ABSTRACT
Clostridium perfringens causes various digestive system disease symptoms in pigs. In the present study, 11 C. perfringens isolates were obtained from diarrheic piglets and 18 from healthy piglets. All of the C. perfringens isolates were shown to be type A using a multiplex PCR assay. The β2 toxin gene was detected in 27/29 C. perfringens isolates, i.e., 81% (9/11) of diarrheic piglets and 100% (18/18) of healthy piglets, and all of the genes had the same sequence. In conclusion, the β2 toxin gene of C. perfringens was distributed widely in Korean piglets regardless of the incidence of diarrhea, and there was no clear relationship with enteric disease. A pulsed-field gel electrophoresis analysis of DNA digested using SmaI demonstrated the non-clonal spread of C. perfringens isolates from piglets.
Keywords: Clostridium perfringens, multiplex PCR, piglet, pulsed-field gel electrophoresis, toxin
Clostridium perfringens is abundant in the soil and in the intestinal contents of animals and humans [6, 10, 15, 23]. However, C. perfringens is a pathogen that induces digestive diseases, such as diarrhea and hemorrhagic and necrotic enteritis in pigs. The pathogenicity of C. perfringens is associated with its major toxins, and C. perfringens isolates are divided into five types (A, B, C, D and E) based on their major toxins (alpha, beta, epsilon and iota toxins) [12, 24]. In South Korea, C. perfringens type A (alpha toxin) is a common isolate from pigs, whereas C. perfringens type C (alpha and beta toxins) is isolated occasionally from pigs with diarrhea and hemorrhagic and necrotic enteritis [27, 28]. However, C. perfringens type A is a ubiquitous pathogen, which can also cause necrotic enteritis and malignant edema [21]. A toxin produced by C. perfringens types A and C, which is referred to as β2 toxin, has a role in the causation of digestive system diseases in horses, pigs, dogs and calves [4, 11]. The β2 toxin sequence shares no significant amino acid sequence homology with C. perfringens β toxin (only 15% identity) or any other known toxin [16]. The β2 toxin is plasmid-borne, at least in some strains, which suggests that there is the potential for mobility and the subsequent transfer of the β2 toxin gene among C. perfringens strains [11, 21, 26]. A high percentage of diarrheic pigs with C. perfringens type A produces the β2 toxin [17, 25]. However, several recent studies have demonstrated the widespread distribution of β2-toxigenic C. perfringens isolates in diseased poultry and healthy calves and pigs [1, 8, 22].
Epidemiological studies of C. perfringens have also been conducted using random amplified polymorphic DNA (RAPD), multi-locus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE) [2, 5, 14, 18,19,20]. PFGE was used for subtyping to provide information on the molecular relatedness of isolates for comparison with other analytical methods [19]. C. perfringens has been isolated from many pigs in South Korea, but no major studies have been performed to characterize the strains or their epidemiology.
Thus, the present study investigated the distribution of alpha (cpa gene), beta (cpb gene), epsilon (etx gene), iota (iap gene), β2 (cpb2 gene) toxins and enterotoxin (cpe gene) in C. perfringens isolates from diarrheic and healthy piglets in South Korea. The genetic characteristics of C. perfringens isolates from diarrheic piglets were also compared using a PFGE assay.
Diarrheal fecal samples were collected from pigs with diarrheal diseases by the Optifarm Solution Co. (Optifarm Co., Osong, Korea) of the Animal Appraisal Organization. Four-hundred diarrheal fecal samples were obtained from seven provinces (Gyeonggi, Chungbuk, Chungnam, Jeonbuk, Jeonnam, Gyeongbuk and Gyeongnam) throughout South Korea between 2009 and 2011. The Animal and Plant Quarantine Agency also collected 136 fecal samples from healthy pigs in six provinces (Gyeonggi, Gyeongnam, Chungnam, Gangwon, Jeonbuk and Jeju) for diarrheal disease monitoring during 2010.
All of the fecal samples were collected aseptically, inoculated into brain heart infusion (BHI) agar (Difco, Beckton, Dickinson and Co., Sparks, MD, U.S.A.) containing 5% sheep blood and incubated anaerobically at 37°C for 24 hr. Two to three presumptive colonies with characteristic dual hemolytic zones were identified by biochemical tests and 16S rRNA sequencing. The genomic DNA was extracted from C. perfringens using a Genomic DNA Extraction Kit (Intron Co., Seongnam, South Korea) and stored at −20°C until use.
