Abstract
Atypical hemolytic uremic syndrome (aHUS) is usually characterized by uncontrolled complement activation. The recent discovery of loss-of-function mutations in DGKE in patients with aHUS and normal complement levels challenged this observation. DGKE, encoding diacylglycerol kinase-ε, has not been implicated in the complement cascade but hypothetically leads to a prothrombotic state. The discovery of this novel mechanism has potential implications for the treatment of infants with aHUS, who are increasingly treated with complement blocking agents. In this study, we used homozygosity mapping and whole-exome sequencing to identify a novel truncating mutation in DGKE (p.K101X) in a consanguineous family with patients affected by thrombotic microangiopathy characterized by significant serum complement activation and consumption of the complement fraction C3. Aggressive plasma infusion therapy controlled systemic symptoms and prevented renal failure, suggesting that this treatment can significantly affect the natural history of this aggressive disease. Our study expands the clinical phenotypes associated with mutations in DGKE and challenges the benefits of complement blockade treatment in such patients. Mechanistic studies of DGKE and aHUS are, therefore, essential to the design of appropriate therapeutic strategies in patients with DGKE mutations.
Hemolytic uremic syndrome (HUS) is characterized by thrombosis of the small vessels, resulting in microangiopathic hemolytic anemia, renal failure, and thrombocytopenia.1 In contrast to classical HUS, which is secondary to infection with specific Escherichia coli serotypes, atypical HUS (aHUS) is caused by genetic or autoimmune factors involved in the complement system.2 Up to 65% of patients progress to ESRD or die within 1 year after diagnosis.3 Most identified disease-causing mutations in aHUS affect genes encoding complement factors, such as CFH, CFB and CFI, and C3 and MCP, leading to uncontrolled activation of the complement cascade at the cell surface level.2 Consequently, treatment aimed to block complement cascade activation has been proven effective.4
Recently, two studies independently identified mutations in DGKE (Online Mendelian Inheritance in Man 15008), encoding diacylglycerol kinase-ε, in families with aHUS and membranoproliferative GN.5,6 The patients in these studies did not show abnormal complement levels, suggesting that DGKE does not play a role in complement activation but causes a thrombotic phenotype by influencing endothelial cells and platelets function.7 The absence of overt complement activation in such patients suggested that treatment with complement inhibitors, such as eculizumab, would be inefficacious. In this study, we expand the clinical phenotype of DGKE-associated diseases to hypocomplementemic aHUS.
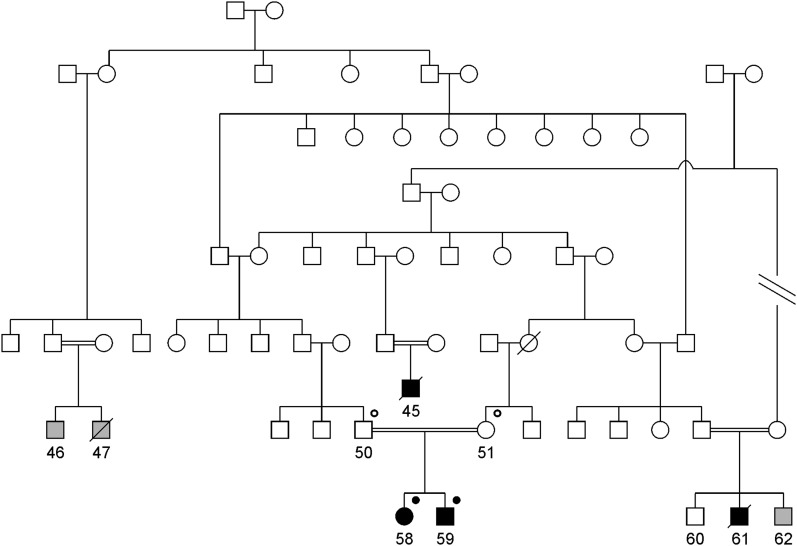
We studied a family spanning multiple generations with at least four recognized loops of consanguinity (Figure 1). The family originates from Linosa, a remote island in the Mediterranean Sea that is inhabited by about 450 persons and characterized by a high rate of endogamy. There were four individuals (45, 58, 59, and 61) affected by thrombotic microangiopathy and three individuals with an unknown phenotype (46, 47, and 62). Individual 45 was reported by history to have had thrombotic microangiopathy with heavy proteinuria at young age and died in childhood. Individual 61 was diagnosed with biopsy-proven aHUS after an episode of acute hemolytic anemia and renal failure after corrective surgery for congenital cataract in 2000 at the age of 1 year. The patient died at the age of 15 months. Individual 47 died in childhood from an unclear illness; individual 62 was subjected to corrective surgery for congenital cataract at the age of 6 months at Gaslini Hospital in 2003. He was found to have high lactate dehydrogenase (579 U/L), low haptoglobin (3 mg/dl), and low serum C3 (69 mg/dl), but because renal function was normal and there was no anemia or proteinuria, he was not subjected to treatment. Four individuals (50, 51, 58, and 59) were available for this study.
