Abstract
Objective
Overexpression of checkpoint kinase 1 (CHK1) is associated with poorer patient outcome and therapeutic resistance in multiple tumor models. Inhibition of CHK1 has been proposed as a strategy to increase the effectiveness of chemotherapeutic agents, especially in p53-deficient tumors. In this study, we evaluated the effects of a novel CHK1 inhibitor, MK-8776, in combination with pemetrexed (PMX) on cell proliferation and survival in a panel of p53 mutant non-small cell lung cancer (NSCLC) cell lines.
Methods
We examined CHK1 expression in 442 resected lung adenocarcinoma specimens using Affymetrix U133A gene expression arrays. We correlated CHK1 mRNA expression with patient survival, tumor differentiation and genomic complexity. We evaluated CHK1 levels in NSCLC cell lines and identified four p53 mutant cell lines with variable CHK1 expression (H1993, H23, H1437 and H1299) based on publicly available gene expression data. We confirmed differential CHK1 mRNA and CHK1 protein levels by qRT-PCR, ELISA, Western Blot analysis (WB) and immunohistochemistry. We examined cell line sensitization to PMX in response to CHK1 inhibition with MK-8776 using WST-1 and clonogenic survival assays.
Results
We found that elevated CHK1 expression in primary lung adenocarcinomas correlates with poor tumor differentiation and significantly worse patient survival. Tumors with elevated CHK1 mRNA levels have a higher number of gene mutations and DNA copy number gain or amplifications. CHK1 inhibition by MK-8776 enhances sensitivity of NSCLC cell lines to PMX. CHK1 mRNA and protein expression are variable among NSCLC cell lines, and cells expressing higher levels of CHK1 protein are more sensitive to the CHK1 inhibition by MK-8776 as compared to low CHK1 expressing cells.
Conclusions
These findings suggest that CHK1 levels may not only serve as a biomarker of poor prognosis in surgically-resected lung adenocarcinomas, but could also be a predictive marker for CHK1 inhibitor sensitivity, pending in vivo and clinical confirmation.
Keywords: CHK1, Lung, Chemosensitivity, NSCLC, Patient survival, Genomic complexity
1. Introduction
Lung cancer is the number one cause of cancer-related deaths in the United States with over 220,000 new cases each year with an overall 5-year survival of 16% [1]. The best chemotherapeutic treatments are often limited by dose-related toxicities [2]. Emerging targeted therapies might be the key in improving survival in lung cancer patients and new oncogenic pathways with novel targets have been identified [3,4]. EGFR-mutated tumors and tumors containing ALK fusion genes are examples of identifiable subgroups of NSCLC’s yet include only small percentages of NSCLC’s [5,6].
The CHK1 pathway has been shown to contribute to therapy resistance [7,8] and overall cell survival by activating DNA damage responses, including G2 arrest and homologous recombination repair (HRR) [9]. CHK1 is being evaluated as a novel target for cancer therapy and there are a number of CHK1 inhibitors in early clinical development [10]. Inhibition of CHK1 increases chemo- and radiation therapy sensitivity in multiple tumor models, including lung [7–14]. Sensitization by CHK1 inhibition appears to be tumor cell selective and preferential toward p53 mutant tumors. The prevailing model is that tumor cells with p53 mutations, which do not arrest in G1 in response to DNA damage, will be selectively sensitized by CHK1 inhibition, and proceed through G2 to mitotic death, while normal cells will be protected from CHK1 inhibition by their other intact checkpoints [15–17].
We have examined gene expression data from 442 resected lung adenocarcinomas, previously published by Shedden et al. [18], and found that CHK1 was one of the top genes that were elevated in patients with the poorest outcomes. CHK1 expression correlated with tumors’ differentiation state and genomic complexity. We have evaluated the effects of a novel CHK1 inhibitor, MK-8776 [14,19] in combination with pemetrexed (PMX), a current clinically prescribed anti-metabolite chemotherapy, on cell proliferation and clonogenic survival in a panel of p53 mutant NSCLC cell lines. We hypothesized that the level of CHK1 mRNA or protein expression, and in turn, CHK1 function may influence the response of NSCLC to PMX treatment by CHK1 inhibition.
2. Materials and methods
2.1. Correlation analysis of genomic data
Affymetrix U133A gene expression array data from 442 lung adenocarcinomas [18] were normalized using Robust Multi-array Average (RMA) method [20]. Normalized DNA copy number values of 371 lung adenocarcinomas from SNP250K StyI arrays were used to assess copy number changes [21]. Somatic mutations of 623 human genes in 180 lung adenocarcinomas were examined to correlate mutation and CHK1 [22] and gene expression data from 79 NSCLC lung cell lines was used to identify cell models [23]. Pearson correlation was used for the correlation analysis of gene expression and gene mutations or copy number changes. Student’s t-test was used for gene expression in different clinical variables. Kaplan–Meier survival curve with log-rank test was used for survival analysis. Lung adenocarcinomas and normal tissues, used as an independent validation set, were collected with informed consent after approval from University of Michigan Institutional Review Board and Ethics Committee and stored in −80 °C until use.
2.2. Cell culture and reagents
H1993, H23, H1437 (adenocarcinomas) and H1299 (large cell carcinoma) cells were obtained from American Type Culture Collection (Manassas) and grown in RPMI1640 supplemented with 10% FBS. Pemetrexed (Eli Lilly Company) was dissolved in PBS. MK-8776 (Merck) was dissolved in DMSO.
