Abstract
In this report, we present a new method for sensitive detection of short DNA sites in single cells with single base resolution. The method combines peptide nucleic acid (PNA) openers as the tagging probes, together with isothermal rolling circle amplification (RCA) and fluorescence-based detection, all performed in a cells-in-flow format. Bis-PNAs provide single base resolution, while RCA ensures linear signal amplification. We applied this method to detect the oncoviral DNA inserts in cancer cell lines using a flow-cytometry system. We also demonstrated quantitative detection of the selected signature sites within single cells in microfluidic nano-liter droplets. Our results show single-nucleotide polymorphism (SNP) discrimination and detection of copy-number variations (CNV) under isothermal non-denaturing conditions. This new method is ideal for many applications in which ultra-sensitive DNA characterization with single base resolution is desired on the level of single cells.
INNOVATION
In this report we describe a novel isothermal method based on peptide nucleic acid (PNA) and rolling circle amplification (RCA) for ultrasensitive detection and quantitation of short specific DNA sites in single cells with single base resolution in a cells-in-flow format. In comparison to a Flow-FISH technique which was previously used to analyze in flow short DNA sequences in chromosomes, our technique does not require any pre-treatment to denature cellular DNA such as high temperatures and high concentrations of form amide. Moreover, we simplified the flow component by adapting our PNA-RCA method to accommodate a one-step nanoscale isothermal assay using a droplet-based microfluidic system. In this method, single cells and assay specific reagents are all encapsulated within single droplets, such that each cell is assayed in a separate reaction vessel of reduced volume, so that mixing and reaction rates are enhanced and contamination is minimized or eliminated.
INTRODUCTION
The biological and medical importance of genomic variation that underlies susceptibility to various common diseases or responses to drugs has become the main motivation for the development of novel diagnostic methods for genetic analysis1–3. Genomic DNA analysis requires sample pre-treatment processes, such as extraction and purification of the target nucleic acids, and after nucleic acid amplification, sequencing and detection. These labor-intensive and time-consuming processes usually result in a high cost for diagnosis. In addition, the accuracy of the diagnosis may be affected because of potential contamination from manual processing. In parallel, advances in microfluidics technology have yielded numerous miniaturized systems which are being deployed for a number of biomedical applications, including the rapid diagnosis of genetic diseases. These miniaturized systems provide innovative tools that significantly enhance the diagnostic sensitivity of DNA detection methods while lowering sample and reagent consumption, thereby increasing throughput and sensitivity and decreasing cost and time of analysis4,5. Microfluidic droplet technology is particularly advantageous when single-cell/single-molecule analysis is required6–8, of particular importance in understanding cell population heterogeneity effects.
In this paper, we present an approach for sensitive detection of short DNA sequences in single mammalian cells with single-base resolution. In this approach, we combined peptide nucleic acid (PNA)-based DNA recognition of short sequences, and signal enhancement provided by the rolling circle amplification (RCA) method, with a cells-in-flow format to identify small genetic variations in individual DNA molecules. To reduce the cost and volume of reagents as well as to perform single-cell analysis, we integrated the PNA-RCA assay into nanoliter volume microfluidic droplets.
DNA-mimicking peptide nucleic acids (PNAs) have proven to be a convenient tool for research and diagnostic assays9–11. PNAs are a prominent class of artificial nucleic acid analogs with a peptide-like backbone onto which nucleobases are grafted in a designed sequence12. It was shown that cationic pyrimidine bis-PNAs exhibit strong and selective binding to duplex DNA creating a local opening of a chosen genomic DNA target site. The displaced dsDNA strand becomes accessible for Watson-Crick pairing with a DNA oligomer that makes possible the formation of an unusual PNA-DNA construct known as a PD-loop structure13–15. The PD-loop formation is exceedingly sequence-specific, owing to the simultaneous formation of Watson-Crick and Hoogsteen base-pairs14,15. Importantly, this phenomenon is limited to the pre-selected 20–25-bp-long site, while the remaining DNA structure preserves its double-helical formation. The PD-loop construct allows for amplification and fluorescent detection of a variety of genetic markers for diagnostics (see schematic in Fig. 1)16,17.