Multiplex PCR was performed to detect the toxin genes, as described previously [3]. The β2 toxin gene (cpb2 gene) of C. perfringens was amplified using a primer set (5′-GAAAGGTAATGGAGAA-3′ and 5′-GCAGAATCAGGATTTT-3′), as described previously [13]. After confirmation of the DNA fragments on a 1% agarose gel, they were extracted from the gel and purified using a Gel Extraction kit (Qiaquick, Qiagen, Valencia, CA, U.S.A.), before sequencing by the Sanger method using an ABI Prism 373 Genetic Analyzer (Applied Biosystems, Foster City, CA, U.S.A.). The cpb2 gene nucleotide sequence data generated in the present study and data obtained from NCBI GenBank were analyzed using CLUSTALW 1.8X, which generated .aln and .nxs files. The .aln files were converted into .meg files using MEGA 4.1. A neighbor-joining (NJ) tree was constructed using Kimura’s two-parameter model, assuming uniform rates of change among sites.
PFGE was performed according to a published method [19] with a small modification using 40U SmaI (MBI Fermentas, Hanover, MD, U.S.A.), and the digested genomic DNA fragments were separated using a CHEF-DR III system (Bio-Rad, Richmond, CA, U.S.A.) at 12°C for 20 hr with an initial pulse for 5 sec and a final pulse for 40 sec at 6 V/cm at an angle of 120°, where a lambda DNA marker (Sigma) was employed as a DNA molecular weight marker. The DNA bands were stained with ethidium bromide, and the DNA banding patterns were analyzed using BioNumerics software (Applied Maths, Kortrijk, Belgium) to calculate the Dice correlation coefficients and to generate a dendrogram, where clustering was performed using the unweighted pair-group method with arithmetic means (UPGMA).
Among the 400 diarrheic fecal samples, C. perfringens alone was isolated from 11 samples without other pathogens (i.e., enterotoxigenic E. coli, Salmonella spp., transmissible gastroenteritis virus, porcine epidemic diarrhea virus and rotavirus). C. perfringens isolates (n=11) were obtained from three regions, as follows: Gyeonggi (n=3), Gyeongnam (n=2) and Chungnam (n=6) (Table 1). Eighteen C. perfringens isolates were obtained from healthy piglets without diarrhea. These isolates (n=18) were obtained from six regions, as follows: Gyeonggi (n=2), Gyeongnam (n=3), Chungnam (n=6), Gangwon (n=1), Jeonbuk (n=3) and Jeju (n=3) (Table 1). Table 1 shows the distributions of the six toxin genes (cpa, cpb, etx, iap, cpb2 and cpe) among the C. perfringens isolates (n=29). All of the C. perfringens isolates (n=11) from diarrheic samples were confirmed as type A, and the cpb2 gene was present in 81% of the isolates (9/11), whereas the cpe gene was not detected in any of the strains. The 18 C. perfringens isolates from healthy piglets were all confirmed as type A, and they possessed the cpb2 gene.
Table 1. Results of the multiplex PCR analysis of the toxin genes* from 29 Clostridium perfringens isolates from piglets.
