Abstract
Rabies post-exposure prophylaxis (PEP) currently comprises administration of rabies vaccine together with rabies immunoglobulin (RIG) of either equine or human origin. In the developing world, RIG preparations are expensive, often in short supply, and of variable efficacy. Therefore, we are seeking to develop a monoclonal antibody cocktail to replace RIG. Here, we describe the cloning, engineering and production in plants of a candidate monoclonal antibody (E559) for inclusion in such a cocktail. The murine constant domains of E559 were replaced with human IgG1κ constant domains and the resulting chimeric mouse-human genes were cloned into plant expression vectors for stable nuclear transformation of Nicotiana tabacum. The plant-expressed, chimeric antibody was purified and biochemically characterized, was demonstrated to neutralize rabies virus in a fluorescent antibody virus neutralization assay, and conferred protection in a hamster challenge model.
Keywords: rabies, post-exposure prophylaxis, RIG, monoclonal antibody, Nicotiana tabacum
Rabies is a zoonotic disease caused by rabies virus (RABV), the type member of the Lyssavirus genus, and is responsible for >55 000 deaths per annum [1] largely in the developing world [2–4], where transmission usually occurs following the bite of an infected dog. If left untreated, the virus progressively infects surrounding neurons and propagates in the central nervous system leading, almost invariably, to death. The disease can be prevented by post-exposure prophylaxis (PEP), which consists of administration of inactivated RABV vaccine together with passive antibody therapy [5–7]. In passive antibody therapy, rabies immunoglobulin (RIG), derived either from immunized human (HRIG) or equine (ERIG) sources [8–11], is infiltrated into the wound site.
However, in the developing world, these serum-derived antibodies often suffer from drawbacks including limited availability, batch-to-batch variation, high cost, contamination with blood-borne adventitious agents, and/or risk of adverse reactions [12]; for these reasons, the World Health Organization (WHO) encourages the development and evaluation of alternative biologics for RIG replacement [13]. One such alternative is offered by monoclonal antibodies (mAbs) that are capable of neutralizing a wide range of RABV isolates [12, 14–18]. Rabies neutralizing antibodies are directed against the viral glycoprotein, and several studies have demonstrated that rabies-specific mAbs can protect rodents after RABV challenge [18–23].
However, given the unique epitope specificity of individual mAbs compared to polyclonal antiserum, any mAb-based product designed to replace RIG would ideally comprise a defined cocktail of RABV-neutralizing mAbs that would provide coverage against a broad range of RABV isolates, minimize the potential for viral escape and have a potency comparable to that of RIG. The low production costs, ability of plants to assemble and modify multimeric proteins such as mAbs, and ease of scalability make plants a viable platform for production of mAbs to replace RIG [24, 25].
Several groups have characterized RABV-neutralizing mAbs [14, 17, 25–30], and the World Health Organization Rabies Collaborating Centers (WHO RCCs) identified 5 murine mAbs [15], with 4 (E559.9.14, M727-5-1, M777-16-3 and 1112-1) recognizing antigenic site II of the glycoprotein and 1 (62-71-3) recognizing antigenic site I [31].
Amongst the mAbs identified by the WHO RCCs that recognize antigenic site II, E559 exhibited the broadest virus neutralization spectrum and greatest potency [15, 32] and therefore represents an important candidate mAb for inclusion in a RIG-replacement cocktail. In this study, we describe the cloning and sequences of the murine E559 antibody heavy and light chains, engineering of a chimeric mouse-human version of E559, expression in tobacco, and characterization of the purified, tobacco-derived, chimeric mAb in terms of in vitro virus neutralization and in vivo protection.
MATERIALS AND METHODS
Cell Lines, Viruses and Plasmids
Hybridoma cell line E559.9.14 [15, 32], expressing murine IgG1κ mAb E559, was kindly provided by Dr Thomas Müller (WHO Collaborating Centre for Rabies Surveillance and Research, Friedrich-Loeffler-Institute, Germany). Cells were cultured at 37°C, under a 5% CO2 atmosphere in CD hybridoma medium (Life Technologies) supplemented with 10% (v/v) heat-inactivated, fetal bovine serum (Life Technologies) and 2 mM L-glutamine (Sigma, UK). For mAb production, the cells were adapted to serum-free conditions.
Lyssavirus strains used included challenge virus standard (CVS) [ATCC VR-959], derived from the original Pasteur virus [33] and animal-derived isolates, as well as RV61, isolated from a person bitten by a dog.
The pL32 and pTRAk.2 plasmids used for plant transformation are described in detail in the online Supplementary Materials.
Agrobacterium tumefaciens strain LBA4404 was purchased from Invitrogen UK. A. tumefaciens strain GV3101::pMP90RK was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (Leibniz Institute, Germany).
Cloning of Full-length Murine E559 IgG
Total RNA from hybridoma cell line E559.9.14 was isolated from 1 × 106 cells using the RNeasy Mini kit (Qiagen). First strand complementary DNA (cDNA) was prepared using the Omniscript RT kit (Qiagen) with oligo-(dT)15 as the primer.
Using the first strand cDNA as template, the murine γ1 heavy chain gene was amplified using primers FR1γ and 932 (see online Supplementary Table 1 for a description of oligonucleotide primers). The murine κ light chain gene was amplified using primers FR1κ and 933. The murine γ1 heavy chain and κ light chain amplicons were digested with XhoI and EcoRI and ligated into binary vector pL32 restricted with the same enzymes.
Cloning of Chimeric Mouse-human E559 IgG
The cloning of the chimeric (mouse-human) heavy (χE559H) and light (χE559L) chain genes, and the codon-optimised versions of these genes, is described in detail in the online Supplementary Materials.
Generation and Screening of Transgenic Nicotiana tabacum Plants
The generation of transgenic plants is described in the online Supplementary Materials. For screening of plants by Western blotting and enzyme-linked immunosorbent assay (ELISA), leaf discs were excised from leaves using the lid of a 1.5 mL Eppendorf tube as a punch. Leaf discs were homogenized using a plastic pestle in 300 µL of PBS, centrifuged at 20 000 × g for 3 minutes, and the supernatant collected for analysis. Total soluble protein content of the supernatant was measured using the bicinchoninic acid (BCA) protein assay kit (Pierce, UK).
Purification of mAbs
For purification of the hybridoma-derived mAb (E559Hyb), hybridoma E559.9.14 cells were grown for 7 days in serum-free conditions, centrifuged (1000 × g, 10 minutes, 4°C) to pellet the cells, and the supernatant filtered (0.2 µm) and applied to an anti-mouse IgG1 (heavy chain specific)-agarose (Sigma, UK) affinity column.