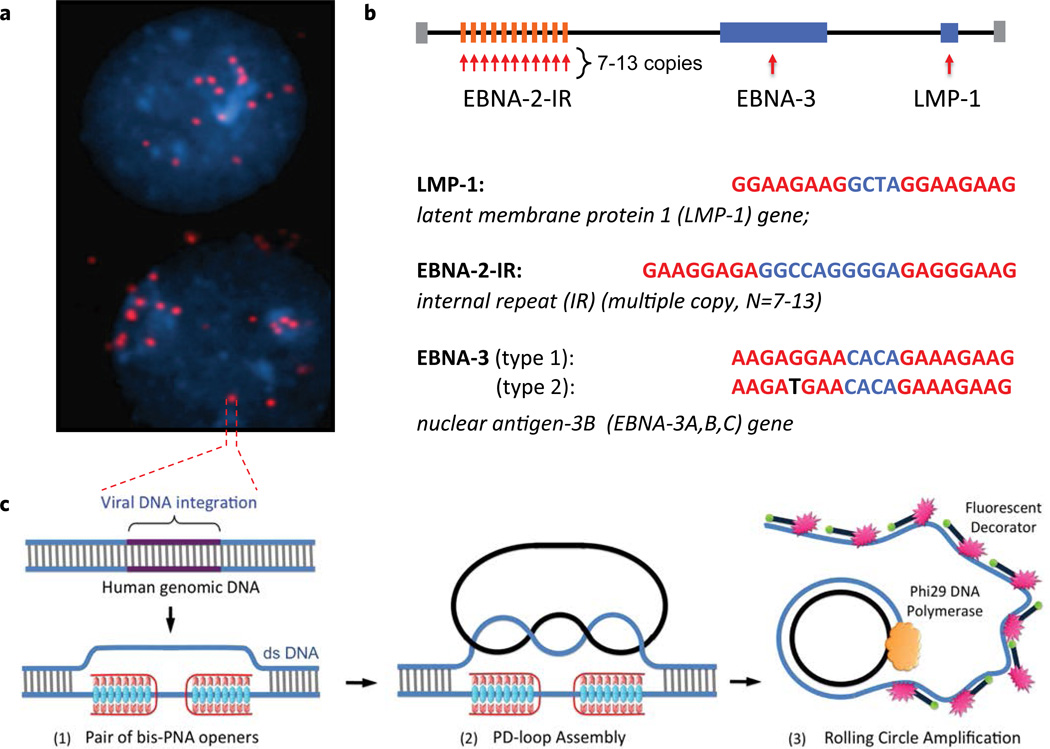
Figure 1.
The PNA-RCA method for detection of short specific DNA target site at the single cell. (a) Multiple fluorescent spots were observed in the BC-1 cells (EBV-positive) when probe corresponding to the LMP-1 gene encoding major transforming protein of Epstein-Barr Virus was applied. The fluorescent signals were acquired separately using two filter sets: DAPI for DNA and Cy3 for labeled RCA product. (b) Target sites in the EBV genomic DNA used in this study. Sequences targeted by PNAs are shown in red. EBNA-3(G)-EBV type 1 signature sites within the EBNA-3 gene differ by a single nucleotide (SNP) from EBNA-3(T)-EBV type 2 in the PNA binding sequence. Mismatch is shown in black. (c) Schematic depiction of the PNA-RCA method for sensitive and specific targeting of unique sequences on non-denatured human genomic DNA. (I) The PNA openers specifically bind to two closely located homopurine DNA sites that are separated by several mixed purine-pyrimidine bases and locally open the double-stranded DNA. (II) The opened region serves as a target for hybridization and ligation of an oligonucleotide probe to form a PD-loop. (III) The small circle on duplex DNA serves as a template for isothermal RCA reaction, which yields a long, single-stranded amplicon that contains thousands of copies of the target sequence. For the fluorescence-based detection, a fluorophore-tagged decorator probes are hybridized to the RCA product.
Previously, we showed that the PNA-RCA approach can be used for fluorescent in situ detection of short single-copy DNA sequences within the bacteria18,19 and more recently in human cells20. Here we demonstrate that this approach can be used in a cells-in-flow format where we are able to detect single base changes. Both DNA opening with PNA probes and RCA amplification require physiologic temperatures, under which cells remain intact and viable, thus obviating the necessity of isolating genomic DNA, a step required for other DNA analyses.