2.3. Quantitative real time PCR
RNA from cell lines was isolated and purified using RNeasy Mini Kits (Qiagen) according to manufacturer’s protocol and reverse transcribed using High Capacity cDNA Transcription Kit (Applied Biosystems). CHK1 transcripts were quantified by quantitative real-time PCR (qRT-PCR) using Platinum SYBR Green qRT-PCR SuperMix-UDG (Invitrogen) in a Rotor-Gene 3000 thermocycler (Corbett Life Science). Relative expression levels were normalized to β-actin expression using 2−ΔΔCt analysis [24]. The qRT-PCR was also used for CHK1 mRNA expression in 101 lung adenocarcinomas and 12 normal tissues, an independent validation set. The demographics of these patients are provided in Table 1. Primer sequences were as follows: ACTB (forward): 5′-ATGCAGAAGGAGATCACTGC-3′; ACTB (reverse): 5′-TCATAGTCCGCCTAGAAGCA-3′; CHK1 (forward): 5′-CGGTGGAGTCATGGCAGTGCCC-3′; CHK1 (reverse): 5′-TCTGGACAGTCTACGGCACGCTTCA-3′.
Table 1.
Clinical characteristics of 101 lung adenocarcinomas in the validation set.
| Variable | Number |
|---|---|
| Age average (years) | 67.0 ± 9.6 |
| Gender | |
| Female | 53 (52.5%) |
| Male | 48 (47.5%) |
| Stage | |
| Stage I | 59 (58.4%) |
| Stage II | 16 (15.8%) |
| Stage III | 26 (25.7%) |
| Differentiation | |
| Well | 28 (27.7%) |
| Moderate | 38 (37.6%) |
| Poor | 34 (33.7%) |
| Deceased (at 5 years) | 44 (43.6%) |
| Alive | 57 (56.9%) |
| Median survival (months) | 28.8 |
2.4. CMA construction and immunohistochemistry
Formalin-fixed, paraffin-embedded blocks (FFPE) of 48 cell lines were arrayed into a cell-line microarray (CMA) using previously described methodology [25]. Immunohistochemical staining was performed on the DAKO Autostainer (DAKO) using DAKO Envision + polymerized HRP and diaminobenzadine as the chromogen. Sections of deparaffinized CMA were microwave-treated for epitope retrieval in 10 mM Tris buffer pH9/1 mM EDTA and incubated overnight with a CHK1 antibody (clone EP691Y, Abcam). Appropriate negative (no primary antibody) and positive controls (breast cancer) were stained in parallel. The immunoreactivity was scored by a three-tier (negative, low- (1+) and high-positive (2+)) modification of the normal grading scheme previously described by Wang et al. [26]. In our analysis, negative meant <10% staining (either true negative or a faint blush). Low was classified as 11–50% staining of the cell population, and high was 51–100%.
2.5. Proliferation analyses
Cell proliferation was measured using cell proliferation reagent WST-1 (Roche Applied Science) according to the manufacturer’s instructions. Cells were seeded in 96-well flat-bottomed microplates and incubated for 24 h. At this time point, defined as T0 h, cells were treated with MK-8776 (T = 0–24 h) at 500 nM concentration. Spectrophotometric quantification of cell proliferation was measured at T72 and normalized to T0. Viability of cells was expressed as the relative percent absorbance of treated versus non-treated cells. For cell count experiment, cells were plated in 24 well plates at the cell concentration as for WST-1 assay and treated as described above. At T72 cells were trypsinized and counted. Cell counts were normalized to T0.
2.6. Chemosensitization
Chemosensitization was measured using WST-1 reagent. At T0 cells were treated with graded concentrations of PMX (T = 0–24 h) followed by the CHK1 inhibitor MK-8776 at 500 nM concentration (T = 24–48 h). After exposure to both drugs, cells were washed and cultured in drug-free medium for 24 h (T = 48–72 h). Data was analyzed using Microsoft Excel 2010 and GraphPad Prism version 5.01 software. Chemosensitization was confirmed by clonogenic survival assay as previously described [9,27], with slight modifications.
2.7. Small interfering RNA
Cells were transfected with SMARTpool CHK1 or non-target (NT) control pool siRNAs (Dharmacon) using Lipofectamine RNAiMAX transfection reagent (Invitrogen) according to the manufacturer’s protocol. For sensitization assays, cells were treated with 10 nmol/L siRNA (CHK1 or NT) and Lipofectamine in the presence of 10% FBS for 24 h and exposed to graded concentrations of PMX for an additional 24 h. Following treatment, cells were cultured in drug-free medium (T = 24 h). Chemosensitization was measured using WST-1 reagent. Cells that received combination treatment of MK-8776 and CHK1 siRNA were cultured with MK-8776 at 750 nM concentration (T = 24 h) following treatment with PMX.
2.8. Immunoblotting and CHK1 ELISA
Cell pellets were lysed and immunoblotted as previously described [28]. Proteins were detected with pS345 CHK1, pS296 CHK1 (Cell Signaling), CHK1 (Abcam), CDC25A (Santa Cruz Biotechnology), and GAPDH (Millipore). PathScan CHK1 Sandwich ELISA kit (Cell Signaling) was used according to manufacturer’s protocol.