Figure 1.
Structure of the pedigree analyzed in the current study. Square symbols represent men, and circles represent women. Consanguineous unions are represented by double horizontal lines. Black-filled symbols represent affected individuals, white symbols represent unaffected individuals, and gray-filled symbols represent phenotype unknown. Black-filled dot superscript represents DGKE mutation in homozygous state; black open dot superscript represents DGKE mutation carriers (heterozygous state).
The index patient (individual 58) presented at 10 months of age with acute renal failure, proteinuria, hematuria, elevated lactate dehydrogenase, low haptoglobin levels, and low platelets (Table 1). Serum complement C3 was 66 mg/dl (normal values=84–192 mg/dl), suggesting a clinical diagnosis of HUS. Serum complement C4 was within normal range (15 mg/dl; normal values=10–42 mg/dl). Plasma infusion was started at 10 ml/kg per day for 2 weeks with improvement of clinical symptoms and laboratory abnormalities (C3 levels increased to 111 mg/dl). Renal biopsy, performed 3 months after presentation because of persistent proteinuria, was consistent with the diagnosis of subacute HUS (Figure 2).
Table 1.
Clinical characteristics of individuals 58 and 59 at presentation and last follow-up
| Individual 58 (Girl) | Individual 59 (Boy) | |||
|---|---|---|---|---|
| Presentation | Last Follow-Up | Presentation | Last Follow-Up | |
| Age (yr) | 0.8 | 5.3 | 0.7 | 3.4 |
| SCr (mg/dl) | 1.5 | 0.29 | 11.4 | 0.30 |
| Platelet count (×109/L) | 64 | 279 | 30 | 201 |
| LDH (IU/L) | 1923 | 414 | 2736 | 551 |
| Haptoglobin | <2 | 46 | <2 | <2 |
| C3 (mg/dl) | 66 | 79 | 86 | 78 |
| Proteinuria | Yes | Yes | Yes | No |
| Hematuria | Yes | Yes | Yes | Yes |
Reference values for C3: 84–192 mg/dl. Proteinuria is defined as a protein-to-creatinine ratio greater than 0.2 mg/mg on a morning urine sample. Hematuria is defined as greater than 3 red blood cells/high power field on urinalysis. SCr, serum creatinine; LDH, lactic dehydrogenase; C3, serum complement C3 level.
Figure 2.
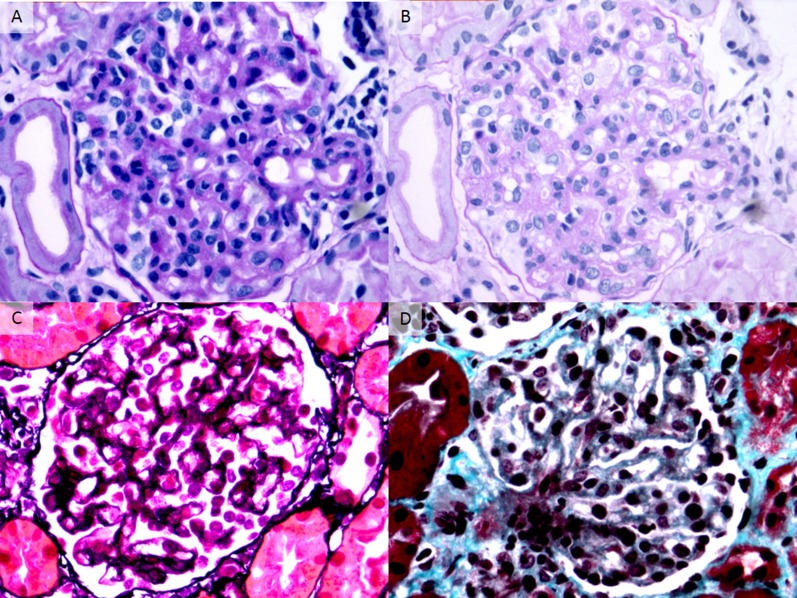
Renal histology studies in individual 58. Representative glomeruli show features of subacute thrombotic microangiopathy. (A) There is global glomerular endothelial cell swelling and mild hypercellularity producing a picture of endotheliosis. No intracapillary fibrin thrombi are identified (periodic acid–Schiff, ×400). (B) The same glomerulus in a deeper level shows irregular thickening and narrow duplication of glomerular basement membranes, which are segmentally associated with mesangial interposition (periodic acid–Schiff, ×400). (C) There is duplication of many glomerular basement membranes (producing double contours) without associated mesangial hypercellularity. The podocytes appear swollen (Jones methenamine silver, ×400). (D) The glomerular capillary lumina are segmentally expanded owing to dissolution of the mesangial matrix, consistent with mesangiolysis. Some of the adjacent glomerular capillary walls appear thickened with duplication of glomerular basement membranes (Masson trichrome, ×400). Immunofluorescence revealed glomerular staining for IgM and fibrin/fibrinogen, with negativity for IgG, IgA, C1q, C3, and C4 (not shown).
