Abstract
Circadian rhythms govern a wide variety of physiological and metabolic functions in most organisms. At the heart of these regulatory pathways in mammals is the clock machinery, a remarkably coordinated transcription-translation system that relies on dynamic changes in chromatin states. Recent findings indicate that regulation also goes the other way, as specific elements of the clock can sense changes in the cellular metabolism. Understanding in full detail the intimate links between cellular metabolism and the circadian clock machinery will provide not only crucial insights into system physiology but also new avenues toward pharmacological intervention of metabolic disorders.
A staggering number of our biological functions are controlled by the circadian clock. Whether we feed, rest, sleep or exercise, the 24 h–based cycle that governs our life has intimate links with our body’s metabolism. Is the endogenous clock present in most organisms, regulating their metabolism, or do variations in their metabolic cycles feed back on the clock? To what extent do changes in our rhythmic life patterns—for example, restricted feeding or altered sleep-wake cycles—coordinately impinge on unique molecular gears common to the body’s clock and metabolism?
The anatomical center of the circadian clock consists of a small region of the brain (approximately 20,000 neurons) known as the suprachiasmatic nucleus (SCN, see Table 1). As circadian rhythms control activities as disparate as the sleep-wake wake cycle, hormone secretion, glucose metabolism, memory formation and changes in body temperature, the finding that there are autonomous oscillations in many tissues of the body constituted a crucial discovery1-3. Indeed, the picture that emerges from numerous studies indicates that a network of interconnected peripheral oscillators operates in the body, with the SCN playing the part of conductor4,5. Central to the core clock mechanism are the Clock and Bmal1 (also known as Arntl) genes, which encode basic-helix–loop–helix (bHLH)-PAS transcription activators that heterodimerize and induce the expression of the genes for two main oscillators, period (Per) and cryptochrome (Cry), which contain E-box elements (CACGTG) in their promoters6,7. Additional genes with promoters containing E-box, D-box and retinoic acid responsive element (RRE) consensus sequences are also clock regulated and make up a large body of circadian clock–controlled genes (CCGs). The Per and Cry genes encode other elements of the core clock machinery that in the classical view function to form heterodimeric complexes that translocate to the nucleus and inhibit Clock–Bmal1-mediated transcription through direct protein-protein interactions8-12. Thus, at its core, circadian rhythmicity is supported by intracellular transcriptional and translational feedback loops that perpetuate oscillations in gene expression. Such an explanation is deceivingly simple; auxiliary proteins necessarily impinge on circadian core components by such mechanisms as methylation, acetylation, phosphorylation and ubiquitination, keeping the core clock strictly timed and yet remarkably plastic, able to adapt to changes in its environment13-18. MicroRNAs and RNA binding proteins that affect RNA stability also participate in timekeeping, providing further complexity to the clock machinery19,20. Moreover, cells cope with the spectacular task of rhythmically synthesizing 10–15% of their transcripts21-23, an undertaking that can be achieved only by a powerful and cyclic mechanism of chromatin remodeling. This is likely to be obtained, at least in part, through the histone acetyltransferase (HAT) activity of the master regulator Clock24.
Table 1.
List of common circadian terms and their definitions
| Term | Definition |
|---|---|
| Circadian | A modifier referring to the approximately 24-h nature of an event. The word ‘circadian’ is derived from the Latin roots circa (about) and diem (day). |
| Entrainment | The process of clock synchronization. For example, light ‘entrains’ the biological clock. |
| Zeitgeber | German word meaning ‘time giver’. An external cue (such as light or food) that entrains the circadian clock. |
| Suprachiasmatic nucleus (SCN) | A small region of the brain consisting of bilateral nuclei that coordinately function to synchronize circadian rhythms in other tissues. |
| Circadian clock–controlled gene (CCG) | A gene expressed in a circadian-dependent manner, usually under the control of a promoter that contains E-box, D-box or RRE elements. |
| Brain and muscle Arnt-like protein-1 (Bmal1) | The mammalian bHLH-PAS transcription factor that dimerizes with Clock to activate gene transcription. |
| CLOCK | A bHLH-PAS transcription factor that dimerizes with BMAL1 to activate promoters that contain E boxes (CACGTG). |
| Cryptochrome proteins (Cry) | Transcriptional repressors that dimerize with Per to inhibit Clock–Bmal1-mediated gene transcription. In plants and invertebrates, these function as light-responsive flavoproteins. |
| Neuronal PAS domain protein 2 (Npas2) | A transcription factor similar to Clock and highly expressed in the forebrain. Npas2 dimerizes with Bmal1 to activate gene transcription. |
| Period homolog proteins (Per) | PAS domain–containing proteins that dimerize with TIM (in Drosophila) or Cry (in mammals) to inhibit Clock–Bmal1-induced gene transcription. |
There is increasing evidence that a smoothly running endogenous clock is crucial for energy balance in organisms from cyanobacteria to mammals25-28. Internal and external clock resonance seems to be energetically favorable for an organism. For example, Arabidopsis thaliana with mismatching internal and environmental periods experience a reduction in leaf chlorophyll content, reduced growth and an increase in mortality29. The alignment of endogenous rhythms with the environment allows coordination of physiology with predictable zeitgebers (Table 1). There may be several reasons for this advantage in mammals, including the prevention of aging and dementia pathologies by the efficient removal of DNA damage–induced by oxidative stress29-34. The recent discovery that the activity of sirtuin-1 (Sirt1), a longevity-associated protein belonging to a family of NAD+-activated histone deacetylases14, oscillates in a circadian fashion broadens our knowledge about the communication between the circadian clock and metabolism, but it also reveals a void in our understanding about the molecular support for such interplay14,15. Sirt1 (the mammalian ortholog of yeast Sir2) shows an oscillation in activity, impinging back on the circadian clock by altering Bmal1 acetylation and CLOCK–BMAL1-induced gene transcription14,35. The discovery of metabolite oscillations during the yeast metabolic cycle26,36,37 combined with evidence of circadian sirtuin activity allows speculation as to whether metabolites such as NAD+ themselves serve a preponderant role in the cellular link between metabolism and the circadian clock (Fig. 1).
