Abstract
Background
A number of studies have investigated the relationship between fibroblast growth factor receptor1 (FGFR1) gene copy number and survival in non-small cell lung cancer (NSCLC) patients. However, conclusions reported by different parties seem to be inconsistent, especially regarding the differences among different histopathologic subtypes. To derive a more precise estimate of the prognostic significance of FGFR1 gene copy number, we have reviewed published studies and carried out a meta-analysis.
Methods
The meta-analysis was conducted in accordance with PRISMA guidelines. The required data for estimation of individual hazard ratios (HRs) for survival were extracted from the publications and an overall HR was calculated.
Results
We identified 6 eligible studies, all dealing with NSCLC. The global quality score ranged 32.5-80%, with a median of 53.33%. For FGFR1 amplification in three studies including differed according to histological type, the overall RR was 0.86 which 95% confidence interval (CI) was 0.75 to 0.99 and P value was 0.048. Combined HR for the six evaluable studies was 1.17 (95% CI: 0.95 to 1.43). In the subgroup of squamous cell lung cancer (SQCC), the combined HR was 1.24 (95% CI: 0.89 to 1.73). For the Asian populations’ studies, the combined HR was 1.67 (95% CI: 1.1 to 2.52).
Conclusions
FGFR1 amplification significantly was more frequent in SQCC. FGFR1 was not associated with poorer survival in patients with NSCLC. Furthermore studies will be needed in terms of survival implications.
Keywords: Lung cancer, meta-analysis, fibroblast growth factor receptor1 (FGFR1), prognosis, survival, systemic review
Introduction
Lung cancer is the most common cause of cancer death in the world (1). Due to the lack of specific symptoms, most of lung cancer patients are in the mid or late stage when they were diagnosed. Although diagnostic approaches, treatment techniques and surgical levels towards lung cancer have been improved greatly in recent years, most lung cancer patients still have bad prognosis with survival rate of five years fluctuating around 15% (2). Therefore, it was of great significance in treatment selection and patient survival rate increase to look for factors relevant to lung cancer prognosis.
Fibroblast growth factor receptor1 (FGFR1) recently has become a hot topic in the research of cancer driver gene. In 2010, Weiss et al. (3), a German scientist, found out that this gene had a large number of amplification in squamous carcinoma specimen. In many subsequent studies, researchers analyzed the role FGFR1 amplification played during lung cancer prognosis; however, conclusions reported by different parties seem to be inconsistent, especially regarding the differences among different histopathologic subtypes. Some believe that squamous cell lung cancer (SQCC) patients with FGFR1 amplification have poor prognosis; others believe that FGFR1 expression was not related to prognosis of SQCC or non-small cell lung cancer (NSCLC). We apply Meta-analysis method to the past research results as a comprehensive quantitative analysis in order to evaluate the effects of FGFR1 amplification on NSCLC prognosis.
Material and methods
Identification and selection of relevant studies
Criteria for eligibility of a study to the meta analysis were: (I) to deal with NSCLC only; (II) to evaluate the correlation between FGFR1 gene copy number and patient survival and analyse FGFR1 in the primary tumour (not in metastatic tissue); (III) to be published as a full paper in the English language literature. Studies published in abstract form were excluded; (IV) to find providing sufficient information, such as P value and survival curve, for the estimation of hazard ratio (HR) and 95% confidence interval (CI); (V) measurement methods, including reverse transcription-polymerase chain reaction (RT-PCR), fluorescent in situ hybridization (FISH) and immunohistochemistry (IHC).
Search for studies was performed using the electronic database PubMed, Embase, Web of Science and Google Scholar until March 31, 2014. The search strategy included the keywords or title combined “lung cancer”, “lung carcinoma”, “FGFR1”, “Fibroblast Growth Factor Receptor 1”, “survival”, and “prognosis”. All studies matching the eligibility criteria were mentioned previously. Two investigators (Wen Yang and Yan-Wen Yao) independently deal with the relevant studies’ data. Any incongruity was settled through discussion until a consensus was reached.