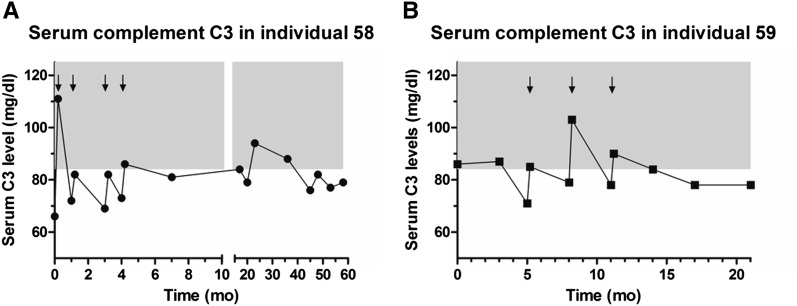
Plasma infusion was conducted every month in the first 1 year after diagnosis and continued every 3 months afterward. Interestingly, despite transitory improvement with plasma infusions, C3 levels remained low during follow-up (Figure 3A). Renal function recovered shortly after diagnosis, but proteinuria and microhematuria persisted during follow-up (Table 1). At 5 years of age, she has normal renal function and is treated daily with a combination of an angiotensin-converting enzyme inhibitor and an angiotensin receptor blocker for proteinuria and every 3 months with plasma infusion.
Figure 3.
Longitudinal data on serum complement C3 levels. Individual 58 (A) and individual 59 (B). Arrows indicate hospital admissions for which there were data on serum complement C3 levels pre- and postplasma infusions. Treatment with plasma infusions is shown to maintain complement levels within normal ranges (84–192 mg/dl; gray area) only for a limited period of time after administration, indicate chronic active complement activation and consumption.
The younger brother (individual 59) of the index patient developed acute renal failure, proteinuria, hemolysis, increased BP, and low platelet count at the age of 8 months after an exanthematous viral infection (sixth disease). Serum complement C3 level was at the lower limit of the normal range (86 mg/dl; normal values=84–192 mg/dl). Serum C4 was within normal range. Because of the similarity in clinical presentation with his older sibling, plasma infusion was initiated. Similar to his sibling, C3 remained below the normal level during follow-up (Figure 3B, Table 1). At 3 years of age, renal function is still normal, proteinuria is absent, and there is persistent microhematuria. The patient remains dependent on plasma infusion (performed every 3 months), and he is not on pharmacological treatment during the plasma-free intervals. The absence of proteinuria at follow-up in patient 59 suggests that early aggressive and continuous treatment with plasma infusion can significantly change the natural history of disease and prevent podocyte damage in patients with DGKE mutations. Both parents are healthy, with no evidence of hemolysis or significant complement consumption at repeated laboratory tests. The average C3 complement fraction level, measured using the same assay, in 426 children without overt immunologic diseases was 114.1 mg/dl (median, 110.5; 95% confidence interval, 111.9 to 116.3).