Figure 1.
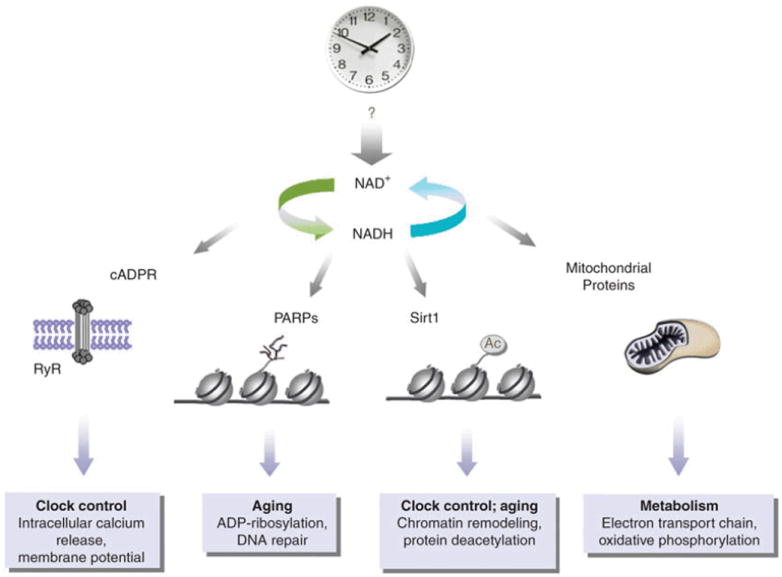
The interplay between the circadian clock and cellular metabolism occurs at various levels. Central to cellular metabolism is the synthesis of NAD+, a process influenced by the circadian clock. Variable levels of NAD+ may influence numerous intracellular pathways. The small molecule cyclic ADP-ribose (cADPR) is produced from NAD+ by ADP-ribosyl cyclases and contributes to circadian oscillations in a ryanodine receptor (RyR)-dependent manner79. Metabolites also have an attractive role in epigenetic control84. NAD+ modulates the activity of chromatin-associated enzymes, such as PARP-1 and Sirt1. The activity of the NAD+-dependent histone deacetylase Sirt1 oscillates and participates in chromatin remodeling at circadian genes. There is no evidence to date for a circadian function of PARP-1, but its role in DNA damage repair64 and the reported interplay with Sirt1 (ref. 66) may be predictive of a circadian role. Finally, the central role of NAD+-activated mitochondrial proteins in energy metabolism begs the question of how they might contribute to circadian control.
EdSumm: The mammalian circadian clock controls many biological functions, including metabolic activity. In this Perspective, the authors present recent literature and discuss the two-way relationship between the clock and metabolism, with NAD+ playing a central part in their integration.
Physiological evidence linking circadian rhythms and metabolism
Accumulating physiological evidence indicates that circadian rhythms and energy state are tightly linked. First of all, rodents that lack expression of circadian clock genes or that express mutant variants of clock genes have abnormal metabolic phenotypes. For example, the Clock mutant mouse (ClockΔ19) on a BALB/c and C57BL/6J background is obese and shows signs of abnormal lipid and glucose metabolism, revealing the importance of a transcriptionally active Clock protein for both normal body weight regulation and glucose and lipid homeostasis27,38. Interestingly, and possibly revealing the epistatic effect of yet unidentified factors, ClockΔ19 mice on an ICR genetic background also show abnormal lipid distribution, although the effect seems to be protection from weight gain on a high fat diet due to decreased fat absorption39. What is clear is that mutations in the Clock gene affect metabolism in each genetic background tested, but in different ways. Bmal1−/− mice also show metabolic abnormalities with impaired insulin responsiveness and reduced gluconeogenesis40. Similarly, mice that lack Bmal1 expression in the liver show fasting-phase hypoglycemia and excessive glucose clearance in response to insulin, probably resulting from the loss of the circadian glucose transporter-2 (Glut2) protein28. These altered metabolic states are somewhat expected, as nuclear hormone receptors, many of which participate directly in fatty acid and glucose metabolism, undergo circadian oscillations38.