| Clinical sign | Strain No. | Province | Date | Major toxin genes | Other toxin genes | ||||
|---|---|---|---|---|---|---|---|---|---|
| cpa | cpb | etx | iap | cpb2 | cpe | ||||
| Diarrhea | KVCC-BA1200001 | Gyeonggi | Jan. 2009 | + | – | – | – | + | – |
| KVCC-BA1200002 | Gyeongnam | Feb. 2009 | + | – | – | – | – | – | |
| KVCC-BA1200010 | Gyeonggi | Dec. 2009 | + | – | – | – | + | – | |
| KVCC-BA1200015 | Gyeongnam | Oct. 2010 | + | – | – | – | + | – | |
| KVCC-BA1200017 | Chungnam | Jul. 2011 | + | – | – | – | + | – | |
| KVCC-BA1200018 | Chungnam | Aug. 2011 | + | – | – | – | + | – | |
| KVCC-BA1200020 | Gyeonggi | Aug. 2011 | + | – | – | – | + | – | |
| KVCC-BA1200021 | Chungnam | Sep. 2011 | + | – | – | – | + | – | |
| KVCC-BA1200022 | Chungnam | Sep. 2011 | + | – | – | – | + | – | |
| KVCC-BA1200024 | Chungnam | Sep. 2011 | + | – | – | – | + | – | |
| KVCC-BA1200025 | Chungnam | Oct. 2011 | + | – | – | – | – | – | |
| No diarrhea | KVCC-BA1200285 | Gyeongnam | Mar. 2010 | + | – | – | – | + | – |
| KVCC-BA1200286 | Gyeongnam | Mar. 2010 | + | – | – | – | + | – | |
| KVCC-BA1200288 | Gangwon | Mar. 2010 | + | – | – | – | + | – | |
| KVCC-BA1200289 | Gyeongnam | Mar. 2010 | + | – | – | – | + | – | |
| KVCC-BA1200290 | Gyeonggi | Mar. 2010 | + | – | – | – | + | – | |
| KVCC-BA1200291 | Gyeonggi | Mar. 2010 | + | – | – | – | + | – | |
| KVCC-BA1200292 | Jeju | Mar. 2010 | + | – | – | – | + | – | |
| KVCC-BA1200293 | Jeju | Mar. 2010 | + | – | – | – | + | – | |
| KVCC-BA1200295 | Jeju | Mar. 2010 | + | – | – | – | + | – | |
| KVCC-BA1200297 | Chungnam | Apr. 2010 | + | – | – | – | + | – | |
| KVCC-BA1200298 | Chungnam | Apr. 2010 | + | – | – | – | + | – | |
| KVCC-BA1200299 | Chungnam | Apr. 2010 | + | – | – | – | + | – | |
| KVCC-BA1200300 | Chungnam | Apr. 2010 | + | – | – | – | + | – | |
| KVCC-BA1200303 | Chungnam | Apr. 2010 | + | – | – | – | + | – | |
| KVCC-BA1200306 | Chungnam | Apr. 2010 | + | – | – | – | + | – | |
| KVCC-BA1200308 | Jeonbuk | Apr. 2010 | + | – | – | – | + | – | |
| KVCC-BA1200325 | Jeonbuk | Apr. 2010 | + | – | – | – | + | – | |
| KVCC-BA1200326 | Jeonbuk | Apr. 2010 | + | – | – | – | + | – | |
*The cpa, cpb, etx, iap, cpb2 and cpe genes encode the alpha, beta, epsilon, iota, β2 and enterotoxin toxins, respectively.
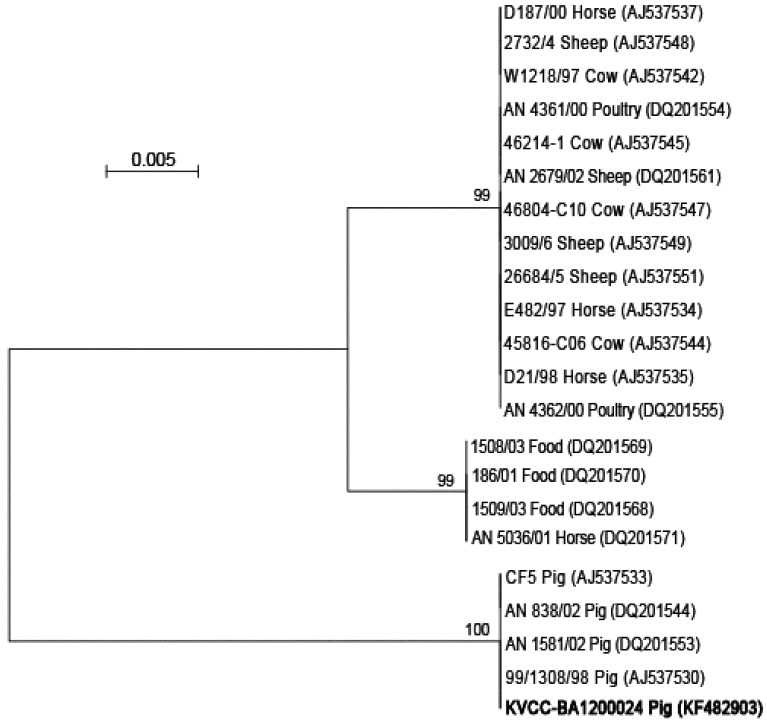
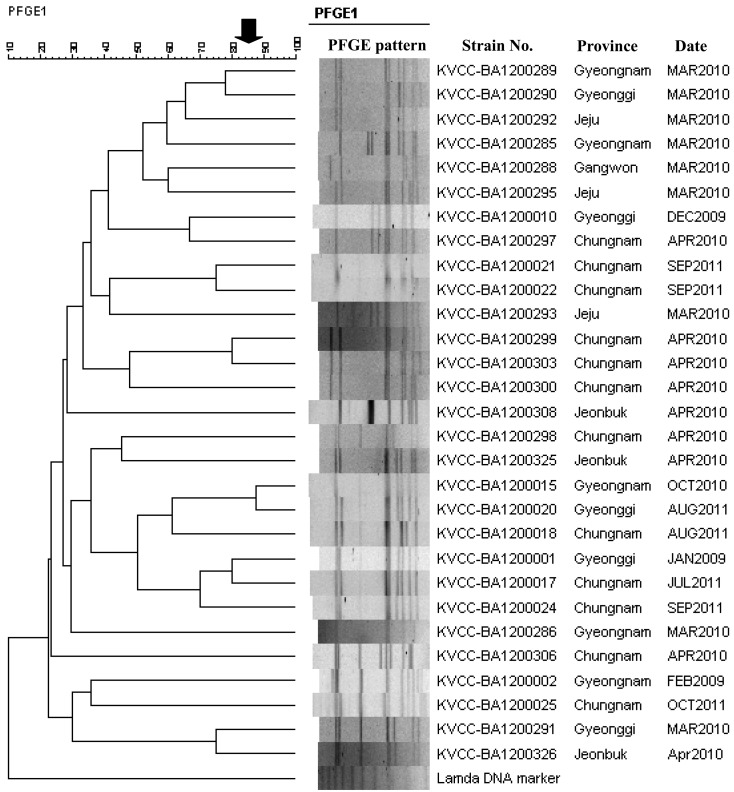
Sequence analysis of the 478 nucleotides of the cpb2 gene showed that the 27 cpb2 genes from Korean isolates all shared the same sequence. To determine the level of homology, the cpb2 genes were compared with those of isolates from animals (cow, horse, pig, poultry and sheep) and food deposited in the NCBI GenBank, which showed that the cpb2 gene (KVCC-BA1200024; GenBank accession no. KF482903) of the Korean piglet isolates