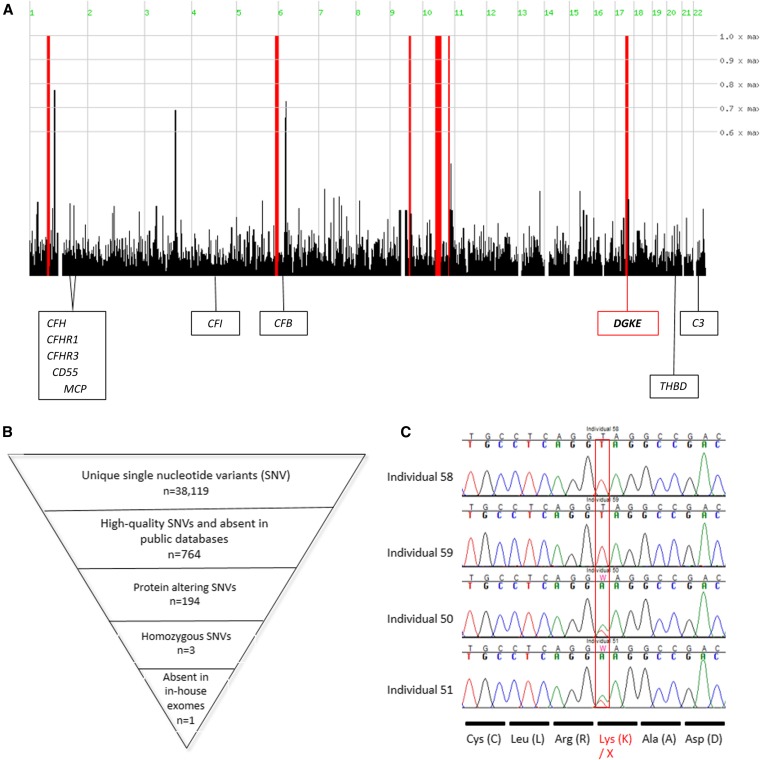
Homozygosity mapping identified six regions across the genome (Figure 4A) that were homozygous in individual 58 and heterozygous in her parents (50 and 51). These regions collectively spanned 64.7 Mb (approximately 2.3% of the genome, including 658 transcriptional units) (Supplemental Table 1). Exome sequencing performed on individual 58 resulted in an average depth of 51.5X, with 95.51% of bases covered at a depth of 4X or higher. We identified 38,119 unique single nucleotide variants (SNVs) (Figure 4B). Of these SNVs, 764 (500 SNVs and 264 indels) SNVs were of high quality and absent from the Single Nucleotide Polymorphism Database, 1000 Genome, and National Heart, Lung, and Blood Institute Exome Variant Server databases. Among those SNVs, 194 (169 SNVs and 24 indels) SNVs were protein-altering variants (truncating, missense, or predicted to affect splicing), and only 3 SNVs were present in homozygous state. Two variants (a deletion of an A 2 bp upstream of exon 4 of PRB3 and a missense p.D2115V in MUC4) were not under any of the homozygosity peaks but they present at high frequency in 45 exomes from IgA nephropathy patients available in the laboratory that were processed at the same time as our patients. The third variant, a homozygous mutation c.301 A>T, resulted in a premature termination at the aminoacidic position 101 (p.K101X) in DGKE. This gene is located within the distal 290 kb of the region of homozygosity on chromosome 17q21–22. Sanger sequencing confirmed that individuals 58 and 59 were homozygous for the p.K101X mutation and that their parents, 50 and 51, were heterozygous for this variant (Figure 4C). Linkage analysis using an autosomal recessive model using the p.K101X mutation as a marker8 resulted in an logarithm of odds score of 2.4 at the DGKE locus, supporting gene localization for this family. Finally, this variant, absent in >6500 publicly available controls, was absent in an additional 118 ethnically and geographically matched controls. DGKE mutations were recently associated to noncomplement-mediated forms of aHUS and membranoproliferative GN,5,6 suggesting that this form of hypocomplementemic thrombotic microangiopathy is allelic to the forms previously reported.
Figure 4.
Identification of a truncating mutation in DGKE. (A) Genome-wide plot for homozygosity. Red bars indicate areas of significant homozygosity in individual 58 with heterozygosity in the parents (individuals 50 and 51). The six regions span 64.7 Mb and encompass 658 transcriptional units. The localization of the complement genes involved in aHUS are displayed; only DGKE is included under an area of significant homozygosity. (B) Schematic representation of the filtering pipeline for exome sequencing data that resulted in only one deleterious homozygous variant included in the chromosome 17 homozygosity region (DGKE p.K101X). (C) Sanger sequencing of DGKE p.K101X in individuals 50, 51, 58, and 59 shows the mutation in homozygosity in the affected patients 58 and 59 and the mutation in heterozygous state in their parents, patients 50 and 51.