The plant-expressed chimeric antibody (χE559P) was purified using Protein A/G agarose as described elsewhere [34]. In the case of the plant-expressed murine E559 (muE559P), an anti-mouse IgG1 (heavy chain specific)-agarose (Sigma, UK) affinity column was used instead.
Column fractions were analyzed on Coomassie stained SDS-PAGE gels. Fractions containing the antibody were pooled, dialyzed against phosphate-buffered saline (PBS), and stored in aliquots at −20°C. Dialyzed material was analyzed by ELISA and SDS-PAGE to determine the concentration, purity, and integrity of the mAb.
Samples destined for animal challenge studies were purified using MabSelect SuRe protein A chromatography on a 5 mL HiTrap column (GE Healthcare). In addition to affinity purification, samples were further purified using Capto Q (GE Healthcare) in flow through mode and polished using ceramic hydroxyapatite (CHT; BioRad Laboratories). All chromatography steps were conducted on an Akta Avant 150 operated via Unicorn 6.0 software.
Antibody concentrations were determined using a sandwich ELISA, by capturing samples with a heavy-chain specific reagent and detection with a light chain specific reagent. Commercially available human IgG1κ (The Binding Site, UK) and mouse IgG1κ (Sigma, UK) were used as concentration standards.
Deglycosylation Using PNGaseF
The deglycosylation protocol using PNGaseF is described in detail in the online Supplementary Materials.
Glycan Analysis of the Plant-derived mAb E559
A glycoproteomic analysis was undertaken by in-gel digestion of S-carbamidomethylated sample and analysis by reverse-phase electrospray ionization mass spectrometry (RP-ESI-MS), as described elsewhere [35]. Tandem MS results were also subjected to Mascot MS/MS ion search (Matrix Science Ltd, London, UK; http://www.matrixscience.com).
Enzyme-Linked Immunosorbent Assay
ELISA for detection of antibody heavy or light chains is described in detail in the online Supplementary Materials.
SDS-PAGE and Western Blotting
Polyacrylamide gel electrophoresis (PAGE) and Western blotting protocols are described in detail in the online Supplementary Materials.
Modified Fluorescent Antibody Virus Neutralization (mFAVN) Assay
Live virus experiments were performed using a modified form of the fluorescent antibody virus neutralization (FAVN) assay described for CVS-11 [36, 37] and described in more detail in the online Supplementary Materials. OIE positive (OIE+) and OIE negative (OIE−) reference sera were included as controls. Virus was considered neutralized if the neutralization titer was >0.5 IU/mL [36].
Hamster Challenge Studies
Four groups of Syrian hamsters were included in the experiment. The challenge and treatment schedule was as follows: Group 1 (uninfected control) comprised 4 hamsters that did not receive any viral inoculum or biologics treatment. Group 2 (4 animals) and groups 3 and 4 (each comprising 9 animals) were all inoculated with 50 µL of 1 × 106 TCID50/mL of a RABV laboratory strain, Challenge Virus Standard CVS (at day 0) intramuscularly and treated subsequently (at day 1) with either PBS (group 2), or with 22.5 IU/kg of either undiluted commercial HRIG (Rabigam [150 IU/mL], National Bioproducts Institute, Pinetown, South Africa) (group 3) or χE559P mAb (group 4). Biologics (groups 3 and 4) and PBS (group 2) were administered in the gastrocnemius muscle in 50 µL volumes to simulate passive immunization in PEP treatment. No rabies vaccine was administered. The hamsters were observed twice daily over 28 days for any symptoms associated with RABV infection. Brain tissues were collected from animals to confirm rabies virus infection for all those hamsters that succumbed during the observation period and assessed for the presence of lyssavirus antigen using the fluorescent antibody test (FAT) [38]. All hamsters surviving for up to 28 days post-infection were killed with isoflurane and tested for rabies as described above. The animal experimental protocols, animal caging and care, as well as end point for the experiments were approved by the Animal Ethics Committee for the use of living vertebrates for research, diagnostic procedures, and product development (Agricultural Research Council-Onderstepoort Veterinary Institute, South Africa).
RESULTS
Cloning of Antibody Heavy and Light Chain Genes From Hybridoma E559.9.14
The murine immunoglobulin γ1 heavy and κ light chain genes expressed by the E559.9.14 hybridoma were amplified by polymerase chain reaction, using first strand cDNA as template. The deduced amino acid sequences of the E559 heavy and light chain genes are presented in Figure 1. Highlighted are important features, such as the complementarity determining regions [39] and the presence of potential N-linked glycosylation sites within the CH2 domain and the light chain VL domain.
Figure 1.
Sequences and mass spectrometry analysis of E559. Deduced amino acid sequences of the heavy chain variable domain (VH), the heavy chain constant region domains (CH1, Hinge, CH2, and CH3), the light chain variable domain (VL), and the light chain constant domain (CL) of E559. Complementarity determining regions (CDRs), as defined by Kabat et al [39], are highlighted in bold and underlined. Amino acids encoded by the primers used for amplification are shown in bold italics. Potential N-linked glycosylation sites are double-underlined. Peptides identified by mass spectrometry analysis of the 25 kDa and 27 kDa isoforms of the E559Hyb light chain are shown aligned below the corresponding VL and CL sequences (see text).
