Abstract
Organisms require an appropriate balance of stability and reversibility in gene expression programs, to maintain cell identity or to enable responses to stimuli; epigenetic regulation is integral to this dynamic control. Post-translational modification of histones by methylation is an important and widespread type of chromatin modification that is known to influence biological processes in the context of development and cellular responses. We provide a broad overview of how histone methylation is regulated and leads to biological outcomes, to evaluate how histone methylation contributes to stable or reversible control. The importance of maintaining or reprogramming histone methylation appropriately is illustrated by links to disease and aging, or possibly transmission of traits across generations.
Introduction
Genetic information encoded in DNA is largely identical in every cell of a eukaryote. Yet cells in different tissues and organs can have widely different gene expression patterns and exhibit specialized functions. Gene expression patterns in different cell types need to be appropriately induced and maintained and also to respond to developmental and environmental changes; inappropriate expression leads to disease. In eukaryotes, the chromatin state – the packaging of DNA with histone proteins - is believed to contribute to control of gene expression. Histone post-translational modifications (PTMs) include phosphorylation, acetylation, ubiquitinylation, methylation and others1,2, and these modifications are thought to contribute to control of gene expression through influencing chromatin compaction or signaling to other protein complexes. Therefore, an appropriate balance of stability and dynamics in histone PTMs is necessary for accurate gene expression.
Histone methylation occurs on all basic residues: arginines3, lysines4 and histidines5. Lysines can be mono (me1)4, di(me2)6, or tri(me3)7 methylated on their ε amine group, arginines can be mono(me1)3, symmetrically dimethylated (me2s), or asymemetrically dimethylated(me2a) on their guanidinyl group8, and histidines have been reported to be monomethylated8,9 although this methylation appears to be rare and has not been further characterized. The most extensively studied histone methylation sites include histone H3 lysine 4 (H3K4), H3K9, H3K27, H3K36, H3K79 and H4K20. Sites of arginine methylation include H3R2, H3R8, H3R17, H3R26 and H4R3. However, many other basic residues throughout the histone proteins H1, H2A, H2B, H3 and H4 have also been recently identified as methylated by mass spectrometry and quantitative proteomic analyses (2, reviewed in 10). The functional effects and the regulation of the newly identified methylation events remain to be determined.
In general, methyl groups are believed to turnover more slowly than many other PTMs and histone methylation was originally thought to be irreversible3. The discovery of a histone H3 lysine 4 (H3K4) demethylase, LSD1 (Lysine Specific Demethylase 1, also known as KDM1A), revealed that histone methylation is in fact reversible11. Now, a plethora of methyltransferases and demethylases have been identified that mediate the addition and removal of methyl groups from different lysine residues on histones. Depending on the biological context, some methylation events may need to be stably maintained (for example, methylation involved in the inheritance through mitosis of a silenced heterochromatin state) whereas others may have to be amenable to change (for example, when cells differentiate or respond to environmental cues). Indeed, methylation at different lysine residues on histones has been shown to display differential turnover rates12. Importantly, the diverse array of methylation events provides exceptional regulatory power. A current model suggests that methylated histones are recognized by chromatin effector molecules (“readers”), causing the recruitment of other molecules to alter the chromatin and/or transcription states13.
To understand the dynamic regulation of and by histone methylation, it is useful to take a holistic view of regulation of and by this chromatin modification. Here we aim to draw together key points – rather than providing comprehensive coverage - regarding how histone methylation is established, reversed or maintained across cell divisions, or possibly even across generations. We describe the principles of how methyl marks might be converted into biological outcome and examples of that demonstrate the importance of appropriate establishment or maintenance of methylation, through considering when methylation regulation goes awry in cancer, mental retardation and aging. Throughout, we refer readers to literature that considers each of these topics in more depth.
Regulation of histone methylation
Histone methyltransferases and demethylases
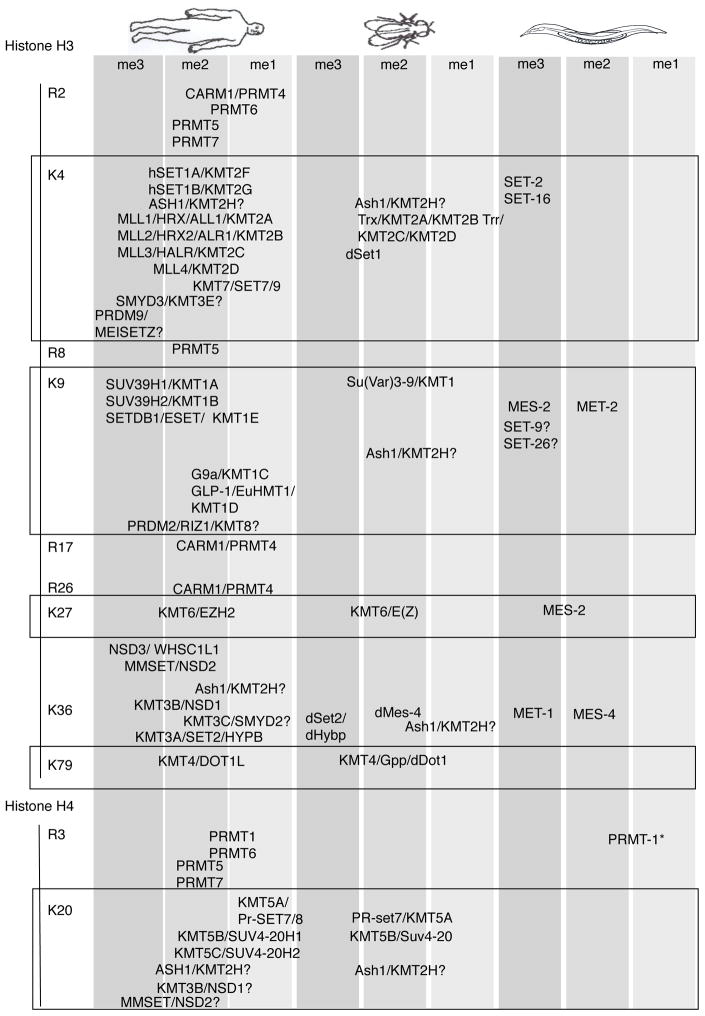
Three families of enzymes have been identified thus far that catalyze the addition of methyl groups donated from S-adenosyl methionine4 to histones. The SET domain containing proteins14 and Dot1 like proteins15 have been shown to methylate lysines and members of the PRMT family have been shown to methylate arginines16 (Table 1). These histone methyltransferases have been shown to methylate histones incorporated in chromatin14, free histones and non-histone proteins17. Calmodulin-lysine N-methyltransferase, a non-SET domain containing protein, has been shown to methylate calmodulin and might have the potential to methylate histones as well18.
Table 1. Histone methyltransferases.
The histone methyltransferases for various lysine and arginine residues are indicated over the specific residues that they have been shown to methylate. Histone methyltransferases, that methylate multiple residues or methylate residues to varying degrees are written over each modification that has been reported. ‘?’ indicates results have not been independently replicated. *: PRMT-1 was shown to mono and asymmetrically dimethylate Histone H4 but the specific residue, which it methylates has not yet been identified. References for individual methyltransferases can be found in Supplementary Information.
|
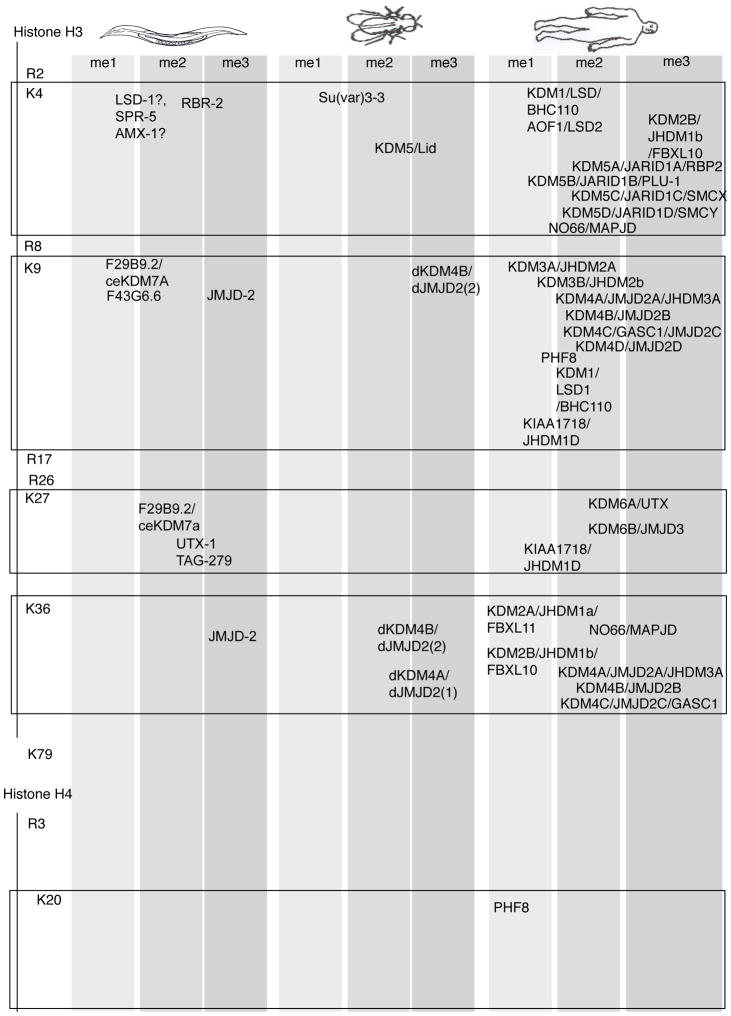
Two families of demethylases have been identified thus far that demethylate methyl-lysines. These are the amine oxidases11 and Jumonji C (JmjC) domain containing, iron-dependent dioxygenases19–21 (Table 2). These enzymes are highly conserved from yeast to humans and demethylate histone and non-histone substrates. Arginine demethylases remain more elusive. Although an initial report suggested that one of the JmjC domain proteins, JMJD6, demethylates arginines22, a more recent study indicates that the main function of JMJD6 is hydroxylation of an RNA splicing factor23. Mono-methyl arginines have also been shown to be converted by PADI4 (also known as PAD4) to citrulline24,25. However, PADI4 is not an arginine demethylase as it works on both methylated and unmethylated arginine24.
