Abstract
In Structural Genomics projects, virtual high-throughput ligand screening can be utilized to provide important functional details for newly determined protein structures. Using a variety of publicly available software tools, it is possible to computationally model, predict, and evaluate how different ligands interact with a given protein. At the Center for Structural Genomics of Infectious Diseases (CSGID) a series of protein analysis, docking and molecular dynamics software is scripted into a single hierarchical pipeline allowing for an exhaustive investigation of protein-ligand interactions. The ability to conduct accurate computational predictions of protein-ligand binding is a vital component in improving both the efficiency and economics of drug discovery. Computational simulations can minimize experimental efforts, the slowest and most cost prohibitive aspect of identifying new therapeutics.
Keywords: Protein, Ligand, High-throughput screening, Docking, Molecular modeling
1 Introduction
In the context of structural genomics (SG), identification of bound ligands can provide many benefits. A bound ligand can increase the stability of crystal packing to provide a higher resolution structure, provide hydrogen bonding interactions to anchor a highly flexible loop region, and/or provide important functional evidence for proteins of unknown function. As structural genomics initiatives move toward more specialized goals (i.e., centers for structural genomics of infectious disease, tuberculosis, biology) identification of ligand bound structures can play an even bigger role: function prediction and validation or early stage drug discovery efforts.
Identifying ligands for co-crystallization experiments in structural genomics requires a different strategy than for a concerted drug discovery effort. The latter is characterized by a high-degree of knowledge about the protein target, its biochemical mechanism, and substrates. This information is used to highly tailor an effort to identify an optimum ligand in order to alter a specific mechanism, most likely through inhibition of the mechanism. The structural genomics effort, by design of target selection, is characterized by a significantly reduced amount of information about the protein. In some circumstances, a newly determined structure will represent the first three-dimensional model of the protein. Any additional protein-ligand interaction data that is generated can provide valuable context for increasing the biological impact of the structure.
In many structural genomics efforts, the program’s throughput does not allow for significant effort or resources to be allocated to further biological experimentation beyond structure determination. This includes the significant amount of time that may be required to obtain new protein crystals with bound ligands, collect data, and refine models, in addition to the time necessary to analyze small molecule compound databases, synthesize compounds, and optimize solubility. Therefore, the introduction of computational approaches to increase efficiency, reduce costs, and improve success of ligand identification for protein targets is a pragmatic approach undertaken by many structural genomics efforts.
At the CSGID a series of protein analysis, docking and molecular dynamics software packages have been combined into a single hierarchical pipeline allowing for an exhaustive investigation of protein-ligand interactions. The APPLIED (Analysis Pipeline for Protein-Ligand Interactions and Experimental Determination) pipeline allows for the evolutionary analysis of protein binding sites with cheminformatics obtained from petascale computational docking experiments to create a high-quality library of datasets of protein-ligand interaction. Such libraries provide global scale analysis of protein domain-small molecule interactions that can be used to provide insights on protein function, predict ligand interactions, and perform early stage computer aided drug discovery.
2 Materials
2.1 Software
The methodologies utilized in the APPLIED Pipeline use the following software packages:
DOCK 6, University of California, San Francisco [1].
AUTODOCK, The Scripps Research Institute [2].
NAB (Nucleic Acid Builder) [3].
CHARMM (Chemistry at HARvard Macromolecular Mechanics), Harvard University [4].
SurfaceScreen, Argonne National Laboratory [5].
Falkon, Argonne National Laboratory [6].
Swift, Argonne National Laboratory [7].
The software is organized into a pipeline using a series of scripts written in the PERL and PYTHON scripting languages. The pipeline is implemented and operates on “Intrepid” an IBM BlueGene/P supercomputer located at the Advanced Leadership Computing Facility (ALCF) at Argonne National Laboratory. Access to Intrepid is provided through the Department of Energy’s INCITE (Innovative and Novel Computational Impact on Theory and Experiment) program.
2.2 Databases
The APPLIED pipeline uses publicly available three-dimensional protein structure data from the Protein Data Bank (PDB) [8]. The ZINC [9] database of commercially available compounds is used for virtual screening.
