Abstract
Bile salts play crucial roles in allowing the gastrointestinal system to digest, transport and metabolize nutrients. They function as nutrient signaling hormones by activating specific nuclear receptors (FXR, PXR, Vitamin D) and G-protein coupled receptors [TGR5, sphingosine-1 phosphate receptor 2 (S1PR2), muscarinic receptors]. Bile acids and insulin appear to collaborate in regulating the metabolism of nutrients in the liver. They both activate the AKT and ERK1/2 signaling pathways. Bile acid induction of the FXR-α target gene, small heterodimer partner (SHP), is highly dependent on the activation PKCζ, a branch of the insulin signaling pathway. SHP is an important regulator of glucose and lipid metabolism in the liver. One might hypothesize that chronic low grade inflammation which is associated with insulin resistance, may inhibit bile acid signaling and disrupt lipid metabolism. The disruption of these signaling pathways may increase the risk of fatty liver and non-alcoholic fatty liver disease (NAFLD). Finally, conjugated bile acids appear to promote cholangiocarcinoma growth via the activation of S1PR2.
Keywords: Bile acids, Sphingosine 1-phosphate receptor 2, Insulin, PKCζ, Glucose metabolism, Liver
1. Introduction
In the past, bile salts were considered to be just detergent molecules that were required for the solubilization of cholesterol in the gall bladder, promoting digestion of dietary lipids and stimulating absorption of lipids, cholesterol and fat-soluble vitamins in the intestines (1). Bile salts were also known to stimulate bile flow, promote cholesterol secretion from the liver, and to have antibacterial properties. However, in 1999, three independent laboratories reported that bile acids were natural ligands for the farnesoid X receptor (FXR-α)(2–4). The recognition that bile acids activated specific nuclear receptors started a renaissance in the field of bile acid research. Since 1999, bile acids have been reported to activate other nuclear receptors (preganane X receptor, vitamin D receptor), G protein coupled receptors [TGR5, sphingosine-1-phosphate receptor 2 (S1PR2), muscarinic receptor 2 (M2)] and cell signaling pathways (JNK 1/2, AKT, and ERK 1/2)(5, 6). Deoxycholic acid (DCA), a secondary bile acid, has also been reported to activate the epidermal growth factor receptor (EGFR) (7). It is now clear that bile acid function as hormones or nutrient signaling molecules that help to regulate glucose, lipid, lipoprotein, energy metabolism and inflammatory responses (5, 6). The role of bile acid-mediated signaling pathways in nonalcoholic fatty liver diseases has been discussed in several excellent reviews (8–12). In this brief review, we will focus on how the insulin signaling pathway and FXR-α cross-talk to regulate hepatic nutrient metabolism.
2. Enterohepatic Circulation of Bile Acids
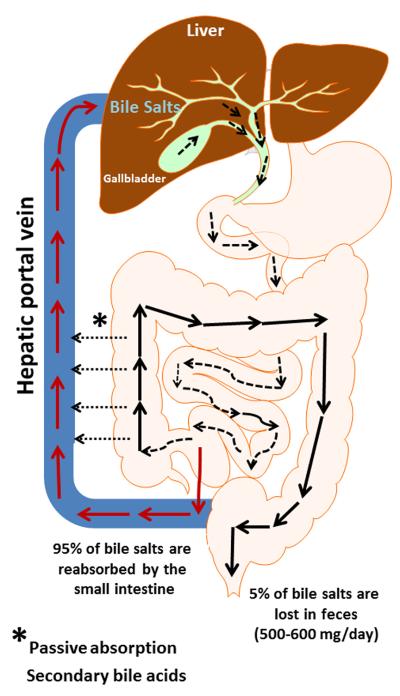
Bile acids are synthesized from cholesterol in liver hepatocytes, conjugated to either glycine or taurine and actively secreted via ABC transporters on the canalicular membrane into biliary bile. Conjugated bile acids are often referred to as bile salts. Bile acid synthesis represents a major out-put pathway of cholesterol from the body. Bile acids are actively secreted from hepatocytes via the bile salt export protein (BSEP, ABCB11) along with phospholipids by ABCB4 and with cholesterol by ABCG5/ABCG8 in a fairly constant ratio under normal conditions(13, 14). Bile acids are detergent molecules and form mixed micelles with cholesterol and phospholipids which help keep cholesterol in solution in the gall bladder. Eating stimulates the gall bladder to contract emptying its contents into the small intestines. Bile salts are crucial for the solubilization and absorption of cholesterol and lipids as well as lipid soluble vitamins (A, D, E, and K). They activate pancreatic enzymes and form mixed micelles with lipids in the small intestines promoting their absorption. Bile acids are efficiently recovered from the intestines, primarily the ileum, by the apical sodium dependent transporter (ASBT). Bile acids are secreted from ileocytes, on the basolateral side, by the organic solute OSTα/OSTβ transporter(15). Secondary bile acids, formed by 7α-dehydroxylation of primary bile acids by anaerobic gut bacteria, can be passively absorbed from the large bowel or secreted in the feces. Absorbed bile acids return to the liver via the portal blood where they are actively transported into hepatocytes primarily via the sodium taurocholate cotransporting polypeptide (NTCP, SLC10A1)(16). Bile acids are again actively secreted from the hepatocyte into bile which stimulates bile flow and the secretion of cholesterol and phospholipids. Bile acids undergo enterohepatic circulation several times each day (Figure 1). During their enterohepatic circulation approximately 500–600 mg/day are lost via fecal excretion and must be replaced by new bile acid synthesis in the liver. The bile acid pool size is tightly regulated as excess bile acids can be highly toxic to mammalian cells.
Figure 1. Enterohepatic circulation of bile acids.
Bile acids are synthesized and conjugated mainly to glycine or taurine in hepatocytes. Bile acids travel to the gall bladder for storage during the fasting state. During digestion, bile acids travel to the duodenum via the common bile duct. 95% of the bile acids delivered to the duodenum are absorbed back into blood within the ileum and circulate back to the liver through the portal vein. 5% of bile acids are lost in feces.