To demonstrate the diagnostic potential of this method, we addressed a clinically relevant problem of detecting cancer-associated herpes-virus in a human cell line, which carries oncoviral DNA inserts within their genomes. Chromosomal integration of these viruses might lead to genetic abnormalities causing malignant transformation of the infected cells21–25. Detecting these viruses in patients with lymphoid diseases and related disorders is of significant clinical importance, as such illnesses are frequently treated differently than their morphologically similar, but non-oncovirus-associated malignancies. Specifically, we focused on Epstein-Barr Virus (EBV) that infects and transforms human B-lymphocytes with an average of 2 to 30 integrated copies or viral episomes per cell, depending on the cell line21. The pathogenesis of EBV-positive lymphomas in man varies in their association with types 1 and 2 EBV26,27. It has been shown28 that EBV type 2 infections are less likely to cause tumors, and if developed, those tumors have a longer incubation period compared to EBV type 1. Even though EBV types 1 and 2 are very homologous, the viral genomes have polymorphisms in the latent genes EBNA2, 3A, 3B, and 3C29–31 and these differences can be used for their identification. To show that our cell-in-flow PNA-RCA method can discriminate SNPs, we performed EBV classification by selecting the signature sites within EBNA-3 gene that has a single G/T mutation for type1/type2, respectively).
We also demonstrated that the cell-in-flow PNA-RCA method could be adapted to a nanoscale assay format. Recently, microfluidic droplet systems have attracted significant interest as they allow picoor nanoliter volumes of samples and reagents to form extremely high-density microreactors32–36. In our work, cells and their specific reagents are all encapsulated within single droplets, such that each cell is assayed in a separate reaction vessel of reduced volume, in which mixing and reaction rates are enhanced and contamination is eliminated. Using this droplet-based microfluidic system, we visualized viral genomes in infected cell and quantified oncoviral DNA in single cells. We conclude that this assay can be applied to identify both the type and multiplicity of viral infection in human cells.
MATERIAL AND METHODS
Cell lines
We performed experiments on well-characterized cell lines that are available from the American Type Culture Collection (ATCC). Cells were propagated according to ATCC protocol. For assays of EBV, we used BC-1 (CRL-2230) B-cell derived from human virus-associated malignancies, AIDS-related body-cavity-based lymphoma cell line and CESS (TIB-190), which are EBV-positive B lymphoblastic cell line derived from patients with myelomonocytic leukemia. As controls for these experiments, we used two EBV-negative human cell lines, Ramos (CRL-1596) B-lymphocyte and CCRF-CEM cells (CCL-119) T-lymphoblast.
Signature site design
We selected a set of target sites, which exhibit maximal diagnostic potential. The PD-loop sites were selected using Human Herpesvirus 4 (EBV) genome sequences available from the Genomes Database (http://www.ncbi.nlm.nih.gov/genomes/GenomesGroup.cgi?taxid=10292). PNAs were purchased from Panagene Inc. PNA-binding sites with different sequences between them were selected. We determine the target site as an arbitrary sequence of nucleobases (up to 10 bp) flanked on both sides by the binding sites for PNA openers, each of which must consist of a short (7–10 bp-long) homopurine–homopyrimidine tract. Thus, the length of the entire site is about 20–30 bp. Statistically, one such site suitable for PD-loop formation is expected per several hundred base pairs of DNA sequence, on average14. The padlock probe is designed in such a manner that its termini are complementary to the displaced strand of the DNA target. Upon circularization, it becomes topologically linked to the target DNA. All oligonucleotide probes used in this study were purchased from Integrated DNA Technologies, Inc. (Coralville, IA). Sequences of padlock oligomers are given in Table 1.
Table 1.