3 Methods
3.1 APPLIED Pipeline Overview
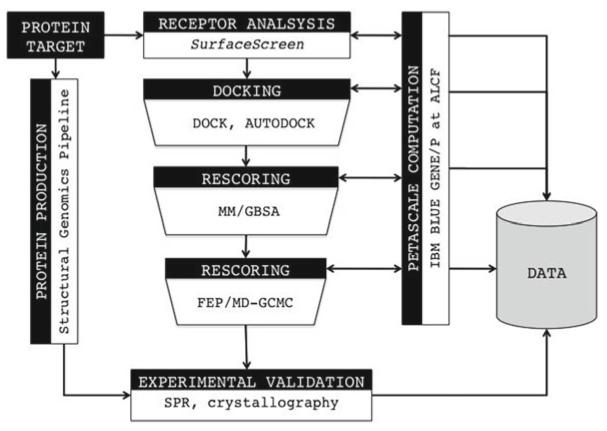
At CSGID virtual ligand screening is driven by the multistage, computational APPLIED pipeline (see Fig. 1). The automated pipeline is a data driven workflow for the rapid transformation of knowledge from initial target characterization into the prediction and validation of small compound binding affinity. All steps require large-scale computation using distributed tools to harness high-performance computing resources for efficient calculations. Computational results are used to drive experimental studies in CSGID’s high-throughput protein structure determination pipeline.
Fig. 1.
The APPLIED pipeline combines automated binding site identifi cation, docking and rescoring into a singular workfl ow to investigate protein-ligand interactions for structural genomics targets
Given a target with an existing three-dimensional structure, automated binding site identification and analysis is conducted using the SurfaceScreen methodology [5, 10, 11]. Based on comparison to a library of binding sites, SurfaceScreen identifies surfaces sharing structural and physicochemical properties, thereby uncovering the most probable active site. The active site is propagated down the pipeline for massively parallel docking simulations using mixed strategies to develop a complete cheminformatics profile of the pocket.
In the language of molecular modeling, ligand screening can be separated into two loosely defined steps, “docking” and “scoring” [12]. The docking step aims to predict the preferred orientation and conformation of the ligand molecule bound to the protein receptor (the ligand “pose”), and the scoring step aims to predict the binding affinity of the ligand for a given ligand orientation. While docking can proceed successfully via heuristic simplifications, the shortcomings of ligand screening approaches stem from the approximate scoring functions. The fundamental principles controlling ligand binding are relatively well understood, but scoring often relies on extremely simplified approximations in order to achieve the computational efficiency needed to handle large databases [13-15]. Nonetheless, to have any predictive and practical value, scoring must reflect the binding free energies with sufficient accuracy.
Arguably, physics-based approaches such as molecular mechanics-generalized born surface area (MM/GBSA) and free energy perturbation molecular dynamics (FEP/MD) simulations represent the most accurate approach to quantitatively characterize the binding free energy of small ligands with macromolecules. These physics-based methods can naturally handle the influence of solvent and dynamic flexibility [14], and indeed, previous studies indicate that they are often more reliable than simpler scoring schemes [16, 17].
In the APPLIED pipeline, after the initial docking poses are generated, compounds are “funneled” into highly parallelized implementations of these complex rescoring methods. The top ranked 10,000 molecules are rescored using MM-GBSA methodology [18]. The FEP/MD-GCMC (molecular dynamics free energy perturbation-grand canonical Monte Carlo) [19] method is then used to rescore the top 100 compounds based on free energy binding estimations. A full run through the pipeline requires over 500,000 computing hours, but has been efficiently scaled for optimal performance on the BlueGene/P.
The pipeline currently docks against the aggregated ZINC library of commercially available compounds [20] (over 21 million in release 12), allowing the easy purchase of compounds and minimizing the need for chemical synthesis capabilities.