Because of the significantly different serologic phenotype in our patients compared with the recently reported patients with DGKE mutations, we explored the possibility that additional functional variants in genes involved in the innate immunity might account for the low serum complement.9 No rare homozygous or heterozygous variants were found in the genes previously associated with complement-mediated forms of aHUS, such as CFH, CFB, CFI, C3, or MCP and other genes of the complement system (Supplemental Table 2), and no other rare deleterious variants were found in the regions of homozygosity. Importantly, all family individuals were homozygous for the wild-type allele at the rs6677604 locus, a single nucleotide polymorphism that is in nearly perfect linkage disequilibrium with a deletion of CFHR3 and CFHR1.10 Deletions of these genes (and CFHR4) are associated with production of autoantibodies to complement factor H and can result in low serum complement.11 Deletion of CFHR4 was excluded by the presence of single nucleotide polymorphisms in the heterozygous state in this gene (Supplemental Table 2). Finally, anti-factor H autoantibodies were absent in all four individuals.12 Taken together, these data strongly implicate the p.K101X mutation in DGKE as the most likely single cause of the hypocomplementemic phenotype of this form of microangiopathy. The mechanism by which mutations in DGKE result in complement activation is unclear. A possibility is that the effect of DGKE on protein kinase C activation would lead to a downregulation of vascular endothelial growth factor receptor 2, which in turn, would result in reduced expression of decay-accelerating factor and consequent activation of complement at the endothelial cells surface.13
The advent of next generation sequencing boosted gene discovery for Mendelian diseases and helped in solving the pathogenesis of numerous familial and sporadic genetic nephropathies.14–16 Although this technology helped to formulate accurate diagnosis and improve knowledge in pathophysiological and therapeutic aspects of many diseases, there are increasing examples in which deep resequencing of human genomes or exomes showed that many genes can harbor mutations with pleiotropic phenotypic effect that confounded traditional gene identification approaches.17,18 In a recent report, Boyer et al.18 performed whole-exome sequencing in families with autosomal dominant FSGS without extrarenal manifestations to surprisingly identify segregating mutations in LMX1B, a gene implicated in the nail patella syndrome.19,20 This phenotypic expansion suggests that the traditional approaches to gene identification for Mendelian diseases, based on strict partitioning of phenotypes before candidate gene or genome-wide studies, might require a more agnostic approach to identify causal deleterious mutations and then classify diseases based on the underlying molecular defect.
Here, using homozygosity mapping coupled to exome sequencing, we identified a previously unreported homozygous truncating mutation in DGKE in a consanguineous family with a form of aHUS characterized by hypocomplementemia and dependence from plasma infusion. The good response to plasma infusion in our patients together with the observation that at least one patient from the study by Lemaire et al.5 had disease recurrence while on treatment with eculizumab suggest that aggressive plasma infusion treatment may significantly change the natural history of this disease.
Our findings expand the phenotypic spectrum of DGKE-related diseases to hypocomplementemic aHUS and have potential implications for treatment. The availability of genome-wide high-density genotyping and whole-exome sequencing allowed us to exclude other genetic causes of low complement level, such as deletions of the CFHR1, CFHR3, and CFHR4 or mutations in other genes involved in aHUS or complement regulation. Notably, Dgke-null mice did not show overt renal disease or a thrombotic phenotype,21 indicating that the exact role of DGKE mutations in aHUS and the complement system still remains to be elucidated.
This family originates from a remote island in the Mediterranean Sea called Linosa, which has only 450 inhabitants and is characterized by a high rate of endogamy. Linosa is a small (2.1 mi2) volcanic island located in the Mediterranean Sea 101 miles south of Sicily. The island has no airport and can be reached only by boat. The presence of multiple individuals affected by aHUS in this family suggests that the p.K101X mutations might have a high prevalence on this island. The common environmental exposure, the reduced gene pool, and the high mutation frequency make this population ideal to study the natural history of this disease and design appropriate functional studies and therapeutic strategies.
Concise Methods
Patients and Families
Patient recruitment was performed at G. Gaslini Institute in Genoa, Italy, between 2008 and 2011. We collected peripheral blood samples for DNA isolation from the proband, her parents, and her brother. The Institutional Review Board for Columbia University and local ethic review committees in Genoa approved the study protocol.