3. Results
3.1. CHK1 mRNA expression correlates with patient survival, tumor differentiation and genomic complexity in lung cancer
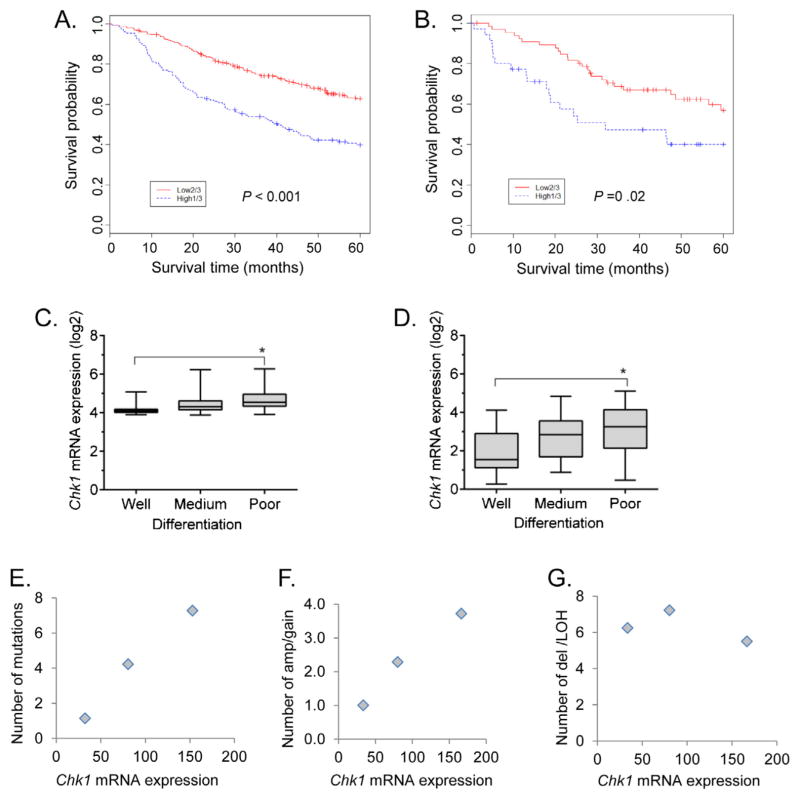
Analysis of gene expression of 442 lung adenocarcinomas [18] revealed that CHK1 is one of the panel of variably-expressed genes significantly associated with poor survival. Patients with high CHK1 mRNA expressing tumors demonstrated an overall poorer 5 year survival by log-rank test (hazard ratio (HR) 2.13; 95% confidence interval (CI) 1.6–2.8; p < 0.001) (Fig. 1A) and univariate Cox model (HR 2.18, 95% CI 1.69–2.81, p < 0.001). A multivariate Cox model adjusted with age, gender and stage indicated that CHK1 mRNA is independently related to survival (HR 2.39; 95% CI 1.8–3.1; p < 0.001). These results were verified with an independent cohort of 101 lung adenocarcinomas using qRT-PCR method, showing a similar difference in 5 year survival by log-rank test (HR 2.06, 95% CI 1.13–3.76, p = 0.02) (Fig. 1B). Further analysis of the gene expression dataset of 442 lung adenocarcinomas revealed that CHK1 mRNA expression was significantly higher in poorly differentiated compared to well differentiated tumors, p = 0.0001 (Fig. 1C). These results were also verified with an independent cohort of 101 lung adenocarcinomas, showing a similar correlation between level of differentiation and CHK1 mRNA expression, p = 0.0003 (Fig. 1D).
Fig. 1.
Relationship of CHK1 mRNA expression with patient survival, tumor differentiation and genomic complexity. (A) Analysis of gene expression of 442 lung adenocarcinomas using Kaplan–Meier survival curve shows that patients with high CHK1 mRNA expressing tumors demonstrate an overall poorer 5 year survival, comparing the higher 1/3 patients vs lower 2/3 patients based on CHK1 mRNA levels (log-rank test, p < 0.001). (B) This was verified in an independent cohort of patients with 101 lung adenocarcinomas with CHK1 mRNA verified by RT-PCR (p = 0.02). (C) Tumors with higher CHK1 mRNA expression are more likely to display a poorly-differentiated phenotype as compared to low CHK1 expressing tumors (p < 0.0001) in 442 lung adenocarcinomas. (D) This was verified in the same independent cohort of patients with 101 lung adenocarcinomas (p = 0.0003). CHK1 mRNA expression in lung adenocarcinomas also correlates with an increased number of genomic alterations, specifically gene mutations or amplifications. Tumors were classified into high, medium and low groups based on CHK1 mRNA expression (CHK1 mRNA >100, 100–50, and <50 as cutoff for these 3 groups). (E) Higher CHK1 mRNA expression correlated with an increased number of gene mutations in 54 lung adenocarcinomas (Pearson correlation, r = 0.993, p < 0.001, the mean mutation ±SD are 7.28 ±10.4, 4.23 ±3.7 and 1.14 ±0.99 for high, medium and low groups, respectively). (F) In a separate data-set of 93 lung adenocarcinomas, higher CHK1 mRNA correlated with DNA amplification/gain (Pearson correlation, r = 0.99, p < 0.001, mean amp/gain ±SD are 3.73 ±3.2, 2.29 ±2.1, and 1.0 ±0.76 for high, medium and low groups, respectively). (G) CHK1 mRNA levels do not correlate with gene deletions or LOH for the same 93 tumors.