The levels of the metabolic hormones glucagon, insulin, ghrelin, leptin and corticosterone are also reported to oscillate in a circadian fashion, as is that of peroxisome proliferator–activated receptor-γ (PPARγ) coactivator-1α (PGC-1α an essential activator of gluconeogenesis in the nutrient-deprived state41-43. PGC-1α seems to be dually regulated by circadian processes: it is both rhythmically expressed and it is deacetylated by Sirt1 in an NAD+-dependent fashion44,45. As is the case with Sirt1, PGC-1α feeds back on the circadian clock, affecting metabolic rate, body temperature and daily locomotion patterns. These PGC-1α–mediated changes in physiology may be due to alterations in Bmal1 transcription, as PGC-1α and RAR-related orphan receptor-α (RORα) coactivate the Bmal1 promoter43.
Finally, restricted feeding is a powerful zeitgeber, one of a limited number of cues that can entrain circadian rhythms in non-SCN tissues but which probably depends on signaling to the dorsomedial hypothalamus46. Some evidence suggests that sterol regulatory element–binding protein-1 (SREBP-1) activation and HMG-CoA reductase expression in the liver, both of which participate in sterol metabolism, may serve a central role in restricted feeding-induced phase shifting47. Because the endogenous circadian clock has an intrinsic ability to anticipate changes in its environment, restricted feeding-induced phase shifting provides convincing evidence for the codependence of circadian and metabolic processes.
Chromatin remodeling by histone acetylation seems to be a key molecular pathway in the translation of the metabolic message to the circadian clock24. The recent findings that another histone deacetylase, HDAC3, interacts with the co-repressor NCOR1 to regulate circadian behavior, insulin signaling and energy expenditure48 underscore the critical role that epigenetic regulation has in circadian and metabolic physiology.
Connections between circadian rhythmicity and energy balance are poignantly apparent in aging studies. Although neurodegenerative diseases and aging frequently coincide with disrupted circadian rhythms and fragmented sleep patterns49, perhaps the Bmal1 −/− mice present the most convincing evidence linking an aging phenotype to disrupted circadian rhythmicity50. These mutant mice show numerous signs of early aging and have a reduced lifespan. Tissues from Bmal1 −/− mice demonstrating an age-dependent reduction in size also show high accumulation of reactive oxygen species (ROS), consistent with the idea that Bmal1 participates in the oxidative stress response51.
Mitochondria are subject to high levels of ROS production, an inexorable byproduct of respiration. As a result, mitochondrial proteins are central targets for oxidative damage and, when damaged, may contribute to aging disorders such as neurodegeneration. ROS-induced damage of mitochondrial DNA, which inevitably leads to reduced respiration efficiency and a further increase in the level of intracellular ROS production, is thought to be central to the aging process, an idea known as the ‘mitochondrial theory of aging’52. Accumulating ROS in the brain probably lead to the age-related neurodegenerative pathologies of Alzheimer’s and Parkinson’s disease. Although still debated53, there is much evidence to back up this theory, particularly in pathologies associated with neurodegeneration54. Interestingly, nicotinamide mononucleotide adenylyl-transferase-1 (Nmnat1), an enzyme involved in the biosynthesis of NAD+, protects against axonal degeneration in Wallerian degeneration slow mice (Wlds). These mice have a spontaneous mutation that increases activation of the Nmnat1 protein, resulting in elevated NAD+ levels and consequent Sirt1-dependent protection against axonal injury55,56. The discovery that Sirt1 activity oscillates suggests that there may be a link between neuroprotection, redox state and circadian rhythmicity14. This association is also supported in Drosophila melanogaster, where the administration of superoxide radical–producing compounds, such as paraquat, results in reduced clock gene cycling in peripheral tissues and a forkhead box O protein (FOXO)-dependent sensitivity to oxidative stress in the central pacemaker32. These studies help bring into focus the importance of circadian control in metabolism and aging, but raise questions as to what fundamentally fuels their interaction at the cellular level.
NAD+, central to both metabolism and circadian rhythmicity?
During the conversion of energy from food to ATP, cells harness energy from the oxidation of food molecules to drive reactions that are required for normal cell function but that may be energetically unfavorable. At the crossroad of cellular metabolism is the mitochondrion, host to the ATP-generating process of oxidative phosphorylation. In the mitochondria, the carrier molecules such as ATP and NAD+ are used in oxidation-reduction reactions that are crucial for the maintenance of energy balance. In the case of NAD+, the acquisition of energy-rich electrons and a hydrogen atom from substrate molecules to form reduced NADH promotes subsequent biosynthesis by the release of hydride ions from reduced NAD to donor molecules. NAD+ is derived from niacin and operates as a coenzyme for multiple cellular dehydrogenases (such as those necessary for the β-oxidation of fatty acids or for the Krebs cycle), at which time its reduction to NADH precedes its subsequent oxidation by the respiratory chain. Therefore, the role of NAD+ as a hydrogen carrier is paramount for the production and maintenance of energy stores.