Genotyping, Mapping, and Sequencing
Genomic DNA was purified from peripheral blood cells using standard procedures. Genome-wide genotyping was conducted on individuals 50, 51, and 58 using the Omni1-quad chips (Illumina). Genotype data were processed for quality controls using PLINK software22 as previously described.23 Homozygosity mapping was performed using the Homozygositymapper program (http://www.homozygositymapper.org/) as previously described.14 Whole-exome sequencing was performed on individual 58 using an Illumina HiSeq 2000 as previously described.15
Web Resources
PubMed: http://www.ncbi.nlm.nih.gov/pubmed; Online Mendelian Inheritance in Man: http://www.ncbi.nlm.nih.gov/omim; the Single Nucleotide Polymorphism Database: http://www.ncbi.nlm.nih.gov/SNP; 1000 Genome Project: www.1000genomes.org/; Exome Variant Server: http://evs.gs.washington.edu/EVS/; Homozygositymapper: http://www.homozygositymapper.org/.
Disclosures
None.
Supplementary Material
Acknowledgments
We thank the patients and their family members for participating in the study.
R.W. is supported by a grant from Fonds NutsOhra Zorgsubsidies (Project Number 1101-058) and a stipend of the Ter Meulen Fund, Royal Netherlands Academy for Arts and Sciences (2012/225). This study was supported by National Human Genome Research Institute (NHGRI) Centers for Mendelian Genomics Grant HG006504 (to R.P.L.) and the Fondazione Malattie Renali nel Bambino (to G.M.G.). S.S.-C. is supported by American Heart Association Grant in Aid 13GRNT14680075.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2013080886/-/DCSupplemental.
References
- 1.Loirat C, Frémeaux-Bacchi V: Atypical hemolytic uremic syndrome. Orphanet J Rare Dis 6: 60, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Noris M, Remuzzi G: Atypical hemolytic-uremic syndrome. N Engl J Med 361: 1676–1687, 2009 [DOI] [PubMed] [Google Scholar]
- 3.Fremeaux-Bacchi V, Fakhouri F, Garnier A, Bienaimé F, Dragon-Durey MA, Ngo S, Moulin B, Servais A, Provot F, Rostaing L, Burtey S, Niaudet P, Deschênes G, Lebranchu Y, Zuber J, Loirat C: Genetics and outcome of atypical hemolytic uremic syndrome: A nationwide French series comparing children and adults. Clin J Am Soc Nephrol 8: 554–562, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Legendre CM, Licht C, Muus P, Greenbaum LA, Babu S, Bedrosian C, Bingham C, Cohen DJ, Delmas Y, Douglas K, Eitner F, Feldkamp T, Fouque D, Furman RR, Gaber O, Herthelius M, Hourmant M, Karpman D, Lebranchu Y, Mariat C, Menne J, Moulin B, Nürnberger J, Ogawa M, Remuzzi G, Richard T, Sberro-Soussan R, Severino B, Sheerin NS, Trivelli A, Zimmerhackl LB, Goodship T, Loirat C: Terminal complement inhibitor eculizumab in atypical hemolytic-uremic syndrome. N Engl J Med 368: 2169–2181, 2013 [DOI] [PubMed] [Google Scholar]
- 5.Lemaire M, Frémeaux-Bacchi V, Schaefer F, Choi M, Tang WH, Le Quintrec M, Fakhouri F, Taque S, Nobili F, Martinez F, Ji W, Overton JD, Mane SM, Nürnberg G, Altmüller J, Thiele H, Morin D, Deschenes G, Baudouin V, Llanas B, Collard L, Majid MA, Simkova E, Nürnberg P, Rioux-Leclerc N, Moeckel GW, Gubler MC, Hwa J, Loirat C, Lifton RP: Recessive mutations in DGKE cause atypical hemolytic-uremic syndrome. Nat Genet 45: 531–536, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ozaltin F, Li B, Rauhauser A, An SW, Soylemezoglu O, Gonul II, Taskiran EZ, Ibsirlioglu T, Korkmaz E, Bilginer Y, Duzova A, Ozen S, Topaloglu R, Besbas N, Ashraf S, Du Y, Liang C, Chen P, Lu D, Vadnagara K, Arbuckle S, Lewis D, Wakeland B, Quigg RJ, Ransom RF, Wakeland EK, Topham MK, Bazan NG, Mohan C, Hildebrandt F, Bakkaloglu A, Huang CL, Attanasio M: DGKE variants cause a glomerular microangiopathy that mimics membranoproliferative GN. J Am Soc Nephrol 24: 377–384, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quaggin SE: DGKE and atypical HUS. Nat Genet 45: 475–476, 2013 [DOI] [PubMed] [Google Scholar]