3. Synthesis of Primary and Secondary Bile Acids
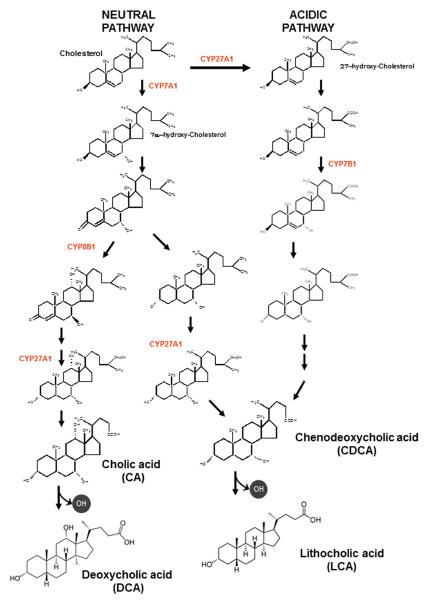
There are two pathways of bile acid synthesis in the liver, the neutral pathway and the acidic pathway (Figure 2). The neutral pathway is believed to be the major pathway of bile acid synthesis in man under normal physiological conditions. The neutral pathway is initiated by cholesterol 7α-hydroxylase (CYP7A1) which is the rate-limiting step in this biochemical pathway. CYP7A1 is a cytochrome P450 monooxygenase and the gene encoding this enzyme is highly regulated by a feed-back repressive mechanism involving the FXR-dependent induction of fibroblast growth factor 15/19 (FGF15/19) by bile acids in the intestines. FGF15/19 binds to the fibroblast growth factor receptor 4 (FGFR4)/β-Klotho complex in hepatocytes activating both the JNK1/2 and ERK1/2 signaling cascades (17–19). Activation of the JNK1/2 pathway has been reported to down-regulate CYP7A1 mRNA in hepatocytes(20). FGFR4 and β-Klotho null mice have increased levels of CYP7A1 and upregulated bile acid synthesis (21, 22). Moreover, treatment of FXR null mice with a specific FXR agonist failed to repress CYP7A1 in the liver(23). These results support an important role of FGF15, synthesized in the intestines by activation of FXR, in the regulation of CYP7A1 and bile acid synthesis in the liver. CYP7A1 has also been reported to be down-regulated by glucagon (24, 25) and pro-inflammatory cytokines (15) and up-regulated by glucose and insulin during the postprandial period (26).
Figure 2. Biosynthetic pathways of bile acids.
Two major pathways are involved in bile acid synthesis. The neutral (or classic) pathway is controlled by cholesterol 7α-hydroxylase (CYP7A1) in the endoplasmic reticulum. The acidic (or alternative) pathway is controlled by sterol 27-hydroxylase (CYP27A1) in mitochondria. The sterol 12α-hydroxylase (CYP8B1) is required to synthesis of cholic acid (CA). The oxysterol 7α-hydroxylase (CYP7B1) is involved in the formation of chenodeoxycholic acid (CDCA) in acidic pathway. The neutral pathway is also able to form CDCA by CYP27A1.
The neutral pathway of bile acid synthesis produces both cholic acid (CA) and chenodeoxycholic acid (CDCA) (Fig. 2). The ratio of CA and CDCA is primarily determined by the activity of sterol 12α-hydroxylase (CYP8B1). The gene encoding CYP8B1 is also highly regulated by bile acids. Bile acids induce the gene encoding small heterodimer partner (SHP) in the liver via activation of the farnesoid X receptor (FXR-α). SHP is an orphan nuclear receptor without a DNA binding domain. It interacts with several transcription factors including hepatocyte nuclear factor 4 (HNF4α) and liver-related homolog-1 (LRH-1) and acts as a dominant negative protein to inhibit transcription. In this regard, a liver specific knockout of LRH-1 completely abolished the expression of CYP8B1, but had little effect on CYP7A1 (27, 28). These results suggest that the interaction of SHP with LRH-1, caused by bile acids, may be the key regulator of hepatic CYP8B1 and the ratio of CA/CDCA.
The acidic or alternative pathway of bile acid synthesis is initiated in the inner membrane of mitochondria by sterol 27-hydroxylase (CYP27A1). This enzyme also has low sterol 25-hydroxylase activity (29, 30). CYP27A1 is capable of further oxidizing the 27-hydroxy group to a carboxylic acid (31). Unlike, CYP7A1, CYP27A1 is widely expressed in various tissues in the body where it may produce regulatory oxysterols (32). Even though CYP27A1 is the initial enzyme in the acidic pathway of bile acid synthesis, it may not be the rate limiting step. The inner mitochondrial membrane is very low in cholesterol content. Hence, cholesterol transport into the mitochondria appears to be a rate limiting step in this pathway (33). In this regard, it has been reported that the expression of the gene encoding steroidogenic acute regulatory (StAR D1) protein, markedly increases (5-fold) the rates of bile acid synthesis in primary hepatocytes via the acidic pathway (32, 34). Over expression of the gene encoding CYP27A1 only minimally increases (<2-fold) bile acid synthesis. The acidic pathway is believed to become more dominant in an individual with cirrhotic liver disease as the neutral pathway is repressed by inflammation (20, 35).
The acidic pathway of bile acid synthesis is now being viewed as an important pathway for generating regulatory oxysterols. For example, 25-hydroxycholesterol and 27-hydroxycholesterol are natural ligands for the liver X receptor (LXR) which is involved in regulating cholesterol and lipid metabolism (35, 36). Moreover, recent studies report that 25-hydroxycholesterol, formed by CYP27A1, can be converted into 5-cholsten-3β-25-diol-3-sulfate in the liver (37). The sulfated 25-hydroxycholesterol is a regulator of inflammatory responses, lipid metabolism and cell proliferation and is located in the liver (36, 38, 39). Recent evidence suggests that sulfated 25-hydroxycholesterol is a ligand for peroxisome proliferator-activated receptor gamma (PPAR), which is a major regulator of inflammation and lipid metabolism (39). The 7α-hydroxylation of oxysterols is catalyzed by oxysterol 7α-hydroxylase (CYP7B1)(40). This biotransformation allows some of these oxysterols to be converted to bile acids. Finally, oxysterols generated in extrahepatic tissues can be transported to the liver and metabolized into bile acids.