Analysis of Hybridoma-derived E559
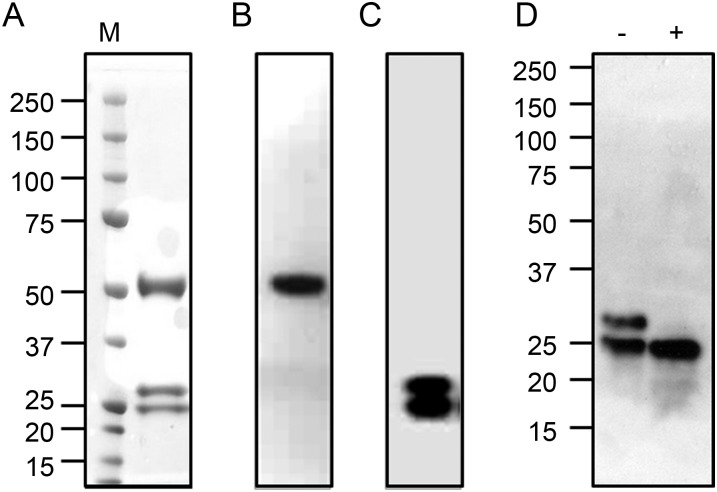
Analysis of the purified murine hybridoma-derived E559 (E559Hyb) by SDS-PAGE under reducing conditions, followed by Coomassie staining, showed the presence of 3 bands with molecular weights of 50 kDa, 27 kDa and 25 kDa (Figure 2A). Western blotting confirmed previous findings [15] that the 50 kDa band corresponded to the heavy chain (Figure 2B), and that the 2 lower molecular weight bands were murine light chains (Figure 2C). The 2 lower molecular weight bands were excised from the gel, treated with trypsin, and analyzed by LC-MS. The panel of peptides generated from each band were nearly identical and in accord with the sequence deduced from the cloned light chain gene (see Figure 1), indicating that these 2 bands are murine κ light chain isoforms. Minor differences in the identified peptides are likely due to differences in the extent of trypsin digestion between the 2 samples. The identification of a potential N-glycosylation site within the VL domain of the light chain (Figure 1) suggested that the difference between the light chain isoforms might be due to the presence of N-linked glycans. E559Hyb was deglycosylated by treatment with PNGaseF. Blotting under reducing conditions shows that after treatment with PNGaseF, the 27 kDa band is lost, leaving only a single band at 25 kDa (Figure 2D), providing evidence that the 27 kDa species is a glycosylated form of the light chain and the 25 kDa band is the aglycosylated species.
Figure 2.
Analysis of hybridoma-derived E559. Hybridoma-derived E559 (E559Hyb) was purified by affinity chromatography and analyzed by SDS-PAGE under reducing conditions, followed by staining with Coomassie Brilliant Blue (A) or blotted to nitrocellulose and probed with HRP-labeled antisera specific for murine γ1 heavy chains (B) or murine κ light chains (C). Purified E559Hyb was also treated with PNGaseF, and proteins were separated by SDS-PAGE under reducing conditions, blotted to nitrocellulose and probed with HRP-labeled light chain-specific antiserum (D). Lane M: molecular weight standards; (−): untreated E559Hyb; (+): PNGaseF-treated E559Hyb. Abbreviation: HRP, horseradish peroxidase.
Characterization of Plant-derived E559
Murine and chimeric (mouse-human) heavy and light chain genes were cloned into the binary vector pL32 and transformed into Agrobacterium tumefaciens. Co-cultivation of Nicotiana tabacum leaf discs with A. tumefaciens strains harboring the recombinant pL32 binary vectors was used to generate transgenic tobacco lines expressing murine heavy (pL32-muE559H), chimeric heavy (pL32-χE559H), murine light (pL32-muE559L), or chimeric light (pL32-χE559L) chains. Several independent plants lines derived from each transformation were screened by ELISA to identify transgenic plants expressing each antibody chain.
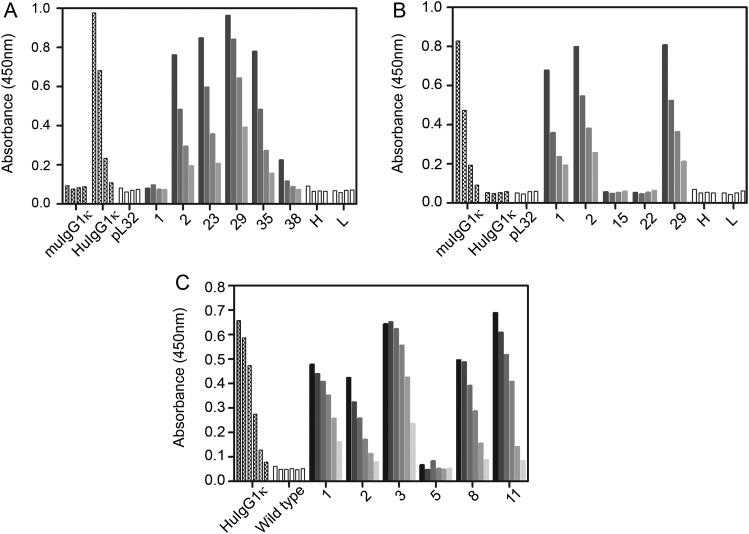
Sexual crossing was used to produce plants lines expressing the fully assembled chimeric E559 (pL32-χE559) or fully assembled murine E559 (pL32-muE559). Plants were analyzed by ELISA for antibody assembly and expression levels. The results from a selected set of plants provide evidence that both the chimeric (Figure 3A) and murine (Figure 3B) antibodies are assembled. Control plants expressing only the heavy chain (pL32-χE559H or pL32-muE559H) or the light chain (pL32-χE559L or pL32-muE559L) did not produce any signal above that of the nonrecombinant pL32 control. In sum, 5 of the 6 plants shown in Figure 3A expressed the chimeric antibody, whereas 3 of the 5 plants shown in Figure 3B expressed the murine antibody.
Figure 3.
ELISA analysis of transgenic plants expressing fully assembled E559 monoclonal antibodies. Leaf discs from selected independent plant lines expressing either (A) chimeric E559 (pL32-χE559), (B) murine E559 (pL32-muE559), or (C) codon-optimised, chimeric E559 (pTRAk-χE559), were extracted in PBS and loaded onto ELISA plates coated with either sheep anti-human IgG1 (panels A and C) or sheep anti-mouse IgG1 (panel B) antisera. Bound antibodies were detected with HRP-labeled antibodies specific for either human κ light chains (A and C) or murine κ light chains (B). Control samples were: isotype-matched, commercially available human IgG1κ (HuIgG1κ) or mouse IgG1κ (muIgG1κ) antibodies; samples from transgenic plant lines expressing only the heavy or light chains of the chimeric E559 (H or L, respectively in panel A); samples from transgenic plant lines expressing only the heavy or light chains of the murine E559 (H or L, respectively, in panel B); a plant line transformed with nonrecombinant binary vector (pL32); and a wild-type (nontransgenic) plant. For panels A and B, plant samples were serially diluted 2-fold, whereas the isotype-matched controls were serially diluted 5-fold. In panel C, all samples and controls were serially diluted 4-fold. Abbreviations: ELISA, enzyme-linked immunosorbent assay; PBS, phosphate-buffered saline.