3.2 Pipeline Architecture
Collectively, the APPLIED pipeline is a hybrid of highly parallel and high-throughput techniques, integrated with an innovative model for parallel scripting at extreme scales, and carefully tuned for the Intrepid BG/P. SurfaceScreen, DOCK, and AUTODOCK leverage the BG/P in a high-throughput computing mode. FEP-REMD/GCMC uses a new, innovative, and highly parallel variant of CHARMM to achieve excellent scaling using MPI.
3.2.1 High-Throughput Computing Mode
SurfaceScreen, DOCK, and AUTODOCK involve many thousands of discrete, loosely coupled computations with significant data exchange taking place via files. An important goal of the pipeline is to ensure that these computations can be performed rapidly and reliably. A set of custom tools was developed to specify and orchestrate the execution of many independent tasks. These tools are based on Swift, a system for the rapid and reliable specification, execution, and management of large-scale computational pipelines [7, 21] and Falkon, a system to efficiently provision cluster resources for long-running workflows composed of short discrete tasks [6].
It has been shown that a loosely coupled approach (e.g., treating application invocations as functions and passing data through the file system) permits effective use of the BG/P for workflows in which applications can be integrated into a larger application as if they were ordinary functions [6, 22]. These middleware components have been extensively tested at scales across the entire BG/P complex and have achieved near-linear speedup on workloads that comfortably encompass the performance characteristics of the pipeline in both file I/O and task duration [6, 22].
3.2.2 Parallel Distributed Replica Mode
One complete molecular dynamics (MD) run of binding free energy calculation consists of one hydration and one binding site calculation each requiring tens of independent FEP windows. It needs to be emphasized that the employment of SSBP/GSBP significantly decreases the size of simulated region (number of simulated atoms), and therefore it is not necessary to invoke a large number of CPUs for a single FEP window. Even with an excellently scaled MD package, like NAMD [23], it is impossible for BG/P to run these tens of FEP windows independently. To take full advantage of BG/P, the Parallel Distributed Replica (REPDSTR) mode is employed to run in a highly efficient parallel/parallel mode. Historically, CHARMM was the first MD package equipped with parallel/parallel mode for free energy calculations.
In REPDSTR mode, each of the underlying multiple I/O controls one FEP window, all the windows run in parallel, and each window is a smaller parallel job that occupies 32 processors (optimized). Thus, the total number of MPI ranks is proportional to the number of FEP windows multiplied by 32. On BG/P, usually each REPDSTR job is run with 2,048 MPI ranks (hydration simulation) or 4,096 MPI ranks (binding site). For a specific binding complex, one is always able to modify the number of FEP windows (replicas) of each interaction type to make the total number of windows 128 or 64, resulting in 4,096 MPI ranks (128 × 32) or 2,048 MPI ranks (64 × 32).
The possible “load balance” problem happening in many parallelized jobs is eliminated naturally due to the similarity of the calculation between these replicas (they all employ CHARMM PERT module and deal with the same structure). Multiple binding complexes with REPDSTR are run so that more racks can be invoked for one single job and accelerate the throughput simultaneously. It needs to be noted that, for a fixed acceptor, the variation of ligand species only causes tiny (~20 atoms) variation in size of simulated binding structure, and therefore the “load balance” problem remains insignificant.
Besides the parallel/parallel structure of the REPDSTR module, another significant advancement is the implementation of replica exchange between these FEP windows. The replica exchange method has been exhaustively proven to be significant in speeding the sampling/convergence of free energy calculations. However, so far those implementations in biological simulations have been limited to relatively small systems and/or a small number of replicas due to lack of efficient parallel/parallel programming and the unavailability of a large number of processors in parallel. With REPDSTR mode and the abundant resource of BG/P, a successful development of the Hamiltonian-exchange scheme for FEP calculation, with infrequent point-to-point message communications (1 per 100 MD steps) between these tens of windows has been implemented. The new replica exchange scheme was proven to be efficient on BG/P (the message communication between FEP windows merely causes ~4 % speed loss compared with normal MD) and significantly increases the convergence/confidence of the free energy calculation.
