Abstract
We demonstrate that ligand hydrophobicity can be used to increase affinity and selectivity of binding between monolayer-protected cationic gold nanoparticles and β– lactoglobulin protein isoforms containing two amino acid mutations.
Selective recognition of proteins is an important aspect in the design of diagnostics1 and separations systems.2 However, the discrimination between proteins with similar structures (e.g. <1% change in amino acid content) presents a difficult challenge that is most commonly addressed using either relatively slow separations techniques2 or through use of antigen-antibody recognition, resulting in stability issues and significantly increasing the overall cost.3 Therefore, development of synthetic materials to rapidly discriminate between proteins with similar structures is important in the design of sensors and media for separations.
Protein–nanoparticle (NP) assemblies provide effective platforms that have successfully been employed in numerous bio-applications including sensing,4 controlling enzymatic behavior,5 protein delivery,6 and producing biocompatible materials.7 Monolayer-functionalized NPs provide a tunable surface for binding with target biomacromolecules, enabling differential NP–protein interactions.8 Selective NP–protein interactions have been utilized as a robust platform for differentiating proteins with different molecular weights, pI values, and sizes.4
The nature of the non-covalent interactions employed for host-guest interactions is an important factor in designing nanoparticles for selective protein binding. In recent studies, we have demonstrated protein isoform differentiation through purely electrostatic interactions in solution9 and on densely-packed immobilized nanoparticle surfaces.10 In this communication, we employ the interplay of hydrophobicity and electrostatics to increase both the affinity and selectivity between isoforms of protein recognition using nanoparticle receptors. These studies indicate that charge and hydrophobicity can work synergistically to provide particles with enhanced recognition capabilities.
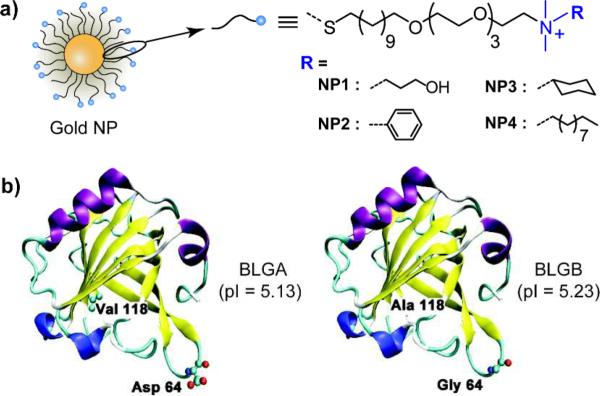
We used monolayer protected cationic gold NPs (core diameter ~2 nm) for differentially binding the proteins. The chemical structures of the monolayer ligands used are shown in Fig. 1a, covering a range of different hydrophobicities of the ligand headgroups. These NPs feature a tetra(ethylene glycol) spacer to isolate the effect of the headgroups, as well as to minimize denaturation of the bound protein.11 Additionally, the charge (zeta potential ~20 mV) and the size (diameter ~10 nm) of the particles are controlled to set the surface hydrophobicity as the only variable parameter. We selected β-lactoglobulin (BLG) as a model protein that has two isoforms: BLG variant A (BLGA) and BLG variant B (BLGB). They differ only in two amino acids: Val 118 (BLGA) → Ala 118 (BLGB) and Asp 64 (BLGA) → Gly 64 (BLGB) (Fig. 1b). Their physiochemical properties such as size, geometry (dumbbell shape), and pI (~5.2) are similar,12 making the discrimination of these proteins challenging. BLG isoforms are known to exhibit multimeric states within certain pH ranges.13 To avoid complex binding phenomena, we carried out the experiments at pH 5.5, where both the isoforms show dimeric structures.14
Fig. 1.
(a) Chemical structures of the cationic gold NPs with different hydrophobicities (NP1 – NP4). (b) The secondary structure of BLGA and BLGB monomers displaying the two different amino acids at position 64 and 118. Color scheme for the secondary structure: purple - α-helix, yellow - β-sheet, blue - 310-helix, cyan - turn, and white - coil.