Using a commercially available human IgG1κ as an ELISA standard, the best expression level of the chimeric E559 (χE559P) was calculated as 1.8 mg/kg of fresh leaf weight (0.04% of total soluble protein), whereas the best yield achieved from the plant-derived, murine E559 (muE559P) was 1.2 mg/kg of fresh leaf weight (0.03% of total soluble protein).
As an alternative expression strategy, codon-optimised versions of the chimeric E559 heavy and light chain genes were cloned into expression cassettes arranged in tandem (head-to-tail orientation) in plant transformation vector pTRAk.2. Co-cultivation of N. tabacum leaf discs with an A. tumefaciens strain harboring the recombinant pTRAk.2 was used to generate transgenic tobacco lines, pTRAk-χE559, which were analyzed by ELISA for antibody assembly and yield (Figure 3C). The best yield of plant-derived chimeric E559 (χE559P) was determined to be 280 mg/kg of fresh leaf weight, approximately 150-fold greater than the nonoptimized, chimeric antibody expressed using the pL32 vector.
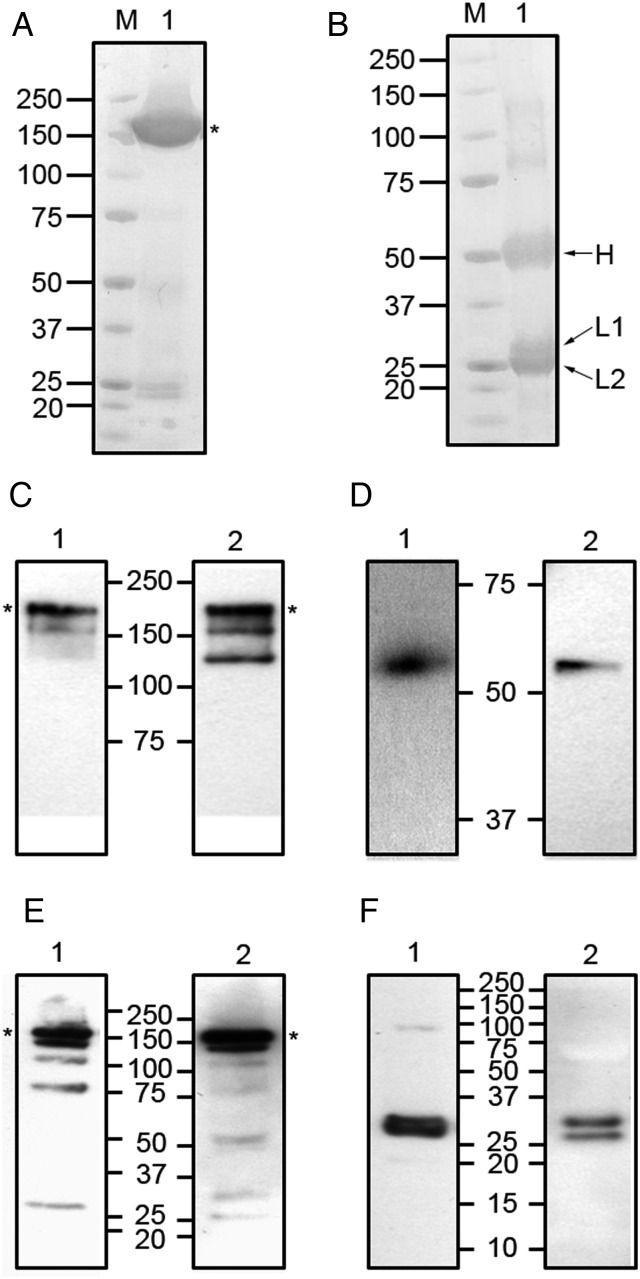
The purified χE559P was analyzed by Coomassie staining under nonreducing and reducing conditions. A nonreducing gel (Figure 4A) showed a predominant high molecular weight band (indicated by the asterisk) at the expected size for the fully assembled antibody and, despite the presence of some minor low molecular weight bands, indicates a high degree of purity was achieved using the single-step (protein A/G) purification. The reducing gel (Figure 4B) shows the heavy chain (indicated by H) migrating at the expected position. As previously observed for the hybridoma-derived E559 (Figure 2A), the plant-derived χE559P also comprises 2 isoforms of the light chain (indicated by L1 and L2). Additional higher molecular weight species in the reducing gel most likely represent incompletely reduced antibody.
Figure 4.
Gel and Western blotting analysis of purified χE559P. Purified, plant-derived chimeric E559 (χE559P) was analyzed by Coomassie staining of polyacrylamide gels under nonreducing (panel A) and reducing (panel B) conditions, and by Western blotting under nonreducing (panels C and E) and reducing (panels D and F) conditions. For Western blotting, the nitrocellulose membranes were probed with HRP-conjugated antibodies specific for heavy chains (panels C and D), or with HRP-conjugated antibodies specific for light chains (panels E and F). The χE559P samples (lane 1) were probed with human-specific reagents, whereas E559Hyb samples (lane 2) were probed with murine-specific reagents. Abbreviations: HRP, horseradish peroxidase; M, molecular weight standards. Asterisks indicate the positions of the fully assembled antibodies.
The purified χE559P was also analyzed by Western blotting, alongside purified E559Hyb. Figure 4C shows the results of a nonreducing blot, detected with antisera specific for human (lane 1) or mouse (lane 2) heavy chains. Both χE559P and E559Hyb samples have a high molecular weight band migrating at the expected position for the fully assembled antibody (indicated by the asterisk), with some additional lower molecular weight bands, representing either assembly intermediates or proteolytic fragments [34]. Under reducing conditions (Figure 4D), both samples showed a single band at approximately 55 kDa, corresponding to the expected size for free heavy chains.
Blotting of the light chains under nonreducing conditions (Figure 4E) revealed the fully assembled antibody for both χE559P (lane 1) and E559Hyb (lane 2) as well as some additional lower molecular weight species. Under reducing conditions (Figure 4F), E559Hyb (lane 2) showed 2 bands, corresponding to the 2 glycoform variants, and χE559P (lane 1) also showed 2 light chain species. The lower band in χE559P corresponded in size to the lower band in E559Hyb, indicating that this is also an aglycosylated form of the light chain. The higher band in χE559P had a slightly faster mobility compared to the glycosylated isoform of E559Hyb, and this probably reflects differences in the N-linked glycan structures between plants and mammals.