Table 2. Histone demethylases.
The histone demethylases for various lysine and arginine residues are indicated over the specific residues that they have been shown to demethylate. Histone demethylases, which demethylate multiple residues or to varying degrees are written over each modification that has been reported. ‘?’ indicates results have not been independently replicated. References for individual demethylases can be found in Supplementary Information.
|
The diversity of chromatin-associated and non-chromatin-associated substrates poses an important challenge for our understanding of the mechanisms by which these enzymes execute their biological functions.
How are the enzymes recruited to their genomic destinations?
Determining how and when methyltransferases and demethylases are recruited to specific histone targets is an important area of current research. Specific DNA sequences have been identified that are responsible for the recruitment of several histone-modifying enzymes. Some of the best-studied examples are the Drosophila Trithorax Group Response Elements (TREs) and Polycomb Group (PcG) Response Elements (PREs), which direct recruitment of TrX (a H3K4 methyltransferase complex) and PcG proteins (the PRC2 complex catalyzes H3K27 trimethylation), respectively, possibly through specific DNA binding transcription factors that recognize these regulatory elements 26–28. In human cells, a DNA sequence that enhances PcG binding has been identified and this shows some similarity to Drosophila PREs29, suggesting that at least some aspect of the PcG recruitment mechanism is conserved.
Long non-coding RNAs (lncRNAs) have also been proposed to play a targeting role by binding to certain methyltransferases and demethylases and directing them to specific genomic locations. lncRNAs have been shown to bind to members of the PRC2 complex30–33, the H3K9 methyltransferase G9a33,34 and the H3K4 methyltransferase complex member WDR535. Particular lncRNAs may influence multiple methyl marks. For instance, the human lncRNA HOTAIR has been shown to bind to PRC2 and to a complex containing the demethylase LSD1, suggesting that this lncRNA might coordinate recruitment of an H3K27 methyltransferase and an H3K4 demethylase to lead to efficient repression of specific loci32. However the roles of lncRNAs in vivo remain unclear. A deletion of mouse Hotair had no effect on Hox gene expression or H3K27me3 levels36, suggesting either functional redundancy or that the function of this lncRNA may have diverged between humans and mice. The identification of over 1,000 lncRNAs37 in mammals, together with the proposal that lncRNAs act both in cis and in trans, suggest that lncRNAs might account for a large amount of the specific targeting of methyl modifying proteins38. The mechanisms require further investigation. Chemical tagging of lncRNAs and knockouts of lncRNAs will facilitate an in vivo examination of endogenous protein partners and also help to validate real methylation targets.
Small non-coding RNAs also play a role in directing chromatin modifications, including histone methylation. For example, in Schizosaccharomyces pombe, the RNA interference (RNAi) machinery is required for establishing and/or maintaining centromeric heterochromatin, which is characterized by H3K9 methylation39–41. The RNAi machinery has also been linked to H3K9 methylation levels and heterochromatin in Arabidopsis thaliana42 and mammals43, suggesting some evolutionary conservation. In Caenorhabditis elegans dsRNA-triggered knock-down of specific genes leads decreased transcription in addition to increased H3K9me3 of the target gene44. In addition, the RNAi machinery has recently been linked to the induction of H3K27me3 and heterochromatin changes during X chromosome inactivation in mice45. However, the RNAi machinery might not solely be associated with inactivating transcription as a recent report in Drosophila shows that Dicer 2 and Argonaute 2 are associated with mostly euchromatic loci and affect global transcription46.
DNA methylation also seems to have a role in directing histone methylation47. For example the A. thaliana H3K9 methyltransferase SUVH4 binds methylated DNA48 and mutation of the methyl-DNA binding domain of the Arabidopsis H3K9 methyltransferase SUVH5 decreases H3K9me249. There is also evidence that histone methylation can influence DNA methylation, with these two marks reinforcing each other to establish repressive chromatin environments. This topic is beyond the scope of this review and we therefore refer readers to relevant reviews50,51.
Collaborative and antagonistic relationships among different histone marks
It is important to note that histone methylation marks do not appear in isolation. Methylation can occur at multiple different sites on the same histone, but some histone marks are mutually exclusive. For instance, in mammals and yeast, dimethylation of H3R2 (by PRMT6 in mammals) is prevented by H3K4me3 and conversely H3R2me2a prevents H3K4 methylation52,53. Similarly, phosphorylation of H3S10 prevents H3K9 methylation in vitro14. Combinatorial histone marks can also alter the recognition and binding of methyl binding proteins. Phosphorylation of H3S10 during the M phase of the cell cycle prevents the H3K9me3 binding protein heterochromatin protein 1 (HP1) from binding to doubly modified histone tails54. Some PTMs on histone tails can recruit methyl modifying proteins and can play a role in determining their substrate specificities. For example, PHF8 binds to H3K4me2/3 and demethylates H3K9me2 but its related enzyme KIAA1718 is directed towards demethylating H3K27me2 upon binding H3K4me2/355. PTMs on one histone tail also influence other histone tails in trans. For example, H2B monoubiquitylation is necessary for H3K4 methylation and H3K79 methylation56–58. In yeast, this relationship might be explained by a component of the COMPASS complex (the yeast homologue of the mammalian MLL complex), Swd2, which interacts with chromatin in an H2B-monoubiquitination-dependent manner59 and also interacts with the H3K79 methyltransferase Dot159. Histone PTMs can therefore play an important role in the recruiting of methyl-modifying enzymes to specific genomic locations60 and in some cases determine their substrate specificities.
Is the enzymatic activity of methyl modifying enzymes important for their function?
When considering the action of methyl modifying enzymes it is important to bear in mind recent findings that suggest some of these proteins have functions independent of their enzymatic activities. For instance, although the enzymatic activity of the H3K27 demethylase UTX has been shown to be important for zebrafish posterior development61 and for muscle specific gene expression during myogenesis62, it has also been shown to regulate T-box family member-dependent gene expression in a demethylase-independent manner63. Similarly, the related H3K27 demethylase JMJD3 has been reported to function in ways that are dependent and independent of its enzymatic activity63.
Regulation by histone methylation
How is methylation recognized?
Recognition of methylated histones is accomplished by proteins with methyl binding domains13 including PHD fingers64,65, WD40 repeats66, CW domains67, PWWP domains68, ankyrin repeats69, and proteins of the Royal superfamily. This superfamily includes proteins with chromodomains70,71, double chromodomains72, chromo barrels73, Tudor domains74, Double/tandem tudor domains75,76, and MBT repeats77. Some proteins containing these domains - including BHC8078, TRIM2479, and UHRF180 - also recognize unmethylated lysine and arginine residues on histones and methylation inhibits their interaction with histones. Addition of the methyl moieties increases the positive charge and hydrophobicity of lysine and arginine, thus facilitating their interactions with proteins hydrophobic properties. Indeed, aromatic cages have been found in several methyl binding proteins, which enable direct interaction with methylated arginine74,81 and lysine residues75,82. Exceptions include the ADD domain of ATRX, which recognizes H3K9me3 via a composite pocket distinct from the aromatic cages discussed above83–85.
The effects of methylation are context dependent
The location of the methyl lysine residue on a histone tail and the degree of methylation (me1, me2 or me3,) have been associated with differential gene expression status. For example, H3K4me3 is generally associated with active transcription86,87 or with genes that are poised for activation, whereas H3K27me3 is associated with repressed chromatin. H3K4me1 is often associated with enhancer function88 whereas H3K4me3 is linked to promoter activity. H3K79me2 is important for cell cycle regulation whereas H3K79me3 is linked to the Wnt signaling pathway89,90.
However, there are instances where the same modifications can be associated with opposing activities, such as transcriptional activation and repression. H3K4me2 and H3K4me3 are illustrative of this point; generally these marks are associated with transcriptional activation but they can also be associated with transcriptional repression64,91. Probably, the change in activity is due to different effector proteins. For instance, when H3K4me2 or me3 marks are bound by the PHD domain containing co-repressor protein inhibitor of growth family member 2 (ING2) they are associated with transcriptional repression64 through the stabilization of a histone deacetylase complex. We propose that the “reader” proteins that recognize specific histone modifications are important components in determining the function of modifications.
Combined marks can also have different roles to the same marks appearing in isolation. Although H3K4me3 and H3K27me3 are marks associated with active and repressive transcription, respectively, when present together they appear to play a role in poising genes for transcription91. Combinatorial histone modifications are efficiently recognized by proteins with multiple domains to effect specific outcomes. For instance, the chromatin regulator tripartite motif-containing 24 (TRIM24) has a PHD and a bromodomain, which recognize unmethylated H3K4 and acetylated H3K23 on the same histone tail79. This finding suggests that proteins that have multiple histone binding domains are ideally suited to incorporate the information from multiple histone modifications to ensure specific biological outcomes, in the case of TRIM24 this binding leads to estrogen-dependent gene activation.
Combinatorial action of methyl-modifying enzymes is also context specific. Coordinated addition of histone methyl-marks that are generally associated with transcriptional activation and removal of marks that are generally associated with transcriptional repression is thought to occur. For instance, in mammals, the H3K27 demethylase UTX has been shown to associate with the H3K4 methyltransferase complex MLL2/392; presumably adding the ‘activating’ mark (H3K4me3) and removing the ‘inhibitory’ mark (H3K27me3) achieves optimal transcriptional activation. Similarly the T-box transcription factors can bring together the H3K4 methyltransferase complex subunit RbBp5 and the H3K27 demethylase JMJD393. For efficient repression, the H3K4me3 demethylases have been shown to physically associate with the repressive Polycomb Group (PcG) proteins (which methylate and bind to H3K27me3) and H3K9 methyltransferases94,95. In addition, some of these repressive complexes also contain histone deacetylases96, suggesting that coordinated methylation regulation and deacetylation occurs to efficiently repress target genes. These findings suggest coordinated and reciprocal relationships may help to form and possibly maintain a stable methylation pattern when and where appropriate.
Roles of methylation in transcriptional processes
The majority of the associations between histone methylation status and transcription are based on correlations between gene expression level and genome wide or locus-specific ChIP studies86,87,97. However, several studies have begun to address the role of histone modifications at specific stages of transcription. It appears that histone methylation plays a role in many levels of transcriptional regulation from chromatin architecture to specific loci regulation through the recruitment of cell-specific transcription factors and interaction with initiation and elongation factors (Box 1). In addition, histone methylation influences RNA processing (Box 2).