PD-sites, PNAs, and circularizable ODN probes.
| PD-sites* | PNAs** | Circularizable linear oligonucleotide (ODN) padlock probes*** |
|---|---|---|
|
LMP-1: GGAAGAAGGCTAGGAAGAAG |
PNA1: H-Lys2-JJTTJTTJ- (eg1)3-CTTCTTCC-Lys-NH2 |
5′-pTAGGAAGAAG-TCACGGAATGGTTACTTGCACAGCCAGCCAGCC TCACGGAATGGTTACTTGCACAGC-GGAAGAAGGC-3′ |
|
EBNA-3: AAGAGGAACACAGAAAGAAG (type 1) |
PNA2: H-Lys2-TTJTJJTT- (eg1)3-TTCCTCTT-Lys-NH2 |
5′-pCAGAAAGAAG-TCACGGAATGGTTACTTGCACAGCCAGCCAGCC TCACGGAATGGTTACTTGCACAGC-AAGAGGAACA-3′ |
| AAGATGAACACAGAAAGAAG (type 2) |
PNA3: H-Lys-CTTCTTTC- (eg1)3-JTTTJTTJ-Lys2-NH2 |
|
|
EBNA-2 IR: GAAGGAGAGGCCAGGGGAGAGGGAAG (multiple copy, N = 7 – 13) |
PNA4: H-Lys-TCTCCTTC- (eg1)3-JTTJJTJT-Lys2-NH2 |
5′-pGGGGAGAGGGAAG-TCACGGAATGGTTACTTGCACAGCCAGC CAGCCTCACGGAATGGTTACTTGCACAGC-GAAGGAGAGGCCA-3′ |
|
PNA5: H-Lys-CTTCCCTC- (eg1)3-JTJJJTTJ-Lys2-NH2 |
||
| RCA primer | 5′-GTGAGGCTGCTGGCTG-3′ | |
| Decorator probe | CY3-TCACGGAATGGTTACTTGCACAGC-biotin |
Binding sites are underlined;
eg1, Lys, and J denote the bis-PNA linker segment, the amino acid lysine, and the nucleobase pseudoisocytosine;
terminal sequences covering PD-site are bolded; decorator binding sites are underlined; primer-binding sites are bolded and italicized.
Flow cytometry
Approximately 106 cells in 100 µl were washed with PBS, followed by centrifugation and resuspension in 100 µl of fixation solution (4% formaldehyde in PBS). Cells were fixed for 15 minutes on ice and rinsed three times in a sodium-phosphate buffer (pH 6.8). Then a standard protocol used earlier for in situ detection has been applied with slight modification20. Specifically, at each step cells were spin down and supernatant with excess of probes and reagents was removed. Step 1. Binding of PNA openers: cells were incubated at 45°C for 4 h with the corresponding PNAs (2 µM final concentration) in 10mM Na-phosphate buffer (pH 6.8) containing 0.1mM EDTA. Step 2. Probe circularization: cells with PNA-opened DNA samples and padlock oligonucleotide probes (10 µM) were incubated with 10U of T4 DNA ligase for 2 h at 37°C in 0.5X T4 DNA Ligase Reaction Buffer. Step 3. The RCA was performed by adding 100 µl of the reaction mixture containing 2 µM of primer, 10U of phi29 DNA polymerase (New England Biolabs), 200 µM dNTPs, in 1X phi29 DNA polymerase buffer. In addition, the reaction mixture contained 20 µM of fluorescently labeled decorator probe (linear oligonucleotides with fluorophores at their termini). The RCA reaction was performed for 2 h at 37°C. The cells were then washed twice in PBS buffer. Cells were analyzed using BD FACSCalibur™ Flow Cytometer with Sorting Option. Additionally, concentrated cell suspension have been dropped onto a clean slide, air-dried, and stained by DAPI (4,6–diamidino–2–phenylindole), a fluorescent dye specific for DNA, and inspected by fluorescent microscopy.
Microfluidic device fabrication
Microfluidic flow chambers were fabricated by soft lithography. Negative photo resist SU-8 2025 or SU-8 2100 (MicroChem, Newton, MA) was deposited onto clean silicon wafers to a thickness of 150 µm and patterned by exposure to UV light through a transparency photomask (CAD/Art Services, Bandon, OR). The Sylgard 184 PDMS (Dow Corning, Midland, MI) was mixed with crosslinker (ratio 10:1), poured onto the photoresist patterns, degassed thoroughly, and cured for at least 1 h at 65°C. The PDMS devices were peeled off the wafer and bonded to glass slides after oxygen-plasma activation of both surfaces. To improve the wetting of the channels with mineral oil in the presence (1 w/w%) of the surfactant (span80), the microfluidic channels were pre-treated with Aquapel (PPG Industries, Pittsburgh, PA) by filling the channels with the solution as received and then flushing with air. Polyethylene tubing with an inner diameter of 0.38mm and an outer diameter of 1.09mm (Becton Dickinson, Franklin Lakes, NJ) connected the channels to the syringes. Syringes were used to load the fluids into the devices and syringe pumps controlled the flow rates.