Initially, we studied the effect of NP structures on NP–BLG complexation using dynamic light scattering (DLS). Table 1 lists the sizes of the complexes between the different NPs and BLGA/BLGB. The sizes of both the BLGA and BLGB proteins were quite similar (~5 nm) and consistent with reported values.15 Likewise, all the NPs have similar hydrodynamic sizes when tested independently. As the NPs bind with BLGA/BLGB, an increase in size is observed for all the cases evidencing the formation of NP-protein conjugates/aggregates. Although some differences in the size of the conjugates can be observed, the polydispersity of the complexes made assessment of affinity impossible. As such, a more detailed thermodynamically analysis is required to assess correctly these phenomena.
Table 1.
Hydrodynamic diameter (% number) of the gold NPs, proteins, and their complexes in 5 mM sodium phosphate buffer (pH 5.5)
| Unbound (nm) | Complex (nm) |
||
|---|---|---|---|
| BLGA | BLGB | ||
| NP1 | 9.8 ± 2.1 | 37.4 ± 22.6 | 50.1 ± 34.2 |
| NP2 | 6.9 ± 1.7 | 44.2 ± 20.3 | 43.6 ± 22.6 |
| NP3 | 8.0 ± 2.4 | 25.7 ± 12.5 | 40.3 ± 18.1 |
| NP4 | 7.5 ± 2.4 | 34.8 ± 20.6 | 33.3 ± 19.7 |
| BLGA | 4.8 ± 0.7 | – | – |
| BLGB | 4.7 ± 0.7 | – | – |
Quantitative analysis of protein-particle interaction was obtained using isothermal titration calorimetry (ITC).16 Using this technique we were able to determine the stoichiometry and affinity of the particle-protein pairs as well as obtain thermodynamic parameters for the binding processes.
The ITC experiments were performed by titrating the NPs into solutions of each protein at 25°C. Depending on the NP, different thermodynamic profiles were obtained (see ESI†). The complexation of BLGA/BLGB with NP1 is exothermic in nature, while NP2 – NP4 binding is an endothermic process. The thermograms were fitted to a one-site binding model that assumes equivalent and non-cooperative NP-BLG binding (see ESI†). Binding stoichiometries (n) and thermodynamic parameters such as binding constants (Kb) and enthalpy changes (ΔH) were obtained from the isothermal curve fitting analysis. The Gibbs free energy changes (ΔG) and entropy changes (ΔS) were calculated by using the standard thermodynamic equations: ΔG = –RTlnKb and ΔG = ΔH – TΔS, where R denotes the universal gas constant and T denotes the absolute temperature. The thermodynamic quantities for each NP–protein complex are summarized in Table 2. It is observed that the BLGA/NP complexes feature significantly higher stoichiometries than that of BLGB/NP (Table 2). Moreover, the ratio of protein to NP varied dramatically depending on the ligand structure of the NPs. Presumably, different stoichiometries arise from the differential binding affinity as a result of various favorable interactions (vide infra). Another important observation is the enthalpy and entropy changes of the complexation processes that depend on the monolayer structures on the NPs. While NP2 – NP4 bind with the protein variants in an entropy-driven process, NP1 features enthalpy driven binding with negative ΔH and ΔS values. These enthalpy or entropy driven processes can be explained by the overall complexation equation (3) that is a combination of two simultaneous processes described in equation (1) and (2).17
| (1) |
| (2) |
| (3) |
where, H2ON, H2OB, and H2ON-B are the water associated with the NP, BLG protein and the NP-BLG complexes respectively.
Table 2.
Thermodynamic parameters for the NP–protein complexation, and the octanol–water partition coefficients of the NPs
| NP | Calculated log Poct | Protein | n * | Kb × 105 (M−1) | ΔH [kcal mol−1] | ΔS [kcal (mol K)−1] | −ΔG [kcal mol−1] |
|---|---|---|---|---|---|---|---|
| NP1 | 0.94 | BLGA | 7.7 ± 0.6 | 5.3 ± 0.1 | −165.8 ± 0.8 | −0.58 ± 0.002 | 7.8 ± 0.01 |
| BLGB | 5.6 ± 0.6 | 3.4 ± 0.1 | −78.3 ± 0.5 | −0.29 ± 0.001 | 7.53 ± 0.02 | ||
| NP2 | 1.95 | BLGA | 16.7 ± 3.3 | 8.4 ± 0.7 | 270.7 ± 3.7 | 0.88 ± 0.01 | 8.07 ± 0.05 |
| BLGB | 8.3 ± 0.8 | 4.6 ± 0.8 | 253.1 ± 8.1 | 0.82 ± 0.03 | 7.70 ± 0.1 | ||
| NP3 | 3.77 | BLGA | 9.1 ± 0.8 | 4.6 ± 0.4 | 10200 ± 60 | 34.2 ± 0.2 | 7.71 ± 0.05 |
| BLGB | 3.8 ± 0.5 | 2.0 ± 0.9 | 9900 ± 660 | 33.2 ± 2.2 | 7.15 ± 0.29 | ||
| NP4 | 5.04 | BLGA | 5.3 ± 0.3 | 1.4 ± 0.7 | 13200 ± 440 | 44.3 ± 1.5 | 6.92 ± 0.32 |
| BLGB | 4.5 ± 0.2 | 1.1 ± 0.6 | 13000 ± 410 | 43.6 ± 1.4 | 6.76 ± 0.36 | ||
n denotes the stoichiometry of [protein] to [NP] based on monomeric state of the protein.