In addition to its redox roles, NAD+ also serves as a substrate for ADP ribosylation. When NAD+ is depleted by sustained increases in the activity of the NAD+-dependent ribosylating enzyme poly(ADP-ribose) polymerase (PARP)-1, cell death can ensue57-59. Mitochondrially localized PARP activity is induced by DNA damage and is thought to contribute to cell death during periods of intense oxidative stress60. Vitamin B3 (consisting of the NAD+ precursors nicotinamide and nicotinic acid) can be particularly useful in preventing NAD+ turnover, as nicotinamide inhibits PARP activity61. The nicotinamide phosphoribosyltransferase enzyme NAMPT (which mediates the biosynthesis of NAD+ from nicotinamide in mammals) is expressed in the mitochondria, among other cellular loci, and as a sirtuin activator can also protect cells from PARP-mediated cell death. Restoration of mitochondrial NAD+, specifically, seems to rescue cells from the PARP-mediated apoptosis that normally occurs when PARP-1 depletes cytosolic pools of NAD+, a rescue that probably depends on one of the sirtuin proteins (for example, Sirt3) that localizes to the mitochondria62.
Not all NAD+ functions are restricted to the mitochondria. On the contrary, its role in both PARP and sirtuin regulation can be cytosolic or nuclear63. The fact that the NAMPT protein, for example, can localize to the cytoplasm, nucleus and mitochondria suggests that microdomains of NAD+ are probably important for stimuli-specific responses64. In fact, it has been proposed that increasing the nuclear availability of NAD+ specifically promotes longevity in yeast in a Sir2-dependent manner65. The Sir2 protein seems to control aging in yeast by mediating transcriptional silencing at telomeres66. The biosynthesis of NAD+ in mammals relies on the activity of the rate-limiting enzyme NAMPT, whose activation seems to be sufficient to drive Sir2-mediated changes in gene expression and activity67. Importantly, recent data reveal that NAD+ levels oscillate with a 24-h cycle, a rhythm that is driven by the circadian clock68,69. The Clock–Bmal1 activator complex regulates Nampt expression, in conjunction with Sirt1, which thereby contributes to the cyclic synthesis of its own coenzyme. Thus, the circadian clock is directly implicated in controlling the intracellular levels of critical metabolites, generating an interlocking of the transcriptional feedback clock loop with the enzymatic feedback loop of the NAD+-salvage pathway69 (Fig. 2).
Figure 2.
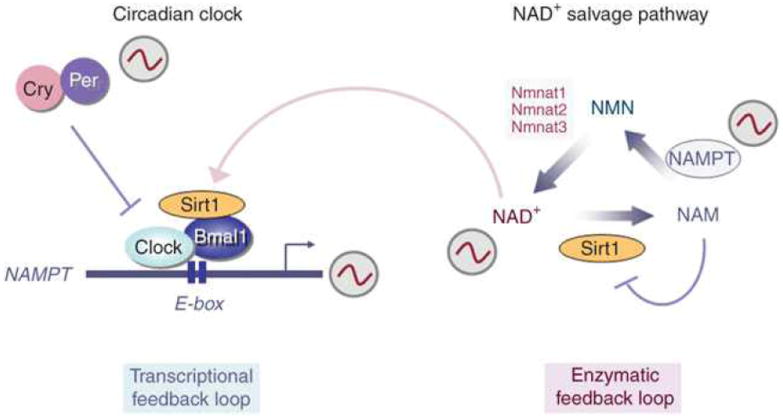
By activating Sirt1, NAD+ conjoins two feedback loops necessary for cross-talk between the circadian clock and metabolite production. The NAD+-salvage pathway is important for regulating intracellular NAD+ levels. After the conversion of nicotinamide (NAM) into nicotinamide mononucleotide (NMN) by NAM phosphoribosyl transferase (NAMPT), NMN is further modified into NAD+ by the nicotinamide mononucleotide adenylyl transferases (Nmnat1,-2 and -3). Whereas NAM inhibits Sirt1 activity, NAD+-activated Sirt1 feeds back into the NAD+-salvage pathway by directly regulating Nampt gene expression in a Clock–Bmal1-dependent manner69. By this mechanism, NAD+ conjoins the two feedback loops, contributing to the fine tuning necessary for achieving energy balance.
EdSumm: The mammalian circadian clock controls many biological functions, including metabolic activity. In this Perspective, the authors present recent literature and discuss the two-way relationship between the clock and metabolism, with NAD+ playing a central part in their integration.
