Abstract
Unc-51-Like Kinase 1 (ULK1) is regarded as a central role in autophagy. Although the details of how ULK1 triggers autophagy are obscure, the relationship between ULK1 expression and the diagnosis and prognosis of cancer patients may guide the clinical practice and scientific research. The aim of this study was to investigate and compare the expression level of ULK1 in 86 paired esophageal squamous cell carcinoma (ESCC) and paracancerous tissues, and to examine the effect of ULK1 expression on the prognosis of ESCC patients. ULK1 was primarily expressed in cytoplasm, but was rarely seen in nucleus. The levels of cytoplasmic ULK1 in ESCC tissues were higher than those in paracancerous tissue (P < 0.01) and significantly associated with lymph node metastasis (LNM) (P = 0.025). Survival analysis showed that patients with low expression of cytoplasmic ULK1 had worse survival time than those with high expression of cytoplasmic ULK1 (hazard ratio = 1.754, 95% confidence interval: 1.022-3.010, P = 0.041), which disappeared after adjustment for TNM stages and LNM (P = 0.319). In conclusion, ULK1 might play an important role in the occurrence and development of ESCC and represent a potential prognostic biomarker for ESCC patients. However, the precise impact of ULK1 on predicting the prognosis of patients with ESCC requires further investigation.
Keywords: Esophageal squamous cell carcinoma, autophagy, UNC51-like kinase 1, survival, biomarker
Introduction
Esophageal cancer is a common malignant tumor in China. Ferlay et al reported that more than 80% of esophageal cancer patients come from developing countries, and China ranks fourth in the world in terms of the incidence of esophageal cancer [1]. In recent years, the incidence of esophageal cancer in China declined from 1998 to 2007 [2]. Despite this encouraging result, esophageal cancer remains a point of major concern because of China’s immense population base. Moreover, the poor sensitivity of esophageal cancer to radiotherapy and chemotherapy not only results in hematogenous metastasis and local infiltration in the early stage, but also reduces the five-year survival rate of patients. Therefore, prevention of esophageal cancer is an arduous task.
The formation and development of malignant tumors remains an active area of research, and there is growing focus on the role of autophagic behavior in cancer. Autophagy occurs during normal physiological processes and serves in cells as a defense mechanism against adverse conditions. Both excessive and insufficient autophagy can lead to various diseases [3]. Previous studies showed that change in autophagic activity is closely related to the occurrence and development of cancer [4]. Suzuki et al. [5] found that at least 36 autophagy-related genes (ATG) exist in yeast. Notably, ATG1 has two homologs in mammals: uncoordinated 51-Like kinases 1 and 2 (ULK1 and ULK2) [6]. ULK1 is known to be a key factor in cellular autophagy [7-9]. Rapamycin acceptor complex (TORC1) and amp-activated protein kinase (AMPK) are highly sensitive to lack of nutrients and energy, and directly act on ULK1 [10,11]. ULK1 then activates the VPS34 complex by causing Beclin-1 phosphorylation to induce autophagy [12].
The detailed mechanism by which autophagy occurs is being actively studied around the world, and the relationship between ULK1 expression and the diagnosis and prognosis of cancer has received increasing attention in recent years. Recent findings show that ULK1 expression can be used as a biomarker of poor prognosis for breast, liver, and esophageal cancers [13-15]. The present study analyzed 86 cases of esophageal squamous cell carcinoma (ESCC) by tissue microarray technology to further investigate whether ULK1 can serve as a biomarker for the diagnosis and prognosis of ESCC.
Materials and methods
Patients
A total of 86 patients with histologically confirmed ESCC were recruited. Immediately after collection, all samples were fixed in 10% neutral formaldehyde solution, conventionally dehydrated, paraffin embedded, and then stored for later use. ESCC patients had no history of cancer treatment before tumor removal. All patients gave written informed consent before enrolling in the study, and the study was approved by the Ethical Committees of Taizhou People’s Hospital and National Engineering Center for Biochip at Shanghai.