Data extraction
HRs and their 95% CIs were used to combine the data. When these information were described in text or tables, we acquired these values directly from two articles (3,4). When these statistical variables were not given directly in the article, they were calculated from survival curves in four articles (5-8) using the methods which reported by Parmar et al. (9). We extracted data on characteristics of studies and patients, measurements, results and so on. In particular, in each report we recorded the first author, country of origin, year of publication, number of patients analyzed, staging of tumor, method of FGFR1 gene copy number detection, cutoff value, histology, number of patients of FGFR1 gene amplification, time of follow-up, and OS data. The primary outcome of the meta-analysis was overall survival.
Quality assessment
Two investigators (Wen Yang and Yan-Wen Yao) independently assessed the quality of the selected studies using the European lung cancer working party quality scale for biological prognostic factors for lung cancer (10). This tool comprises four quality parameters: scientific design, laboratory methodology, generalizability and results analysis. Each category had a maximum score from 8 to 10 points, and the overall score was 40 points. The final scores were expressed as percentages, ranging from 0 to 100%. The Higher values were obtained, the better methodological quality was indicated. The scores were compared and a consensus value for each item was reached in meetings of all investigators needed to be present.
Statistical analysis
Survival data from each study were analyzed in terms of the HRs and 95% CI directly or calculated by Kaplan-Meier curves as previously described by Parmar et al. (9) and Tierney et al. (11).
Statistical heterogeneity was measured using the chi squared Q test and the I2 statistic. Significant heterogeneity was determined at a P value less than 0.10. I2 was used to quantify inconsistencies, where a value more than 50% indicates visible heterogeneity. When visible heterogeneity was observed, the random effects model was used. When no visible heterogeneity was observed, the fixed effects model was used for meta-analysis (12). The individual HR estimates were combined into an overall HR, which less than 1 implied a poor survival for the group with FGFR1 gene amplification by convention. This impact of FGFR1 gene copy number on survival was considered as statistically significant if the 95% CI for the overall HR did not overlap 1.
We assessed the subgroup additionally, including the histological type (NSCLC or SQCC), country of origin (Asian or non-Asian). The slection of the model of subgroup analysis was based on the convention, as previously described. Publication bias including funnel plot and Egger’s test was performed. By convention, an observed P value less than 0.05 implied a great statistical significance for summary HR and publication biases. Survival rates on the graphical representation of the survival curves were extracted by Engauge Digitizer version 5.0. HRs and their 95% CI were calculated by STATA version 11.0.
Results
Eligible studies and characteristics
Ten publications (3-8,13-16), published between 1997 and 2013, were selected. They all reported the prognostic value for survival of FGFR1 gene copy number in lung cancer patients, assessing FGFR1 gene amplification in the primary tumour. One publication (13) of these studies was excluded because the histological type of cohort was small cell lung cancer (SCLC). One publication (14) was excluded because the cohort of patients were NSCLC with brain metastases. Two publications (15,16) was excluded because the technique of the detection was immature. There were six publications eligible for the meta-analysis finally (3-8). The total number of included patients was 1,128, ranging from 100 to 264 patients per study (median number =188). The main characteristics of the six eligible publications are described in Table 1. In the six studies included in the analysis, gene copy number of FGFR1 was evaluated by FISH in five (3-5,7,8) by RT-PCR in one (6). Using cut off values for overexpression chosen by each author, 166 (15.69%) of the 1,128 patients in this meta-analysis had FGFR1 amplification. Comprehensive analysis for overall FGFR1 amplification in three studies including differed according to histological type, the overall RR was 0.9 which 95% CI was 0.85 to 0.96 and P value was 0.001 (Table 2 and Figure 1). The outcome showed that significantly more frequent in SQCC.