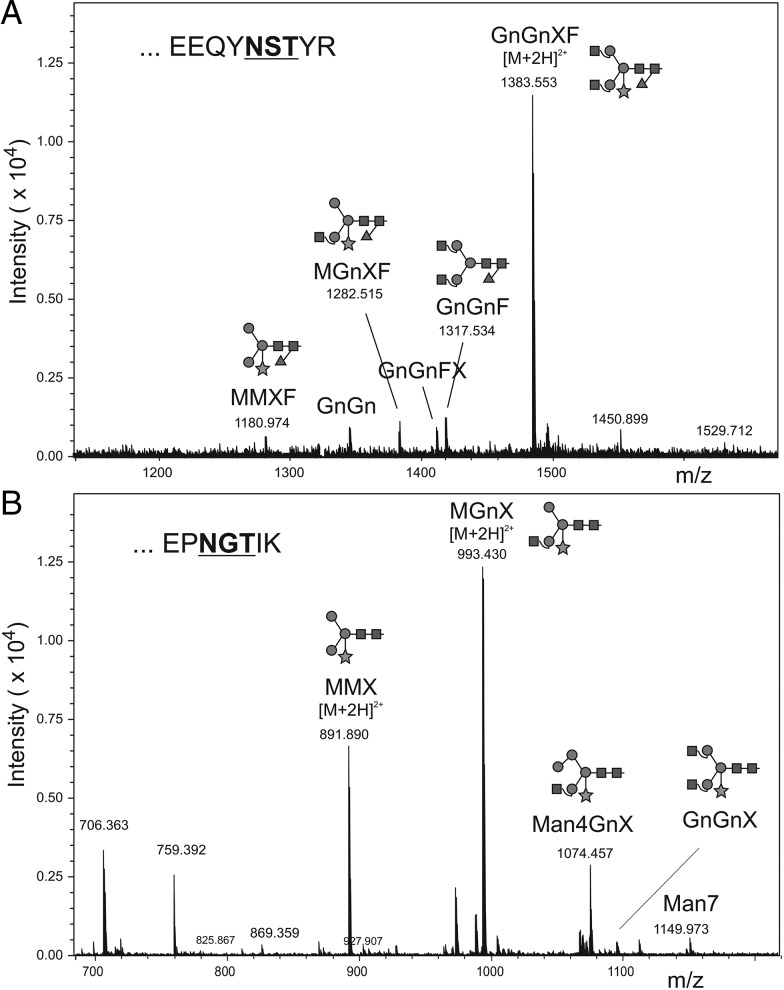
Glycoproteomic Analysis
Sequence analysis of heavy and light chains of mAb E559 predicted the presence of 2 potential N-linked glycosylation sites, a conserved site in the antibody Fc region, and one in the VL domain. The plant-derived antibody was subjected to glycoproteomic analysis by RP-ESI-MS (Figure 5). Glycopeptides comprising the Fc glycosylation site EEQFNSTFR (Figure 5A) and the VL glycosylation site EPNGTIK (Figure 5B) were identified (N-linked glycosylation sites are underlined). The glycan analysis revealed that χE559P heavy chain displayed glycan compositions typical of plant glycoproteins, with predominantly complex type glycans containing xylose and fucose (GnGnXF), which are presumed to be the β1,2-linked xylose residues attached to the β-linked mannose and the α1,3-fucose residue linked to the Asn-linked N-acetyl-glucosamine. The light chain glycosylation pattern (MGnX and MMX) was also largely typical of plant glycoproteins, except for the lack of the α1,3-fucose residue linked to the Asn-linked N-acetyl-glucosamine. Tandem MS results were subjected to Mascot MS/MS ion search, which confirmed the sample to contain essentially mAb E559.
Figure 5.
Glycan analysis of χE559P. Purified, plant-derived chimeric E559 was analyzed by in-gel digestion of S-carbamidomethylated sample and RP-ESI-MS. Deconvoluted spectra of the glycopeptide elution region of the Fc glycopeptide (A) and light chain glycopeptide (B). Masses correspond to oligomannosidic and complex-type structures. Abbreviations: F, fucose; Gn, N-acetylglucosamine; M, mannose; RP-ESI-MS, reverse-phase electrospray ionization mass spectrometry; X, xylose.
In vitro Neutralization
The hybridoma-derived E559 (E559Hyb) and both plant-derived antibodies (muE559P and χE559P) were tested for neutralization of a diverse panel of lyssavirus species and strains using the mFAVN assay. The results (Table 1) show that both plant-derived antibodies mirrored the hybridoma-derived antibody in terms of breadth of neutralization. Representative viruses from phylogroups I and II [5, 40] were assayed for their ability to be neutralized by the antibodies. All tested phylogroup I viruses, covering the type species member (classical RABV), Duvenhage virus, European bat lyssavirus types 1 and 2, and Australian bat lyssavirus, were neutralized by all 3 antibodies and, except for Duvenhage virus, also by the OIE+ control. No neutralization was observed for the phylogroup II viruses tested (Lagos bat virus and Mokola virus).
Table 1.
Virus Neutralizing Activity of Plant-derived Antibodies
| Phylogroup | Lyssavirus Species (Genotype) | Virus Reference No. | Animal of Origin | Country of Origin | OIE+ | muE559P | χE559P | E559Hyb |
|---|---|---|---|---|---|---|---|---|
| I | RABV (1) | CVS | Standard stock | n/a | + | + | + | + |
| RV51 | Fox | USA | + | + | + | + | ||
| RV61 | Human ex dog | UK (ex India) | + | + | + | + | ||
| RV108 | Bat | Chile | + | + | + | + | ||
| RV410 | Mongoose | South Africa | + | + | + | + | ||
| RV437 | Raccoon | Estonia | + | + | + | + | ||
| RV1237 | Deer | Yugoslavia | + | + | + | + | ||
| II | LBV (2) | RV1 | Bat (E. helvum) | Nigeria | − | − | − | − |
| MOK (3) | RV4 | Shrew (Crocidura spp.) | Nigeria | − | − | − | − | |
| I | DUVV (4) | RV131 | Bat (N. thebaica) | Zimbabwe | − | + | + | + |
| EBLV1 (5) | RV9 | Bat (E. serotinus) | Germany | + | + | + | + | |
| EBLV2 (6) | RV1781 | Bat (M. daubentonii) | UK | + | + | + | + | |
| ABLV (7) | RV634 | Fruit bat | Australia | + | + | + | + |
A modified fluorescent antibody virus neutralization (mFAVN) assay was used to compare the virus neutralizing activity of plant-derived chimeric E559 (χE559P), plant-derived murine E559 (muE559P), hybridoma-derived murine E559 (E559Hyb) and pooled dog reference sera from immunized animals (OIE+) against different lyssaviruses. Virus abbreviations: ABLV, Australian bat lyssavirus; CVS, challenge virus standard; DUVV, Duvenhage virus; EBLV1, European bat lyssavirus type 1; EBLV2, European bat lyssavirus type 2; LBV, Lagos bat virus; MOK, Mokola virus. Virus was considered neutralized if the neutralization titer was >0.5 IU/mL [36]. (+) indicates neutralization, (−) indicates no neutralization.