The primary bile acids CA and CDCA are converted into deoxycholic acid (DCA) and lithocholic acid (LCA), respectively, by a small population of intestinal anaerobic bacteria (41, 42). Bile acid 7α-dehydroxylation occurs via a multistep biochemical pathway found in a few species of the genus Clostridium(41). However, bile acids must be deconjugated by bile salt hydrolase before 7α-dehydroxylation occurs. Bile salt hydrolase is found in a large number of different intestinal bacteria. The levels of DCA in the bile acid pool of humans can vary from 1% to over 50% as the human liver is unable to convert DCA back to CA (41). As the amount of DCA is increased in the bile acid pool there is an increase in the hydrophobicity and toxicity to mammalian cells. The amount of DCA in the bile acid pool in man is determined primarily by levels and activity of bile acid 7α-dehydroxylating gut bacteria and intestinal transit time (43). A Western diet appears to increase the levels of bile acid 7α-dehydroxylating bacteria in the intestines and is associated with an increase in DCA in bile (44). The composition of the human bile acid pool is important as bile acids are now known to be regulatory molecules which vary in their ability to activate different nuclear receptors and G-protein coupled receptors (GPCRs)(6). Hence, the bile acid pool composition can regulate the physiology of cells in the gastrointestinal system by differentially regulating different nuclear receptor and GPCRs and by direct toxicity to mammalian cells.
4. The secondary bile acid 7-oxolithocholic acid is reduced by host 11β-hydroxysteroid dehydrogenase 1
A number of gut bacterial have bile acid 7α-hydroxysteroid dehydrogenase activity which can generate 7-oxolithocholic acid (7-oxo-LCA) from chenodeoxycholic acid (45). 7-Oxo-LCA that is absorbed from the gut is reduced to chenodeoxycholic acid and small amounts of ursodeoxycholic acid by 11β-hydroxysteroid dehydrogenase 1 (11β-HSDH-1) in the liver (46, 47). There are two known isoforms of 11β-HSDH in humans and rodents, 11β-HSDH-1 and 11β-HSDH2. 11β-HSDH-2 has been reported to primary oxidize the 11β-hydroxy group of cortisol converting it to cortisone, essentially inactivating this glucocorticoid. In contrast, 11β-HSDH-1 appears to primarily function to reduce cortisone to cortisol producing an active glucocorticoid(48). Because 11β-HSDH-1 specifically reduces 7-oxo-LCA and this secondary bile acid accumulates in 11β-HSDH null mice, it has been proposed that 7-oxo-LCA may be a useful serum marker for 11β-HSDH-1 deficiency. Moreover, 7-oxo-LCA, or analogs, might function as competitive inhibitors of 11β-HSDH-1 which could be useful in treating various metabolic diseases.
5. Bile Acids Vary in their Ability to Activate Nuclear Receptors and GPCRs
Bile acids can activate several different nuclear receptors (FXR, PXR and Vitamin D) and GPCRs (TGR5, S1PR2, and [M2] Muscarinic receptor). The ability of different bile acids to activate FXR-α occurs in the following order CDCA>LCA = DCA>CA; for the pregnane X receptor (PXR) LCA>DCA>CA and the vitamin D receptor, 3-oxo-LCA>LCA>DCA>CA (49). LCA is the best activator of PXR and the vitamin D receptor which correlates with the hydrophobicity and toxicity of this bile acid toward mammalian cells. Activation of PXR and the vitamin D receptor induces genes encoding enzymes which metabolize LCA into a more hydrophilic and less toxic metabolite (49). These nuclear receptors appear to function in the protection of cells from hydrophobic bile acids. In contrast, FXR-α appears to play a much more extensive role in the body by regulating bile acid synthesis, transport, and enterohepatic circulation. Moreover, FXR-α also participates in the regulation of glucose, lipoprotein and lipid metabolism in the liver as well as a suppressor of inflammation in the liver and intestines (6). FXR null mice rapidly develop liver cancer suggesting FXR is a tumor suppressor (50, 51). Bile acids and FXR participate in the regulation of the level and composition of the intestinal microbiome by regulating antibacterial defenses in the small intestines and by direct effects on gut bacteria (52, 53).
TGR5, also referred to as membrane-type bile acid receptor (M-BAR), was the first GPCR to be reported to be activated by bile acids in the order LCA>DCA>CDCA>CA(54). TGR5 is a Gαs type receptor which activates adenyl cyclase activity increasing the rate of the synthesis of c-AMP (55). TGR5 is widely expressed in human tissues including: intestinal neuroendocrine cells, gall bladder, spleen, brown adipose tissue, macrophages and cholangiocytes, but not hepatocytes (55). TGR5 may play a role on various physiological processes in the body. TGR5 appears to be important in regulating energy metabolism. It has been postulated that bile acids may activate TGR5 in brown adipose tissue, activating type 2 iodothyroxine deiodinase, leading to increased levels of thyroid hormone and stimulation of energy metabolism (56). Moreover, TGR5 has been reported to promote the release of glucagon-like peptide-1 release from neuroendocrine cells which increases insulin release in the pancreas (57). These results suggest that TGR5 may play a role in glucose homeostasis in the body. TGR5 is a potential target for drug development for treating type 2 diabetes and other metabolic disorders (55, 58).
Taurine conjugated bile acids have been reported to activate specific muscarinic receptors (59). There are five different muscarinic receptors (M1 to M5) which are differentially expressed in various tissues in the body. Lithocholytaurine has been reported to activate the M3 muscarinic receptor on gastric chief cells stimulating the secretion of pepsinogen (60). Muscarinic receptors may play an important role in colon cancer. Muscarinic receptors are over expressed in colon cancer cells and activation stimulates proliferation, migration and invasion (61). Moreover, activation of muscarinic receptors stimulates matrix metalloproteinase 1-dependent invasion of colon cancer cells (62). However, the role of taurine conjugated bile acids in activating muscarinic receptors to promote colon cancer is unclear as most bile acids found in feces are unconjugated.
6. Interplay of Sphingosine 1-phosphate Receptor 2, Insulin and FXR in Regulating Hepatic Metabolism
Both unconjugated and conjugated bile acids activate the insulin signaling (AKT) and ERK1/2 pathways in hepatocytes (63). Interesting, insulin and bile acids both activated glycogen synthase activity to a similar extent in primary rat hepatocytes. Moreover, the addition of both insulin and bile acids to the culture medium resulted in an additive effect on activation of glycogen synthase activity in primary hepatocytes. Infusion of taurocholate (TCA) into the chronic bile fistula rat rapidly activated the AKT and ERK1/2 signaling pathway and glycogen synthase activity (63). In addition, there was a rapid down-regulation of the gluconeogenic genes, PEP carboxykinase (PEPCK) and glucose-6-phophatase (G-6-Pase) and a marked up-regulation of SHP mRNA in these sample livers (64). These results suggest that TCA functions much like insulin to regulate hepatic glucose metabolism both in vitro and in vivo.