To determine the potential basis for variable CHK1 expression between different primary lung tumors, we evaluated CHK1 mRNA expression and gene mutations in 54 lung adenocarcinomas [18,22]. Tumors were divided into three groups based on their CHK1 mRNA expression as either low <50 (n = 7), medium 50–100 (n = 22) or high >100 (n = 25). Tumors with high CHK1 mRNA expression contain a higher number of gene mutations, p < 0.001 (Fig. 1E). We then evaluated a second cohort of 93 lung adenocarcinomas, again dividing them into three groups based on CHK1 mRNA expression, low (n = 8), medium (n = 35) and high (n = 50), and examined for copy number change as determined by SNP arrays [21]. Higher CHK1 mRNA expression correlated with a higher number of DNA copy number gain or amplifications, p < 0.001 (Fig. 1F). CHK1 mRNA did not correlate with the number of DNA deletions or loss of heterozygosity (LOH) events (Fig. 1G). These results suggest that increasing CHK1 expression may be a survival mechanism in tumors for coping with genetic gains/amplifications or mutations and is associated with a less differentiated phenotype.
Using the same 54 lung adenocarcinomas, we correlated CHK1 levels to specific gene mutations and found that CHK1 mRNA is higher in p53 mutated tumor as compared to wild type p53 tumors (p = 0.01) (Fig. S1). CHK1 mRNA levels were not different between EGFR or KRAS mutated samples and wild type samples (p > 0.05) (Fig. S1, EGFR box plot not shown). From Shedden’s 442 lung adenocarcinomas [18], CHK1 mRNA is higher in stage 3 tumors (vs stage 1, t-test, p = 0.002), and also higher in current smokers (vs never smoking, t-test, p = 0.001) (Fig. S2).
Supplementary material related to this article can be found, in the online version, at http://dx.doi.org/10.1016/j.lungcan.2013.09.010.
3.2. NSCLC cell lines have variable CHK1 mRNA and protein expression
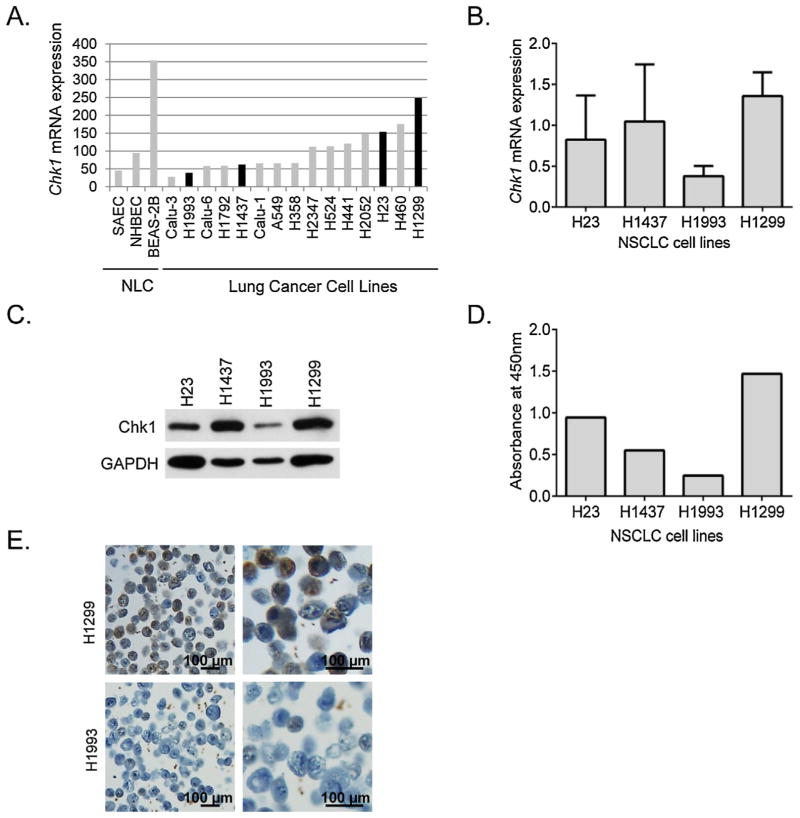
To examine whether the level of CHK1 is associated with response to CHK1 inhibition, we identified four p53 mutant cell lines with variable levels of CHK1 expression based on publically-available gene expression data [23] (Fig. 2A). We confirmed CHK1 mRNA expression by qRT-PCR and correlated to CHK1 protein expression by Western Blot (WB), ELISA, and immunohistochemistry. QRT-PCR revealed slight differences in CHK1 mRNA expression when compared to the gene array data (Fig. 2B), with H1437 demonstrating high expression of CHK1 (relative to β-actin). Western Blot (Fig. 2C) and ELISA (Fig. 2D) confirmed CHK1 expression differences, with high levels of protein seen in H1299 and H1437 cells, intermediate levels in H23 cells, and the lowest in H1993 cells. IHC performed on a CMA confirmed the relative CHK1 expression seen by qRT-PCR with representative samples from H1299 and H1993 showing high and low expression, respectively of nuclear localized CHK1 protein (Fig. 2E).