Box 1. Histone methylation regulation is important for transcriptional control.
An interesting and untested hypothesis is that histone methylation could influence transcription by bringing physically separate regions of chromatin close together through chromosomal looping163 (figure panel A). This could include enhancer and promoter regions or, in the case of repressive interactions, insulator elements164. However histone methylation might be a consequence of chromosomal looping. For instance, PcG proteins can regulate H3K27 methylation of distal sites after initial recruitment to a specific site165. Whether chromosomal looping is a cause or a consequence of transcriptional regulation remains to be determined.
Histone modifications can affect the higher-order chromatin structure directly166 or indirectly through recruiting chromatin remodeling complexes167,168. For example: the BPTF component of the chromatin remodeling complex NuRF contains a PHD finger that recognizes H3K4me365; DPF3, a component of the BAF complex, contains a double PHD finger that interacts with methylated histones169; and the chromodomains of CHD proteins also bind to methylated histones72,170,171. In yeast, H3K36me3 can recruit a histone deacetylase to indirectly affect transcription172.
Inaccessible chromatin domains can be “opened” by so-called “pioneering factors”173, which are sequence-specific DNA-binding transcription factors (such as FOXA1 and GATA4) (Figure panel B). After binding of the pioneering factors (PF), DNA methylation and histone modifications could participate in making the chromatin more accessible for other transcription factors, the pre-initiation complex (PIC) and RNA polymerase II (RNAPII)174.
Certain histone methylation patterns (such as stretches of chromatin marked by high density of H3K4 and H3K79 methylation) also appear to be necessary for binding of transcription factors (Figure panel C), as highlighted by a study demonstrating that histone modifications affect MYC binding to promoters in humans175 (presumably by providing a euchromatic environment, which facilitates sequence-specific binding). More work needs to be done to determine how widespread the role of histone modifications is in setting up local regions with specific histone modification signatures that are either conducive or antagonistic to the stable localization of DNA-binding factors, histone modifying enzymes and effector proteins.
It is still unclear whether the recruitment of chromatin remodeling machinery to sites of transcription176 enables more efficient transcription and/or is necessary for elongation to begin. The PHD finger of TAF3, a component of the transcription factor II D complex (which itself is an essential component of the RNA pol II preinitiation complex) binds to H3K4me3177, suggesting that the RNA pol II machinery communicates directly with histone methylation to regulate transcription. The phosphorylation status of Pol II (which determines whether Pol II is in the initiation or elongation phase) regulates the binding of different chromatin modifying proteins to Pol II; these proteins methylate H3K4 or H3K36 or demethylate H3K27 to potentially facilitate transcription initiation or elongation (elongation shown in Figure panel D).
Despite advancements in deciphering the role of histone methylation in transcriptional control there is still a lot of uncertainty regarding the order of events, which are beginning to be deciphered176 but are still far from clear.
Box 2. The role of methylation in RNA splicing.
Histone methylation has been implicated in the control of RNA splicing. Intriguingly, the average exon length of many eukaryotic species is similar to the length of DNA wrapped around one nucleosome178, whereas intron length varies greatly. The association of the splicing factor U2 snRNP with chromatin is enhanced by H3K4 trimethylation179,180. This appears to be mediated by U2 snRNP interacting with the H3K4 binding proteins Chd1 and Sgf29 in human cell lines179,180. Also, recent global ChIP-seq analyses in C. elegans, mice and humans show that exons are enriched for H3K36me3 compared to introns and alternatively spliced exons have lower levels of H3K36me3 than constitutively spliced exons181.
In vitro assays have shown that the rate of transcriptional elongation can affect splicing. Since H3K36me3 can recruit a histone deacetylase complex175, which represses transcription, a kinetic model for splicing has been proposed in which the histone methylation can affect the rate of transcription and thus influence splicing182. In addition, several studies have shown that knockdown of SETD2 (a H3K36 methyltransferase) affects alternative splicing183, potentially due to the interaction of the splicing regulator polypyrimidine tract binding protein 1 (PTB1) with the H3K36me3 binding protein, MRG1573,179,184. More recent studies have shown that global splicing inhibitors can lead to repositioning of H3K36me3 and impaired recruitment of SETD2, suggesting that splicing also regulates H3K36 trimethylation183,185. These results suggest that there may be two-way communication between histone methylation and splicing; additional work is necessary to fully understand the functional consequence of this communication.
Biological importance of methylation dynamics
Histone methylation dynamics is known to play important roles in many biological processes including cell cycle regulation, DNA damage and stress response, development and differentiation (reviewed in 60,98–101). The importance of the tight regulation of histone methylation is illustrated by emerging links to disease and aging.
Histone methylation and cancer
Several lines of evidence suggest that aberrant histone methylation is likely to play a role in cancer (see review 102,103). Initial studies showed that changes in global levels of certain histone methylation events are correlated with increased cancer recurrence and poor survival (Table 3). Although it remains to be determined whether these changes are causal, they nevertheless might be developed into potential biomarkers for drug discovery and for diagnosis or prognosis. More recent studies provide increasing genetic evidence that suggests histone methylation events play a causal role in tumorigenesis.
Table 3. Global changes in histone methylation marks have been reported in various types of cancers.
Correlative studies report altered histone methylation patterns in specific cancer types189–197. In some of these cases multiple methyl marks have been shown to change in concert highlighting the importance of combinatorial modifications effect on biological functions.
| Cancer type | Methyl mark | Consequence | Reference |
|---|---|---|---|
| Prostate cancer | ↓ H3K4me2 | Higher recurrence | 1 |
| ↓ H4R2me2 | Higher recurrence | 1 | |
| Lung cancer | ↓ H3K4me2 | Poorer survival | 2, 3 |
| Kidney cancer | ↓ H3K4me2 | Poorer survival | 3 |
| Breast cancer | ↓ H3K4me2 | Poorer survival | 5 |
| ↓ H3K27me3 | Poorer survival | 7 | |
| ↓ H4R3me2 | Worse clinical outcomes | 5 | |
| ↓ H4K20me3 | Worse clinical outcomes | 5 | |
| Pancreatic cancer | ↓ H3K4me2 | Poorer survival | 4 |
| ↓ H3K9me2 | Poorer survival | 4 | |
| ↓ H3K27me3 | Poorer survival | 7 | |
| Gastric adenocarcinoma | ↑ H3K9me3 | Poorer survival | 6 |
| Ovarian cancer | ↓ H3K27me3 | Poorer survival | 7 |
| Lymphomas | ↓ H4K20me3 | Associated with | 8 |
| Colon adenocarcinomas | ↓ H4K20me3 | Associated with | 8 |
Mutations in or altered expression of histone methyl-modifiers and methyl binding proteins correlate with increased incidence of a variety of different cancers (reviewed by 102,103). For example, the H3K27me3 methyltransferase is upregulated in a number of cancers, including prostate cancer104, breast cancer105 and lymphomas106. Importantly, activating point mutations in EZH2 have recently been identified that are associated with B cell lymphomas106, in agreement with the idea that EZH2 is oncogenic. Consistent with this, somatic inactivating point mutations in the H3K27 demethylase UTX were found in a variety of human cancers107. However, EZH2 does not always work as an oncogene; mutations that cause a loss of methyltransferase activity of EZH2 have been identified in myeloidysplastic syndromes108, suggesting that EZH2 functions as a tumor suppressor in that cancer type. The dual role of EZH2 as an oncogene or a tumor suppressor highlights the context-dependent nature of oncogenes and tumor suppressor genes and raises the possibility that H3K27me3 may have alternative functions in different cell types.
Importantly, methyltransferases and demethylases are found at sites of chromosomal translocations (Box 3) and several of these chromosomal fusion proteins have been shown to have tumorigenic properties109–111. Such gene fusions could either lead to aberrant deposition or removal of histone methylation (if the fusion results in mistargeting of the methyltransferase or demethylase), or inappropriate recruitment of other proteins (such as transcription factors) by the mutant methyl binding domain.
Box 3. Chromosomal Translocations of Methyltransferases and Demethylases.
Methyltransferases and demethylases are found at sites of chromosomal translocations in several reported types of cancer, suggesting that regulation of histone methylation could play a causal role in tumorigenesis. For a comprehensive review on histone modifications and cancer please see 102,103.
A notable example of a methyltransferase implicated in tumorigenesis is the H3K4 methyltransferase MLL1, which is a frequent target of chromosomal translocations in acute myeloid leukemia (AML), acute lymphoblastic leukaemia (ALL) or mixed lineage leukaemia (MLL). It is unclear whether it is the misregulation of the methyltransferase activity of MLL1 or the activity of the new fusion proteins that is tumorigenic. Fusion of MLL and AF9 causes cell proliferation and increased HoxA9 expression and mutation of this fusion protein’s DNA binding or recruitment domains prevented leukemic transformations111. Interestingly, MLL1 has roles in proliferation of normal hematopoietic stem cells186, which may be the cells of origin for some leukemias.
The gene encoding the H3K36 methyltransferase NSD1 has also been shown to be a site of translocations in various cancers. A translocation between nucleoporin 98 (NUP98) and NSD1 occurs in 5% of childhood acute myeloid leukemia’s187. The fusion protein, which includes the PHD and SET domains of NSD1, is sufficient to cause AML in mice, coincident with increases in H3K36me and decreases in H3K27me3 at specific genomic loci109. While the wildtype fusion protein permits myeloid progenitor proliferation, point mutations in the SET domain of this fusion protein do not, suggesting that the methyltransferase activity of this fusion protein is essential for its tumorigenic potential109.
In certain leukemias NUP98 is fused to the demethylase JARID1A; the fusion protein contains the PHD domain but not the demethylase domain of JARID1A188. This fusion protein and an artificial fusion of the PHD domain of PHF23 to NUP98 induce AML. Mutations in the PHD domain that eliminate its H3K4me3 binding abolish its leukemic transformative potential110. These results suggest that it is the mis-localization of NUP98 via the PHD domain rather than the mis-regulation of JARID1A, which causes the leukemia.