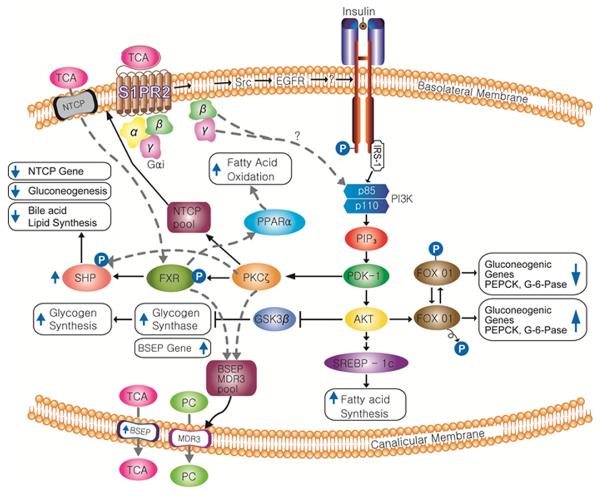
Conjugated bile acids activate the insulin signaling (AKT) and ERK 1/2 pathways via a pertussis toxin sensitive mechanism in primary hepatocytes and in vivo (63, 64). These results suggest that conjugated bile acids activate a Gαi coupled GPCR in hepatocytes. The addition of pertussis toxin or expression dominant negative Gαi in primary rat hepatocytes blocked the ability of TCA to activate the AKT signaling pathway and to down-regulate PEPCK and G-6-Pase. Surprisingly, inhibiting PI3 kinase with Wortmannin, but not an AKT or ERK1/2 chemical inhibitor, markedly decreased the ability of TCA to induce SHP mRNA. Moreover, inhibiting PKCζ with a chemical inhibitor or shRNA markedly inhibited TCA induction of SHP and the bile salt exporter (ABCB11), both FXR target genes (64). PKCζ is activated by phosphoinositide-dependent protein kinase-1 (PDK-1), a branch pathway of the insulin signaling pathway. Chemical inhibitors of other isoforms of PKC had no effect on the induction of SHP by TCA in primary hepatocytes. It has been reported that PKCζ phosphorylates FXR-α (65) and may allow for its activation of target gene expression. In contrast, phosphorylation of FXR-α by AMPK inhibits the ability of FXR to induce target genes (66). PKCζ has been reported to be important for the translocation of the bile acid transporters NTCP (SLC10A1) and BSEP (ABC B11) to the basolateral and canalicular membranes, respectively (65, 67). Finally, it has been recently reported that PKCζ phosphorylates SHP allowing both to translocate to the nucleus and down-regulate genes via epigenetic mechanisms (68). In total, these results all suggest that the insulin signaling pathway is an important regulator of FXR-α activation and bile acid signaling in the liver (Figure 3).
Figure 3. Interrelationship between sphingosine 1-phosphate receptor 2 and the insulin signaling pathway in regulating hepatic nutrient metabolism.
S1PR2, sphingosine 1-phosphate receptor 2; Src, Src Kinase; EGFR, epidermal growth factor receptor; PPARα, peroxisome proliferator-activated receptor alpha; NTCP, Na+/taurocholate cotransporting polypeptide; BSEP, bile salt export pump; PC, phosphotidylcholine; PECK, phosphoenolpyruvate carboxykinase; G6Pase, glucose-6-phosphatase; PDK1, phosphoinositide-dependent protein kinase 1; AKT, protein kinase B; SREBP, sterol regulatory element-binding protein; PKCζ, protein kinase C zeta; FXR, farnesoid x receptor; SHP, small heterodimeric partner; MDR3, phospholipid transporter (ABCB4); GSK3β, glycogen synthase kinase 3 beta.
The identity of the major Gαi GPCR activating the AKT and ERK1/2 signaling pathway in primary hepatocytes was recently reported to be the S1PR2 by screening lipid-activated GPCRs expressed in HEK-293 cells. All conjugated bile acids tested activated the S1PR2. Moreover, JTE-013, a S1PR2 chemical antagonist or a recombinant lentivirus encoding S1PR2 shRNA markedly inhibited the activation of ERK1/2 and AKT in primary hepatocytes and in the chronic bile fistula rat (69).
The activation of the insulin signaling pathway and FXR-α appear to collaborate in the coordinate regulation of glucose, bile acid and lipid metabolism in the liver. SHP, an FXR target gene, is an important pleotropic regulator of multiple metabolic pathways in the liver (Figure 3). The S1PR2 appears to be an important regulator of hepatic lipid metabolism as S1PR2 null mice rapidly (2 weeks) develop overt fatty livers on a high fat diet as compared to wild-type mice (unpublished data). It is well established that inflammation and the synthesis of inflammatory cytokines i.e. TNFα inhibit insulin signaling by activation of the JNK 1/2 signaling pathway which phosphorylates insulin receptor substrate 1 (70). Inflammation is believed to be an important factor in the development of type 2 diabetes and fatty liver disease. A Western diet is correlated with low grade chronic inflammation and insulin resistance. Inhibition of the insulin signaling pathway may decrease the ability of bile acids to activate FXR-α, induce SHP and other FXR target genes leading to an increased risk of fatty liver and non-alcoholic fatty liver disease (NAFLD) (55).
6.1 Conjugated bile acids stimulate cholangiocarcinoma growth via the S1PR2
Cholangiocarcinoma (CCA) is an often fatal cancer of the biliary tract and its occurrence is associated with chronic cholestasis and elevation of conjugated primary bile acids in the liver and serum. Bile duct obstruction has been reported to promote the growth of CCA. In vitro, conjugated but not unconjugated bile acids promote the growth of CCA (71, 72). In recent studies, Lui R et al. reported that the S1PR2 is highly expressed in both rat and human CCA lines and in human CCA tissues (73). TCA activated the AKT and ERK1/2 signaling pathways in both human and rat CCA cells in culture. Moreover, TCA stimulated CCA cell proliferation, migration and invasion was strongly inhibited by JTE-013, a chemical antagonist of the S1PR2, and by a lentiviral shRNA directed against S1PR2. Finally, FXR-α and ASBT were significantly down-regulated in CCA as compared to normal cholangiocytes. These data suggest that conjugated bile acids promote the invasive growth of CCA via the S1PR2. S1PR2 and FXR may represent new targets for treating CCA.