Table 1. Characteristics of the study populations.
| First author | Year | Histology | Stage | N pts | Positive N | Positive rate | Method | HR estimation | Cut off | Results |
|---|---|---|---|---|---|---|---|---|---|---|
| Weiss | 2010 | SQCC | I-III | 155 | 15 | 9.68% | FISH | Survival curve | Copy number >4 | NS |
| Rebecca | 2012 | SQCC | I-IV | 226 | 37 | 16.37% | FISH | Survival curve | FGFR1/CEP8 ratio ≥2.2 | NS |
| Kim | 2012 | SQCC | I-III | 262 | 34 | 12.98% | FISH | HR | Copy number >9 | Poor prognosis |
| Sasaki | 2012 | NSCLC | I-IV | 100 | 32 | 32.00% | RT-PCR | Survival curve | Copy number >4 | NS |
| Tran | 2013 | NSCLC | I-IV | 264 | 37 | 14.02% | Dual-Colour ISH | survival curve | FGFR1/CEP8 ratio ≥2.0 | NS |
| Craddock | 2013 | SQCC | I-IV | 121 | 11 | 9.09% | FISH | HR | Copy number >5 | NS |
N pts, number of patients; HR, hazard ratio; NSCLC, non-small-cell lung cancer; SQCC, squamous cell lung cancer; FISH, fluorescence in situ hybridization; PCR, polymerase chain reaction; NS, non-significant.
Table 2. Incidence rate in different histologic type.
| Author | Non-amplification |
Amplification |
|||||
|---|---|---|---|---|---|---|---|
| SQCC | Non-SQCC | Sum | SQCC | Non-SQCC | Sum | ||
| Weiss | 140 | 77* | 217 | 15 | 0 | 15 | |
| Rebecca | 37 | – | 37 | 189 | – | 189 | |
| Kim | 228 | – | 228 | 34 | – | 34 | |
| Sasaki | 38 | 30 | 68 | 27 | 5 | 32 | |
| Tran | 25 | 139 | 164 | 6 | 24 | 30 | |
| Craddock | 99 | – | 99 | 22 | – | 22 | |
SQCC, squamous cell lung cancer. *, Patients with Non-SQCC were exclude for survival analysis.
Figure 1.
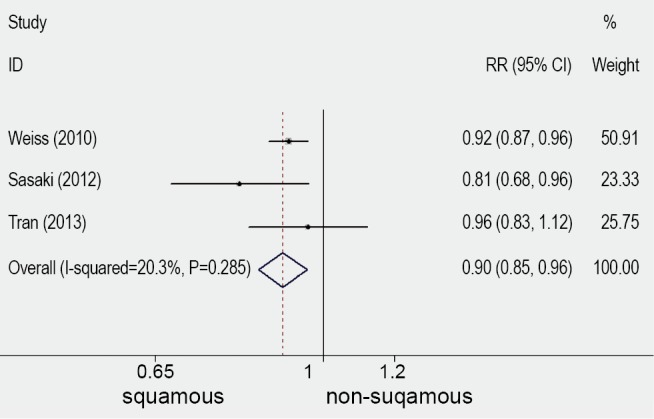
Analysis for overall FGFR1 amplification in three studies including differed according to histological type. FGFR1, fibroblast growth factor receptor1.
Qualitative assessment
We assessed the quality of the selected studies using the European lung cancer working party quality scale for biological prognostic factors for lung cancer. Overall, the global quality score ranged 32.5-80%, with a median of 53.33% (Table 3).
Table 3. Results of the methodological assessment by the European Lung Cancer Working Party score.