clustered with the European enteritis pig group in the phylogenetic tree (Fig. 1). Figure 2 shows the DNA banding patterns of the C. perfringens isolates obtained by PFGE analysis. PFGE of the C. perfringens isolates generated 7–10 DNA fragments, which ranged in size from 48.5 to 485 kb. There was low similarity between the C. perfringens isolates from healthy and diarrheic piglets, which indicates that C. perfringens exhibits genetic variation in pig farms.
Fig. 1.
Phylogenetic relationships among the β2 toxin gene sequences. The trees were constructed using the neighbor-joining method with Mega 4.1 based on 1,000 bootstrap replicates. The cpb2 gene (KF482903) identified in Korean piglet isolates is indicated in bold.
Fig. 2.
Pulsed-field gel electrophoresis profiles of Clostridium perfringens isolates from piglets, which were analyzed using the BioNumerics program (Applied Maths) to generate a dendrogram based on the Dice coefficients.
The alpha and beta toxins produced by C. perfringens cause porcine digestive system diseases, such as diarrhea or necrotizing enterocolitis [8]. The β2toxin, which is comparable to β toxin, is also known to cause these diseases [9]. Therefore, the β2 toxin gene has been investigated actively in European countries, the USA and African countries [14]. Bueschel et al. reported that β2 was present in 11.1% of C. perfringens isolates from healthy pigs, whereas the majority of isolates (90.9%) from pigs with enteritis or diarrhea were positive for β2, which strongly suggests that this gene has a significant association with diarrhea in piglets [4].
In the present study, 29 C. perfringens isolates (11 from diarrheic and 18 from healthy piglets) were detected where only cpa was present as a major toxin. Nine positive isolates for the cpb2 gene were also obtained from diarrheic piglets and 18 from healthy piglets. Interestingly, β2-toxigenic C. perfringens was isolated frequently from healthy piglets in South Korea, which contrasts with previous reports from the U.S.A. and Europe. Recently, Farzan et al. reported that the presence of the cpb2 gene in the intestinal contents of normal and diarrheic piglets in the USA did not differ significantly, although further work is required to confirm whether the presence of the cpb2 gene in C. perfringens has a pathogenic role during enteric infections of piglets [7].
In conclusion, the β2 toxin gene of C. perfringens did not have a clear relationship with enteric disease. However, the β2 toxin gene of C. perfringens might be toxigenic in healthy piglets, and it could cause enteric pathogenicity. The PFGE analysis of 29 C. perfringens isolates from piglets showed that they grouped into 28 separate clusters based on a criterion of 85% similarity, which suggests that C. perfringens is not geographically restricted and that it is widely distributed.