7. Summary and Future Directions
There appears to be extensive interplay between bile salts and insulin signaling in the regulation of nutrient metabolism in both the intestines and liver. Bile salts play a key role in the solubilization and absorption of nutrients from the intestines. The absorption of nutrients stimulates the secretion of insulin from the pancreas. Moreover, bile acids may also stimulate the secretion of insulin by activating TGR5 in intestinal neuroendocrine cells resulting in the secretion of glucagon-like peptide-1. In the liver, bile salts and insulin both activate the AKT and ERK1/2 signaling pathways which yields a stronger signal than either alone. The activation of PKCζ, a branch of the insulin signaling pathway, is required for the optimal induction of FXR target genes and the regulation of the cellular location of bile acid transporters (Figure 3).
Perhaps an important future direction of bile acid signaling research would be to determine to what extent inflammation affects bile acid signaling and hepatic metabolism. Moreover, it is currently unclear what physiological role activation of the ERK1/2 by bile acids and insulin has on hepatic nutrient metabolism. The elucidation of epigenetic mechanisms of gene regulation in the liver by bile acids may be important for regulating nutrient metabolism during the feed/fast cycle. Finally, the role of the S1PR2 and conjugated bile acids in the growth of cholangiocytes and CCA is another important direction for research in this area. The study of bile acids as hormones continues to be a growing and expanding field of biomedical research.
Highlights.
Bile acids are important signaling molecules.
Bile acids can activate nuclear receptors and GPCRs.
Bile acid-mediated signaling pathways play important roles in lipid and glucose metabolism.
Dysregulation of bile acid-mediated signaling pathways contributes to various metabolic diseases.
Acknowledgments
Financial Support: The work was supported by A.D. Williams Award (to HZ), National Institutes of Health (NIH) Grant R01 DK-057543 to PH and HZ. This study is also partially supported by VA Merit Award 1BX0013828-01 to PH; 1I01BX001390 to HZ.
List of Abbreviations
- ASBT
apical sodium dependent transporter
- AKT
protein kinase B
- BSEP
bile salt export protein (ABCB11)
- CA
cholic acid
- CCA
cholangiocarcinoma
- CDCA
chenodeoxycholic acid
- CYP7A1
cholesterol 7α-hydroxylase
- CYP7B1
oxysterol 7α-hydroxylase
- CYP27A1
sterol 27-hydroxylase
- CYP8B1
12α-hydroxylase
- DCA
deoxycholic acid
- EGFR
epidermal growth factor receptor
- ERK1/2
extracellular signal-regulated kinase
- FGF15/19
fibroblast growth factor 15/19
- FXR
farnesoid x receptor
- G-6-Pase
glucose-6-phosphatase
- GCA
glycocholic acid
- GDCA
glycodeoxycholic acid
- GPCR
G-protein coupled receptor
- HNF4a
hepatocyte nuclear factor 4
- LCA
lithocholic acid
- LRH-1
liver-related homolog-1
- LXR
liver X receptor
- M1–5
muscarinic receptor 1–5
- NAFLD
non-alcoholic fatty liver disease
- NTCP
sodium taurocholate cotransporting polypeptide
- P13K
phosphatidylinositol-3-kinase
- PEPCK
PEP carboxykinse
- PXR
pregnane X receptor
- S1P
sphingosine 1-phosphate
- S1PR2
sphingosine 1-phosphate receptor 2
- SHP
small heterodimer partner
- TCA
taurocholate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vlahcevic ZRH, D.M, Hylemon PB. Physiology and pathophysiology of enterohepatic circulation of bike acids. In: Zakim F, Boyer T, editors. Hepatology: A Textbook of Liver Disease. W. B. Sanders Co; Philadelphia: 1996. pp. 376–471. [Google Scholar]
- 2.Makishima M, Okamoto AY, Repa JJ, Tu H, Learned RM, Luk A, Hull MV, et al. Identification of a nuclear receptor for bile acids. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- 3.Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA, Stimmel JB, et al. Bile acids: natural ligands for an orphan nuclear receptor. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- 4.Wang H, Chen J, Hollister K, Sowers LC, Forman BM. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol Cell. 1999;3:543–553. doi: 10.1016/s1097-2765(00)80348-2. [DOI] [PubMed] [Google Scholar]
- 5.Schaap FG, Trauner M, Jansen PL. Bile acid receptors as targets for drug development. Nat Rev Gastroenterol Hepatol. 2014;11:55–67. doi: 10.1038/nrgastro.2013.151. [DOI] [PubMed] [Google Scholar]
- 6.de Aguiar Vallim TQ, Tarling EJ, Edwards PA. Pleiotropic roles of bile acids in metabolism. Cell Metab. 2013;17:657–669. doi: 10.1016/j.cmet.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qiao L, Studer E, Leach K, McKinstry R, Gupta S, Decker R, Kukreja R, et al. Deoxycholic acid (DCA) causes ligand-independent activation of epidermal growth factor receptor (EGFR) and FAS receptor in primary hepatocytes: inhibition of EGFR/mitogen-activated protein kinase-signaling module enhances DCA-induced apoptosis. Mol Biol Cell. 2001;12:2629–2645. doi: 10.1091/mbc.12.9.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Claudel T, Trauner M. Adiponectin, bile acids, and burnt-out nonalcoholic steatohepatitis: new light on an old paradox. Hepatology. 2013;57:2106–2109. doi: 10.1002/hep.26340. [DOI] [PubMed] [Google Scholar]
- 9.Fuchs M. Non-alcoholic Fatty liver disease: the bile Acid-activated farnesoid x receptor as an emerging treatment target. J Lipids. 2012;2012:934396. doi: 10.1155/2012/934396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Jadhav K, Zhang Y. Bile acid receptors in non-alcoholic fatty liver disease. Biochem Pharmacol. 2013;86:1517–1524. doi: 10.1016/j.bcp.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eguchi A, Povero D, Alkhouri N, Feldstein AE. Novel therapeutic targets for nonalcoholic fatty liver disease. Expert Opin Ther Targets. 2013;17:773–779. doi: 10.1517/14728222.2013.789502. [DOI] [PubMed] [Google Scholar]