3.3 Approaches
3.3.1 Stage 1: Receptor Surface Analysis
The SurfaceScreen methodology attempts to optimize two components, global shape and local physicochemical texture, for discovering the similarity between surfaces [5, 10, 11]. Utilizing global shape and local physicochemical texture, the protein surfaces are shape matched against libraries of annotated surfaces extracted from the PDB. The Delauney triangulation and alpha-shape methods are used to accurately decompose and describe pockets and voids in protein structure [24-27].
SurfaceScreen has proven useful in identifying distant functional relationships between proteins void of sequence or structural homology. By focusing on a library of binding sites, the method is used to screen for homologous binding sites. In the pipeline, similar binding surfaces are run in parallel to the target surface to predict cross reactivity reactions (i.e., side effects) or identify species-specific inhibitors (see Fig. 1). For example, one can automatically eliminate compounds with high-predicted binding affinity for human proteins while screening for inhibitors against bacterial homologs.
3.3.2 Stage 2: Initial Docking Pose
Once regions of receptor surfaces are identified via SurfaceScreen, it is relatively straightforward to perform docking and scoring of a large database of ligands. The mixed success of different docking methods against a particular target has inspired the integration of two different docking applications in the pipeline: DOCK and AUTODOCK.
Both software packages have been ported and optimized to run on the BlueGene/P architecture. During docking, both the active site residues and ligands are allowed to be flexible to achieve “induced fit” docking. The top ranked 5,000 compounds, as evaluated by each application’s internal scoring function, are passed along for re-scoring.
3.3.3 Stage 3: MM/GBSA Re-Scoring
A molecular mechanics (MM) potential function based on the Generalized Born (BG) and surface area (SA) approximation is employed for further refinement of initial docking poses and calculation of binding energies [28]. MM/GBSA relies on more complex, physically realistic models for solvation, electrostatic interactions, and conformational change and has been shown to outperform most docking program’s internal scoring functions [16, 17].
The calculation of MM/GBSA energies involves minimization with a conjugate gradient method, molecular dynamics (MD) simulation (Langevin at constant temperature), another minimization round, and a final energy evaluation. Implementation of this method in the pipeline allows both the ligand and selected residues within the receptor-binding pocket to be flexible. Since rescoring is applied to all ligands output from the prior docking runs, it allows one to universally evaluate/rank/sort the results from both DOCK and AUTODOCK. Modules from the molecular modeling software Nucleic Acid Builder (NAB) is used to drive the MM-GBSA scoring procedures [3].
3.3.4 Stage 4: FEP/MD Rescoring
The equilibrium binding constant Kb for the process corresponding to the association of a ligand L to a protein P + L → LP, can be expressed as a sequence of well-defined steps that can be calculated from free energy perturbation (FEP) MD simulations [29-31]. Furthermore, biasing potentials restraining the translation, orientation, and conformation of the ligand can help enhance the convergence of the calculations [29-36]. Such an FEP procedure can provide correct results as long as the effect of all the restraining potentials is rigorously taken into account and unbiased.
FEP/MD methods are challenging and ambitious at the present time and a certain level of skepticism exists on the role of such computationally demanding methods. However, rescoring based on FEP/MD simulations for protein-ligand interactions has the potential to become a powerful tool in drug discovery and optimization [31, 32, 34, 37]. Nonetheless, despite outstanding developments in simulation methodologies, carrying out brute-force FEP/MD calculations of large macromolecular assemblies surrounded by explicit solvent molecules often remain prohibitive. For this reason, it is necessary to seek ways to decrease the computational cost of FEP/MD calculations while keeping them accurate.
An attractive strategy to decrease the cost of FEP/MD computations consists of simulating a small number of explicit atoms in the vicinity of the region of interest, while representing the influence of the surrounding with an effective “boundary potential” [38-41]. This is reasonable because binding specificity is often dominated by local interactions in the vicinity of the ligand, while the remote regions of the receptor contribute in only an average manner. The method used in the present study is called the Generalized Solvent Boundary Potential (GSBP). GSBP includes both the solvent-shielded static field from the distant atoms of the macromolecule and the reaction field from the dielectric response of the solvent acting on the atoms of the simulation region.