These findings suggest that loss and/or gain of function through translocation events can cause cancer, although the possibility that some of the translocation events may represent “passenger” translocations cannot be excluded. It is interesting to note that in some cases, such as NSD1 proteins in leukemia, cancer progression is dependent on the methylase/demethylase activity of these proteins. In other cases, such as the NUP98-JARID1A fusion, the capacity to bind to a certain methyl mark through the PHD domain may cause progression by inappropriate targeting of a fusion protein to novel genomic locations.
These links between methyl-modifying enzymes and cancer indicate that an appropriate balance between stable methylation and dynamic methylation of histones is essential for cancer prevention. Methyl-modifying and methyl-binding proteins are thus candidate pharmacological targets for cancer and possibly other human diseases.
Histone methyation and Intellectual disability
A variety of histone modifiers have been implicated in intellectual disability syndromes112, supporting the concept that appropriate regulation of histone modifications during nervous system development is essential for brain function. For example, truncations in the histone methyltransferase NSD1 have been found in 77% of patients with Sotos syndrome, which is associated with intellectual disability113. Additionally, 9q subtelomere deletion syndrome (9qSTDS) is caused by mutations in the H3K9 methyltransferase EHMT1 and haploinsufficiency of EHMT1 is thought to cause intellectual disability114. Mice that are heterozygous for Ehmt1 display autistic-like features115 suggesting that EHMT1 has a conserved role in regulating normal neural function.
The role of histone modifiers in cognitive disorders is supported by studies of X-linked mental retardation (XLMR). At least seven proteins that have been identified as mutated in XLMR are potential methyl modifying enzymes or methyl binding proteins, including: MECP2116 (a methyl CpG binding protein), JARID1C/SMCX95,117 (an H3K4 demethylase), PHF8118–120 (an H3K9/H4K20 demethylase), BCOR121 (an ankyrin repeat containing which complexes with Polycomb Group proteins), ATRX83–85 (an H3K9me3 binding protein), PHF6122 (a PHD containing protein), and BRWD3123 (a bromo and WD repeat containing protein). Detailed molecular mechanisms for how these proteins regulate cognitive function remain largely unknown. In the case of MECP2, its disruption leads to Rett syndrome and global changes in neuronal chromatin structure124 which accompany global changes in histone methylation patterns. ATRX has been shown to bind to H3K9me3 and to play a role in chaperoning variant histones to telomeres83–85,125–127. Exactly what ATRX does at regions of heterochromatin (such as telomeric and pericentromeric heterochromatin) is unclear. However, the activity of ATRX probably involves remodeling of heterochromatin structure through the helicase activity of this protein as some mutations in ATRX that cause ATRX syndrome have recently been shown to disrupt its ATP hydrolysis activity128. Interestingly, a recent study identified a patient with severe intellectual disability who carries duplication of both MeCP2 and ATRX, suggesting a possible functional link between these two proteins in regulating cognition129.
We speculate that the above XLMR gene products regulate cognition via different molecular mechanisms, such as regulation of a specific gene expression signature that is important for cognition, of heterochromatin structure and function and/or of the balance between heterochromatin and euchromatin. It is also possible that all of these proteins regulate a common process that influences human cognition, but that they do so via different paths. Mouse models with point mutations that eliminate specific binding or enzymatic activity of proteins implicated in intellectual disability will be powerful tools to determine the impact of chromatin modifications, through using the battery of cognitive analysis tools available for mice.
Histone methylation and Aging
Histone methylation and methyl modifying proteins have recently been shown to play a role in the regulation of organismal lifespan and tissue aging. Loss of the appropriate balance between stable and dynamic methyl marks in adult stem cells may contribute to the decline of individual tissue function with age130. Evidence that global histone methylation levels change with age raise the possibility that global acquisition or loss of stable methyl marks could contribute to organismal aging. Specifically, H4K20me3 levels increase with age in rat livers131 but H3K27me3 levels show a decrease in somatic tissues with age in C. elegans132. Heterochromatin seems to decrease in cells from older individuals133 or in cells from patients with the premature aging disease, the Hutchinson-Gilford progeria syndrome (HGPS)134. Cells from individuals with HGPS also show decreased H3K27me3 on the inactive X chromosome as well as a decrease in the level of the H3K27 trimethyltransferase EZH2135. These results suggest that a global decrease in heterochromatin, and the concomitant mis-regulation and mis-expression of many genes that are silenced in young healthy individuals, could play a causal role in aging.
Several recent studies have shown that manipulations of histone methyltransferases136–138 and demethylases132,137,139,140 alter lifespan in C. elegans and Drosophila. Knockdown of the methyltransferases set-2, set-4, set-6, set-9, set-15, set-26 and blmp-1, and demethylases rbr-2, lsd-1, T26A5.5 and utx-1 each affected longevity in C. elegans132,136,137,139,141. It was further shown that knockdown or mutation of the H3K4 methyltransferase SET-2 and the H3K4 methylation complex components ASH-2 and WDR-5 extends C. elegans lifespan predominantly through influencing the activity of these components in the germline137. Consistent with these findings, over-expression of the H3K4me3 demethylase RBR-2 extends worm lifespan whereas mutation or knockdown of RBR-2 reverts the long lifespan caused by mutation or knockdown of members of the H3K4 methyltransferase complex137. The Drosophila homologue of RBR-2, Lid, has also been shown to slow aging140, suggesting that H3K4me3 regulatory proteins are conserved regulators of longevity. Also, knockdown or deletion of one copy of the H3K27me3 demethylase UTX-1 was sufficient to extend worm lifespan in a germline independent manner132,139. Interestingly, a recent report in Drosophila showed that heterozygous mutation of E(Z), a member of the H3K27 trimethyltransferase PRC2 complex also extends longevity138. This apparent contradiction of mutations in a H3K27 methyltransferase and in a demethylase both extending lifespan could be due to specific regulation of longevity in different tissues, cells or organisms, perhaps depending on the loci targeted by these enzymes. Methyltransferases and demethylases could be potential targets of small molecules to slow aging and prevent diseases. However, these links to aging need to be investigated in higher organisms. Histone methyl marks have not yet been shown to play a direct role in regulating longevity. In lower organisms, this could be addressed by investigating the impact on longevity of mutating a given lysine residue to an unmethylatable amino acid, although caution would have to be exercised in the interpretation of these results since altering the lysine residue by mutation is not equivalent to the loss of modification.
Inheritance of histone methylation marks
The examples of cancer and aging indicate the importance of maintaining the correct patterns of methylation throughout an organism’s lifetime. Therefore, there need to be mechanisms to ensure the stability of methylation through cell divisions. By contrast, most methyl marks need to be ‘reset’ in the germline, but some recent evidence suggests stability of methylation across generations is possible.
Inheritance through cell divisions
Initial in vitro and ex vivo studies on the SV40 replication fork showed that histones were not present immediately after replication and nucleosomes reassociated with the DNA a short distance (225–285 nucleotides) from the branch point 142. A subsequent in vitro study using a hybrid bacteriophage and eukaryotic DNA replication system demonstrated that the entire histone octamer could be segregated to one of the newly replicated daughter strands without being dissociated from the DNA143. More recently, SILAC quantitative mass spectrometry analysis using temporally labeled histones suggests that H3.1-H4 tetramers are segregated conservatively whereas new H2A and H2B histones are incorporated into new DNA144. Similarly in S. cerevisiae analysis using inducible expression of tagged histone H3 revealed that there was predominantly conservative inheritance of whole nucleosomes but that actively transcribed genes did contain both new and old H3–H4 dimers145.
Moreover, the mechanism by which PTMs are transmitted seems inherently problematic. If histones are indeed maintained through replication on one daughter strand, how can histones with the same PTMs be reintroduced to the appropriate DNA loci on the other strand? If histones are removed during DNA replication and then replaced with new histones after replication the problem of reintroducing the same PTMs on appropriate histones is doubled. If histone dimers are split to each daughter strand in a semi-conservative manner then marks would only have to be reintroduced on the other dimers; recent papers suggest this is a rare event144,145 but inheritance of histone methylation is also rare.
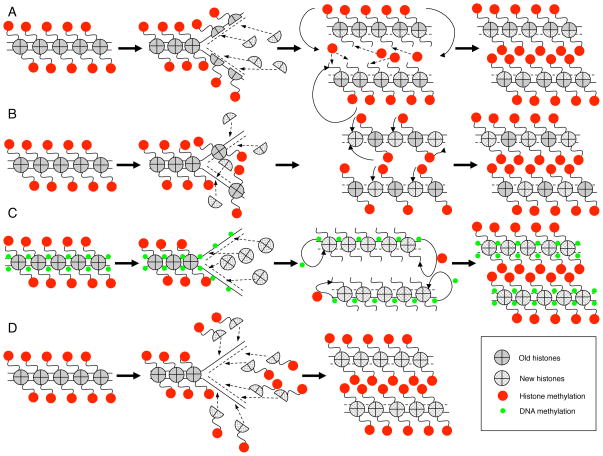
Several current theories exist to explain the propagation of histone PTMs96,146,147 (Figure 1). First, histones on newly synthesized DNA could be immediately modified to maintain an epigenetic memory. Interestingly, studies showing that MLL is retained on mitotic chromatin148 and that PcG proteins remain bound to chromatin during DNA replication149 provide some evidence in support of this hypothesis. Another theory19 suggests that histones are replicated in a semi-conserved manner (Figure 1a); each daughter chromosome would get one half of the histone octamer and then PTMs on the newly introduced histones using the old histones as a template. Alternatively, entire histone octamers could be segregated to each of the daughter strands and the PTMs of new histones could be informed by neighboring marked histones (Figure 1b). Another idea is that DNA methylation could help maintain histone PTM modifications by recruiting methyl modifiers146 (Figure 1c). However it is unknown whether DNA methylation always correlates with inherited histone methylation and DNA methylation has not yet been observed in quantifiable amounts in C. elegans150, which display inheritance of histone methylation marks.
Figure 1. Models for inheritance of histone methylation marks.
a) Semiconservative replication of histones (and their marks). The half of the histone octamer, which is inherited (and therefore appropriately marked), could inform the cell machinery to mark the newly added histone proteins. b) Entire histone octamers are segregated to each of the daughter strands in alternating fashion and the newly synthesized histones could be informed by the neighboring parental histone marks. c) DNA methylation acts as a signal to methylate specific new histones. d) Deposition of histones containing pre-existing modifications. Previously labeled free histones could be recruited to important sites where an epigenetic memory is needed and integrated into newly synthesized DNA.