- 8.Gudbjartsson DF, Jonasson K, Frigge ML, Kong A: Allegro, a new computer program for multipoint linkage analysis. Nat Genet 25: 12–13, 2000 [DOI] [PubMed] [Google Scholar]
- 9.Degn SE, Jensenius JC, Thiel S: Disease-causing mutations in genes of the complement system. Am J Hum Genet 88: 689–705, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gharavi AG, Kiryluk K, Choi M, Li Y, Hou P, Xie J, Sanna-Cherchi S, Men CJ, Julian BA, Wyatt RJ, Novak J, He JC, Wang H, Lv J, Zhu L, Wang W, Wang Z, Yasuno K, Gunel M, Mane S, Umlauf S, Tikhonova I, Beerman I, Savoldi S, Magistroni R, Ghiggeri GM, Bodria M, Lugani F, Ravani P, Ponticelli C, Allegri L, Boscutti G, Frasca G, Amore A, Peruzzi L, Coppo R, Izzi C, Viola BF, Prati E, Salvadori M, Mignani R, Gesualdo L, Bertinetto F, Mesiano P, Amoroso A, Scolari F, Chen N, Zhang H, Lifton RP: Genome-wide association study identifies susceptibility loci for IgA nephropathy. Nat Genet 43: 321–327, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore I, Strain L, Pappworth I, Kavanagh D, Barlow PN, Herbert AP, Schmidt CQ, Staniforth SJ, Holmes LV, Ward R, Morgan L, Goodship TH, Marchbank KJ: Association of factor H autoantibodies with deletions of CFHR1, CFHR3, CFHR4, and with mutations in CFH, CFI, CD46, and C3 in patients with atypical hemolytic uremic syndrome. Blood 115: 379–387, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dragon-Durey MA, Loirat C, Cloarec S, Macher MA, Blouin J, Nivet H, Weiss L, Fridman WH, Frémeaux-Bacchi V: Anti-Factor H autoantibodies associated with atypical hemolytic uremic syndrome. J Am Soc Nephrol 16: 555–563, 2005 [DOI] [PubMed] [Google Scholar]
- 13.Mason JC, Lidington EA, Yarwood H, Lublin DM, Haskard DO: Induction of endothelial cell decay-accelerating factor by vascular endothelial growth factor: A mechanism for cytoprotection against complement-mediated injury during inflammatory angiogenesis. Arthritis Rheum 44: 138–150, 2001 [DOI] [PubMed] [Google Scholar]
- 14.Sanna-Cherchi S, Burgess KE, Nees SN, Caridi G, Weng PL, Dagnino M, Bodria M, Carrea A, Allegretta MA, Kim HR, Perry BJ, Gigante M, Clark LN, Kisselev S, Cusi D, Gesualdo L, Allegri L, Scolari F, D’Agati V, Shapiro LS, Pecoraro C, Palomero T, Ghiggeri GM, Gharavi AG: Exome sequencing identified MYO1E and NEIL1 as candidate genes for human autosomal recessive steroid-resistant nephrotic syndrome. Kidney Int 80: 389–396, 2011 [DOI] [PubMed] [Google Scholar]
- 15.Sanna-Cherchi S, Sampogna RV, Papeta N, Burgess KE, Nees SN, Perry BJ, Choi M, Bodria M, Liu Y, Weng PL, Lozanovski VJ, Verbitsky M, Lugani F, Sterken R, Paragas N, Caridi G, Carrea A, Dagnino M, Materna-Kiryluk A, Santamaria G, Murtas C, Ristoska-Bojkovska N, Izzi C, Kacak N, Bianco B, Giberti S, Gigante M, Piaggio G, Gesualdo L, Kosuljandic Vukic D, Vukojevic K, Saraga-Babic M, Saraga M, Gucev Z, Allegri L, Latos-Bielenska A, Casu D, State M, Scolari F, Ravazzolo R, Kiryluk K, Al-Awqati Q, D’Agati VD, Drummond IA, Tasic V, Lifton RP, Ghiggeri GM, Gharavi AG: Mutations in DSTYK and dominant urinary tract malformations. N Engl J Med 369: 