It is also possible to reduce the computational cost of FEP/ MD simulations and even improve their accuracy by using an intermediate approach combining some aspects of both explicit and implicit solvent treatments [38, 40]. It consists of simulating a small number of explicit solvent molecules in the vicinity of a region of interest, while representing the influence of the surrounding solvent with an effective “solvent boundary potential” [38-41]. Recently a Hamiltonian-exchange scheme in CHARMM based on the Parallel Distributed Replica (REPDSTR) was implemented to form an FEP/REMD/GCMC methodology. Convergence is improved significantly with a reduced cost of computation [42]. This represents a significant breakthrough in how the CHARMM biomolecular simulation package can be utilized on leadership scale machines such as the BlueGene/P.
4 Notes
The APPLIED pipeline implementation is only available on Intrepid at the ALCF. All individual software components are freely available and there is no technical limitation that would preclude replicating the pipeline on other large-scale computing resources.
Acknowledgments
This research used resources of the Argonne Leadership Computing Facility at Argonne National Laboratory, which is supported by the Office of Science of the US Department of Energy under contract DE-AC02-06CH11357. We would like to acknowledge Drs. Devleena Shivakumar, Mike Wilde, Zhao Zhang for valuable discussions and support on computational method development and implementation. The submitted manuscript has been created by UChicago Argonne, LLC, Operator of Argonne National Laboratory (“Argonne”). Argonne, a US Department of Energy Office of Science laboratory, is operated under Contract No. DE-AC02-06CH11357. The US Government retains for itself, and others acting on its behalf, a paid-up nonexclusive, irrevocable worldwide license in said article to reproduce, prepare derivative works, distribute copies to the public, and perform publicly and display publicly, by or on behalf of the Government. This work was in part supported with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contracts No. HHSN272200700058C and HHSN272201200026C and by the National Institute of Heath Grant GM094585.
References
- 1.Lang PT, Brozell SR, Mukherjee S, Pettersen EF, Meng EC, Thomas V, Rizzo RC, Case DA, James TL, Kuntz ID. DOCK 6: combining techniques to model RNA-small molecule complexes. RNA. 2009;15(6):1219–1230. doi: 10.1261/rna.1563609. doi: 10.1261/rna.1563609, rna.1563609 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Macke T, Case DA. Modeling unusual nucleic acid structures. In: Molecular modeling of nucleic acids. American Chemical Society. 1998;682:379–393. [Google Scholar]
- 4.Brooks BR, Brooks CL, 3rd, Mackerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Binkowski TA, Joachimiak A. Protein functional surfaces: global shape matching and local spatial alignments of ligand binding sites. BMC Struct Biol. 2008;8:45. doi: 10.1186/1472-6807-8-45. doi: 10.1186/ 1472-6807-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raicu I, Zhao Y, Dumitrescu C, Foster I, Wilde M. Falkon: a fast and light-weight task execution framework. IEEE/ACM Super Computing. 2007 [Google Scholar]
- 7.Zhao Y, Hategan M, Clifford B, Foster I, von Laszewski G, Raicu I, Stef-Praun T, Wilde M. Swift: fast, reliable, loosely coupled parallel computation; IEEE International workshop on scientific workflows; Salt Lake City, Utah, USA. 2007. [Google Scholar]
- 8.Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Feng Z, Gilliland GL, Iype L, Jain S, Fagan P, Marvin J, Padilla D, Ravichandran V, Schneider B, Thanki N, Weissig H, Westbrook JD, Zardecki C. The Protein Data Bank. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 6 No 1):899–907. doi: 10.1107/s0907444902003451. doi:S0907444902003451 [pii] [DOI] [PubMed] [Google Scholar]