Note: The error values are the standard errors.
The first process of non-covalent interaction between the NP and BLG is an exothermic reaction. However, the second process of water reorganization of the well-defined solvent shell is endothermic in nature with both ΔH and ΔS positive. It is clear that the hydrophobic NPs (NP2 – NP4) release higher amounts of water of solvation from the binding interface, with concomitant positive entropy change (2nd process predominant). On the other hand, the increased hydrophilicity of the hydroxyl functional group in NP1 might involve stronger non-covalent interaction along with other favorable interactions, such as hydrogen bonding. As a result, a negative ΔH is observed (1st process predominant) that is somewhat offset by the unfavorable entropy change.
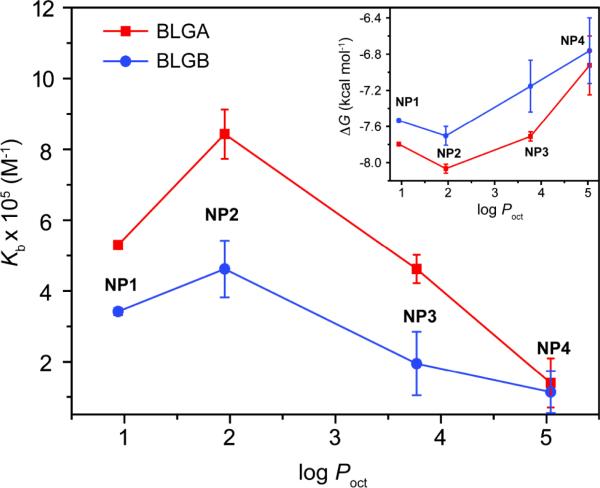
We investigated the quantitative relationship between the hydrophobicity of the ligand headgroups and the binding affinities of the NP–protein dyads. We determined the computed octanol–water partition coefficient (log Poct) of the NP headgroups and used these as a measure of surface hydrophobicity.18 The plot of binding constant (Kb) against log Poct displays an interesting behavior (Fig. 2). As the hydrophobicity of the NPs increases, the binding affinity first increases, and then gradually attenuates. This trend presumably arises from the cooperative effect of hydrophobic and electrostatic interactions. An optimum balance of the two interactions is the key to achieve the maximum binding affinity observed for NP2. In addition, the π–π interactions of the aromatic amino acids on the proteins with the phenyl ring of NP2 might play a role to enhance the affinity. However, differences in binding affinities between BLGA-NP and BLGB-NP vary significantly depending on the NPs. The extra carboxyl group of BLGA (Asp64) together with neighboring negative residues should have higher electrostatic interactions with the NPs possessing lower hydrophobicity, leading to greater differentiation between BLGA and BLGB.9,19 On the other hand, interactions between the quaternary ammonium cation of the NP and anionic residues on BLG are likely to be sterically diminished by the long decyl hydrocarbon chain of NP4, with consequent loss of differentiation between BLGA and BLGB. The Gibbs free energy change against NP hydrophobicity for the NP-protein dyads also showed a similar trend as the binding affinity (Fig. 2 inset). Taken together, these studies demonstrate that a subtle difference in the monolayer structure on the NPs can alter NP–protein affinity significantly.
Fig. 2.
Plot of binding constant (Kb) vs. partition coefficient (log Poct) of the ligand headgroups on the NP monolayer. The inset shows plot of the Gibbs free energy change (ΔG) vs. log Poct. The lines are drawn to guide the eye.