Tissue microarray (TMA)
The TMA contained 86 pairs of ESCC and adjacent normal tissues. Details on sample preparation procedure have been described previously [16,17]. The samples were sliced and stained with hematoxylin and eosin for each donor wax block. A total of 172 sits of ESCC and adjacent tissues were selected under a microscope. A tissue microarray instrument was used to obtain a tissue core with a 1.5 mm diameter from the donor wax block. This tissue core was inserted into an acceptor wax block with 172 lattices and subsequently cut into 4 μm-thick sections. Details on the operation of the tissue microarray instrument have been described in previous studies [16,17]. Each spot of the TMA was further reviewed by pathologist.
Immunohistochemistry (IHC)
The TMA was baked at 63°C for 1 h to dewax and hydrate conventionally. The EDTA heat repair method was performed for 20 min for antigen retrieval. After boiling, the slide was cooled by water for 30 min and then placed in an endogenous peroxidase blocker for 15 min. The slide was removed from the blocker and then washed thrice (1 min each) with phosphate-buffered saline (PBS). Antibody 1 was added and diluted at 1:30 using antibody dilution solution (Dako, CA, USA). It was then stored overnight in a refrigerator at 4°C. The humidor was removed from the refrigerator and placed in a room. Excess Antibody 1 was removed using paper after the humidor reached room temperature. The slide was washed thrice with PBS, immersed in the EnVision™+/HRP rabbit working solution from Dako, incubated for 30 min, and then washed thrice with PBS. The slide was stained with diaminobenzidine and distilled water was used to remove the color. After re-staining with hematoxylin, washing with water, and differentiating, the slide was completely washed with water to restore its color to blue. Each chip was conventionally dehydrated until it became transparent and then sealed with a neutral gum. PBS was used as negative control instead of Antibody 1, and self-control was set for each slide.
Tissue samples with rich cells and easily observable spots were considered valid, whereas those with folded slices, coloring failure, and 14spots without cells were considered invalid or insignificant. Invalid or insignificant spots were removed from the samples. Staining intensity was scored as follows: 0, negative; 1, weak; 2, moderate, 2; 3, strong straining. The percentage of ULK1 positive cells was scored as follows: 0, no staining cell; 1, < 20%; 2, 20% to 75%; 3, ≥ 75%. The total histological scores (Staining intensity × percentage of positive cells) greater than 4 indicated high expression, whereas scores equal to or less than 4 indicated low expression.
Statistical analyses
The SPSS software (IBM, CA, USA) was used for statistical analysis. The difference in the levels of ULK1 between ESCC and adjacent tissues was analyzed using Mann-Whitney test. The χ2 test was performed to analyze the relationship between ULK1 expression and clinical features. The Kaplan-Meier method and log-rank test were used to investigate the relationship between ULK1 expression and survival rate. Cox regression model was used to examine whether ULK1 and clinical features can serve as biomarkers of ESCC. A P value < 0.05 was considered statistically significant.
Results
IHC analyses showed that ULK1 exhibited different expression levels in ESCC and adjacent tissues (Figure 1). ULK1 was principally expressed in the cytoplasm, but is rarely seen in the nucleus of ESCC cells. Therefore, nuclear ULK1 expression was excluded from further analysis. The expression level of cytoplasmic ULK1 was higher in ESCC tissues than in adjacent tissues (P = 0.002). Statistical analysis revealed that cytoplasmic ULK1 expression was significantly related to sex (P = 0.002) (Table 1). Furthermore, low expression of cytoplasmic ULK1 was significantly associated with lymph node metastasis (LNM) (P = 0.038). No difference was observed between other clinical features and cytoplasmic ULK1 expression.
Figure 1.