621–629, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaki M, Airik R, Ghosh AK, Giles RH, Chen R, Slaats GG, Wang H, Hurd TW, Zhou W, Cluckey A, Gee HY, Ramaswami G, Hong CJ, Hamilton BA, Cervenka I, Ganji RS, Bryja V, Arts HH, van Reeuwijk J, Oud MM, Letteboer SJ, Roepman R, Husson H, Ibraghimov-Beskrovnaya O, Yasunaga T, Walz G, Eley L, Sayer JA, Schermer B, Liebau MC, Benzing T, Le Corre S, Drummond I, Janssen S, Allen SJ, Natarajan S, O’Toole JF, Attanasio M, Saunier S, Antignac C, Koenekoop RK, Ren H, Lopez I, Nayir A, Stoetzel C, Dollfus H, Massoudi R, Gleeson JG, Andreoli SP, Doherty DG, Lindstrad A, Golzio C, Katsanis N, Pape L, Abboud EB, Al-Rajhi AA, Lewis RA, Omran H, Lee EY, Wang S, Sekiguchi JM, Saunders R, Johnson CA, Garner E, Vanselow K, Andersen JS, Shlomai J, Nurnberg G, Nurnberg P, Levy S, Smogorzewska A, Otto EA, Hildebrandt F: Exome capture reveals ZNF423 and CEP164 mutations, linking renal ciliopathies to DNA damage response signaling. Cell 150: 533–548, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Al-Romaih KI, Genovese G, Al-Mojalli H, Al-Othman S, Al-Manea H, Al-Suleiman M, Al-Jondubi M, Atallah N, Al-Rodayyan M, Weins A, Pollak MR, Adra CN: Genetic diagnosis in consanguineous families with kidney disease by homozygosity mapping coupled with whole-exome sequencing. Am J Kidney Dis 58: 186–195, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boyer O, Woerner S, Yang F, Oakeley EJ, Linghu B, Gribouval O, Tête MJ, Duca JS, Klickstein L, Damask AJ, Szustakowski JD, Heibel F, Matignon M, Baudouin V, Chantrel F, Champigneulle J, Martin L, Nitschké P, Gubler MC, Johnson KJ, Chibout SD, Antignac C: LMX1B mutations cause hereditary FSGS without extrarenal involvement. J Am Soc Nephrol 24: 1216–1222, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen H, Lun Y, Ovchinnikov D, Kokubo H, Oberg KC, Pepicelli CV, Gan L, Lee B, Johnson RL: Limb and kidney defects in Lmx1b mutant mice suggest an involvement of LMX1B in human nail patella syndrome. Nat Genet 19: 51–55, 1998 [DOI] [PubMed] [Google Scholar]
- 20.Dreyer SD, Zhou G, Baldini A, Winterpacht A, Zabel B, Cole W, Johnson RL, Lee B: Mutations in LMX1B cause abnormal skeletal patterning and renal dysplasia in nail patella syndrome. Nat Genet 19: 47–50, 1998 [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez de Turco EB, Tang W, Topham MK, Sakane F, Marcheselli VL, Chen C, Taketomi A, Prescott SM, Bazan NG: Diacylglycerol kinase epsilon regulates seizure susceptibility and long-term potentiation through arachidonoyl-inositol lipid signaling. Proc Natl Acad Sci U S A 98: 4740–4745, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC: PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanna-Cherchi S, Kiryluk K, Burgess KE, Bodria M, Sampson MG, Hadley D, Nees SN, Verbitsky M, Perry BJ, Sterken R, Lozanovski VJ, Materna-Kiryluk A, Barlassina C, Kini A, Corbani V, Carrea A, Somenzi D, Murtas C, Ristoska-Bojkovska N, Izzi C, Bianco B, Zaniew M, Flogelova H, Weng PL, Kacak N, Giberti S, Gigante M, Arapovic A, Drnasin K, Caridi G, Curioni S, Allegri F, Ammenti A, Ferretti S, Goj V, Bernardo L, Jobanputra V, Chung WK, Lifton RP, Sanders S, State M, Clark LN, Saraga M, Padmanabhan S, Dominiczak AF, Foroud T, Gesualdo L, Gucev Z, Allegri L, Latos-Bielenska A, Cusi D, Scolari F, Tasic V, Hakonarson H, Ghiggeri GM, Gharavi AG: Copy-number disorders are a common cause of congenital kidney malformations. Am J Hum Genet 91: 987–997, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.