The present study indicates that careful monitoring of the distribution and toxin patterns of C. perfringens will be necessary to develop preventive measures to facilitate disease control.
Acknowledgments
We are grateful to Optifarm Solution Laboratories for providing Clostridium perfringens isolates.
REFERENCES
- 1.Badagliacca P., Di. Provvido A., Scattolini S., Pompei G., Di. Giannatale E.2010. Toxin genotyping of Clostridium perfringens strains using a polymerase chain reaction protocol. Vet. Ital. 46: 113–118 [PubMed] [Google Scholar]
- 2.Baker A. A., Davis E., Rehberger T., Rosener D.2010. Prevalence and diversity of toxigenic Clostridium perfringens and Clostridium difficile among swine herds in the midwest. Appl. Environ. Microbiol. 76: 2961–2967. doi: 10.1128/AEM.02459-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baums C. G., Schotte U., Amtsberg G., Goethe R.2004. Diagnostic multiplex PCR for toxin genotyping of Clostridium perfringens isolates. Vet. Microbiol. 100: 11–16. doi: 10.1016/S0378-1135(03)00126-3 [DOI] [PubMed] [Google Scholar]
- 4.Bueschel D. M., Jost B. H., Billington S. J., Trinh H. T., Songer J. G.2003. Prevalence of cpb2, encoding beta2 toxin, in Clostridium perfringens field isolates: correlation of genotype with phenotype. Vet. Microbiol. 94: 121–129. doi: 10.1016/S0378-1135(03)00081-6 [DOI] [PubMed] [Google Scholar]
- 5.Chalmers G., Bruce H. L., Hunter D. B., Parreira V. R., Kulkarni R. R., Jiang Y. F., Prescott J. F., Boerlin P.2008. Multilocus Sequence Typing Analysis of Clostridium perfringens isolates from Necrotic Enteritis outbreaks in Broiler Chicken Populations. J. Clin. Microbiol. 46: 3957–3964. doi: 10.1128/JCM.01548-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fach P., Popoff M. R.1997. Detection of enterotoxigenic Clostridium perfringens in food and fecal samples with a duplex PCR and the slide latex agglutination test. Appl. Environ. Microbiol. 63: 4232–4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farzan A., Kircanski J., Delay J., Soltes G., Songer J. G., Friendship R., Prescott J. F.2013. An investigation into the association between cpb2-encoding Clostridium perfringens type A and diarrhea in neonatal piglets. Can. J. Vet. Res. 77: 45–53 [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrarezi M. C., Cardoso T. C., Dutra I. S.2008. Genotyping of Clostridium perfringens isolated from calves with neonatal diarrhea. Anaerobe 14: 328–331. doi: 10.1016/j.anaerobe.2008.12.001 [DOI] [PubMed] [Google Scholar]
- 9.Garmory H. S., Chanter N., French N. P., Bueschel D., Songer J. G., Titball R. W.2000. Occurrence of Clostridium perfringens β2-toxin amongst animals, determined using genotyping and subtyping PCR assays. Epidemiol. Infect. 124: 61–67. doi: 10.1017/S0950268899003295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gharaibeh S., Al Rifai R., Al-Majali A.2010. Molecular typing and antimicrobial susceptibility of Clostridium perfringens from broiler chickens. Anaerobe 16: 586–589. doi: 10.1016/j.anaerobe.2010.10.004 [DOI] [PubMed] [Google Scholar]
- 11.Gibert M., Jolivet-Reynaud C., Popoff M. R.1997. Beta2 toxin, a novel toxin produced by Clostridium perfringens. Gene 203: 65–73. doi: 10.1016/S0378-1119(97)00493-9 [DOI] [PubMed] [Google Scholar]
- 12.Hatheway C. L.1990. Toxigenic Clostridia. Clin. Microbiol. Rev. 3: 66–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herholz C., Miserez R., Nicolet J., Frey J., Popoff M., Gibert M., Gerber H., Straub R.1999. Prevalence of β2-Toxigenic Clostridium perfringens in Horses with intestinal disorders. J. Clin. Microbiol. 37: 358–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johansson A., Aspan A., Bagge E., BÅverud V., Engström B. E., Johansson K. E.2006. Genetic diversity of Clostridium perfringens type A isolates from animals, food poisoning outbreaks and sludge. BMC Microbiol. 6: 47. doi: 10.1186/1471-2180-6-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joshy L., Chaudhry R., Dhawan B., Kumar L., Das B. K.2006. Incidence and characterization of Clostridium perfringens isolated from antibiotic-associated diarrhoeal patients: a prospective study in an Indian hospital. J. Hosp. Infect. 63: 323–329. doi: 10.1016/j.jhin.2005.12.015 [DOI] [PubMed] [Google Scholar]
