, Maccoss MJ. Anal Chem. 2010;82:10116. doi: 10.1021/ac102179j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arsene CG, Ohlendorf R, Burkitt W, Pritchard C, Henrion A, O'Connor G, Bunk DM, Guttler B. Anal Chem. 2008;80:4154. doi: 10.1021/ac7024738. [DOI] [PubMed] [Google Scholar]
- 22.Ionescu R, Vlasak J. Anal Chem. 2010;82:3198. doi: 10.1021/ac902752e. [DOI] [PubMed] [Google Scholar]
- 23.Brady LJ, Martinez T, Balland A. Anal Chem. 2007;79:9403. doi: 10.1021/ac7017469. [DOI] [PubMed] [Google Scholar]
- 24.Cao J, Gonzalez-Covarrubias V, Straubinger RM, Wang H, Duan X, Yu H, Qu J, Blanco JG. Anal Chem. 2010;82:2680. doi: 10.1021/ac902314m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blumenthal RD. Curr Opin Mol Ther. 2004;6:90. [PubMed] [Google Scholar]
- 26.Urva SR, Balthasar JP. MAbs. 2010;2:67. doi: 10.4161/mabs.2.1.10781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferl GZ, Wu AM, DiStefano JJ., 3rd Ann Biomed Eng. 2005;33:1640. doi: 10.1007/s10439-005-7410-3. [DOI] [PubMed] [Google Scholar]
- 28.Diez C, Barrado E, Marinero P, Sanz M. J Chromatogr A. 2008;1180:10. doi: 10.1016/j.chroma.2007.12.036. [DOI] [PubMed] [Google Scholar]
- 29.Duan X, Young R, Straubinger RM, Page B, Cao J, Wang H, Yu H, Canty JM, Qu J. J Proteome Res. 2009;8:2838. doi: 10.1021/pr900001t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qu J, Straubinger RM. Rapid Commun Mass Spectrom. 2005;19:2857. doi: 10.1002/rcm.2138. [DOI] [PubMed] [Google Scholar]
- 31.Anderson L, Hunter CL. Mol Cell Proteomics. 2006;5:573. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. Mol Cell Proteomics. 2007;6:2212. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fusaro VA, Mani DR, Mesirov JP, Carr SA. Nat Biotechnol. 2009;27:190. doi: 10.1038/nbt.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker SH, Papas BN, Comins DL, Muddiman DC. Anal Chem. 2010;82:6636. doi: 10.1021/ac101227a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whiteaker JR, Zhao L, Abbatiello SE, Burgess M, Kuhn E, Lin C, Pope ME, Razavi M, Anderson NL, Pearson TW, Carr SA, Paulovich AG. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.005645. M110 005645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anderson NL, Anderson NG, Haines LR, Hardie DB, Olafson RW, Pearson TW. J Proteome Res. 2004;3:235. doi: 10.1021/pr034086h. [DOI] [PubMed] [Google Scholar]
- 37.Duan X, Weinstock-Guttman B, Wang H, Bang E, Li J, Ramanathan M, Qu J. Anal Chem. 2010;82:2488. doi: 10.1021/ac902869y. [DOI] [PubMed] [Google Scholar]
- 38.Qu J, Qu Y, Straubinger RM. Anal Chem. 2007;79:3786. doi: 10.1021/ac062184r. [DOI] [PubMed] [Google Scholar]
- 39.Tu C, Li J, Young R, Page BJ, Engler F, Halfon MS, Canty JM, Jr, Qu J. Anal Chem. 2011;83:4802. doi: 10.1021/ac200376m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu H, Straubinger RM, Cao J, Wang H, Qu J. J Chromatogr A. 2008;1210:160. doi: 10.1016/j.chroma.2008.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duan X, Engler FA, Qu J. J Mass Spectrom. 2010;45:1477. doi: 10.1002/jms.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang H, Straubinger RM, Aletta JM, Cao J, Duan X, Yu H, Qu J. J Am Soc Mass Spectrom. 2009;20:507. doi: 10.1016/j.jasms.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Farias SE, Kline KG, Klepacki J, Wu CC. Anal Chem. 2010;82:3435. doi: 10.1021/ac100359p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Urva SR, Yang VC, Balthasar JP. J Immunoassay Immunochem. 2010;31:1. doi: 10.1080/15321810903404772. [DOI] [PubMed] [Google Scholar]
- 45.Haidopoulos D, Konstadoulakis MM, Antonakis PT, Alexiou DG, Manouras AM, Katsaragakis SM, Androulakis GF. Eur J Surg Oncol. 2000;26:742. doi: 10.1053/ejso.2000.0996. [DOI] [PubMed] [Google Scholar]
- 46.Sharkey RM, Goldenberg DM, Murthy S, Pinsky H, Vagg R, Pawlyk D, Siegel JA, Wong GY, Gascon P, Izon DO, et al. Cancer. 1993;71:2082. doi: 10.1002/1097-0142(19930315)71:6<2082::aid-cncr2820710625>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.