In vivo Challenge Studies
The efficacy of the χE559P in post-exposure prophylaxis was examined in hamsters injected with a lethal dose of a laboratory strain of RABV (CVS-11; Table 2). In this in vivo protection assay, all uninfected hamsters (group 1) survived. All hamsters that were infected with challenge virus and received mock PEP in the form of PBS (group 2) died after 14 days. The survival rates for hamsters that received PEP in the form of 22.5 IU/kg of either HRIG (group 3) or χE559P (group 4) was >50% after 14 days, although after 28 days survival dropped to zero and 11% for HRIG and χE559P groups, respectively. None of the groups received vaccine as part of the PEP regimen. The data show that the χE559P antibody is at least as effective as the HRIG.
Table 2.
In vivo Efficacy of χE559P for Postexposure Prophylaxis
| Group (Treatment) | 14 d | 28 d |
|---|---|---|
| Group 1 (Uninfected control) | 4/4 | 4/4 |
| Group 2 (PBS) | 0/4 | 0/4 |
| Group 3 (HRIG) | 5/9 | 0/9 |
| Group 4 (χE559P) | 6/9 | 1/9 |
Four groups of Syrian hamsters were included in the experiment. Group 1 (uninfected control) animals did not receive any viral inoculum or biologics treatment. Groups 2, 3 and 4 were all inoculated with a genotype 1 RABV variant (at day 0) and treated subsequently (at day 1) with either PBS (group 2), HRIG (Rabigam) (group 3), or purified χE559P mAb (group 4). Data are presented as the no. of surviving hamsters/no. of hamsters tested. Abbreviations: HRIG, human rabies immunoglobulin; PBS, phosphate-buffered saline.
DISCUSSION
Current PEP for bites by rabid animals involves the use of blood-derived RIG, which can display batch-to-batch variation and may be of limited availability in case of sudden mass exposures. The concerns arising from the use of blood-derived products could be circumvented, and consistent batches of neutralizing antibodies could be produced in large quantities by adopting an approach based on a cocktail of rabies neutralizing mAbs. To this end, it is envisaged that RIG could be replaced by a mAb cocktail, produced using plants as the expression platform. Two different mAb production platforms in plants have already gained regulatory approval for human trials (Pharma-Planta Consortium, personal communication to J. Ma; [41]), demonstrating that plants are amenable to current Good Manufacturing Practice (cGMP) compliance [42].
We compared the murine hybridoma-derived E559 (E559Hyb) with the same murine antibody produced in N. tabacum (muE559P), as well as a mouse-human chimeric version (χE559P), also expressed in N. tabacum. In vitro testing of virus neutralization demonstrated that all 3 versions of E559 were equivalent, with all 3 neutralizing phylogroup I viruses but not the phylogroup II viruses. This is in accord with previous reports showing that neutralizing antibodies targeting phylogroup I viruses are not effective at neutralizing phylogroup II viruses [40, 43].
E559 has a predicted glycosylation site in the framework region of the VL domain, which appears to be utilized, as 2 forms of the hybridoma-derived light chain (glycosylated and aglycosylated) are observed under reducing conditions, with the higher molecular weight form disappearing after treatment with PNGaseF. Two isoforms were also observed in the plant expressed χE559P. The effect of the VL glycosylation is unknown, as both glycosylated and aglycosylated forms of the light chain were present in the hybridoma and plant preparations used for assessment of antibody functionality.
Purified χE559P was analyzed by mass spectrometry and was shown to be glycosylated with typical plant complex glycan structures. It is well established that plant N-linked glycosylation differs from murine glycosylation [44], due to differences in complex glycan processing in the Golgi compartment. Previous studies have shown that plant-derived mAbs can have different half-lives in animals, compared to mammalian-derived mAbs [25, 45]. Although these differences have been attributed to differences in glycosylation, a more recent study [46] found no difference in the clearance rates of a RABV-neutralizing human mAb expressed in hybridoma cells or plants. The impact on the in vivo half-life of the glycosylation differences between E559Hyb and χE559P will need to be addressed in human trials.
Functionally, χE559P retained neutralization activity and had the same breadth of lyssavirus coverage as E559Hyb. In vivo, the chimeric antibody was as effective as a commercial HRIG product in a hamster challenge model.
The potential for viral escape, and the need to provide protection across a broad range of lyssaviruses, means that a single mAb will probably not be sufficient for a rabies PEP product, and this has been recognized by various groups [15, 17, 18, and 31]. However, the cost of mAbs produced in mammalian cell bioreactors is currently prohibitive for rabies products intended for use in resource-poor settings, so it seems unlikely that products combining 2 or more mAbs produced using such traditional platforms will be commercially viable outside the developed world. Production of RABV-neutralizing mAbs in plants raises hopes that these mAbs will be available in quantities sufficient to meet the needs for PEP in rabies-endemic areas, particularly across the developing world.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. The authors would like to thank Marie Paule Kieny (World Health Organization, Geneva, Switzerland) and Thomas Müller (WHO Collaborating Centre for Rabies Surveillance and Research, Friedrich-Loeffler-Institute, Federal Research Institute for Animal Health, Wusterhausen, Germany) for the E559 hybridoma, Rainer Fischer (Fraunhofer IME, Aachen, Germany) for providing the pTRAk vector, Polymun Scientific Immunbiologische Forschung GmbH (Nussdorfer Lände 11, 1190 Vienna, Austria) for providing the 4E10 sequences and Helen Byers (Medical Biomics Centre, St George's University of London, UK) for assistance with the mass spectrometry analysis of the hybridoma-derived E559 light chain. Thanks also to Derek Healy (AHVLA) and Jacobeth Miyen for help with the FAVN and FAT respectively. Ms Gugulethu Mkhize was instrumental in undertaking initial in vitro and in vivo experiments but has since left the ARC-OVI.