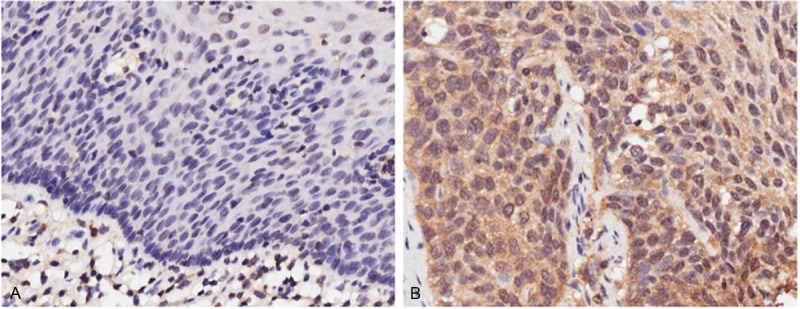
The expression level of ULK1 varies between ESCC and paracanerous tissues. Sections stained for the ULK1 from paracancerous (A) and tissue (B) tissues from ESCC patients. Scale bar = 200 μM.
Table 1.
Relationship between cytoplasmic ULK1 expression and clinicopathological characteristics of ESCC patients
| clinicopathological characteristics | ULK1 expression | P value | |
|---|---|---|---|
|
| |||
| High | Low | ||
| Age (years) | |||
| ≤ 60 | 36 | 20 | 0.635 |
| > 60 | 16 | 12 | |
| Sex | |||
| male | 33 | 30 | 0.002 |
| female | 19 | 2 | |
| Tumor size (cm) | |||
| ≤ 4 | 30 | 16 | 0.494 |
| > 4 | 20 | 16 | |
| Histologic grade | |||
| I | 7 | 5 | 0.812 |
| II | 34 | 22 | |
| III | 11 | 5 | |
| TNM stage | |||
| I+II | 36 | 16 | 0.067 |
| III+IV | 15 | 16 | |
| LNM | |||
| Yes | 16 | 18 | 0.038 |
| No | 35 | 14 | |
Median survival time (MST) of patients was 30 months. During the follow-up period of 61 months, 27 (51.9%) patients with high expression of cytoplasmic ULK1 and 26 (81.3%) patients with low expression of cytoplasmic ULK1 died from the disease. The MST of patients with high expression of cytoplasmic ULK1 was 33 months, and that of patients with low expression of cytoplasmic ULK1 was 22 months (log-rank test, P = 0.036) (Figure 2). Univariate Cox analysis showed that TNM stage III+IV [hazard ratio (HR) = 1.732, 95% confidence interval (CI): 1.312-2.262, P < 0.001], LNM (HR = 2.314, 95% CI: 1.354-3.955, P = 0.002), and low expression of cytoplasmic ULK1 (HR = 1.754, 95% CI: 1.022-3.010, P = 0.041) were associated with poor survival in ESCC patients (Table 2). Multivariate Cox analysis showed that TNM stage is the only independent factor for overall survival (HR = 2.197, 95% CI: 1.138-4.243, P = 0.019).
Figure 2.
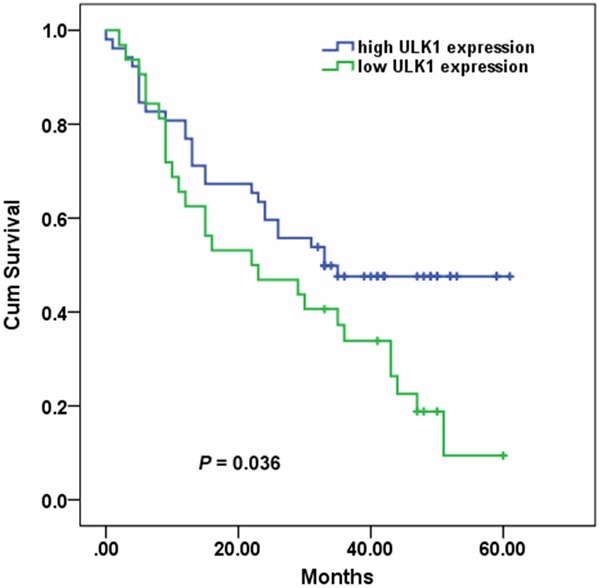
Comparison of overall survival according to cytoplasmic ULK1 expression. Patients with low ULK1 expression had a shorter overall survival (P = 0.036).
Table 2.