Another model suggests that lncRNAs could be inherited and subsequently recruit appropriate histone modifying enzymes to regions which need to maintain an epigenetic memory38,96. Also, since the RNAi machinery can play a role in maintaining heterochromatin (as discussed above)39–41, inherited small interfering RNAs (siRNAs) could be a mechanism of re-establishing histone methylation patterns. Another viable model is that free histones are modified before binding to the newly synthesized DNA (“pre-marked”) (Figure 1d). In this scenario, only a limited number of methylation marks would be inherited or else the cell would need hundreds of unique combinations of modified free histones to ensure faithful inheritance. These models rely on a system in which the cell “knows” which methyl marks it must maintain on newly synthesized DNA and which marks should be erased; how this could be accomplished is unknown. The above-discussed theories are not mutually exclusive and could potentially work together.
Transgenerational epigenetic inheritance
Traits can be inherited in a non-Mendelian fashion by extranuclear nucleic acids, gene conversion, mosaicism, prions and epigenetic transgenerational inheritance, adding to the complexity of phenotypic diversity. Epigenetic inheritance has been implicated in inherited phenotypic differences in model organisms ranging from yeast to rats (for review see 151) and correlative reports suggest transgenerational epigenetic effects occur in humans (reviewed in 151), although the molecular mechanisms have not yet been deciphered. Some transgenerational effects of RNA interference have been shown to be transmitted from generation to generation in C. elegans152–154 raising the possibility that a subclass of small RNAs could - through their reported role in maintaining heterochromatin - be one mechanism of reestablishing appropriate histone methylation patterns in descendants. Indeed dsRNA’s directed against smg-1 led to inherited siRNAs and altered H3K9me3 levels at the smg-1 locus for two generations after removal of C. elegans from the dsRNA44.
Some specific cases of transgenerational epigenetic inheritance seem to involve inherited histone methylation patterns151, although transmission of chromatin modification through the gametes has not been shown conclusively. Histone modifications can be propagated during mitosis and meiosis146,149,155,156, however, histone methylation marks are generally thought to be erased between generations by epigenetic reprogramming157. Nevertheless, possible inheritance of histone methylation levels has been reported in Dictyostelium and C. elegans. Live-cell imaging of single-gene transcription in Dictyostelium suggests that states of high levels of transcription of specific genes can be passed from mother to daughter158. Interestingly, knock out of the H3K4 methyltransferase Set1, the H3K4 methyltransferase complex subunit Ash2 or mutation of H3K4 to alanine eliminated the inheritance of active transcriptional states158. Deletion of the DNA methyltransferase DnmA or the H3K36 methyltransferase Set2 did not have this effect158. Mutations of one of the C. elegans homologues of LSD1, SPR-5, causes progressive sterility159. Early generations of spr-5 mutant worms are phenotypically similar to WT worms. However, over generations, fertility becomes progressively impaired concomitant with an increase in global levels of H3K4me2 in spr-5 mutant worms159,160. Mutations in the C. elegans H3K4 trimethylation complex members SET-2 and WDR-5 extend the longevity137. However, genetically wildtype descendents of these H3K4 trimethylation mutants also have extended longevity, and this persist for 3 additional generations161. The mechanism by which altering this H3K4 trimethylation complex causes an epigenetic memory of extended longevity remains to be deciphered. Although no global decrease in H3K4me3 was detected in descendents of worms with decreased H3K4me3, this does not rule out that specific loci or specific cells could inherit an epigenetic memory of altered H3K4me3 or that the altered H3K4me3 in the parental generation could regulate the production of other RNA or proteins that could be inherited161.
A recent study in Drosophila suggests that transient environmental manipulations can lead to the inheritance of eye color alterations in response to heat shock owing to inheritance of disrupted heterochromatin162. The transcription factor ATF-2 associates with HP1 and is required for heterochromatin formation. Stress induced phosphorylation of ATF-2 causes ATF-2 to dissociate from heterochromatin; this disrupts heterochromatin formation in the parent that is subjected to the stress and its offspring. It will be interesting to determine if inheritance of changes in histone methylation cause this phenomenon. Although correlative, these studies show that modifications of methyltransferases, demethylases or histone methyl interacting proteins have the potential to regulate biological phenotypes across generations. Specific genomic loci might maintain an epigenetic memory through inherited histone methylation patterns. As more examples of non-Mendelian inheritance are described, using these systems to understand how an epigenetic memory can escape erasure between generations will be an exciting field of research.
Perspectives
Chromatin PTMs are essential components of epigenetic regulation. In contrast to DNA CpG methylation, which is prominent only in some higher eukaryotes, histone methylation is present in organisms such as C. elegans and Drosophila in which DNA methylation is largely absent. The N-C bond formed as a result of methylation is quite stable, and yet methylation events are reversible, as evidenced by the plethora of histone demethylases. Therefore we propose that histone methylation functions to confer both stability (in terminal differentiation, for example) and reversibility (in response to stimuli, for example) of gene regulation to an organism. An appropriate balance between stable and dynamic histone methylation is thus necessary to maintain normal biological function.
Methyl modifying enzymes play a crucial role in virtually every aspect of biology and disruption of their function leads to developmental defects, diseases or aging. Many interesting questions remain beyond the further characterization of novel histone methylation marks in biology. Does histone methylation directly affect transcription? This could be tested experimentally by utilizing in vitro transcription assays with chromatinized templates. How are chromatin-modifying enzymes recruited to their target sites? Non-coding transcripts have been shown to recruit Trx and PcG proteins, but are these sufficient to explain the diversity and specificity that we observe? Are other as of yet unidentified mechanisms also involved in controlling the specificity? We expect that large-scale ChIP-seq experiments will begin to define the genomic locations of these enzymes and reveal factors (such as DNA-binding proteins) that are required for their localization.
A major challenge is posed by the realization that methylation also occurs on non-histone proteins; this makes it difficult to unequivocally assign the function of a specific methyl-modifying enzyme to modification of histones and/or non-histone substrates. One of the key first steps will be to define methylomes for various cell types. However, the bottleneck is that antibodies that can bind to methyl groups on a wide range of substrates are not readily available, but they are essential for the identification of methylomes using technologies such as quantitative mass spectrometry (for example, SILAC144). Alternative approaches may be needed. For instance, it may be possible to begin by defining methylation substrates for every methyltransferase in vitro using whole proteome protein arrays. Substantial genomic and genetic investigations are required to understand the mechanism by which methyl-modifying enzymes and readers exert their biological functions. Additionally identifying the complex network of protein-protein, protein-DNA, and protein-RNA interactions, under normal conditions and disease states, will help to delineate the specificity that allows different cells such amazing phenotypic diversity.
Supplementary Material
At-a-glance summary.
Methylation occurs on various basic residues on histones and depending on the degree of methylation and the location of the methylated residue there are different functional outcomes.
Histone methylation is a dynamic process and methyl marks can be added or removed by specific enzymes. Other proteins can recognize and bind methylated residues to effect phenotypic outcomes.
Histone modifying enzymes can be recruited to specific loci by DNA sequence, ncRNA, DNA methylation, or other post-translational marks on histone tails.
Histone methylation can be dynamic or stable through the life of a cell and even through mitosis and meiosis and can, on some occasions, be inherited from parents to children.
Histone methylation helps to explain our phenotypic diversity from cell to cell and when its regulation and the balance between stable and dynamic marks are altered diseases such as cancer and intellectual disability can ensue.
Histone methylation has been shown to play a role in virtually all biological processes from DNA repair, cell cycle, stress response, and transcription to development, differentiation, and aging.
Histone methylation levels change with age and methyltransferases and demethylases have been shown to regulate longevity of several model organisms. In some instances these enzymes have been shown to have a transgenerational effect on lifespan.
Acknowledgments
We thank E. Pollina for critical reading of the manuscript. The work from the Shi lab is supported by grants from the NIH (GM058012, GM071004, and NCI118487). E.L.G. was supported by a Helen Hay Whitney Post-Doctoral fellowship. We apologize for literature omitted due to space limitations.
Glossary Terms
- Epigenetics
Epigenetics refers to heritable changes in gene expression, which occur without alterations in DNA sequence. Unlike genetic changes, which are irreversible, histone methylation marks span the range from highly dynamic (e.g. changing along with the action of the transcription machinery) to stable through cell divisions to emerging evidence that there may be stability across generations
- SILAC
Stable isotope labeling by amino acids in cell culture. Two cell populations (experimental and control) grown in media containing only heavy (e.g. 13C, 15N) or only light forms (e.g. 12C, 14N) of particular amino acids can be compared by quantitative mass spectrometry proteomics
- Trithorax-group (Trx)
Groups of chromatin regulatory proteins which typically act to activate or maintain gene expression
- MLL complex
MLL (mixed-lineage leukemia or myeloid/lymphoid) proteins are the mammalian homologues of Trithorax and contain methyltransferase activity. This protein family is frequently at the site of chromosomal translocations. These proteins are frequently found in multi-protein complexes
- Polycomb Group (PcG)
Chromatin regulatory proteins typically involved in repressing gene expression
- PRC2
Polycomb repressive complex 2 is a polycomb group complex, which trimethylates H3K27. The core PRC2 subunits are Suz12, Eed, and the methyltransferases E(Z)/EZH2 but additional proteins do complex with these core subunits
- Pre-initiation complex
A large complex of proteins necessary for the transcription of protein-coding genes. It helps to appropriately position RNA pol II and orient the DNA in the active site of RNA pol II
- snRNP
small nuclear ribonucleoproteins are RNA protein complexes that, together with other proteins and unmodified pre-mRNA, form a complex where splicing occurs
- symmetrically dimethylated
Arginines can be dimethylated either symmetrically or asymmetrically. Symmetrically dimethylated arginines have methyl groups on each of the two nitrogen’s while asymmetrically dimethylated arginines have two methyl groups on a single nitrogen
- heterochromatin
Tightly packed DNA which is generally inactive
- X chromosome inactivation
Since females have two X chromosomes, X-inactivation by silencing one of the X chromosomes into inactive heterochromatin, prevents female cells from having twice as many X chromosome gene products as males
- RNA interference
Small RNAs which bind to specific messenger RNA molecules or to the promoters of genes can regulate their activity by sequestering, cleaving, or enhancing the target mRNA
- PHD fingers
Plant Homeo Domains are nuclear Zn2+ binding domains ranging from ~50–80 amino acids and typically have a four cysteines, one histidine, three cysteines signature. They bind to both histone and non-histone proteins and in some cases function as an E3 ligase
- WD40 repeats
A short ~40 amino acid domain usually terminating in tryptophan (W) and aspartic acid (D), which forms a circularized beta propeller structure. Can serve as scaffolding protein for multiprotein complexes
- CW domains
A ~45–55 amino acid zinc-binding domain containing at least four cysteine (C) and two tryptophan (W) residues exclusively found in eukaryotes
- PWWP domains
These ~135 amino acid domains have a central core consisting of proline tryptophan tryptophan proline (PWWP) and are found in eukaryotes from yeast to mammals. The PWWP domain has a barrel-like five-stranded structure and a five-helix bundle
- ADD domains
This domain is named after its presence in three proteins ATRX, DNMT3, and DNMT3L, which binds to histone H3. It contains a GATA-lke C2C2 zinc finger and a C4C4 imperfect PHD finger. It contains ~120 amino acids
Biographies
Eric L. Greer: Eric L. Greer graduated from Case Western Reserve University, and obtained a Ph.D. in cancer biology at Stanford University. While a graduate student Eric worked in the laboratory of Dr. Anne Brunet to study the molecular mechanisms that regulate longevity. As a post-doc, in Dr. Yang Shi’s lab, Eric is exploring the mechanisms of transgenerational epigenetic inheritance in the nematode C. elegans. Eric’s work has demonstrated that methyltransferases and demethylases can regulate longevity in the parental generation and can regulate, in a transgenerational manner, the longevity of wildtype descendants.