Financial support. The authors are grateful to the EU-funded Pharma-Planta, ERC Future-Pharma Advanced Grant, and COST (FAO94) projects, the Wellcome Trust, as well as the Hotung Foundation. C. V. D. was funded by the Dr Hadwen Trust and did not participate in experiments involving animals, or cells or tissues from animals or from human embryos. This work was supported by St George's, University of London, UK, and AHVLA, Weybridge, UK.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.World Health Organization. WHO Fact Sheet no. 99. http://web.archive.org/web/20130310122553/http://www.who.int/mediacentre/factsheets/fs099/en/index.html . Accessed 19 February 2014.
- 2.Hampson K, Dobson A, Kaare M, et al. Rabies exposures, post-exposure prophylaxis and deaths in a region of endemic canine rabies. PLoS Negl Trop Dis. 2008;2:e339. doi: 10.1371/journal.pntd.0000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dodet B. The fight against rabies in Africa: from recognition to action. Vaccine. 2009;27:5027–32. doi: 10.1016/j.vaccine.2009.06.030. [DOI] [PubMed] [Google Scholar]
- 4.Fooks AR. Rabies remains a ‘neglected disease. Euro Surveill. 2005;10:211–2. [PubMed] [Google Scholar]
- 5.Both L, Banyard AC, van Dolleweerd C, Horton DL, Ma JK, Fooks AR. Passive immunity in the prevention of rabies. Lancet Infect Dis. 2012;12:397–407. doi: 10.1016/S1473-3099(11)70340-1. [DOI] [PubMed] [Google Scholar]
- 6.Rupprecht CE, Hanlon CA, Hemachudha T. Rabies re-examined. Lancet Infect Dis. 2002;2:327–43. doi: 10.1016/s1473-3099(02)00287-6. [DOI] [PubMed] [Google Scholar]
- 7.Hemachudha T, Laothamatas J, Rupprecht CE. Human rabies: a disease of complex neuropathogenetic mechanisms and diagnostic challenges. Lancet Neurol. 2002;1:101–9. doi: 10.1016/s1474-4422(02)00041-8. [DOI] [PubMed] [Google Scholar]
- 8.Ertl HC. Novel vaccines to human rabies. PLoS Negl Trop Dis. 2009;3:e515. doi: 10.1371/journal.pntd.0000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rupprecht CE, Gibbons RV. Clinical practice: prophylaxis against rabies. N Engl J Med. 2004;351:2626–35. doi: 10.1056/NEJMcp042140. [DOI] [PubMed] [Google Scholar]
- 10.Warrell M. Rabies and African bat lyssavirus encephalitis and its prevention. Int J Antimicrob Agents. 2010;36(Suppl 1):S47–52. doi: 10.1016/j.ijantimicag.2010.06.021. [DOI] [PubMed] [Google Scholar]
- 11.Satpathy DM, Sahu T, Behera TR. Equine rabies immunoglobulin: a study on its clinical safety. J Indian Med Assoc. 2005;103:238. 41–2. [PubMed] [Google Scholar]
- 12.World Health Organization. Geneva: WHO; 2002. WHO consultation on a rabies monoclonal antibody cocktail for rabies post exposure treatment. 23–24 May 2002. World Health Organization, Geneva, Switzerland http://web.archive.org/web/20100525173651/http://www.who.int/rabies/vaccines/en/mabs_final_report.pdf . Accessed 19 February 2014. [Google Scholar]
- 13.World Health Organization. Geneva, Switzerland: 2005. WHO expert consultation on rabies (1st report). WHO technical report series 931. [PubMed] [Google Scholar]
- 14.Bakker AB, Marissen WE, Kramer RA, et al. Novel human monoclonal antibody combination effectively neutralizing natural rabies virus variants and individual in vitro escape mutants. J Virol. 2005;79:9062–8. doi: 10.1128/JVI.79.14.9062-9068.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Müller T, Dietzschold B, Ertl H, et al. Development of a mouse monoclonal antibody cocktail for post-exposure rabies prophylaxis in humans. PLoS Negl Trop Dis. 2009;3:e542. doi: 10.1371/journal.pntd.0000542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Both L, Banyard AC, van Dolleweerd C, Wright E, Ma JK, Fooks AR. Monoclonal antibodies for prophylactic and therapeutic use against viral infections. Vaccine. 2013;31:1553–9. doi: 10.1016/j.vaccine.2013.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goudsmit J, Marissen WE, Weldon WC, et al. Comparison of an anti-rabies human monoclonal antibody combination with human polyclonal anti-rabies immune globulin. J Infect Dis. 2006;193:796–801. doi: 10.1086/500470. [DOI] [PubMed] [Google Scholar]
- 18.Prosniak M, Faber M, Hanlon CA, Rupprecht CE, Hooper DC, Dietzschold B. Development of a cocktail of recombinant-expressed human rabies virus-neutralizing monoclonal antibodies for postexposure prophylaxis of rabies. J Infect Dis. 2003;188:53–6. doi: 10.1086/375247. [DOI] [PubMed] [Google Scholar]
- 19.Dietzschold B, Gore M, Casali P, et al. Biological characterization of human monoclonal antibodies to rabies virus. J Virol. 1990;64:3087–90. doi: 10.1128/jvi.64.6.3087-3090.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enssle K, Kurrle R, Kohler R, et al. A rabies-specific human monoclonal antibody that protects mice against lethal rabies. Hybridoma. 1991;10:547–56. doi: 10.1089/hyb.1991.10.547. [DOI] [PubMed] [Google Scholar]
- 21.Champion JM, Kean RB, Rupprecht CE, et al. The development of monoclonal human rabies virus-neutralizing antibodies as a substitute for pooled human immune globulin in the prophylactic treatment of rabies virus exposure. J Immunol Methods. 2000;235:81–90. doi: 10.1016/s0022-1759(99)00223-9. [DOI] [PubMed] [Google Scholar]
- 22.Hanlon CA, DeMattos CA, DeMattos CC, et al. Experimental utility of rabies virus-neutralizing human monoclonal antibodies in post-exposure prophylaxis. Vaccine. 2001;19:3834–42. doi: 10.1016/s0264-410x(01)00135-9. [DOI] [PubMed] [Google Scholar]
- 23.Muhamuda K, Madhusudana SN, Ravi V. Use of neutralizing murine monoclonal antibodies to rabies glycoprotein in passive immunotherapy against rabies. Hum Vaccin. 2007;3:192–5. doi: 10.4161/hv.3.5.4386. [DOI] [PubMed] [Google Scholar]