Like mitochondrial PARPs, nuclear PARP proteins are also activated following DNA damage, where they recruit factors necessary for subsequent DNA repair, thereby preserving viability and normal cellular function70. Cytosolic and nuclear NAD+ also carries out an important role via its activation of the deacetylase Sirt1, whose expression has been determined to be both cytosolic and nuclear but not mitochondrial. The recent evidence that Sirt1 activity oscillates over a 24-h cycle14 provides a novel mechanism for how the cell might rely on circadian-controlled cycling in energy systems to maintain energy balance. Because Sirt1 is a histone deacetylase, its primary function may be to translate cellular energy states to chromatin remodeling and then into changes in gene expression. Although Sirt1 activity is not confined exclusively by NAD+ levels, Sirt1 has been described to function in part as a redox sensor, using the coenzyme NAD+ to catalyze the acetyl transfers from substrate proteins71. Therefore, central to the redox state of the cell, as a substrate for PARP proteins, and as an activator of the sirtuin family of longevity-associated histone deacetylases, NAD+ maintains a privileged position in the cell, able to monitor both its metabolic state and, possibly, its fate. Intriguingly, a molecular and functional link has been described between Sirt1 and PARP-1 in the control of cell death72.
Cycling proteins or metabolites?
The interplay of circadian rhythmicity and energy balance may result from the fluctuations of proteins required for normal metabolic function, fluctuations in metabolites themselves or both. Some evidence suggests that neither takes center stage, but rather that both may work together to maintain energy balance. Respiration may itself be subject to circadian control; mRNA levels for several complex I proteins seem to oscillate in expression (reviewed in ref.73). In rodents, a circadian oscillation in the expression of approximately 20 respiratory-chain protein subunits in the SCN peaks just before dawn, preceding the circadian peak of neuronal firing when metabolic demands are at their highest23,74. As respiration produces its own metabolites, the cells must find ways to avoid the damaging effects of this necessary activity. In mice, the oscillation of hexokinase and malate dehydrogenase, proteins that utilize glucose or ketone bodies to provide the energy for oxidative phosphorylation, demonstrates the necessity for such compensation23.
Microarray analysis of D. melanogaster fly head RNA also reveals oscillations in numerous detox, stress response and metabolic proteins, some of which are NAD+ or NADP+ activated. These include aldehyde dehydrogenase, oxido reductase and short-branched chain acyl CoA dehydrogenase75. Detoxification enzymes constitute a large proportion of cycling genes in the fly head, accounting for at least six cytochrome P450 family members as well as glutathione S-transferase. Cycling genes are also manifest in the fly body, where many redox-associated genes show circadian oscillations. In keeping with the D. melanogaster microarray data, melatonin-driven glutathione peroxidase production peaks in both the brain and the liver in rodents. This event may be synchronized specifically for scavenging ROS as ROS levels peak just before maximal glutathione peroxidase production73. The expression profile of numerous other metabolism-associated mRNAs, such as those encoding the glucagon receptor, glucokinase, glucagon, Glut2, glucose-6-phosphate transport protein, pyruvate kinase and pyruvate dehydrogenase, show circadian oscillation23,76,77. These observations lend strength to the hypothesis that circadian oscillations of mitochondrial proteins influence the cell’s metabolic state and allow the cell to respond efficiently to DNA damage induced by accumulating oxidative stress. As DNA damage is thought to contribute to neurodegenerative disorders such as Parkinson’s and Alzheimer’s disease, it is reasonable to speculate that the circadian fluctuations of proteins responsible for reducing oxidative stress may help prevent development of these disease states.
Although the circadian activity or expression of metabolic proteins is likely to contribute to energy balance, a recent study of the yeast metabolic cycle in nutrient-limiting conditions reveals that yeast metabolites themselves are actually cyclically produced and that they drive cell division, resulting in a peak of cell division while respiration is at its nadir37. Metabolite cycling seems to be finely tuned. For example, amino acid and nucleotide precursors peak during the oxidative phase, whereas metabolites associated with glycogen metabolism (such as pyruvate and glucose-6-phosphate) reach a maximum during reductive and building phases. Although these cycles are ultradian (with periodicities of less than 24 h), the synchronization required for normal cell cycling suggests that there are similar oscillations in mammalian cells, although they are temporally disparate. Whether cycles such as these occur on an ultradian or a circadian cycle in mammalian tissues has yet to be demonstrated.
The small molecule cyclic ADP-ribose (cADPR), which is produced from NAD+ by ADP-ribosyl cyclases, modulates circadian oscillations in cytosolic calcium release in plant and animal cells in a ryanodine receptor–dependent manner78. It was recently reported to be synthesized in a circadian fashion in some organisms, where it contributes to circadian clock gene expression79. The NAD+ dependency of cADPR production lends further support for the idea that oscillations in metabolites may serve as a prerequisite for circadian modulation of cellular metabolism. Notably, heme oxygenase-2, an enzyme involved in heme degradation, is light inducible. This is relevant considering that both PAS domains of Npas2 (a clock protein that dimerizes with Bmal1 and controls circadian gene expression) were recently reported to bind heme and respond to changes in the cell’s redox state in the form of altered Bmal1 binding80,81. Heme also serves as a ligand for the REV-ERB proteins, an interaction that results in transcriptional repression of Bmal1 and negative regulation of hepatic gluconeogenic genes82,83. The contribution of heme to circadian rhythms is further supported by the fact that the gene controlling the rate-limiting step for heme biosynthesis, aminolevulinate synthase-1 (Alas1), is itself transcriptionally regulated by Bmal1–Npas2 heterodimers84.