- 9.Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG. ZINC: a free tool to discover chemistry for biology. J Chem Inf Model. 2012;52(7):1757–1768. doi: 10.1021/ci3001277. doi: 10.1021/ci3001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Binkowski TA, Adamian L, Liang J. Inferring functional relationships of proteins from local sequence and spatial surface patterns. J Mol Biol. 2003;332(2):505–526. doi: 10.1016/s0022-2836(03)00882-9. [DOI] [PubMed] [Google Scholar]
- 11.Binkowski TA, Joachimiak A, Liang J. Protein surface analysis for function annotation in high-throughput structural genomics pipeline. Protein Sci. 2005;14(12):2972–2981. doi: 10.1110/ps.051759005. doi: 10.1110/ps.051759005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shoichet BK. Virtual screening of chemical libraries. Nature. 2004;432(7019):862–865. doi: 10.1038/nature03197. doi: 10.1038/nature03197, nature 03197 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shoichet BK, Leach AR, Kuntz ID. Ligand solvation in molecular docking. Proteins. 1999;34(1):4–16. doi: 10.1002/(sici)1097-0134(19990101)34:1<4::aid-prot2>3.0.co;2-6. doi: 10.1002/(SICI)1097-0134(19990101)34:1<4::AID-PROT2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 14.Carlson HA, Masukawa KM, Rubins K, Bushman FD, Jorgensen WL, Lins RD, Briggs JM, McCammon JA. Developing a dynamic pharmacophore model for HIV-1 integrase. J Med Chem. 2000;43(11):2100–2114. doi: 10.1021/jm990322h. jm990322h [pii] [DOI] [PubMed] [Google Scholar]
- 15.Schneider G, Bohm HJ. Virtual screening and fast automated docking methods. Drug Discov Today. 2002;7(1):64–70. doi: 10.1016/s1359-6446(01)02091-8. S1359644 601020918 [pii] [DOI] [PubMed] [Google Scholar]
- 16.Price DJ, Jorgensen WL. Computational binding studies of human pp 60c-src SH2 domain with a series of nonpeptide, phosphophenyl-containing ligands. Bioorg Med Chem Lett. 2000;10(18):2067–2070. doi: 10.1016/s0960-894x(00)00401-7. S0960-894X(00)00401-7 [pii] [DOI] [PubMed] [Google Scholar]
- 17.Wesolowski SS, Jorgensen WL. Estimation of binding affinities for celecoxib analogues with COX-2 via Monte Carlo-extended linear response. Bioorg Med Chem Lett. 2002;12(3):267–270. doi: 10.1016/s0960-894x(01)00825-3. S0960894X01008253 [pii] [DOI] [PubMed] [Google Scholar]
- 18.Graves AP, Shivakumar DM, Boyce SE, Jacobson MP, Case DA, Shoichet BK. Rescoring docking hit lists for model cavity sites: predictions and experimental testing. J Mol Biol. 2008;377(3):914–934. doi: 10.1016/j.jmb.2008.01.049. doi: 10.1016/j. jmb.2008.01.049, S0022-2836(08)00096-X [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng Y, Roux B. Computation of binding free energy with molecular dynamics and grand canonical Monte Carlo simulations. J Chem Phys. 2008;128(11):115103. doi: 10.1063/1.2842080. doi: 10.1063/ 1.2842080. [DOI] [PubMed] [Google Scholar]
- 20.Irwin JJ, Shoichet BK. ZINC-a free database of commercially available compounds for virtual screening. J Chem Inf Model. 2005;45(1):177–182. doi: 10.1021/ci049714. doi: 10.1021/ci049714+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Y, Wilde M, Foster I, editors. Workflows for eScience. Springer; London: 2007. Virtual Data Language: a typed workflow notation for diversely structured scientific data. [Google Scholar]
- 22.Raicu I, Zhang Z, Wilde M, Foster I, Beckman P, Iskra K, Clifford B. Towards loosely coupled programming on a petascale system. 2008 IEEE/ACM SuperComputing. [Google Scholar]
- 23.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Binkowski TA, Naghibzadeh S, Liang J. CASTp: computed atlas of surface topography of proteins. Nucleic Acids Res. 2003;31(13):3352–3355. doi: 10.1093/nar/gkg512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang J, Edelsbrunner H, Fu P, Sudhakar PV, Subramaniam S. Analytical shape computation of macromolecules: I. Molecular area and volume through alpha shape. Proteins. 1998;33(1):1–17. [PubMed] [Google Scholar]