Fig. 2.
(A) Gene expression analysis for CHK1 mRNA levels in representative sample of 15 cultured lung cancer cell lines and in nontransformed lung cells (NLC). Based on these results, we selected two apparent under-expressing cell lines, H1993 and H1437, and two over-expressing cell lines, H23 and H1299, for further analysis. (B) Relative expression of CHK1 to β-actin by qRT-PCR in selected NSCLC cell lines. Data shown is the aggregate of three independent assays, and suggests that H1437 is actually a CHK1 over-expressing cell line. CHK1 protein levels in selected NSCLC cell lines by Western Blot (C) and by ELISA (D) confirming the three higher CHK1 expressing cell lines (H23, H1437, and H1299) and the one lower expressing line (H1993). (E) CHK1 expression by immunohistochemistry in two representative cell lines (H1993 and H1299) showing increased nuclear CHK1 protein expression (brown stain) in the H1299 cells compared to the H1993 cells. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. CHK1 inhibition preferentially sensitizes cells expressing higher levels of CHK1
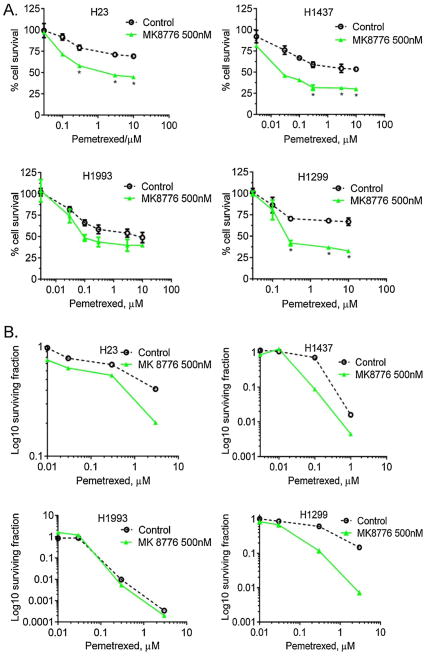
To ascertain the effects of CHK1 inhibition on PMX sensitivity in high versus low CHK1 expressing cells, we assessed the effects of MK-8776 alone on cell viability and proliferation. We found that MK-8876 treatment had no effect on either high or low CHK1 expressing cell lines (data not shown). To determine whether CHK1 inhibition by MK-8776 differentially sensitized to PMX in high versus low CHK1 expressing cells, we treated H23, H1437, H1299 and H1993 cells with graded concentrations of PMX (T = 0–24 h) followed by 24 h treatment with MK-8776 (Fig. 3A). At as low as 0.3 μM of PMX, there was a statistically significant reduction in proliferation in the high expressing CHK1 cells H23 (p = 0.0012), H1437 (p = 0.0006) and H1299 (p = 0.0006), but not in the low CHK1 expressing H1993 cells (p = 0.26). We then evaluated the effects of MK-8776 on PMX sensitivity using clonogenic assays. It confirmed that MK-8776 sensitized high CHK1 expressing cells to PMX as evidenced by a decrease in the surviving fraction (5-fold change in H23, 3.4-fold in H1437, and 8-fold in H1299 as based on IC50; data not shown), but did not sensitize H1993 (Fig. 3B).
Fig. 3.
(A) Chemosensitization to PMX by the CHK1 inhibitor, MK-8776, as measured by WST-1 assay. Cells were treated with graded concentrations of PMX for 24 h (T0–T24 h) followed by 24 h treatment with MK-8776 at 500 nM (T24–T48 h). CHK1 inhibition preferentially sensitized cells expressing higher levels of CHK1 (H23, H1437 and H1299) as compared to low CHK1-expressing cells (H1993). At as low as 0.3 μM PMX concentration there was significant reduction in percent of proliferating cells in H23 (p = 0.0012), H1437 (p = 0.0006) and H1299 (p = 0.0006). No significant change in H1993 cells (p = 0.26). Each experiment was performed using 3 replicates for each drug concentration. All experiments were repeated a minimum of 3 times. (B) Chemosensitization by clonogenic survival assay shown in log scale. Low CHK1-expressing H1993 cells are not sensitized to PMX in combination with MK-8776, whereas over-expressing cell lines are sensitized.