Univariate and multivariate Cox regression analysis of overall survival
| Features | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
|
|
|
|||
| HR (95% CI) | P value | HR (95% CI) | P value | |
| age (years), > 60 vs ≤ 60 | 0.747 (0.424-1.315) | 0.312 | ||
| sex, male v female | 2.188 (1.068-4.481) | 0.032 | ||
| tumor size (cm), > 4 vs ≤ 4 | 0.662 (0.384-1.143) | 0.139 | ||
| tumor differentiation, moderate, poor vs well | 1.007 (0.645-1.571) | 0.977 | ||
| TNM stage, III+IV vs I+II | 1.732 (1.312-2.262) | < 0.001 | 2.197 (1.138-4.243) | 0.019 |
| LNM, yes vs no | 2.314 (1.354-3.955) | 0.002 | 0.585 (0.157-2.177) | 0.424 |
| ULK1, low vs high | 1.754 (1.022-3.010) | 0.041 | 1.334 (0.757-2.353) | 0.319 |
Discussion
Identifying novel biomarkers and verifying their applications in the diagnosis and prognosis of cancers are important for individualized treatment of cancer patients. In this study, we found that the expression of ULK1 in ESCC tissues were significantly increased. Cytoplasmic ULK1 expression was associated with LNM and survival of ESCC patients. Thus, ULK1 might hold potential as a biomarker for ESCC patients.
Autophagy in mammalian cells can be divided into 3 categories: macroautophagy, microautophagy, and chaperone-mediated autophagy. In macroautophagy, the membrane from the endoplasmic reticulum surrounds parts of the cytoplasm to form autophagysome and then to integrate with the lysosome to degrade its contents. In microautophagy, the lysosomal membrane directly covers long-lived proteins and degrades them. In chaperone-mediated autophagy, protein to be degraded becomes physically associated with chaperones and is transferred into the lysosome, where it is enzymatically digested. As a robust degradation mechanism, macroautophagy can degrade and eliminate proteins in the cytoplasm, including whole organelles. Therefore, macroautophagy is very important for the inhibition of tumors [18,19]. However, growing evidence have demonstrated that autophagy is a double-edged sword in the carcinogenesis of various cancers [20,21]. Autophagy can act both as agonist and inhibitor of tumor formation in different types, stages and microenvironments of cancer [20,21].
As the only kinase of the core autophagy machinery for macroautophagy [22], ULK1 is a key factor influencing the occurrence and development of ESCC. Clinical and anatomical evidence indicate that blood supply to the esophagus is not as rich as those to other organs, such as the liver. Therefore, starvation response is larger when cancerous growth occurs in the esophagus, which in turn causes stronger activation of autophagy. This may explain the elevated expression of ULK1 observed in ESCC. Previous studies revealed that high ULK1 expression is related to poor prognosis of patients with hepatocellular carcinoma [15], ESCC [13], and breast cancer [14]. However, in the present study, we found a different effect of ULK1 expression on the prognosis of ESCC patients. This difference may be explained by the different TNM stages of ESCC patients in two studies. The majority of ESCC patients were at TNM stages I (33.3%) and II (33.1%) in previous study [13], whereas those in our study were at TNM stages II (52.9%) and III (35.3%). These findings demonstrate that ULK1 may play different roles at different TNM stages of ESCC, indicating that autophagy has dual function in carcinogenesis and cancer development. In addition to its role in self-conservation and self-protection in normal cells, autophagy also appears to restrict tumor development by selectively destroying tumor cells. However, detailed understanding of the effect of ULK1 on ESCC will require further studies on this topic.
In summary, low expression of cytoplasmic ULK1 correlated with LNM and poor survival in ESCC patients. ULK1 expression might be used to predict clinical outcomes in ESCC patients. Validation of these results in a larger sample size will open doors to its use as an ESCC biomarker in clinical settings.
Acknowledgements
This work was supported by the 12th Five-Year Plan Key Project of Science and Technology, China (grant No. 2013ZX10002007), the Shanghai Committee of Science and Technology, China (grant No. 13440701500), and the Jiangsu Province Science and Technology Support Program, China (grant No. BE2012729).