- 24.Paul M, van Dolleweerd C, Drake PM, et al. Molecular pharming: future targets and aspirations. Hum Vaccin. 2011;7:375–82. doi: 10.4161/HV.7.3.14456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ko KS, Tekoah Y, Rudd PM, et al. Function and glycosylation of plant-derived antiviral monoclonal antibody. P Natl Acad Sci USA. 2003;100:8013–8. doi: 10.1073/pnas.0832472100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schumacher CL, Dietzschold B, Ertl HC, Niu HS, Rupprecht CE, Koprowski H. Use of mouse anti-rabies monoclonal antibodies in postexposure treatment of rabies. J Clin Invest. 1989;84:971–5. doi: 10.1172/JCI114260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Kruif J, Bakker AB, Marissen WE, et al. A human monoclonal antibody cocktail as a novel component of rabies postexposure prophylaxis. Ann Rev Med. 2007;58:359–68. doi: 10.1146/annurev.med.58.061705.145053. [DOI] [PubMed] [Google Scholar]
- 28.Bakker AB, Python C, Kissling CJ, et al. First administration to humans of a monoclonal antibody cocktail against rabies virus: safety, tolerability, and neutralizing activity. Vaccine. 2008;26:5922–7. doi: 10.1016/j.vaccine.2008.08.050. [DOI] [PubMed] [Google Scholar]
- 29.Sloan SE, Hanlon C, Weldon W, et al. Identification and characterization of a human monoclonal antibody that potently neutralizes a broad panel of rabies virus isolates. Vaccine. 2007;25:2800–10. doi: 10.1016/j.vaccine.2006.12.031. [DOI] [PubMed] [Google Scholar]
- 30.Gogtay N, Thatte U, Kshirsagar N, et al. Safety and pharmacokinetics of a human monoclonal antibody to rabies virus: a randomized, dose-escalation phase 1 study in adults. Vaccine. 2012;30:7315–20. doi: 10.1016/j.vaccine.2012.09.027. [DOI] [PubMed] [Google Scholar]
- 31.Both L, van Dolleweerd C, Wright E, et al. Production, characterization, and antigen specificity of recombinant 62-71-3, a candidate monoclonal antibody for rabies prophylaxis in humans. FASEB J. 2013;27:2055–65. doi: 10.1096/fj.12-219964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneider LG, Meyer S. Antigenic determinants of rabies virus as demonstrated by monoclonal antibody. In: Bishop DH, Compans RW, editors. The Replication of Negative Strand Viruses: Developments in Cell Biology. Vol. 7. North Holland: Elsevier; 1981. pp. 947–83. [Google Scholar]
- 33.Kissling RE. Growth of rabies virus in non-nervous tissue culture. Proc Soc Exp Biol Med. 1958;98:223–5. doi: 10.3181/00379727-98-23997. [DOI] [PubMed] [Google Scholar]
- 34.Hehle VK, Paul MJ, Drake PM, Ma JK, van Dolleweerd CJ. Antibody degradation in tobacco plants: a predominantly apoplastic process. BMC biotechnology. 2011;11:128. doi: 10.1186/1472-6750-11-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stadlmann J, Pabst M, Kolarich D, Kunert R, Altmann F. Analysis of immunoglobulin glycosylation by LC-ESI-MS of glycopeptides and oligosaccharides. Proteomics. 2008;8:2858–71. doi: 10.1002/pmic.200700968. [DOI] [PubMed] [Google Scholar]
- 36.Cliquet F, Aubert M, Sagne L. Development of a fluorescent antibody virus neutralisation test (FAVN test) for the quantitation of rabies-neutralising antibody. J Immunol Methods. 1998;212:79–87. doi: 10.1016/s0022-1759(97)00212-3. [DOI] [PubMed] [Google Scholar]
- 37.Brookes SM, Parsons G, Johnson N, McElhinney LM, Fooks AR. Rabies human diploid cell vaccine elicits cross-neutralising and cross-protecting immune responses against European and Australian bat lyssaviruses. Vaccine. 2005;23:4101–9. doi: 10.1016/j.vaccine.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 38.Dean DJ, Abelseth MK, Atanasiu P. The fluorescent antibody test. In: Meslin F-X, Kaplan MM, Koprowski H, editors. Laboratory Techniques in Rabies. 4th ed. Geneva: WHO; 1996. pp. 88–95. [Google Scholar]
- 39.Kabat EA, Wu TT, Perry HM, Gottesman KS, Foeller C. 5th Edition ed. Bethesda, Maryland: National Institutes of Health; 1991. Sequences of proteins of immunological interest. [Google Scholar]
- 40.Badrane H, Bahloul C, Perrin P, Tordo N. Evidence of two Lyssavirus phylogroups with distinct pathogenicity and immunogenicity. J Virol. 2001;75:3268–76. doi: 10.1128/JVI.75.7.3268-3276.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCormick AA, Reddy S, Reinl SJ, et al. Plant-produced idiotype vaccines for the treatment of non-Hodgkin's lymphoma: safety and immunogenicity in a phase I clinical study. Proc Natl Acad Sci U S A. 2008;105:10131–6. doi: 10.1073/pnas.0803636105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whaley KJ, Hiatt A, Zeitlin L. Emerging antibody products and Nicotiana manufacturing. Hum Vaccines. 2011;7:349–56. doi: 10.4161/hv.7.3.14266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horton DL, McElhinney LM, Marston DA, et al. Quantifying antigenic relationships among the lyssaviruses. J Virol. 2010;84:11841–8. doi: 10.1128/JVI.01153-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cabanes-Macheteau M, Fitchette-Laine AC, Loutelier-Bourhis C, et al. N-glycosylation of a mouse IgG expressed in transgenic tobacco plants. Glycobiology. 1999;9:365–72. doi: 10.1093/glycob/9.4.365. [DOI] [PubMed] [Google Scholar]
- 45.Triguero A, Cabrera G, Rodriguez M, et al. Differential N-glycosylation of a monoclonal antibody expressed in tobacco leaves with and without endoplasmic reticulum retention signal apparently induces similar in vivo stability in mice. Plant Biotechnol J. 2011;9:1120–30. doi: 10.1111/j.1467-7652.2011.00638.x. [DOI] [PubMed] [Google Scholar]
- 46.Lee JH, Park DY, Lee KJ, et al. Intracellular reprogramming of expression, glycosylation, and function of a plant-derived antiviral therapeutic monoclonal antibody. PloS one. 2013;8:e68772. doi: 10.1371/journal.pone.0068772. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.