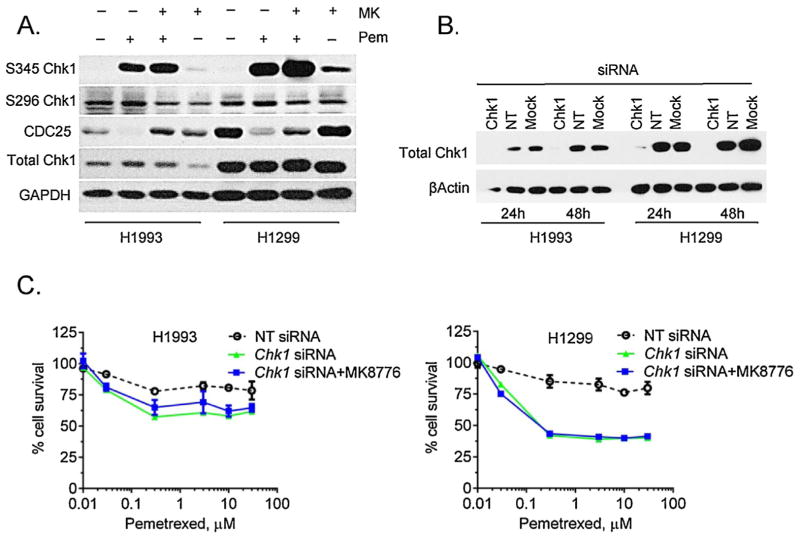
We then evaluated the effect of PMX/MK-8776 treatment on the levels of phosphorylated CHK1 (the S296, autophosphorylation site, and the S345 ATR/ATM-mediated phosphorylation site), as well as CDC25A, which is degraded in response to CHK1 activation and contributes to G2-checkpoint activation in response to DNA damage (Fig. 4A). Consistent with CHK1 inhibition by MK-8776, autophosphorylation at S296 was modestly decreased by MK-8776 both in the presence and absence of PMX in both cell lines. The loss of CDC25A protein in response to PMX was blocked by MK-8776 and is consistent with CHK1 inhibition resulting in CDC25A protein stabilization. ATR/ATM-mediated phosphorylation of CHK1 at S345, as anticipated, was increased in response to PMX or MK-8776 single agent treatments and is likely a reflection of DNA damage [29]. The combination of PMX and MK-8776 further increased S345 CHK1 phosphorylation and the magnitude of this effect was the greatest in H1299 cells.
Fig. 4.
(A) Western Blot analyses of the MK-8776 plus PMX in low and high-expressing CHK1 cell lines. The higher CHK1 expressing line, H1299, displays a greater response in S345 autophosphorylation with combination treatment. Also of note, H1299 appears to have a higher baseline CDC25A expression which is reduced with PMX treatment alone. The addition of a CHK1 inhibitor increases CDC25A expression indicating mitotic entry. (B) Western Blot analysis of CHK1 siRNA treatment showing knockout of CHK1 protein expression at 24 and 48 h. In addition, cells were treated with non-target siRNAs (NT) and Lipofectamine RNAiMAX transfection reagent (Mock). (C) Effect of CHK1 siRNA on proliferation in H1993 and H1299 cell lines. H1299 cells show a greater reduction in proliferation than H1993 cells when treated with CHK1 siRNA vs NT siRNA. The combination of CHK1 siRNA and MK-8776 at 750 nM had no effect compared to siRNA use alone.
To confirm that MK-8776 sensitized cells by selective inhibition of CHK1, we treated H1993 and H1299 cells with CHK1 siRNA and found an almost complete elimination of CHK1 protein at 24 and 48 h (Fig. 4B) post-transfection, with no changes observed in the controls. We then tested the ability of CHK1 siRNA and CHK1 siRNA plus MK-8776 to sensitize to PMX (Fig. 4C). Both treatments produced similar sensitization to PMX, confirming that MK-8776-mediated sensitization is through CHK1 inhibition. The extent of the sensitization to PMX was more pronounced in H1299 cells as compared to H1993, which is consistent with sensitization by MK-8776 alone (Fig. 3A). These results demonstrate that MK-8776 similarly inhibits CHK1 in high versus low CHK1 expressing cells but results in a greater sensitization to PMX in high CHK1 expressing cells.
4. Discussion
In this study, we have shown that CHK1 inhibition by MK-8776 enhances sensitivity of NSCLC cell lines to PMX. CHK1 mRNA and protein expression are variable among NSCLC cell lines, and cells expressing higher levels of CHK1 protein are more sensitive to the CHK1 inhibition as compared to low CHK1 expressing cells. We have also found that higher CHK1 expression in primary lung adenocarcinomas correlates with poor tumor differentiation, higher stage, smoking status, and worse survival. Furthermore, elevated CHK1 expression correlates with increased number of genomic alterations, specifically amplifications or gene mutations.
DNA damage is central to many chemotherapy regimens in lung cancer treatment. The normal cell cycle requires functional p53 to stop cell cycling at the G1-checkpoint. CHK1 regulates the S-phase and G2 checkpoints, preventing cells from entering mitosis. When p53-mutant cancer cells undergo genomic damage in combination with CHK1 inhibition, they lose both checkpoints and progress through the cell cycle with unrepaired DNA damage resulting in preferential killing [16]. CHK1 appears to play a central role in chemotherapy resistance of lung cancers by this mechanism [30]. Inhibiting CHK1 function may increase the effectiveness, and lower the required doses of commonly used lung cancer chemotherapy agents.
The correlation of CHK1 overexpression with decreased survival that we have seen has been also seen in other cancers. In patients with liver cancer, both recurrence-free survival and overall survival were significantly diminished in tumors with CHK1 over-expression [31]. Inhibition of CHK1 in mice bearing p53-deficient triple-negative, poorly-differentiated breast cancer reduced tumor growth, prolonged host survival, and this effect was not seen in more differentiated p53 wild type tumors [32]. CHK1 function is critical for tumor cells with complex genomes to maintain survival [33], because cells with the most complex genetic alterations may require more stringent checkpoints for DNA repair. In our tumor cohort, the subgroup of lung adenocarcinomas with increased number of gene mutations, amplifications or gains demonstrated the highest level of CHK1 expression. Interestingly, CHK1 mRNA did not correlate with LOH or gene deletions. CHK1 levels may reflect an essential role of CHK1 in responding to this type of genomic complexity although this area requires further study [34,35].