Whereas circadian oscillations in the activity of the NAD+-activated Sir2 protein homologs may affect DNA repair or silencing, cell-cycle progression and glucose homeostasis in a manner reflective of the cell’s metabolic state, oscillations in NAD+ or NADP+ levels may provide the explanation for why food remains such a potent zeitgeber. Specifically, high concentrations of the oxidized forms of NAD, NAD+ and NADP+, have been described to inhibit the binding of Clock or Npas2 to Bmal1, preventing E-box binding and subsequent circadian target gene transcription. Conversely, NADH or NADPH have been described to promote dimerization of Bmal1 with Npas2 or Clock, thus contributing to Clock-driven gene transcription85. The profile of circadian gene expression, therefore, is likely to depend directly on the metabolic state of the cell.
Implications for metabolite oscillations on circadian physiology
Whether there are mammalian metabolite oscillations analogous to those of the yeast metabolic cycle is still unclear, but it remains a tantalizing possibility. The fact that cellular demands are met temporally as a function of the cell’s metabolic cycle is likely to be true for all cells, regardless of the organism. In the context of mammalian Sirt1 circadian activity, it seems likely that metabolite oscillations in the coenzyme NAD+ must also occur in a cyclical, circadian manner. If metabolite fluctuations are organized temporally in a circadian manner, what might this mean physiologically? The central functions of NAD+ in DNA repair, gene silencing, the cell cycle and circadian control (Fig. 1) indicate that the consequences of its aberrant regulation could be numerous and physiologically severe. It is conceivable that food restriction impinges on circadian rhythms because it disrupts NAD+-NADH cycling, essentially allowing the redox state of individual cells and tissues to alter rhythmicity. The absence of Clock–Bmal1 dimerization in the presence of increased levels of oxidized NAD is one piece of evidence supporting this idea85. As such, it is easy to imagine sophisticated schemes coordinating SCN-driven rhythms with those of a phase-shifted periphery for drug administration and efficacy. Already there are numerous drugs, perhaps most commonly known within cancer chemotherapeutic strategies, administered following a circadian protocol so that the maximal benefit might be achieved from their use86,87.
Sirtuins are also a powerful example of the extent to which metabolite oscillations might affect physiology as its protein substrates include not only histones but transcription factors and cofactors such p53, FOXO, PGC-1α, NF-κB and PPARγ88. These proteins have been implicated in disease states ranging from the metabolic syndromes and obesity to cancer. Furthermore, as it is the coenzyme for Sirt1, timed production of NAD+ may be important in neuronal and axonal integrity, promoting neuronal survival under conditions of metabolic stress56,89.
As observed throughout circadian biology, peripheral loops and feedback mechanisms are crucial for proper timing. In this sense, NAD+ fits stereotypically what is required in an oscillating system. For example, oxidative phosphorylation requires NADH for ATP production, but then NAD+ provides itself as substrate for PARP-mediated repair to deal with the resulting oxidative stress. Likewise, as coenzyme for Sirt1, NAD+ puts the brakes per se on the system, by slowing the metabolic cycle—or rather by restoring it in preparation for the next cycle of energy production. Deciphering the molecular interplay between these regulators is likely to provide a new and much awaited glimpse of how the circadian clock ‘senses’ cellular metabolism, and vice versa.
Acknowledgments
We thank all the members of our laboratory for stimulating discussions and reading of the manuscript. Work in our laboratory is supported by the US National Institutes of Health (R01 GM081634; R21 AG033888), the Cancer Research Coordinating Committee of the University of California and the Institut National de la Sante et la Recherche Medicale (INSERM U904, France).