- 16.Jost B. H., Billington S. J., Trinh H. T., Bueschel D. M., Songer J. G.2005. Atypical cpb2 genes, Encoding beta2-toxin in Clostridium perfringens isolates of nonporcine origin. Infect. Immun. 73: 652–656. doi: 10.1128/IAI.73.1.652-656.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klaasen H. L., Molkenboer M. J., Bakker J., Miserez R., Hani H., Frey J., Popoff M. R., andvan den Bosch J. F.1999. Detection of β2 toxin gene of Clostridium perfringens in diarrhoeic piglets in the Netherlands and Switzerland. FEMS Immunol. Med. Microbiol. 24: 325–332 [DOI] [PubMed] [Google Scholar]
- 18.Leflon-Guibout V., Pons J. L., Heym B., Nicolas-Chanoine M. H.1997. Typing of Clostridium perfringens strains by use of Random Amplified Polymorphic DNA (RAPD) system in comparison with zymotyping. Anaerobe 3: 245–250. doi: 10.1006/anae.1997.0094 [DOI] [PubMed] [Google Scholar]
- 19.Lin Y. T., Labbe R.2003. Enterotoxigenicity and genetic relatedness of Clostridium perfringens isolates from retail foods in the United States. Appl. Environ. Microbiol. 69: 1642–1646. doi: 10.1128/AEM.69.3.1642-1646.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lukinmaa S., Takkunen E., Siitonen A.2002. Molecular epidemiology of Clostridium perfringens related to food-borne outbreaks of disease in Finland from 1984 to 1999. Appl. Environ. Microbiol. 68: 3744–3749. doi: 10.1128/AEM.68.8.3744-3749.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petit L., Gibert M., Popoff M. R.1999. Clostridium perfringens: toxinotype and genotype. Trends Microbiol. 7: 104–110. doi: 10.1016/S0966-842X(98)01430-9 [DOI] [PubMed] [Google Scholar]
- 22.Schotte U., Truyen U., Neubauer H.2004. Significance of β2-toxigenic Clostridium perfringens infections in animals and their predisposing factors-A Review. J. Vet. Med. B Infect. Dis. Vet. Public Health 51: 423–426. doi: 10.1111/j.1439-0450.2004.00802.x [DOI] [PubMed] [Google Scholar]
- 23.Songer J. G.1996. Clostridial Enteric Disease of Domestic Animals. Clin. Microbiol. Rev. 9: 216–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Titball R. W., Naylor C. E., Basak A. K.1999. The Clostridium perfringens alpha-toxin. Anaerobe 5: 51–64. doi: 10.1006/anae.1999.0191 [DOI] [PubMed] [Google Scholar]
- 25.van Asten A. J., Nikolaou G. N., Grone A.2010. The occurrence of cpb2-toxigenic Clostridium perfringens and the possible role of the β2-toxin in enteric disease of domestic animals, wild animals and humans. Vet. J. 183: 135–140. doi: 10.1016/j.tvjl.2008.11.005 [DOI] [PubMed] [Google Scholar]
- 26.Waters M., Savoie A., Garmory H. S., Bueschel D., Popoff M. R., Songer J. G., Titball R. W., McClane B. A., Sarker M. R.2003. Genotyping and Phenotyping of Beta2-Toxigenic Clostridium perfringens fecal isolates associated with gastrointestinal disease in piglets. J. Clin. Microbiol. 41: 3584–3591. doi: 10.1128/JCM.41.8.3584-3591.2003 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Yeh J. G., Park K. Y., Cho S. K.1993. Studies on the Clostridium perfringens type C infection of pig in Korea. Korea. J. Vet. Res. 33: 419–427 [Google Scholar]
- 28.Yoo H. S., Lee S. U., Park K. Y., Park Y. H.1997. Molecular typing and epidemiological survey of prevalence of Clostridium perfringens types by Multiplex PCR. J. Clin. Microbiol. 35: 228–232 [DOI] [PMC free article] [PubMed] [Google Scholar]