- 12.Adorini L, Pruzanski M, Shapiro D. Farnesoid X receptor targeting to treat nonalcoholic steatohepatitis. Drug Discov Today. 2012;17:988–997. doi: 10.1016/j.drudis.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 13.Trauner M, Boyer JL. Bile salt transporters: molecular characterization, function, and regulation. Physiol Rev. 2003;83:633–671. doi: 10.1152/physrev.00027.2002. [DOI] [PubMed] [Google Scholar]
- 14.Quazi F, Molday RS. Lipid transport by mammalian ABC proteins. Essays Biochem. 2011;50:265–290. doi: 10.1042/bse0500265. [DOI] [PubMed] [Google Scholar]
- 15.Dawson PA, Hubbert ML, Rao A. Getting the mOST from OST: Role of organic solute transporter, OSTalpha-OSTbeta, in bile acid and steroid metabolism. Biochim Biophys Acta. 2010;1801:994–1004. doi: 10.1016/j.bbalip.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doring B, Lutteke T, Geyer J, Petzinger E. The SLC10 carrier family: transport functions and molecular structure. Curr Top Membr. 2012;70:105–168. doi: 10.1016/B978-0-12-394316-3.00004-1. [DOI] [PubMed] [Google Scholar]
- 17.Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG, Luo G, et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2:217–225. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 18.Potthoff MJ, Kliewer SA, Mangelsdorf DJ. Endocrine fibroblast growth factors 15/19 and 21: from feast to famine. Genes Dev. 2012;26:312–324. doi: 10.1101/gad.184788.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cicione C, Degirolamo C, Moschetta A. Emerging role of fibroblast growth factors 15/19 and 21 as metabolic integrators in the liver. Hepatology. 2012;56:2404–2411. doi: 10.1002/hep.25929. [DOI] [PubMed] [Google Scholar]
- 20.Gupta S, Stravitz RT, Dent P, Hylemon PB. Down-regulation of cholesterol 7alpha-hydroxylase (CYP7A1) gene expression by bile acids in primary rat hepatocytes is mediated by the c-Jun N-terminal kinase pathway. J Biol Chem. 2001;276:15816–15822. doi: 10.1074/jbc.M010878200. [DOI] [PubMed] [Google Scholar]
- 21.Yu C, Wang F, Kan M, Jin C, Jones RB, Weinstein M, Deng CX, et al. Elevated cholesterol metabolism and bile acid synthesis in mice lacking membrane tyrosine kinase receptor FGFR4. J Biol Chem. 2000;275:15482–15489. doi: 10.1074/jbc.275.20.15482. [DOI] [PubMed] [Google Scholar]
- 22.Ito S, Fujimori T, Furuya A, Satoh J, Nabeshima Y, Nabeshima Y. Impaired negative feedback suppression of bile acid synthesis in mice lacking betaKlotho. J Clin Invest. 2005;115:2202–2208. doi: 10.1172/JCI23076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kong B, Wang L, Chiang JY, Zhang Y, Klaassen CD, Guo GL. Mechanism of tissue-specific farnesoid X receptor in suppressing the expression of genes in bile-acid synthesis in mice. Hepatology. 2012;56:1034–1043. doi: 10.1002/hep.25740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hylemon PB, Gurley EC, Stravitz RT, Litz JS, Pandak WM, Chiang JY, Vlahcevic ZR. Hormonal regulation of cholesterol 7 alpha-hydroxylase mRNA levels and transcriptional activity in primary rat hepatocyte cultures. J Biol Chem. 1992;267:16866–16871. [PubMed] [Google Scholar]
- 25.Song KH, Chiang JY. Glucagon and cAMP inhibit cholesterol 7alpha-hydroxylase (CYP7A1) gene expression in human hepatocytes: discordant regulation of bile acid synthesis and gluconeogenesis. Hepatology. 2006;43:117–125. doi: 10.1002/hep.20919. [DOI] [PubMed] [Google Scholar]
- 26.Li T, Francl JM, Boehme S, Ochoa A, Zhang Y, Klaassen CD, Erickson SK, et al. Glucose and insulin induction of bile acid synthesis: mechanisms and implication in diabetes and obesity. J Biol Chem. 2012;287:1861–1873. doi: 10.1074/jbc.M111.305789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee YK, Schmidt DR, Cummins CL, Choi M, Peng L, Zhang Y, Goodwin B, et al. Liver receptor homolog-1 regulates bile acid homeostasis but is not essential for feedback regulation of bile acid synthesis. Mol Endocrinol. 2008;22:1345–1356. doi: 10.1210/me.2007-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mataki C, Magnier BC, Houten SM, Annicotte JS, Argmann C, Thomas C, Overmars H, et al. Compromised intestinal lipid absorption in mice with a liver-specific deficiency of liver receptor homolog 1. Mol Cell Biol. 2007;27:8330–8339. doi: 10.1128/MCB.00852-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X, Hylemon P, Pandak WM, Ren S. Enzyme activity assay for cholesterol 27-hydroxylase in mitochondria. J Lipid Res. 2006;47:1507–1512. doi: 10.1194/jlr.M600117-JLR200. [DOI] [PubMed] [Google Scholar]
- 30.Li X, Pandak WM, Erickson SK, Ma Y, Yin L, Hylemon P, Ren S. Biosynthesis of the regulatory oxysterol, 5-cholesten-3beta, 25-diol 3-sulfate, in hepatocytes. J Lipid Res. 2007;48:2587–2596. doi: 10.1194/jlr.M700301-JLR200. [DOI] [PubMed] [Google Scholar]
- 31.Cali JJ, Russell DW. Characterization of human sterol 27-hydroxylase. A mitochondrial cytochrome P-450 that catalyzes multiple oxidation reaction in bile acid biosynthesis. J Biol Chem. 1991;266:7774–7778. [PubMed] [Google Scholar]
- 32.Bjorkhem I. Cerebrotendinous xanthomatosis. Curr Opin Lipidol. 2013;24:283–287. doi: 10.1097/MOL.0b013e328362df13. [DOI] [PubMed] [Google Scholar]