Disclosure of conflict of interest
None.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Zeng HM, Zheng RS, Zhang SW, Zou XN, Li N, Dai Z, Chen WQ. Analysis and prediction of esophageal cancer incidence trend in China. Zhonghua Yu Fang Yi Xue Za Zhi. 2012;46:593–597. [PubMed] [Google Scholar]
- 3.Marino G, Lopez-Otin C. Autophagy: molecular mechanisms, physiological functions and relevance in human pathology. Cell Mol Life Sci. 2004;61:1439–1454. doi: 10.1007/s00018-004-4012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin SK, White E. Tumor suppression by autophagy through the management of metabolic stress. Autophagy. 2008;4:563–566. [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki K, Ohsumi Y. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 2007;581:2156–2161. doi: 10.1016/j.febslet.2007.01.096. [DOI] [PubMed] [Google Scholar]
- 6.Nazarko VY, Zhong Q. ULK1 targets Beclin-1 in autophagy. Nat Cell Biol. 2013;15:727–728. doi: 10.1038/ncb2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong PM, Puente C, Ganley IG, Jiang X. The ULK1 complex: sensing nutrient signals for autophagy activation. Autophagy. 2013;9:124–137. doi: 10.4161/auto.23323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan J, Kuroyanagi H, Tomemori T, Okazaki N, Asato K, Matsuda Y, Suzuki Y, Ohshima Y, Mitani S, Masuho Y, Shirasawa T, Muramatsu M. Mouse ULK2, a novel member of the UNC-51-like protein kinases: unique features of functional domains. Oncogene. 1999;18:5850–5859. doi: 10.1038/sj.onc.1202988. [DOI] [PubMed] [Google Scholar]
- 10.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang S, Li Y, Zhu YH, Wu XQ, Tang J, Li Z, Feng GK, Deng R, Li DD, Luo RZ, Zhang MF, Qin W, Wang X, Jia WH, Zhu XF. Intensive expression of UNC-51-like kinase 1 is a novel biomarker of poor prognosis in patients with esophageal squamous cell carcinoma. Cancer Sci. 2011;102:1568–1575. doi: 10.1111/j.1349-7006.2011.01964.x. [DOI] [PubMed] [Google Scholar]
- 14.Pike LR, Singleton DC, Buffa F, Abramczyk O, Phadwal K, Li JL, Simon AK, Murray JT, Harris AL. Transcriptional up-regulation of ULK1 by ATF4 contributes to cancer cell survival. Biochem J. 2013;449:389–400. doi: 10.1042/BJ20120972. [DOI] [PubMed] [Google Scholar]
- 15.Xu H, Yu H, Zhang X, Shen X, Zhang K, Sheng H, Dai S, Gao H. UNC51-like kinase 1 as a potential prognostic biomarker for hepatocellular carcinoma. Int J Clin Exp Pathol. 2013;6:711–717. [PMC free article] [PubMed] [Google Scholar]
- 16.He C, Jiang H, Geng S, Sheng H, Shen X, Zhang X, Zhu S, Chen X, Yang C, Gao H. Expression of c-Myc and Fas correlates with perineural invasion of pancreatic cancer. Int J Clin Exp Pathol. 2012;5:339–346. [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, He C, He C, Chen B, Liu Y, Kong M, Wang C, Lin L, Dong Y, Sheng H. Nuclear PKM2 expression predicts poor prognosis in patients with esophageal squamous cell carcinoma. Pathol Res Pract. 2013;209:510–515. doi: 10.1016/j.prp.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 18.Xie ZP, Klionsky DJ. Autophagosome formation: Core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 19.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 20.Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Apel A, Zentgraf H, Buchler MW, Herr I. Autophagy-A double-edged sword in oncology. Int J Cancer. 2009;125:991–995. doi: 10.1002/ijc.24500. [DOI] [PubMed] [Google Scholar]
- 22.Dorsey FC, Rose KL, Coenen S, Prater SM, Cavett V, Cleveland JL, Caldwell-Busby J. Mapping the Phosphorylation Sites of Ulk1. J Proteome Res. 2009;8:5253–5263. doi: 10.1021/pr900583m. [DOI] [PubMed] [Google Scholar]