| Weiss | Rebecca | Kim | Sasaki | Tran | Craddock | Average | |
|---|---|---|---|---|---|---|---|
| Scientific design | 3 | 6 | 7 | 6 | 6 | 6 | 5.67 |
| Laboratory methodology | 5 | 5 | 10 | 8 | 5 | 6 | 6.50 |
| Generalizability | 1 | 4 | 10 | 2 | 6 | 6 | 4.83 |
| Results analysis | 4 | 4 | 5 | 4 | 4 | 5 | 4.33 |
| 32.50% | 47.50% | 80.00% | 50.00% | 52.50% | 57.50% | 53.33% |
Meta-analysis
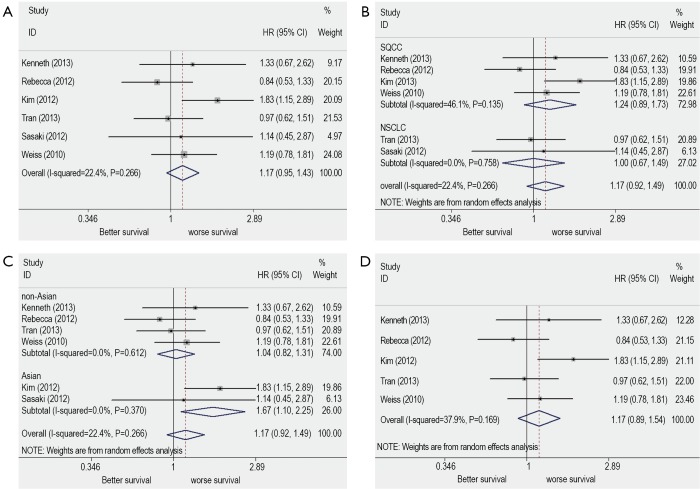
The combined HR for all six eligible studies was 1.17 (95% CI: 0.95 to 1.43) using fixed effects model, indicating that FGFR1 gene copy number had no significant survival impact in patients with NSCLC (Figure 2A). There was no significant heterogeneity between studies (I2=22.4%, P=0.266) (Figure 2A).
Figure 2.
Meta-analysis of hazard ratio (HR) of the effect of FGFR1 amplification on survival in patients with non-small cell lung cancer (NSCLC). (A) All six eligible studies; (B) Subgroup of histological subtypes; (C) Subgroup of country of origin; and (D) five FISH evaluated studies. FGFR1, fibroblast growth factor receptor1; FISH, fluorescence in situ hybridization.
When subgrouped according to histological subtypes, the combined HR for the NSCLC studies was 1 (95% CI: 0.67 to 1.49), the pooled HR for SQCC was 1.24 (95% CI: 0.89 to 1.73) (Figure 2B).
When subgrouped according to country of origin, the combined HR for the Asian populations’ studies was 1.67 (95% CI: 1.1 to 2.52) while for Non-Asian populations’ studies was 1.04 (95% CI: 0.82 to 1.73) (Figure 2C).
The sensitivity analysis showed that omitting any single study did not influence the pooled HR. We conduct another analysis after the literature using RT-PCR was excluded, then the combined HR was 1.17 and the 95% CI was 0.81-1.68 (Figure 2D). The results were consistent with the above-mentioned ones. For publication bias test, a more formal evaluation either using Bgger’s test or Egger’s test also showed no evidence of significant publication bias (Bgger’s test: Z=0.19, P=0.851; Egger’s test: coef. =0.068, P=0.908, 95% CI: –1.47-1.6) (Figure 3). This suggested absence of publication bias in all studies.
Figure 3.
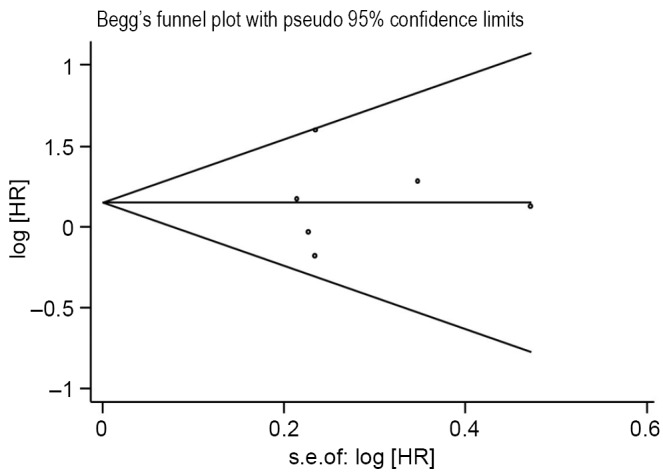
Begg’s funnel plot with pseudo 95% confidence limits.