When we analyzed CHK1 signaling, we found that in response to DNA damage from PMX, there was an increase in phosphorylation of the S345 site, which was more pronounced with CHK1 inhibition. Consistent with CHK1 inhibition, autophosphorylation at S296 was modestly reduced by MK-8776, both in the presence and absence of PMX, indicating inhibition of CHK1 kinase activity. Our results are consistent with previous studies [29], which have suggested that pS345 CHK1 is a marker of DNA damage in response to CHK1 inhibition, while pS296 CHK1 is a marker of CHK1 catalytic activity. Active CHK1 inhibits CDC25 phosphatases, ultimately resulting in cell cycle arrest and cell survival [33]. The reduction of CDC25 in response to PMX was blocked by MK-8776 leading to CDC25 stabilization, which is consistent with CHK1 inhibition. Both the low and high-expressing CHK1 NSCLC cell lines exhibited a similar pattern of CDC25A response, suggesting equivalent inhibition of CHK1 although sensitization by CHK1 inhibition was greater in the high CHK1 expressing cell lines.
Our data implies that total CHK1 levels, based upon protein expression, may serve as a useful biomarker for CHK1 inhibitor sensitivity for lung cancers. Additionally, 60–70% of NSCLC tumors are p53 mutant and p53 sequencing is commonplace in the current clinical treatment of NSCLC. The future testing of NSCLC tumors with p53 sequencing combined with CHK1 expression could allow for characterization of those tumors with poor clinical outcomes and potentially modify current chemotherapy regimens. Further studies to validate total CHK1 as a potential biomarker in NSCLC primary tumors and clinical biopsies are needed in addition to in vivo studies to verify the efficacy of CHK1 inhibitors as sensitizers and also the reliability of CHK1 as a biomarker.
Acknowledgments
The authors would like to thank Drs. Lin Lin and Amy Myers for helpful discussions and suggestions on the experimental design, and Dr. Ernest Nadal for critical review and constructive comments.
Grant support R01CA163895 (Morgan MA).
Abbreviations
- CHK1
checkpoint kinase 1
- LOH
loss of heterozygosity
- NSCLC
non-small cell lung cancer
- PMX
pemetrexed
- RMA
Robust Multi-Array Average
Footnotes
Conflict of interest
The authors have no potential conflict of interest relevant to this article.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J, et al. Eastern Cooperative Oncology Group. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346:92–8. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 3.Bryant CM, Albertus DL, Kim S, Chen G, Brambilla C, Guedj M, et al. Clinically relevant characterization of lung adenocarcinoma subtypes based on cellular pathways: an international validation study. PLoS ONE. 2010;5:e11712. doi: 10.1371/journal.pone.0011712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayes DN, Monti S, Parmigiani G, Gilks CB, Naoki K, Bhattacharjee A, et al. Gene expression profiling reveals reproducible human lung adenocarcinoma subtypes in multiple independent patient cohorts. J Clin Oncol. 2006;24:5079–90. doi: 10.1200/JCO.2005.05.1748. [DOI] [PubMed] [Google Scholar]
- 5.Rosell R, Moran T, Queralt C, Porta R, Cardenal F, Camps C, et al. Spanish Lung Cancer Group. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361:958–67. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 6.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–770. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou J, Chen Z, Malysa A, Li X, Oliveira P, Zhang Y, et al. A kinome screen identifies checkpoint kinase 1 (CHK1) as a sensitizer for RRM1-dependent gemcitabine efficacy. PLoS ONE. 2013;8:e58091. doi: 10.1371/journal.pone.0058091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrassa L, Broggini M, Erba E, Damia G. Chk1, but not Chk2, is involved in the cellular response to DNA damaging agents: differential activity in cells expressing, or not, p53. Cell Cycle. 2004;3:1175–9. [PubMed] [Google Scholar]
- 9.Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, et al. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat Cell Biol. 2005;7:195–201. doi: 10.1038/ncb1212. [DOI] [PubMed] [Google Scholar]
- 10.Dai Y, Grant S. New insights into checkpoint kinase 1 in the DNA damage response signaling network. Clin Cancer Res. 2010;16:376–83. doi: 10.1158/1078-0432.CCR-09-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morgan MA, Parsels LA, Zhao L, Parsels JD, Davis MA, Hassan MC, et al. Mechanism of radiosensitization by the Chk1/2 inhibitor AZD7762 involves abrogation of the G2 checkpoint and inhibition of homologous recombinational DNA repair. Cancer Res. 2010;70:4972–81. doi: 10.1158/0008-5472.CAN-09-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parsels LA, Morgan MA, Tanska DM, Parsels JD, Palmer BD, Booth RJ, et al. Gemcitabine sensitization by checkpoint kinase 1 inhibition correlates with inhibition of a Rad51 DNA damage response in pancreatic cancer cells. Mol Cancer Ther. 2009;8:45–54. doi: 10.1158/1535-7163.MCT-08-0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitchell JB, Choudhuri R, Fabre K, Sowers AL, Citrin D, Zabludoff SD, et al. In vitro and in vivo radiation sensitization of human tumor cells by a novel checkpoint kinase inhibitor, AZD7762. Clin Cancer Res. 2010;16:2076–84. doi: 10.1158/1078-0432.CCR-09-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Montano R, Chung I, Garner KM, Parry D, Eastman A. Preclinical development of the novel Chk1 inhibitor SCH900776 in combination with DNA-damaging agents and antimetabolites. Mol Cancer Ther. 2012;11:427–38. doi: 10.1158/1535-7163.MCT-11-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Z, Xiao Z, Gu WZ, Xue J, Bui MH, Kovar P, et al. Selective Chk11 inhibitors differentially sensitize p53-deficient cancer cells to cancer therapeutics. Int J Cancer. 2006;119:2784–9. doi: 10.1002/ijc.22198. [DOI] [PubMed] [Google Scholar]
- 16.Ma CX, Janetka JW, Piwnica-Worms H. Death by releasing the breaks: CHK1 inhibitors as cancer therapeutics. Trends Mol Med. 2011;17:88–96. doi: 10.1016/j.molmed.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koniaras K, Cuddihy AR, Christopoulos H, Hogg A, O’Connell MJ. Inhibition of Chk1-dependent G2 DNA damage checkpoint radiosensitizes p53 mutant human cells. Oncogene. 2001;20:7453–6. doi: 10.1038/sj.onc.1204942. [DOI] [PubMed] [Google Scholar]
- 18.Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14:822–7. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guzi TJ, Paruch K, Dwyer MP, Labroli M, Shanahan F, Davis N, et al. Targeting the replication checkpoint using SCH 900776, a potent and functionally selective CHK1 inhibitor identified via high content screening. Mol Cancer Ther. 2011;10:591–602. doi: 10.1158/1535-7163.MCT-10-0928. [DOI] [PubMed] [Google Scholar]
- 20.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 21.Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–8. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–75. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou BB, Peyton M, He B, Liu C, Girard L, Caudler E, et al. Targeting ADAM-mediated ligand cleavage to inhibit HER3 and EGFR pathways in non-small cell lung cancer. Cancer Cell. 2006;10:39–50. doi: 10.1016/j.ccr.2006.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Nocito A, Kononen J, Kallioniemi OP, Sauter G. Tissue microarrays (TMAs) for high-throughput molecular pathology research. Int J Cancer. 2001;94:1–5. doi: 10.1002/ijc.1385. [DOI] [PubMed] [Google Scholar]
- 26.Wang S, Saboorian MH, Frenkel E, Hynan L, Gokaslan ST, Ashfaq R. Laboratory assessment of the status of her-2/neu protein and oncogene in breast cancer specimens: Comparison of immunohistochemistry assay with fluorescence in situ hybridisation assays. J Clin Pathol. 2000;53:374–81. doi: 10.1136/jcp.53.5.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawrence TS. Ouabain sensitizes tumor cells but not normal cells to radiation. Int Radiat Oncol Biol Phys. 1998;15:953–8. doi: 10.1016/0360-3016(88)90132-0. [DOI] [PubMed] [Google Scholar]
- 28.Silvers AL, Lin L, Bass AJ, Chen G, Wang Z, Lin J, et al. Decreased selenium-binding protein 1 in esophageal adenocarcinoma results from posttranscriptional and epigenetic regulation and affects chemosensitivity. Clin Cancer Res. 2010;16(7):2009–21. doi: 10.1158/1078-0432.CCR-09-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parsels LA, Qian Y, Tanska DM, Gross M, Zhao L, Hassan MC, et al. Assessment of chk1 phosphorylation as a pharmacodynamic biomarker of chk1 inhibition. Clin Cancer Res. 2011;17:3706–15. doi: 10.1158/1078-0432.CCR-10-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartucci M, Svensson S, Romania P, Dattilo R, Patrizii M, Signore M, et al. Therapeutic targeting of Chk1 in NSCLC stem cells during chemotherapy. Cell Death Differ. 2012;19:768–77. doi: 10.1038/cdd.2011.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong J, Hu K, Yuan Y, Sang Y, Bu Q, Chen G, et al. CHK1 targets spleen tyrosine kinase (L) for proteolysis in hepatocellular carcinoma. J Clin Invest. 2012;122:2165–7. doi: 10.1172/JCI61380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma CX, Cai S, Li S, Ryan CE, Guo Z, Schaiff WT, et al. Targeting Chk1 in p53-deficient triple-negative breast cancer is therapeutically beneficial in human-in-mouse tumor models. J Clin Invest. 2012;122:1541–52. doi: 10.1172/JCI58765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cho SH, Toouli CD, Fujii GH, Crain C, Parry D. Chk1 is essential for tumor cell viability following activation of the replication checkpoint. Cell Cycle. 2005;4:131–9. doi: 10.4161/cc.4.1.1299. [DOI] [PubMed] [Google Scholar]
- 34.Bartek J, Bartkova J, Lukas J. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene. 2007;26:7773–9. doi: 10.1038/sj.onc.1210881. [DOI] [PubMed] [Google Scholar]
- 35.Bartkova J, Hamerlik P, Stockhausen MT, Ehrmann J, Hlobilkova A, Laursen H, et al. Replication stress and oxidative damage contribute to aberrant constitutive activation of DNA damage signalling in human gliomas. Oncogene. 2010;29:5095–102. doi: 10.1038/onc.2010.249. [DOI] [PubMed] [Google Scholar]