Yang Shi: Yang Shi is the Merton Bernfield Professor of Neonatology in the Division of Newborn Medicine at Children’s Hospital in Boston and Professor of the Department of Cell Biology at Harvard Medical School. He received his Ph.D. at New York University and conducted his postdoctoral research at Princeton University. His lab identified the first histone demethylase and continues to investigate the mechanisms that underlie dynamic regulation of histone methylation, proteins which recognize various methylated and unmethylated states of histones, and the impact of this regulation on physiological and pathological condtions.
References
- 1.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 2.Tan M, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–28. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byvoet P, Shepherd GR, Hardin JM, Noland BJ. The distribution and turnover of labeled methyl groups in histone fractions of cultured mammalian cells. Arch Biochem Biophys. 1972;148:558–67. doi: 10.1016/0003-9861(72)90174-9. [DOI] [PubMed] [Google Scholar]
- 4.Murray K. The Occurrence of Epsilon-N-Methyl Lysine in Histones. Biochemistry. 1964;3:10–5. doi: 10.1021/bi00889a003. [DOI] [PubMed] [Google Scholar]
- 5.Fischle W, Franz H, Jacobs SA, Allis CD, Khorasanizadeh S. Specificity of the chromodomain Y chromosome family of chromodomains for lysine-methylated ARK(S/T) motifs. J Biol Chem. 2008;283:19626–35. doi: 10.1074/jbc.M802655200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paik WK, Kim S. E-N-dimethyllysine in histones. Biochemical and biophysical research communications. 1967;27:479–83. [Google Scholar]
- 7.Hempel K, Lange HW, Birkofer L. Epsilon-N-trimethyllysine, a new amino acid in histones. Die Naturwissenschaften. 1968;55:37. doi: 10.1007/BF00593411. [DOI] [PubMed] [Google Scholar]
- 8.Borun TW, Pearson D, Paik WK. Studies of histone methylation during the HeLa S-3 cell cycle. J Biol Chem. 1972;247:4288–98. [PubMed] [Google Scholar]
- 9.Gershey EL, Haslett GW, Vidali G, Allfrey VG. Chemical studies of histone methylation. Evidence for the occurrence of 3-methylhistidine in avian erythrocyte histone fractions. J Biol Chem. 1969;244:4871–7. [PubMed] [Google Scholar]
- 10.Young NL, Dimaggio PA, Garcia BA. The significance, development and progress of high-throughput combinatorial histone code analysis. Cellular and molecular life sciences: CMLS. 2010;67:3983–4000. doi: 10.1007/s00018-010-0475-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi Y, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–53. doi: 10.1016/j.cell.2004.12.012. This study provides evidence of the first histone demethylase demonstrating that histone methylation is reversible and dynamic. [DOI] [PubMed] [Google Scholar]
- 12.Zee BM, et al. In vivo residue-specific histone methylation dynamics. J Biol Chem. 2010;285:3341–50. doi: 10.1074/jbc.M109.063784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–40. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rea S, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–9. doi: 10.1038/35020506. This study identified the first methyltransferase and that different modifications can affect each other. They show that phosphorylation of S10 inhibits K9 methylation. [DOI] [PubMed] [Google Scholar]
- 15.Feng Q, et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Current biology: CB. 2002;12:1052–8. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 16.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell research. 2011;21:381–95. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang J, Berger SL. The emerging field of dynamic lysine methylation of non-histone proteins. Curr Opin Genet Dev. 2008;18:152–8. doi: 10.1016/j.gde.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 18.Magnani R, Dirk LM, Trievel RC, Houtz RL. Calmodulin methyltransferase is an evolutionarily conserved enzyme that trimethylates Lys-115 in calmodulin. Nature communications. 2010;1:43. doi: 10.1038/ncomms1044. [DOI] [PubMed] [Google Scholar]
- 19.Tsukada Y, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–6. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 20.Whetstine JR, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–81. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 21.Cloos PA, et al. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature. 2006;442:307–11. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- 22.Chang B, Chen Y, Zhao Y, Bruick RK. JMJD6 is a histone arginine demethylase. Science. 2007;318:444–7. doi: 10.1126/science.1145801. [DOI] [PubMed] [Google Scholar]
- 23.Webby CJ, et al. Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science. 2009;325:90–3. doi: 10.1126/science.1175865. [DOI] [PubMed] [Google Scholar]
- 24.Cuthbert GL, et al. Histone deimination antagonizes arginine methylation. Cell. 2004;118:545–53. doi: 10.1016/j.cell.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, et al. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306:279–83. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- 26.Chan CS, Rastelli L, Pirrotta V. A Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. Embo J. 1994;13:2553–64. doi: 10.1002/j.1460-2075.1994.tb06545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tillib S, et al. Trithorax- and Polycomb-group response elements within an Ultrabithorax transcription maintenance unit consist of closely situated but separable sequences. Molecular and cellular biology. 1999;19:5189–202. doi: 10.1128/mcb.19.7.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fritsch C, Brown JL, Kassis JA, Muller J. The DNA-binding polycomb group protein pleiohomeotic mediates silencing of a Drosophila homeotic gene. Development. 1999;126:3905–13. doi: 10.1242/dev.126.17.3905. [DOI] [PubMed] [Google Scholar]
- 29.Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell. 2010;140:99–110. doi: 10.1016/j.cell.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsai MC, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pandey RR, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–46. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 34.Nagano T, et al. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–20. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 35.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–4. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schorderet P, Duboule D. Structural and functional differences in the long non-coding RNA hotair in mouse and human. PLoS genetics. 2011;7:e1002071. doi: 10.1371/journal.pgen.1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koziol MJ, Rinn JL. RNA traffic control of chromatin complexes. Curr Opin Genet Dev. 2010;20:142–8. doi: 10.1016/j.gde.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verdel A, et al. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–6. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noma K, et al. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat Genet. 2004;36:1174–80. doi: 10.1038/ng1452. [DOI] [PubMed] [Google Scholar]
- 41.Sugiyama T, Cam H, Verdel A, Moazed D, Grewal SI. RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc Natl Acad Sci U S A. 2005;102:152–7. doi: 10.1073/pnas.0407641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–9. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 43.Fukagawa T, et al. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nature cell biology. 2004;6:784–91. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- 44.Gu SG, et al. Amplification of siRNA in Caenorhabditis elegans generates a transgenerational sequence-targeted histone H3 lysine 9 methylation footprint. Nat Genet. 2012;44:157–64. doi: 10.1038/ng.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ogawa Y, Sun BK, Lee JT. Intersection of the RNA interference and X-inactivation pathways. Science. 2008;320:1336–41. doi: 10.1126/science.1157676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cernilogar FM, et al. Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature. 2011;480:391–5. doi: 10.1038/nature10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bartke T, et al. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010;143:470–84. doi: 10.1016/j.cell.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnson LM, et al. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Current biology: CB. 2007;17:379–84. doi: 10.1016/j.cub.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rajakumara E, et al. A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo. Genes Dev. 2011;25:137–52. doi: 10.1101/gad.1980311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fuks F. DNA methylation and histone modifications: teaming up to silence genes. Curr Opin Genet Dev. 2005;15:490–5. doi: 10.1016/j.gde.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 51.Rountree MR, Selker EU. DNA methylation and the formation of heterochromatin in Neurospora crassa. Heredity. 2010;105:38–44. doi: 10.1038/hdy.2010.44. [DOI] [PubMed] [Google Scholar]
- 52.Guccione E, et al. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature. 2007;449:933–7. doi: 10.1038/nature06166. [DOI] [PubMed] [Google Scholar]
- 53.Kirmizis A, et al. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature. 2007;449:928–32. doi: 10.1038/nature06160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fischle W, et al. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–22. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- 55.Horton JR, et al. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat Struct Mol Biol. 2010;17:38–43. doi: 10.1038/nsmb.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–8. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 57.Krogan NJ, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell. 2003;11:721–9. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 58.Kim J, et al. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–71. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee JS, et al. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell. 2007;131:1084–96. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- 60.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 61.Lan F, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–94. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 62.Seenundun S, et al. UTX mediates demethylation of H3K27me3 at muscle-specific genes during myogenesis. Embo J. 2010;29:1401–11. doi: 10.1038/emboj.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miller SA, Mohn SE, Weinmann AS. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol Cell. 2010;40:594–605. doi: 10.1016/j.molcel.2010.10.028. This is the first study to show that demethylases have biological function independent of their demethylase activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shi X, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–9. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wysocka J, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 66.Margueron R, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–7. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hoppmann V, et al. The CW domain, a new histone recognition module in chromatin proteins. Embo J. 2011;30:1939–52. doi: 10.1038/emboj.2011.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Y, et al. Regulation of Set9-mediated H4K20 methylation by a PWWP domain protein. Mol Cell. 2009;33:428–37. doi: 10.1016/j.molcel.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Collins RE, et al. The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules. Nat Struct Mol Biol. 2008;15:245–50. doi: 10.1038/nsmb.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–20. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 71.Bannister AJ, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–4. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 72.Flanagan JF, et al. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–5. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 73.Zhang P, et al. Structure of human MRG15 chromo domain and its binding to Lys36-methylated histone H3. Nucleic acids research. 2006;34:6621–8. doi: 10.1093/nar/gkl989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang Y, et al. TDRD3 is an effector molecule for arginine-methylated histone marks. Mol Cell. 2010;40:1016–23. doi: 10.1016/j.molcel.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Botuyan MV, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–73. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312:748–51. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- 77.Kim J, et al. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO reports. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lan F, et al. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;448:718–22. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tsai WW, et al. TRIM24 links a non-canonical histone signature to breast cancer. Nature. 2010;468:927–32. doi: 10.1038/nature09542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rajakumara E, et al. PHD finger recognition of unmodified histone H3R2 links UHRF1 to regulation of euchromatic gene expression. Mol Cell. 2011;43:275–84. doi: 10.1016/j.molcel.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sprangers R, Groves MR, Sinning I, Sattler M. High-resolution X-ray and NMR structures of the SMN Tudor domain: conformational variation in the binding site for symmetrically dimethylated arginine residues. Journal of molecular biology. 2003;327:507–20. doi: 10.1016/s0022-2836(03)00148-7. [DOI] [PubMed] [Google Scholar]