References
- 1.Giebultowicz JM. Phil Trans R Soc Lond B. 2001;356:1791–1799. doi: 10.1098/rstb.2001.0960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morse D, Sassone-Corsi P. Trends Neurosci. 2002;25:632–637. doi: 10.1016/s0166-2236(02)02274-9. [DOI] [PubMed] [Google Scholar]
- 3.Yoo SH, et al. Proc Natl Acad Sci USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schibler U, Sassone-Corsi P. Cell. 2002;111:919–922. doi: 10.1016/s0092-8674(02)01225-4. [DOI] [PubMed] [Google Scholar]
- 5.Kalsbeek A, et al. J Biol Rhythms. 2006;21:458–469. doi: 10.1177/0748730406293854. [DOI] [PubMed] [Google Scholar]
- 6.Reppert SM, Weaver DR. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 7.Bell-Pedersen D, et al. Nat Rev Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darlington TK, et al. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- 9.Gekakis N, et al. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 10.Kume K, et al. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 11.Griffin EA, Jr, Staknis D, Weitz CJ. Science. 1999;286:768–771. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- 12.Shearman LP, et al. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- 13.Naidoo N, Song W, Hunter-Ensor M, Sehgal A. Science. 1999;285:1737–1741. doi: 10.1126/science.285.5434.1737. [DOI] [PubMed] [Google Scholar]
- 14.Nakahata Y, et al. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belden WJ, Dunlap JC. Cell. 2008;134:212–214. doi: 10.1016/j.cell.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee C, Weaver DR, Reppert SM. Mol Cell Biol. 2004;24:584–594. doi: 10.1128/MCB.24.2.584-594.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toh KL, et al. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 18.Etchegaray JP, et al. J Biol Chem. 2006;281:21209–21215. doi: 10.1074/jbc.M603722200. [DOI] [PubMed] [Google Scholar]
- 19.Cheng HY, et al. Neuron. 2007;54:813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green CB, Besharse JC. Proc Natl Acad Sci USA. 1996;93:14884–14888. doi: 10.1073/pnas.93.25.14884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akhtar RA, et al. Curr Biol. 2002;12:540–550. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- 22.Duffield GE, et al. Curr Biol. 2002;12:551–557. doi: 10.1016/s0960-9822(02)00765-0. [DOI] [PubMed] [Google Scholar]
- 23.Panda S, et al. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 24.Doi M, Hirayama J, Sassone-Corsi P. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 25.Woelfle MA, Ouyang Y, Phanvijhitsiri K, Johnson CH. Curr Biol. 2004;14:1481–1486. doi: 10.1016/j.cub.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 26.Chen Z, Odstrcil EA, Tu BP, McKnight SL. Science. 2007;316:1916–1919. doi: 10.1126/science.1140958. [DOI] [PubMed] [Google Scholar]
- 27.Turek FW, et al. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamia KA, Storch KF, Weitz CJ. Proc Natl Acad Sci USA. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dodd AN, et al. Science. 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- 30.Sharifian A, Farahani S, Pasalar P, Gharavi M, Aminian O. J Circadian Rhythms. 2005;3:15. doi: 10.1186/1740-3391-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krishnan N, Davis AJ, Giebultowicz JM. Biochem Biophys Res Commun. 2008;374:299–303. doi: 10.1016/j.bbrc.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng X, Yang Z, Yue Z, Alvarez JD, Sehgal A. Proc Natl Acad Sci USA. 2007;104:15899–15904. doi: 10.1073/pnas.0701599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH. Proc Natl Acad Sci USA. 1998;95:8660–8664. doi: 10.1073/pnas.95.15.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Green RM, Tingay S, Wang ZY, Tobin EM. Plant Physiol. 2002;129:576–584. doi: 10.1104/pp.004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Asher G, et al. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 36.Murray DB, Klevecz RR, Lloyd D. Exp Cell Res. 2003;287:10–15. doi: 10.1016/s0014-4827(03)00068-5. [DOI] [PubMed] [Google Scholar]
- 37.Tu BP, et al. Proc Natl Acad Sci USA. 2007;104:16886–16891. doi: 10.1073/pnas.0708365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang X, et al. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 39.Oishi K, et al. FEBS Lett. 2006;580:127–130. doi: 10.1016/j.febslet.2005.11.063. [DOI] [PubMed] [Google Scholar]
- 40.Rudic RD, et al. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sinha MK, et al. J Clin Invest. 1996;97:1344–1347. doi: 10.1172/JCI118551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hansen AP, Johansen K. Diabetologia. 1970;6:27–33. doi: 10.1007/BF00425888. [DOI] [PubMed] [Google Scholar]
- 43.Liu C, Li S, Liu T, Borjigin J, Lin JD. Nature. 2007;447:477–481. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- 44.Rodgers JT, et al. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 45.Li X, et al. Mol Cell. 2007;28:91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 46.Gooley JJ, Schomer A, Saper CB. Nat Neurosci. 2006;9:398–407. doi: 10.1038/nn1651. [DOI] [PubMed] [Google Scholar]