- 26.Liang J, Edelsbrunner H, Fu P, Sudhakar PV, Subramaniam S. Analytical shape computation of macromolecules: II. Inaccessible cavities in proteins. Proteins. 1998;33(1):18–29. [PubMed] [Google Scholar]
- 27.Liang J, Edelsbrunner H, Woodward C. Anatomy of protein pockets and cavities: measurement of binding site geometry and implications for ligand design. Protein Sci. 1998;7(9):1884–1897. doi: 10.1002/pro.5560070905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kollman PA. Free energy calculations: applications to chemical and biochemical phenomena. Chem Rev. 1993;93:2395–2417. [Google Scholar]
- 29.Deng Y, Roux B. Calculation of standard binding free energies: aromatic molecules in the T4 lysozyme L99A mutant. J Chem Theory Comput. 2006;2(5):1255–1273. doi: 10.1021/ct060037v. doi: 10.1021/ct060037v. [DOI] [PubMed] [Google Scholar]
- 30.Roux B, Nina M, Pomes R, Smith JC. Thermodynamic stability of water molecules in the bacteriorhodopsin proton channel: a molecular dynamics free energy perturbation study. Biophys J. 1996;71(2):670–681. doi: 10.1016/S0006-3495(96)79267-6. doi: 10.1016/ S0006-3495(96)79267-6, S0006-3495(96) 79267-6 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J, Deng Y, Roux B. Absolute binding free energy calculations using molecular dynamics simulations with restraining potentials. Biophys J. 2006;91(8):2798–2814. doi: 10.1529/biophysj.106.084301. doi : 10.1529/biophysj.106.084301, S0006-3495(06)71994-4 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boresch S, Tettinger F, Leitgeb M, Karplus M. Absolute binding free energies: a quantitative approach for their calculation. J Phys Chem. 2003;107:9535–9551. [Google Scholar]
- 33.Woo HJ, Dinner AR, Roux B. Grand canonical Monte Carlo simulations of water in protein environments. J Chem Phys. 2004;121(13):6392–6400. doi: 10.1063/1.1784436. [DOI] [PubMed] [Google Scholar]
- 34.Woo HJ, Roux B. Calculation of absolute protein-ligand binding free energy from computer simulations. Proc Natl Acad Sci U S A. 2005;102(19):6825–6830. doi: 10.1073/pnas.0409005102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hermans J, Wang L. Inclusion of loss of translational and rotational freedom in theoretical estimates of free energies of binding. application to a complex of benzene and mutant t4 lysozyme. J Am Chem Soc. 1997;119:2707–2714. [Google Scholar]
- 36.Gilson MK, Given JA, Bush BL, McCammon JA. The statistical-thermodynamic basis for computation of binding affinities: a critical review. Biophys J. 1997;72:1047–1069. doi: 10.1016/S0006-3495(97)78756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simonson T, Archontis G, Karplus M. Free energy simulations come of age: proteinligand recognition. Acc Chem Res. 2002;35:430–437. doi: 10.1021/ar010030m. [DOI] [PubMed] [Google Scholar]
- 38.Beglov D, Roux B. Finite representation of an infinite bulk system: solvent boundary potential for computer simulations. J Chem Phys. 1994;100:9050–9063. [Google Scholar]
- 39.Berkowitz M, McCammon JA. Molecular dynamics with stochastic boundary conditions. Chem Phys Lett. 1982;90:215–217. [Google Scholar]
- 40.Im W, Bernèche S, Roux B. Generalized solvent boundary potential for computer simulations. J Chem Phys. 2001;114(7):2924–2937. [Google Scholar]
- 41.Warshel A, King G. Polarization constraints in molecular dynamics simulation of aqueous solutions: the surface constraint all atom solvent (scaas) model. Chem Phys Lett. 1985;121:127–129. [Google Scholar]
- 42.Jiang W, Hodoscek M, Roux B. Computation of absolute hydration and binding free energy with free energy perturbation distributed replica-exchange molecular dynamics. J Chem Theory Comput. 2009;5:2583–2588. doi: 10.1021/ct900223z. [DOI] [PMC free article] [PubMed] [Google Scholar]