- 82.Nielsen PR, et al. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 2002;416:103–7. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- 83.Iwase S, et al. ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome. Nat Struct Mol Biol. 2011;18:769–76. doi: 10.1038/nsmb.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Eustermann S, et al. Combinatorial readout of histone H3 modifications specifies localization of ATRX to heterochromatin. Nat Struct Mol Biol. 2011;18:777–82. doi: 10.1038/nsmb.2070. [DOI] [PubMed] [Google Scholar]
- 85.Dhayalan A, et al. The ATRX-ADD domain binds to H3 tail peptides and reads the combined methylation state of K4 and K9. Human molecular genetics. 2011;20:2195–203. doi: 10.1093/hmg/ddr107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bernstein BE, et al. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci U S A. 2002;99:8695–700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Santos-Rosa H, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–11. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 88.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–8. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 89.Schulze JM, et al. Linking cell cycle to histone modifications: SBF and H2B monoubiquitination machinery and cell-cycle regulation of H3K79 dimethylation. Mol Cell. 2009;35:626–41. doi: 10.1016/j.molcel.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mohan M, et al. Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom) Genes Dev. 2010;24:574–89. doi: 10.1101/gad.1898410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 92.Lee MG, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–50. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 93.Miller SA, Huang AC, Miazgowicz MM, Brassil MM, Weinmann AS. Coordinated but physically separable interaction with H3K27-demethylase and H3K4-methyltransferase activities are required for T-box protein-mediated activation of developmental gene expression. Genes Dev. 2008;22:2980–93. doi: 10.1101/gad.1689708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee MG, Norman J, Shilatifard A, Shiekhattar R. Physical and functional association of a trimethyl H3K4 demethylase and Ring6a/MBLR, a polycomb-like protein. Cell. 2007;128:877–87. doi: 10.1016/j.cell.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 95.Tahiliani M, et al. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature. 2007;447:601–5. doi: 10.1038/nature05823. [DOI] [PubMed] [Google Scholar]
- 96.Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nature reviews. Genetics. 2010;11:285–96. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Noma K, Allis CD, Grewal SI. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–5. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- 98.Pedersen MT, Helin K. Histone demethylases in development and disease. Trends Cell Biol. 2010;20:662–71. doi: 10.1016/j.tcb.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 99.Nottke A, Colaiacovo MP, Shi Y. Developmental roles of the histone lysine demethylases. Development. 2009;136:879–89. doi: 10.1242/dev.020966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Greenberg RA. Histone tails: Directing the chromatin response to DNA damage. FEBS letters. 2011;585:2883–90. doi: 10.1016/j.febslet.2011.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Eissenberg JC, Shilatifard A. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev Biol. 2010;339:240–9. doi: 10.1016/j.ydbio.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nature reviews. Cancer. 2010;10:457–69. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Albert M, Helin K. Histone methyltransferases in cancer. Seminars in cell & developmental biology. 2010;21:209–20. doi: 10.1016/j.semcdb.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 104.Varambally S, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–9. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 105.Kleer CG, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A. 2003;100:11606–11. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Visser HP, et al. The Polycomb group protein EZH2 is upregulated in proliferating, cultured human mantle cell lymphoma. British journal of haematology. 2001;112:950–8. doi: 10.1046/j.1365-2141.2001.02641.x. [DOI] [PubMed] [Google Scholar]
- 107.van Haaften G, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41:521–3. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ernst T, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42:722–6. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 109.Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nature cell biology. 2007;9:804–12. doi: 10.1038/ncb1608. This is the first study to show that a fusion protein caused by chromosomal translocations has a causal regulation of cancer formation. [DOI] [PubMed] [Google Scholar]
- 110.Wang GG, et al. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 2009;459:847–51. doi: 10.1038/nature08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Milne TA, et al. Multiple interactions recruit MLL1 and MLL1 fusion proteins to the HOXA9 locus in leukemogenesis. Mol Cell. 2010;38:853–63. doi: 10.1016/j.molcel.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Iwase S, Shi Y. Histone and DNA modifications in mental retardation. Progress in drug research. Fortschritte der Arzneimittelforschung. Progres des recherches pharmaceutiques. 2011;67:147–73. doi: 10.1007/978-3-7643-8989-5_8. [DOI] [PubMed] [Google Scholar]
- 113.Kurotaki N, et al. Haploinsufficiency of NSD1 causes Sotos syndrome. Nat Genet. 2002;30:365–6. doi: 10.1038/ng863. [DOI] [PubMed] [Google Scholar]
- 114.Kleefstra T, et al. Further clinical and molecular delineation of the 9q subtelomeric deletion syndrome supports a major contribution of EHMT1 haploinsufficiency to the core phenotype. Journal of medical genetics. 2009;46:598–606. doi: 10.1136/jmg.2008.062950. [DOI] [PubMed] [Google Scholar]
- 115.Balemans MC, et al. Reduced exploration, increased anxiety, and altered social behavior: Autistic-like features of euchromatin histone methyltransferase 1 heterozygous knockout mice. Behavioural brain research. 2010;208:47–55. doi: 10.1016/j.bbr.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 116.Amir RE, et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–8. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 117.Iwase S, et al. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–88. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 118.Kleine-Kohlbrecher D, et al. A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation. Mol Cell. 2010;38:165–78. doi: 10.1016/j.molcel.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Qi HH, et al. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature. 2010;466:503–7. doi: 10.1038/nature09261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Liu W, et al. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature. 2010;466:508–12. doi: 10.1038/nature09272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ng D, et al. Oculofaciocardiodental and Lenz microphthalmia syndromes result from distinct classes of mutations in BCOR. Nat Genet. 2004;36:411–6. doi: 10.1038/ng1321. [DOI] [PubMed] [Google Scholar]
- 122.Lower KM, et al. Mutations in PHF6 are associated with Borjeson-Forssman-Lehmann syndrome. Nat Genet. 2002;32:661–5. doi: 10.1038/ng1040. [DOI] [PubMed] [Google Scholar]
- 123.Field M, et al. Mutations in the BRWD3 gene cause X-linked mental retardation associated with macrocephaly. American journal of human genetics. 2007;81:367–74. doi: 10.1086/520677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Skene PJ, et al. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol Cell. 2010;37:457–68. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Goldberg AD, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–91. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci U S A. 2010;107:14075–80. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010;24:1253–65. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mitson M, Kelley LA, Sternberg MJ, Higgs DR, Gibbons RJ. Functional significance of mutations in the Snf2 domain of ATRX. Human molecular genetics. 2011;20:2603–10. doi: 10.1093/hmg/ddr163. [DOI] [PubMed] [Google Scholar]
- 129.Honda S, et al. Concomitant microduplications of MECP2 and ATRX in male patients with severe mental retardation. Journal of human genetics. 2011 doi: 10.1038/jhg.2011.131. [DOI] [PubMed] [Google Scholar]
- 130.Pollina EA, Brunet A. Epigenetic regulation of aging stem cells. Oncogene. 2011;30:3105–26. doi: 10.1038/onc.2011.45. [DOI] [PubMed] [Google Scholar]
- 131.Sarg B, Koutzamani E, Helliger W, Rundquist I, Lindner HH. Postsynthetic trimethylation of histone H4 at lysine 20 in mammalian tissues is associated with aging. J Biol Chem. 2002;277:39195–201. doi: 10.1074/jbc.M205166200. [DOI] [PubMed] [Google Scholar]