- 47.Brewer M, Lange D, Baler R, Anzulovich A. J Biol Rhythms. 2005;20:195–205. doi: 10.1177/0748730405275952. [DOI] [PubMed] [Google Scholar]
- 48.Alenghat T, et al. Nature. 2008;456:997–1000. doi: 10.1038/nature07541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barnard AR, Nolan PM. PLoS Genet. 2008;4:e1000040. doi: 10.1371/journal.pgen.1000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gorbacheva VY. Proc Natl Acad Sci USA. 2005;102:3407–3412. doi: 10.1073/pnas.0409897102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pang CY, Ma YS, Wei YU. Front Biosci. 2008;13:3661–3675. doi: 10.2741/2957. [DOI] [PubMed] [Google Scholar]
- 53.Fukui H, Moraes CT. Trends Neurosci. 2008;31:251–256. doi: 10.1016/j.tins.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khusnutdinova E, et al. Ann NY Acad Sci. 2008;1147:1–20. doi: 10.1196/annals.1427.001. [DOI] [PubMed] [Google Scholar]
- 55.Mack TG, et al. Nat Neurosci. 2001;4:1199–1206. doi: 10.1038/nn770. [DOI] [PubMed] [Google Scholar]
- 56.Araki T, Sasaki Y, Milbrandt J. Science. 2004;305:1010–1013. doi: 10.1126/science.1098014. [DOI] [PubMed] [Google Scholar]
- 57.Cipriani G, et al. J Biol Chem. 2005;280:17227–17234. doi: 10.1074/jbc.M414526200. [DOI] [PubMed] [Google Scholar]
- 58.Ha HC, Snyder SH. Proc Natl Acad Sci USA. 1999;96:13978–13982. doi: 10.1073/pnas.96.24.13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kauppinen TM, Swanson RA. Neuroscience. 2007;145:1267–1272. doi: 10.1016/j.neuroscience.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 60.Du L, et al. J Biol Chem. 2003;278:18426–18433. doi: 10.1074/jbc.M301295200. [DOI] [PubMed] [Google Scholar]
- 61.Virág L, Szabo C. Pharmacol Rev. 2002;54:375–429. doi: 10.1124/pr.54.3.375. [DOI] [PubMed] [Google Scholar]
- 62.Yang H, et al. Cell. 2007;130:1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. J Biol Chem. 2007;282:6823–6832. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- 64.Kitani T, Okuno S, Takeuchi M, Fujisawa H. J Neurochem. 2003;86:77–85. doi: 10.1046/j.1471-4159.2003.01817.x. [DOI] [PubMed] [Google Scholar]
- 65.Anderson RM, et al. J Biol Chem. 2002;277:18881–18890. doi: 10.1074/jbc.M111773200. [DOI] [PubMed] [Google Scholar]
- 66.Oberdoerffer P, Sinclair DA. Nat Rev Mol Cell Biol. 2007;8:692–702. doi: 10.1038/nrm2238. [DOI] [PubMed] [Google Scholar]
- 67.Revollo JR, Grimm AA, Imai S. J Biol Chem. 2004;279:50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- 68.Ramsey KM, et al. Science. 2009 Mar 19; doi: 10.1126/science.1171641. published online. [DOI] [Google Scholar]
- 69.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Science. 2009 Mar 12; doi: 10.1126/science.1170803. published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schreiber V, Dantzer F, Ame JC, de Murcia G. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 71.Sauve AA, Wolberger C, Schramm VL, Boeke JD. Annu Rev Biochem. 2006;75:435–465. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- 72.Kolthur-Seetharam U, Dantzer F, McBurney MW, de Murcia G, Sassone-Corsi P. Cell Cycle. 2006;5:873–877. doi: 10.4161/cc.5.8.2690. [DOI] [PubMed] [Google Scholar]
- 73.Langmesser S, Albrecht U. Chronobiol Int. 2006;23:151–157. doi: 10.1080/07420520500464437. [DOI] [PubMed] [Google Scholar]
- 74.Aujard F, Herzog ED, Block GD. Neuroscience. 2001;106:255–261. doi: 10.1016/s0306-4522(01)00285-8. [DOI] [PubMed] [Google Scholar]
- 75.Ceriani MF, et al. J Neurosci. 2002;22:9305–9319. doi: 10.1523/JNEUROSCI.22-21-09305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.la Fleur SE, Kalsbeek A, Wortel J, Fekkes ML, Buijs RM. Diabetes. 2001;50:1237–1243. doi: 10.2337/diabetes.50.6.1237. [DOI] [PubMed] [Google Scholar]
- 77.la Fleur SE, Kalsbeek A, Wortel J, van der Vliet J, Buijs RM. J Neuroendocrinol. 2001;13:1025–1032. doi: 10.1046/j.1365-2826.2001.00717.x. [DOI] [PubMed] [Google Scholar]
- 78.Guse AH, et al. Nature. 1999;398:70–73. doi: 10.1038/18024. [DOI] [PubMed] [Google Scholar]
- 79.Dodd AN, et al. Science. 2007;318:1789–1792. doi: 10.1126/science.1146757. [DOI] [PubMed] [Google Scholar]
- 80.Ben-Shlomo R, et al. Chronobiol Int. 2005;22:455–471. doi: 10.1081/CBI-200062353. [DOI] [PubMed] [Google Scholar]
- 81.Dioum EM, et al. Science. 2002;298:2385–2387. doi: 10.1126/science.1078456. [DOI] [PubMed] [Google Scholar]
- 82.Raghuram S, et al. Nat Struct Mol Biol. 2007;14:1207–1213. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yin L, et al. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- 84.Kaasik K, Lee CC. Nature. 2004;430:467–471. doi: 10.1038/nature02724. [DOI] [PubMed] [Google Scholar]
- 85.Rutter J, Reick M, Wu LC, McKnight SL. Science. 2001;293:510–514. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- 86.Lévi F, Filipski E, Iurisci I, Li XM, Innominato P. Cold Spring Harb Symp Quant Biol. 2007;72:465–475. doi: 10.1101/sqb.2007.72.030. [DOI] [PubMed] [Google Scholar]
- 87.Eriguchi M, et al. Biomed Pharmacother. 2003;57:S92–S95. [Google Scholar]
- 88.Guarente L. Nature. 2006;444:868–874. doi: 10.1038/nature05486. [DOI] [PubMed] [Google Scholar]
- 89.Qin W, et al. J Biol Chem. 2006;281:21745–21754. doi: 10.1074/jbc.M602909200. [DOI] [PubMed] [Google Scholar]