We also observed from Table 2 that an enthalpy-entropy compensation process was operative similar to many host-guest systems.20 The physical significance of enthalpy-entropy compensation was determined from the linear correlation using the relation TΔS = αΔH + TΔS0, where α is the slope and TΔS0 is the intercept. A linear relationship was obtained for these thermodynamic quantities with a correlation coefficient of 0.999 (Fig. S2). The near-unit slope (α = 1.00) suggests that significant conformational changes occurred at the interaction interface,21 similar to that found in other flexible systems such as protein–protein interactions,20 demonstrating the biomimetic nature of the protein-particle recognition process.
Conclusions
In summary, we have demonstrated that electrostatic and hydrophobic interactions can be employed to tune NP-protein affinity. Using BLGA/BLGB protein isoforms as a testbed, we observed that thermodynamic parameters for complexation between BLGA/BLGB and the NPs depend primarily on the relative hydrophobicities of the ligands present on the NPs. Notably, the maximum binding affinity was achieved through proper balance of electrostatics and hydrophobicity. These studies demonstrate the ability of functionalized NPs to achieve selective binding with subtly different proteins, a starting point in engineering particles with high selectivities required for applications such as biosensing.
Supplementary Material
Acknowledgements
The authors are grateful to the NIH (GM077173 and EB014277), the Fundamental Research Funds for the Central Universities, and the China Scholarship Council for financial support of this work.
Footnotes
Electronic Supplementary Information (ESI) available: Experimental details, ITC, and DLS analyses. See DOI: 10.1039/b000000x/
Notes and references
- 1.a Hanash S. Nature. 2003;422:226–232. doi: 10.1038/nature01514. [DOI] [PubMed] [Google Scholar]; b Pulido R, van Huijsduijnen RH. FEBS J. 2008;275:848–866. doi: 10.1111/j.1742-4658.2008.06250.x. [DOI] [PubMed] [Google Scholar]
- 2.a Chang WWP, Hobson C, Bomberger DC, Schneider LV. Electrophoresis. 2005;26:2179–2186. doi: 10.1002/elps.200410283. [DOI] [PubMed] [Google Scholar]; b Lecoeur M, Gareil P, Varenne A. J. Chromatogr. A. 2010;1217:7293–7301. doi: 10.1016/j.chroma.2010.09.043. [DOI] [PubMed] [Google Scholar]; c Guiochon G. J. Chromatogr. A. 2007;1168:101–168. doi: 10.1016/j.chroma.2007.05.090. [DOI] [PubMed] [Google Scholar]; d Saxena A, Tripathi BP, Kumar M, Shahi VK. Adv. Colloid Interface Sci. 2009;145:1–22. doi: 10.1016/j.cis.2008.07.004. [DOI] [PubMed] [Google Scholar]; e Vissers JPC. J. Chromatogr. A. 1999;856:117–143. doi: 10.1016/s0021-9673(99)00692-5. [DOI] [PubMed] [Google Scholar]
- 3.Murphy GP, Elgamal AA, Su SL, Bostwick DG, Holmes EH. Cancer. 1998;83:2259–2269. [PubMed] [Google Scholar]
- 4.a You CC, Miranda OR, Gider B, Ghosh PS, Kim IB, Erdogan B, Krovi SA, Bunz UHF, Rotello VM. Nat. Nanotechnol. 2007;2:318–323. doi: 10.1038/nnano.2007.99. [DOI] [PubMed] [Google Scholar]; b Aili D, Selegard R, Baltzer L, Enander K, Liedberg B. Small. 2009;5:2445–2452. doi: 10.1002/smll.200900530. [DOI] [PubMed] [Google Scholar]; c De M, Rana S, Akpinar H, Miranda OR, Arvizo RR, Bunz UHF, Rotello VM. Nat. Chem. 2009;1:461–465. doi: 10.1038/nchem.334. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Rana S, Singla AK, Bajaj A, Elci SG, Miranda OR, Mout R, Yan B, Jirik FR, Rotello VM. ACS Nano. 2012;6:8233–8240. doi: 10.1021/nn302917e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a You CC, De M, Han G, Rotello VM. J. Am. Chem. Soc. 2005;127:12873–12881. doi: 10.1021/ja0512881. [DOI] [PubMed] [Google Scholar]; b Wu Z, Zhang B, Yan B. Int. J. Mol. Sci. 2009;10:4198–4209. doi: 10.3390/ijms10104198. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Knecht LD, Ali N, Wei Y, Hilt JZ, Daunert S. ACS Nano. 2012;6:9079–9086. doi: 10.1021/nn303308v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a Jiang W, Kim BYS, Rutka JT, Chan WCW. Nat. Nanotechnol. 2008;3:145–150. doi: 10.1038/nnano.2008.30. [DOI] [PubMed] [Google Scholar]; b Ghosh P, Yang X, Arvizo R, Zhu Z, Agasti SS, Mo Z, Rotello VM. J. Am. Chem. Soc. 2010;132:2642–2645. doi: 10.1021/ja907887z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu M, Qian L, Briñas RP, Lymar ES, Hainfeld JF. Angew. Chem., Int. Ed. 2007;46:5111–5114. doi: 10.1002/anie.200701180. [DOI] [PubMed] [Google Scholar]
- 8.Moyano DF, Rotello VM. Langmuir. 2011;27:10376–10385. doi: 10.1021/la2004535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen K, Xu Y, Rana S, Miranda OR, Dubin PL, Rotello VM, Sun L, Guo X. Biomacromolecules. 2011;12:2552–2561. doi: 10.1021/bm200374e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Y, Engel Y, Yan Y, Chen K, Moyano DF, Dubin PL, Rotello VM. J. Mat. Chem. B. 2013;1:5230–5234. doi: 10.1039/C3TB20377H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rana S, Yeh Y, Rotello VM. Curr. Opin. Chem. Biol. 2010;14:828–834. doi: 10.1016/j.cbpa.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sawyer L, Kontopidis G. Biochim. Biophys. Acta., Protein Struct. Mol. Enzymol. 2000;1482:136–148. doi: 10.1016/s0167-4838(00)00160-6. [DOI] [PubMed] [Google Scholar]
- 13.Qin BY, Bewley MC, Creamer LK, Baker EN, Jameson GB. Protein Sci. 1999;8:75–83. doi: 10.1110/ps.8.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.a Majhi PR, Ganta RR, Vanam RP, Seyrek E, Giger K, Dubin PL. Langmuir. 2006;22:9150–9159. doi: 10.1021/la053528w. [DOI] [PubMed] [Google Scholar]; b Verheul M, Pedersen JS, Roefs SP, de Kruif KG. Biopolymers. 1999;49:11–20. doi: 10.1002/(SICI)1097-0282(199901)49:1<11::AID-BIP2>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 15.Gao JY, Dubin PL. Biopolymers. 1999;49:185–193. doi: 10.1002/(SICI)1097-0282(199902)49:2<185::AID-BIP6>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 16.a Cedervall T, Lynch I, Lindman S, Berggard T, Thulin E, Nilsson H, Dawson KA, Linse S. Proc. Natl. Acad. Sci. USA. 2007;104:2050–2055. doi: 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]; b De M, You CC, Srivastava S, Rotello VM. J. Am. Chem. Soc. 2007;129:10747–10753. doi: 10.1021/ja071642q. [DOI] [PubMed] [Google Scholar]
- 17.De M, Miranda OR, Rana S, Rotello VM. Chem. Commun. 2009:2157–2159. doi: 10.1039/b900552h. [DOI] [PubMed] [Google Scholar]
- 18.a Ghose AK, Crippen GM. J. Comput. Chem. 1986;7:565–577. [Google Scholar]; b Rana S, Yu X, Patra D, Moyano DF, Miranda OR, Hussain I, Rotello VM. Langmuir. 2012;28:2023–2027. doi: 10.1021/la204017z. [DOI] [PubMed] [Google Scholar]
- 19.Xu Y, Mazzawi M, Chen K, Sun L, Dubin PL. Biomacromolecules. 2011;12:1512–1522. doi: 10.1021/bm101465y. [DOI] [PubMed] [Google Scholar]
- 20.Houk KN, Leach AG, Kim SP, Zhang X. Angew. Chem., Int. Ed. 2003;42:4872–4897. doi: 10.1002/anie.200200565. [DOI] [PubMed] [Google Scholar]
- 21.Gattuso G, Nepogodiev SA, Stoddart JF. Chem. Rev. 1998;98:1919–1958. doi: 10.1021/cr960133w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.