- 132.Maures TJ, Greer EL, Hauswirth AG, Brunet A. The H3K27 demethylase UTX-1 regulates C. elegans lifespan in a germline-independent, insulin-dependent manner. Aging Cell. 2011;10:980–90. doi: 10.1111/j.1474-9726.2011.00738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Scaffidi P, Misteli T. Lamin A-dependent nuclear defects in human aging. Science. 2006;312:1059–63. doi: 10.1126/science.1127168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Scaffidi P, Misteli T. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nature medicine. 2005;11:440–5. doi: 10.1038/nm1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Shumaker DK, et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc Natl Acad Sci U S A. 2006;103:8703–8. doi: 10.1073/pnas.0602569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.McColl G, et al. Pharmacogenetic analysis of lithium-induced delayed aging in Caenorhabditis elegans. J Biol Chem. 2008;283:350–7. doi: 10.1074/jbc.M705028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Greer EL, et al. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature. 2010;466:383–7. doi: 10.1038/nature09195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Siebold AP, et al. Polycomb Repressive Complex 2 and Trithorax modulate Drosophila longevity and stress resistance. Proc Natl Acad Sci U S A. 2010;107:169–74. doi: 10.1073/pnas.0907739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jin C, et al. Histone demethylase UTX-1 regulates C. elegans life span by targeting the insulin/IGF-1 signaling pathway. Cell metabolism. 2011;14:161–72. doi: 10.1016/j.cmet.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 140.Li L, Greer C, Eisenman RN, Secombe J. Essential functions of the histone demethylase lid. PLoS genetics. 2010;6:e1001221. doi: 10.1371/journal.pgen.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ni Z, Ebata A, Alipanahiramandi E, Lee SS. Two SET domain containing genes link epigenetic changes and aging in Caenorhabditis elegans. Aging Cell. 2011 doi: 10.1111/j.1474-9726.2011.00785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sogo JM, Stahl H, Koller T, Knippers R. Structure of replicating simian virus 40 minichromosomes. The replication fork, core histone segregation and terminal structures. Journal of molecular biology. 1986;189:189–204. doi: 10.1016/0022-2836(86)90390-6. [DOI] [PubMed] [Google Scholar]
- 143.Bonne-Andrea C, Wong ML, Alberts BM. In vitro replication through nucleosomes without histone displacement. Nature. 1990;343:719–26. doi: 10.1038/343719a0. [DOI] [PubMed] [Google Scholar]
- 144.Xu M, et al. Partitioning of histone H3-H4 tetramers during DNA replication-dependent chromatin assembly. Science. 2010;328:94–8. doi: 10.1126/science.1178994. This paper uses SILAC to show that histone dimers can be passaged in a conserved manner through mitosis. [DOI] [PubMed] [Google Scholar]
- 145.Katan-Khaykovich Y, Struhl K. Splitting of H3-H4 tetramers at transcriptionally active genes undergoing dynamic histone exchange. Proc Natl Acad Sci U S A. 2011;108:1296–301. doi: 10.1073/pnas.1018308108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Martin C, Zhang Y. Mechanisms of epigenetic inheritance. Curr Opin Cell Biol. 2007;19:266–72. doi: 10.1016/j.ceb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 147.Moazed D. Mechanisms for the inheritance of chromatin States. Cell. 2011;146:510–8. doi: 10.1016/j.cell.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Blobel GA, et al. A reconfigured pattern of MLL occupancy within mitotic chromatin promotes rapid transcriptional reactivation following mitotic exit. Mol Cell. 2009;36:970–83. doi: 10.1016/j.molcel.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Francis NJ, Follmer NE, Simon MD, Aghia G, Butler JD. Polycomb proteins remain bound to chromatin and DNA during DNA replication in vitro. Cell. 2009;137:110–22. doi: 10.1016/j.cell.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Simpson VJ, Johnson TE, Hammen RF. Caenorhabditis elegans DNA does not contain 5-methylcytosine at any time during development or aging. Nucleic acids research. 1986;14:6711–9. doi: 10.1093/nar/14.16.6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Youngson NA, Whitelaw E. Transgenerational epigenetic effects. Annual review of genomics and human genetics. 2008;9:233–57. doi: 10.1146/annurev.genom.9.081307.164445. [DOI] [PubMed] [Google Scholar]
- 152.Vastenhouw NL, et al. Gene expression: long-term gene silencing by RNAi. Nature. 2006;442:882. doi: 10.1038/442882a. [DOI] [PubMed] [Google Scholar]
- 153.Grishok A, Tabara H, Mello CC. Genetic requirements for inheritance of RNAi in C. elegans. Science. 2000;287:2494–7. doi: 10.1126/science.287.5462.2494. [DOI] [PubMed] [Google Scholar]
- 154.Rechavi O, Minevich G, Hobert O. Transgenerational Inheritance of an Acquired Small RNA-Based Antiviral Response in C. elegans. Cell. 2011 doi: 10.1016/j.cell.2011.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hall IM, et al. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–7. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- 156.Karachentsev D, Sarma K, Reinberg D, Steward R. PR-Set7-dependent methylation of histone H4 Lys 20 functions in repression of gene expression and is essential for mitosis. Genes Dev. 2005;19:431–5. doi: 10.1101/gad.1263005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hajkova P, et al. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452:877–81. doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Muramoto T, Muller I, Thomas G, Melvin A, Chubb JR. Methylation of H3K4 Is required for inheritance of active transcriptional states. Current biology: CB. 2010;20:397–406. doi: 10.1016/j.cub.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 159.Katz DJ, Edwards TM, Reinke V, Kelly WG. A C. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell. 2009;137:308–20. doi: 10.1016/j.cell.2009.02.015. This is the first paper to show that there is a potential inheritance of histone methylation marks through generations in multicellular organisms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Nottke AC, et al. SPR-5 is a histone H3K4 demethylase with a role in meiotic double-strand break repair. Proc Natl Acad Sci U S A. 2011;108:12805–10. doi: 10.1073/pnas.1102298108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Greer EL, et al. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature. 2011;479:365–71. doi: 10.1038/nature10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Seong KH, Li D, Shimizu H, Nakamura R, Ishii S. Inheritance of stress-induced, ATF-2-dependent epigenetic change. Cell. 2011;145:1049–61. doi: 10.1016/j.cell.2011.05.029. [DOI] [PubMed] [Google Scholar]
- 163.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–11. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 164.Deng W, Blobel GA. Do chromatin loops provide epigenetic gene expression states? Curr Opin Genet Dev. 2010;20:548–54. doi: 10.1016/j.gde.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kahn TG, Schwartz YB, Dellino GI, Pirrotta V. Polycomb complexes and the propagation of the methylation mark at the Drosophila ubx gene. J Biol Chem. 2006;281:29064–75. doi: 10.1074/jbc.M605430200. [DOI] [PubMed] [Google Scholar]
- 166.Shogren-Knaak M, et al. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–7. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 167.Suganuma T, Workman JL. Signals and combinatorial functions of histone modifications. Annual review of biochemistry. 2011;80:473–99. doi: 10.1146/annurev-biochem-061809-175347. [DOI] [PubMed] [Google Scholar]
- 168.Bell O, Tiwari VK, Thoma NH, Schubeler D. Determinants and dynamics of genome accessibility. Nature reviews. Genetics. 2011;12:554–64. doi: 10.1038/nrg3017. [DOI] [PubMed] [Google Scholar]
- 169.Lange M, et al. Regulation of muscle development by DPF3, a novel histone acetylation and methylation reader of the BAF chromatin remodeling complex. Genes Dev. 2008;22:2370–84. doi: 10.1101/gad.471408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Pray-Grant MG, Daniel JA, Schieltz D, Yates JR, 3rd, Grant PA. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–8. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- 171.Sims RJ, 3rd, et al. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J Biol Chem. 2005;280:41789–92. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Li B, et al. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007;316:1050–4. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- 173.Cirillo LA, et al. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–89. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 174.Serandour AA, et al. Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome research. 2011;21:555–65. doi: 10.1101/gr.111534.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Guccione E, et al. Myc-binding-site recognition in the human genome is determined by chromatin context. Nature cell biology. 2006;8:764–70. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- 176.Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–92. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Vermeulen M, et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 178.Zhu L, et al. Patterns of exon-intron architecture variation of genes in eukaryotic genomes. BMC genomics. 2009;10:47. doi: 10.1186/1471-2164-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Vermeulen M, et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142:967–80. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 180.Sims RJ, 3rd, et al. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28:665–76. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kolasinska-Zwierz P, et al. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41:376–81. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.de Almeida SF, et al. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat Struct Mol Biol. 2011;18:977–83. doi: 10.1038/nsmb.2123. [DOI] [PubMed] [Google Scholar]
- 184.Luco RF, et al. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kim S, Kim H, Fong N, Erickson B, Bentley DL. Pre-mRNA splicing is a determinant of histone H3K36 methylation. Proc Natl Acad Sci U S A. 2011;108:13564–9. doi: 10.1073/pnas.1109475108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.McMahon KA, et al. Mll has a critical role in fetal and adult hematopoietic stem cell self-renewal. Cell stem cell. 2007;1:338–45. doi: 10.1016/j.stem.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 187.Cerveira N, et al. Frequency of NUP98-NSD1 fusion transcript in childhood acute myeloid leukaemia. Leukemia: official journal of the Leukemia Society of America, Leukemia Research Fund, U K. 2003;17:2244–7. doi: 10.1038/sj.leu.2403104. [DOI] [PubMed] [Google Scholar]
- 188.van Zutven LJ, et al. Identification of NUP98 abnormalities in acute leukemia: JARID1A (12p13) as a new partner gene. Genes, chromosomes & cancer. 2006;45:437–46. doi: 10.1002/gcc.20308. [DOI] [PubMed] [Google Scholar]
- 189.Lin YH, et al. Global reduction of the epigenetic H3K79 methylation mark and increased chromosomal instability in CALM-AF10-positive leukemias. Blood. 2009;114:651–8. doi: 10.1182/blood-2009-03-209395. [DOI] [PubMed] [Google Scholar]
- 190.Manuyakorn A, et al. Cellular histone modification patterns predict prognosis and treatment response in resectable pancreatic adenocarcinoma: results from RTOG 9704. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2010;28:1358–65. doi: 10.1200/JCO.2009.24.5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Elsheikh SE, et al. Global histone modifications in breast cancer correlate with tumor phenotypes, prognostic factors, and patient outcome. Cancer research. 2009;69:3802–9. doi: 10.1158/0008-5472.CAN-08-3907. [DOI] [PubMed] [Google Scholar]
- 192.Barlesi F, et al. Global histone modifications predict prognosis of resected non small-cell lung cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2007;25:4358–64. doi: 10.1200/JCO.2007.11.2599. [DOI] [PubMed] [Google Scholar]
- 193.Park YS, et al. The global histone modification pattern correlates with cancer recurrence and overall survival in gastric adenocarcinoma. Annals of surgical oncology. 2008;15:1968–76. doi: 10.1245/s10434-008-9927-9. [DOI] [PubMed] [Google Scholar]
- 194.Seligson DB, et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–6. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- 195.Fraga MF, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 196.Wei Y, et al. Loss of trimethylation at lysine 27 of histone H3 is a predictor of poor outcome in breast, ovarian, and pancreatic cancers. Molecular carcinogenesis. 2008;47:701–6. doi: 10.1002/mc.20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Seligson DB, et al. Global levels of histone modifications predict prognosis in different cancers. The American journal of pathology. 2009;174:1619–28. doi: 10.2353/ajpath.2009.080874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.