- 33.Pandak WM, Ren S, Marques D, Hall E, Redford K, Mallonee D, Bohdan P, et al. Transport of cholesterol into mitochondria is rate-limiting for bile acid synthesis via the alternative pathway in primary rat hepatocytes. J Biol Chem. 2002;277:48158–48164. doi: 10.1074/jbc.M205244200. [DOI] [PubMed] [Google Scholar]
- 34.Ren S, Hylemon PB, Marques D, Gurley E, Bodhan P, Hall E, Redford K, et al. Overexpression of cholesterol transporter StAR increases in vivo rates of bile acid synthesis in the rat and mouse. Hepatology. 2004;40:910–917. doi: 10.1002/hep.20382. [DOI] [PubMed] [Google Scholar]
- 35.Axelson M, Sjovall J. Potential bile acid precursors in plasma--possible indicators of biosynthetic pathways to cholic and chenodeoxycholic acids in man. J Steroid Biochem. 1990;36:631–640. doi: 10.1016/0022-4731(90)90182-r. [DOI] [PubMed] [Google Scholar]
- 36.Ren S, Ning Y. Sulfation of 25-hydroxycholesterol regulates lipid metabolism, inflammatory responses, and cell proliferation. Am J Physiol Endocrinol Metab. 2014;306:E123–130. doi: 10.1152/ajpendo.00552.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ren S, Hylemon P, Zhang ZP, Rodriguez-Agudo D, Marques D, Li X, Zhou H, et al. Identification of a novel sulfonated oxysterol, 5-cholesten-3beta, 25-diol 3-sulfonate, in hepatocyte nuclei and mitochondria. J Lipid Res. 2006;47:1081–1090. doi: 10.1194/jlr.M600019-JLR200. [DOI] [PubMed] [Google Scholar]
- 38.Xu L, Bai Q, Rodriguez-Agudo D, Hylemon PB, Heuman DM, Pandak WM, Ren S. Regulation of hepatocyte lipid metabolism and inflammatory response by 25-hydroxycholesterol and 25-hydroxycholesterol-3-sulfate. Lipids. 2010;45:821–832. doi: 10.1007/s11745-010-3451-y. [DOI] [PubMed] [Google Scholar]
- 39.Xu L, Shen S, Ma Y, Kim JK, Rodriguez-Agudo D, Heuman DM, Hylemon PB, et al. 25-Hydroxycholesterol-3-sulfate attenuates inflammatory response via PPARgamma signaling in human THP-1 macrophages. Am J Physiol Endocrinol Metab. 2012;302:E788–799. doi: 10.1152/ajpendo.00337.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yantsevich AV, Dichenko YV, Mackenzie F, Mukha DV, Baranovsky AV, Gilep AA, Usanov SA, et al. Human steroid and oxysterol 7alpha-hydroxylase CYP7B1: substrate specificity, azole binding and misfolding of clinically relevant mutants. FEBS J. 2014 doi: 10.1111/febs.12733. [DOI] [PubMed] [Google Scholar]
- 41.Ridlon JM, Hylemon PB. A potential role for resistant starch fermentation in modulating colonic bacterial metabolism and colon cancer risk. Cancer Biol Ther. 2006;5:273–274. doi: 10.4161/cbt.5.3.2728. [DOI] [PubMed] [Google Scholar]
- 42.Ridlon JM, Ikegawa S, Alves JM, Zhou B, Kobayashi A, Iida T, Mitamura K, et al. Clostridium scindens: a human gut microbe with a high potential to convert glucocorticoids into androgens. J Lipid Res. 2013;54:2437–2449. doi: 10.1194/jlr.M038869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas LA, Veysey MJ, Bathgate T, King A, French G, Smeeton NC, Murphy GM, et al. Mechanism for the transit-induced increase in colonic deoxycholic acid formation in cholesterol cholelithiasis. Gastroenterology. 2000;119:806–815. doi: 10.1053/gast.2000.16495. [DOI] [PubMed] [Google Scholar]
- 44.McGarr SE, Ridlon JM, Hylemon PB. Diet, anaerobic bacterial metabolism, and colon cancer: a review of the literature. J Clin Gastroenterol. 2005;39:98–109. [PubMed] [Google Scholar]
- 45.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 46.Penno CA, Morgan SA, Vuorinen A, Schuster D, Lavery GG, Odermatt A. Impaired oxidoreduction by 11beta-hydroxysteroid dehydrogenase 1 results in the accumulation of 7-oxolithocholic acid. J Lipid Res. 2013;54:2874–2883. doi: 10.1194/jlr.M042499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Odermatt A, Da Cunha T, Penno CA, Chandsawangbhuwana C, Reichert C, Wolf A, Dong M, et al. Hepatic reduction of the secondary bile acid 7-oxolithocholic acid is mediated by 11beta-hydroxysteroid dehydrogenase 1. Biochem J. 2011;436:621–629. doi: 10.1042/BJ20110022. [DOI] [PubMed] [Google Scholar]
- 48.Morris DJ, Latif SA, Brem AS. An alternative explanation of hypertension associated with 17alpha-hydroxylase deficiency syndrome. Steroids. 2014;79:44–48. doi: 10.1016/j.steroids.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 49.Makishima M, Lu TT, Xie W, Whitfield GK, Domoto H, Evans RM, Haussler MR, et al. Vitamin D receptor as an intestinal bile acid sensor. Science. 2002;296:1313–1316. doi: 10.1126/science.1070477. [DOI] [PubMed] [Google Scholar]
- 50.Kim I, Morimura K, Shah Y, Yang Q, Ward JM, Gonzalez FJ. Spontaneous hepatocarcinogenesis in farnesoid X receptor-null mice. Carcinogenesis. 2007;28:940–946. doi: 10.1093/carcin/bgl249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Su H, Ma C, Liu J, Li N, Gao M, Huang A, Wang X, et al. Downregulation of nuclear receptor FXR is associated with multiple malignant clinicopathological characteristics in human hepatocellular carcinoma. Am J Physiol Gastrointest Liver Physiol. 2012;303:G1245–1253. doi: 10.1152/ajpgi.00439.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Islam KB, Fukiya S, Hagio M, Fujii N, Ishizuka S, Ooka T, Ogura Y, et al. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology. 2011;141:1773–1781. doi: 10.1053/j.gastro.2011.07.046. [DOI] [PubMed] [Google Scholar]
- 53.Inagaki T, Moschetta A, Lee YK, Peng L, Zhao G, Downes M, Yu RT, et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci U S A. 2006;103:3920–3925. doi: 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kawamata Y, Fujii R, Hosoya M, Harada M, Yoshida H, Miwa M, Fukusumi S, et al. A G protein-coupled receptor responsive to bile acids. J Biol Chem. 2003;278:9435–9440. doi: 10.1074/jbc.M209706200. [DOI] [PubMed] [Google Scholar]