Discussion
FGFR is a transmembrane protein of receptor tyrosine kinase which includes FGFR1, FGFR2, FGFR3 and FGFR4 (17). FGFRs signal transmission is necessary to normal cell growth and differentiation, participating with physiological processes of angiogenesis, embryogenesis, bone formation, and wound healing, and is quite close to tumor genesis and progression. Among numerous tumors, such as breast cancer, bladder cancer, prostate cancer, NSCLC, FGFR activating mutations or ligand/receptor overexpression causes signal to be continuously activated which is not only closely linked to tumor genesis, progression and poor prognosis but also plays an important role in tumor angiogenesis, tumor invasion and metastasis (18,19). Therefore, FGFR, being closely related to tumor patients’ prognosis, is commonly known as the key target of anti-tumor.
There were six publications eligible for the meta-analysis. The heterogeneity test show I2=22.4% (P=0.266), which indicated no obvious heterogeneity. Then analyzed with fixed effect model, combined HR was 1.17 and 95% CI was 0.95 to 1.43, indicating FGFR1 amplification was irrelevant to NSCLC prognosis and effect size was similar to most of research results. In the former part of this paper, we mentioned that FGFR1 amplification was abundant in SQCC, while in other types of NSCLC amplification was rare. So was FGFR1 amplification related to its prognosis on patients of SQCC? Therefore, we conduct subgroup analysis to the data and receive results showing that subgroup combined HR of SQCC was 1.24 and 95% CI was 0.89 to 1.73, indicating influences of FGFR1 amplification on prognosis of SQCC do not have statistical significance. In racial distinctions, we divide the included people into Asian subgroup and non-Asian subgroup for analysis. Asian subgroup was of combined HR 1.67 and 95% CI was 1.1 to 2.52, while non-Asian subgroup was of combined HR 1.04 and 95% CI was 0.82 to 1.31. Results from race subgroup analysis show that FGFR1 amplification showed poor NSCLC prognosis among Asian people; however, researches included were small and there were only two subgroups with research targets of Asian people of total number of 362, so there is a possibility of bias. We need more researches to prove the phenomenon.
Meta-analysis is a quantitative analysis method based on previous research works, greatly influenced by the quality of previous research materials. We refer to the literature quality evaluation standard applied by European lung cancer working party quality scale for biological prognostic factors for lung cancer. Due to the limitations of Meta-analysis itself, there were various biases during the analysis process. The most common bias is publication bias. We adopt Begg’s funnel plot and Egger’s plot to evaluate publication bias with results showing that bias coefficient was 0.068, 95% CI was 1.47 to 1.6, P=0.908. The 95% CI covers 0 point and P was more than 0.05, indicating publication bias was of no statistical significance. Certainly, it is avoidable that there was omission of literatures in other languages and some negative results being excluded into the analysis, as well as test results undergoing were not to be published yet. The above-mentioned could also become the cause of possible publication bias.
In aspect of technical bias, it is mainly about test methods. In the past, most will adopt RT-PCR method in semi-quantitative gene detection which however imposes higher requirements on sample source and could not verify whether RNA tissue sample ground and extracted is fully a tumor tissue. As the development and sophistication of FISH technology in recent years, PT-PCR application in gene amplification detection is gradually out of historical stage. FISH detection can be conducted by paraffin-embedded tissue and have high correctness in aneuploidy amplification detection of low resolution and high resolution chromosome, with good sensitivity and specificity and objective interpretation of results, making this method become the golden standard of gene amplification detection. Among six literatures included in this paper, out of five adopts FISH technology for detection (incl. Dual-Colour ISH) and one adopts RT-PCR detection. In order to eliminate effects of different detection method on statistical results, we conduct another analysis when the literature using RT-PCR is excluded. The results showed that the combined HR was 1.17 and 95% CI was 0.81 to 1.68, indicating FGFR1 amplification is not related to NSCLC prognosis. The results were consistent with the above-mentioned ones.
In conclusion, this study conducts statistical analysis to relevant literatures through classic Meta-analysis method. Results show that FGFR1 has no statistical significance to both NSCLC and SQCC prognosis. Due to small numbers of literatures included and existence of various biases, it is necessary for more research, more detailed data and more standard detection technologies to prove that in order to receive more convincing results.
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- 1.Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014. CA Cancer J Clin 2014;64:9-29 [DOI] [PubMed] [Google Scholar]
- 2.Wistuba II, Gelovani JG, Jacoby JJ, et al. Methodological and practical challenges for personalized cancer therapies. Nat Rev Clin Oncol 2011;8:135-41 [DOI] [PubMed] [Google Scholar]
- 3.Weiss J, Sos ML, Seidel D, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med 2010;2:62ra93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim HR, Kim DJ, Kang DR, et al. Fibroblast growth factor receptor 1 gene amplification is associated with poor survival and cigarette smoking dosage in patients with resected squamous cell lung cancer. J Clin Oncol. 2013;31:731-7 [DOI] [PubMed] [Google Scholar]
- 5.Heist RS, Mino-Kenudson M, Sequist LV, et al. FGFR1 amplification in squamous cell carcinoma of the lung. J Thorac Oncol 2012;7:1775-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sasaki H, Shitara M, Yokota K, et al. Increased FGFR1 copy number in lung squamous cell carcinomas. Mol Med Rep 2012;5:725-8 [DOI] [PubMed] [Google Scholar]
- 7.Tran TN, Selinger CI, Kohonen-Corish MR, et al. Fibroblast growth factor receptor 1 (FGFR1) copy number is an independent prognostic factor in non-small cell lung cancer. Lung Cancer 2013;81:462-7 [DOI] [PubMed] [Google Scholar]
- 8.Craddock KJ, Ludkovski O, Sykes J, et al. Prognostic value of fibroblast growth factor receptor 1 gene locus amplification in resected lung squamous cell carcinoma. J Thorac Oncol 2013;8:1371-7 [DOI] [PubMed] [Google Scholar]
- 9.Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med 1998;17:2815-34 [DOI] [PubMed] [Google Scholar]
- 10.Steels E, Paesmans M, Berghmans T, et al. Role of p53 as a prognostic factor for survival in lung cancer: a systematic review of the literature with a meta-analysis. Eur Respir J 2001;18:705-19 [DOI] [PubMed] [Google Scholar]
- 11.Tierney JF, Stewart LA, Ghersi D, et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 2007;8:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DerSimonian R, Laird N.Meta-analysis in clinical trials. Control Clin Trials 1986;7:177-88 [DOI] [PubMed] [Google Scholar]
- 13.Schultheis AM, Bos M, Schmitz K, et al. Fibroblast growth factor receptor 1 (FGFR1) amplification is a potential therapeutic target in small-cell lung cancer. Mod Pathol 2014;27:214-21 [DOI] [PubMed] [Google Scholar]
- 14.Preusser M, Berghoff AS, Berger W, et al. High rate of FGFR1 amplifications in brain metastases of squamous and non-squamous lung cancer. Lung Cancer 2014;83:83-9 [DOI] [PubMed] [Google Scholar]
- 15.Volm M, Koomägi R, Mattern J, et al. Prognostic value of basic fibroblast growth factor and its receptor (FGFR-1) in patients with non-small cell lung carcinomas. Eur J Cancer 1997;33:691-3 [DOI] [PubMed] [Google Scholar]
- 16.Mano Y, Takahashi K, Ishikawa N, et al. Fibroblast growth factor receptor 1 oncogene partner as a novel prognostic biomarker and therapeutic target for lung cancer. Cancer Sci 2007;98:1902-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang LY, Edenson SP, Yu YL, et al. A natural kinase-deficient variant of fibroblast growth factor receptor 1. Biochemistry 1996;35:10134-42 [DOI] [PubMed] [Google Scholar]
- 18.Ford MD, Cauchi J, Greferath U, et al. Expression of fibroblast growth factors and their receptors in rat glomeruli. Kidney Int 1997;51:1729-38 [DOI] [PubMed] [Google Scholar]
- 19.Nguyen HB, Estacion M, Gargus JJ. Mutations causing achondroplasia and thanatophoric dysplasia alter bFGF-induced calcium signals in human diploid fibroblasts. Hum Mol Genet 1997;6:681-8 [DOI] [PubMed] [Google Scholar]