- 55.Duboc H, Tache Y, Hofmann AF. The bile acid TGR5 membrane receptor: From basic research to clinical application. Dig Liver Dis. 2014;46:302–312. doi: 10.1016/j.dld.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watanabe M, Houten SM, Mataki C, Christoffolete MA, Kim BW, Sato H, Messaddeq N, et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439:484–489. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- 57.Kumar DP, Rajagopal S, Mahavadi S, Mirshahi F, Grider JR, Murthy KS, Sanyal AJ. Activation of transmembrane bile acid receptor TGR5 stimulates insulin secretion in pancreatic beta cells. Biochem Biophys Res Commun. 2012;427:600–605. doi: 10.1016/j.bbrc.2012.09.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tiwari A, Maiti P. TGR5: an emerging bile acid G-protein-coupled receptor target for the potential treatment of metabolic disorders. Drug Discov Today. 2009;14:523–530. doi: 10.1016/j.drudis.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 59.Raufman JP, Cheng K, Zimniak P. Activation of muscarinic receptor signaling by bile acids: physiological and medical implications. Dig Dis Sci. 2003;48:1431–1444. doi: 10.1023/a:1024733500950. [DOI] [PubMed] [Google Scholar]
- 60.Raufman JP, Zimniak P, Bartoszko-Malik A. Lithocholyltaurine interacts with cholinergic receptors on dispersed chief cells from guinea pig stomach. Am J Physiol. 1998;274:G997–1004. doi: 10.1152/ajpgi.1998.274.6.G997. [DOI] [PubMed] [Google Scholar]
- 61.Cheng K, Chen Y, Zimniak P, Raufman JP, Xiao Y, Frucht H. Functional interaction of lithocholic acid conjugates with M3 muscarinic receptors on a human colon cancer cell line. Biochim Biophys Acta. 2002;1588:48–55. doi: 10.1016/s0925-4439(02)00115-1. [DOI] [PubMed] [Google Scholar]
- 62.Raufman JP, Cheng K, Saxena N, Chahdi A, Belo A, Khurana S, Xie G. Muscarinic receptor agonists stimulate matrix metalloproteinase 1-dependent invasion of human colon cancer cells. Biochem Biophys Res Commun. 2011;415:319–324. doi: 10.1016/j.bbrc.2011.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fang Y, Studer E, Mitchell C, Grant S, Pandak WM, Hylemon PB, Dent P. Conjugated bile acids regulate hepatocyte glycogen synthase activity in vitro and in vivo via Galphai signaling. Mol Pharmacol. 2007;71:1122–1128. doi: 10.1124/mol.106.032060. [DOI] [PubMed] [Google Scholar]
- 64.Cao R, Cronk ZX, Zha W, Sun L, Wang X, Fang Y, Studer E, et al. Bile acids regulate hepatic gluconeogenic genes and farnesoid X receptor via G(alpha)i-protein-coupled receptors and the AKT pathway. J Lipid Res. 2010;51:2234–2244. doi: 10.1194/jlr.M004929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Frankenberg T, Miloh T, Chen FY, Ananthanarayanan M, Sun AQ, Balasubramaniyan N, Arias I, et al. The membrane protein ATPase class I type 8B member 1 signals through protein kinase C zeta to activate the farnesoid X receptor. Hepatology. 2008;48:1896–1905. doi: 10.1002/hep.22431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lien F, Berthier A, Bouchaert E, Gheeraert C, Alexandre J, Porez G, Prawitt J, et al. Metformin interferes with bile acid homeostasis through AMPK-FXR crosstalk. J Clin Invest. 2014;124:1037–1051. doi: 10.1172/JCI68815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sarkar S, Bananis E, Nath S, Anwer MS, Wolkoff AW, Murray JW. PKCzeta is required for microtubule-based motility of vesicles containing the ntcp transporter. Traffic. 2006;7:1078–1091. doi: 10.1111/j.1600-0854.2006.00447.x. [DOI] [PubMed] [Google Scholar]
- 68.Seok S, Kanamaluru D, Xiao Z, Ryerson D, Choi SE, Suino-Powell K, Xu HE, et al. Bile acid signal-induced phosphorylation of small heterodimer partner by protein kinase Czeta is critical for epigenomic regulation of liver metabolic genes. J Biol Chem. 2013;288:23252–23263. doi: 10.1074/jbc.M113.452037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Studer E, Zhou X, Zhao R, Wang Y, Takabe K, Nagahashi M, Pandak WM, et al. Conjugated bile acids activate the sphingosine-1-phosphate receptor 2 in primary rodent hepatocytes. Hepatology. 2012;55:267–276. doi: 10.1002/hep.24681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Copps KD, White MF. Regulation of insulin sensitivity by serine/threonine phosphorylation of insulin receptor substrate proteins IRS1 and IRS2. Diabetologia. 2012;55:2565–2582. doi: 10.1007/s00125-012-2644-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang H, Li TW, Peng J, Tang X, Ko KS, Xia M, Aller MA. A mouse model of cholestasis-associated cholangiocarcinoma and transcription factors involved in progression. Gastroenterology. 2011;141:378–388. 388, e371–374. doi: 10.1053/j.gastro.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sirica AE, Zhang Z, Lai GH, Asano T, Shen XN, Ward DJ, Mahatme A, et al. A novel “patient-like” model of cholangiocarcinoma progression based on bile duct inoculation of tumorigenic rat cholangiocyte cell lines. Hepatology. 2008;47:1178–1190. doi: 10.1002/hep.22088. [DOI] [PubMed] [Google Scholar]
- 73.Liu RZ, R, Zhou X, Liang X, Campbell DJ, Zhang X, Zhang LY, Shi R, Wang G, Pandak WM, Sirica AE, Hylemon PB, ZHou H. Conjugated Bile Acids Promote Cholangiocarcinoma Cell Invasive Growth via Activation of Sphingosine 1-phosphate Receptor 2. Hepatology. 2014 doi: 10.1002/hep.27085. DOI: 10.1002/hep.27085. [DOI] [PMC free article] [PubMed] [Google Scholar]