The generation of monodisperse droplets in a microchannel through shearing flow at a flow-focusing zone is illustrated in Fig. 3a. Three perpendicular inlet channels form a nozzle. The center stream contains a suspension of cells (105 cells/ml) and the RCA reaction mix, while the two opposing side streams contain the oil phase. Individual syringe pumps control the flow rate of oil and cell suspension. To form droplets, the flow-rate ratio of water to oil is adjusted to the Q w=Q o = 0:5 (Q w= 1 µl per min and Q o = 2 µl per min). The generated droplet volume is ≈1.8 nl, corresponding to a spherical-drop diameter of 150 µm.
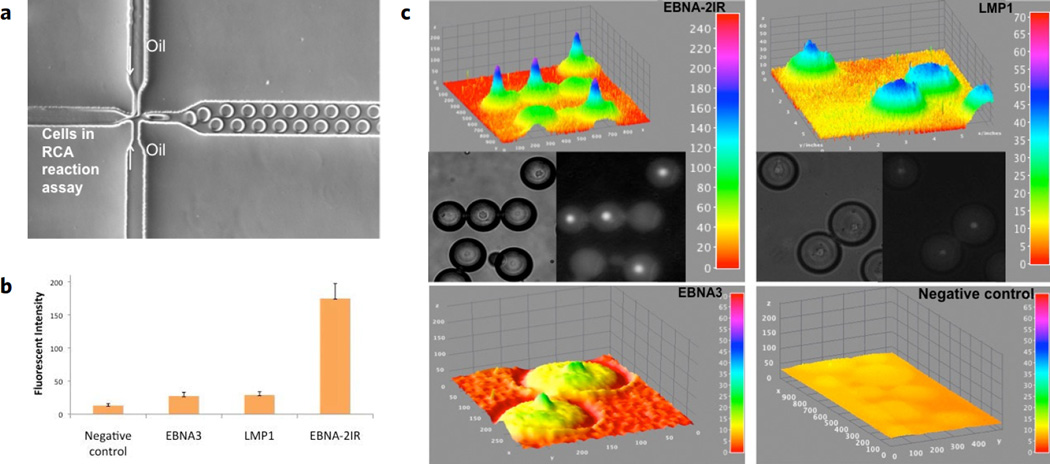
Figure 3.
Copy-number variation (CNV) detection by the microfluidic droplet PNA-RCA method. (a) A microfluidic platform consisted of droplet generation chip and an in-channel incubation chamber. The generation of monodisperse droplets is illustrated in a microchannel through shearing flow at a flow-focusing. (b) Quantitation of oncoviral DNA target sites within BC-1 cell line via the PNA-RCA assay in droplets. Fluorescent intensities were recorded for the single copy genes LMP-1 and EBNA-3 and for the multiple copies EBNA-2 IR repeats target sites. (c) Representative droplet images (DIC – left panel, CY3 – right panel) and the fluorescence intensity distributions from these images are plotted as 3D surface plots in ImageJ (top panel, the color code at the right indicates fluorescence intensities). 3D intensity profiles (z-axis) were obtained from a single cell within each droplet at the end of amplification process.
Image analysis
Fluorescence images of droplets were captured on a Zeiss 200 Axiovert microscope using an AxioCAM MRm digital camera. For the cell studies, fluorescence signals were captured separately using appropriate filter set for Cy3. Image processing and analysis was conducted using ImageJ software.
RESULTS
Principle of the method
The schematic of the PNA-RCA method is shown in Fig. 1c. The PNA-oligomers spontaneously penetrates double-stranded DNA (dsDNA) at two closely located sites through simultaneous formation of Watson-Crick and Hoogsteen base pairs12,15. The PNAs bind to one of the two DNA strands of a target sequence, leaving the opposite strand displaced. The opened site is 20–30 nucleotides long, while the remainder of the DNA structure preserves the overall double-helical form (Fig. 1c,(1)). To form a PD-loop, a linear oligonucleotide (ODN) was designed in such a manner that its termini, separated by a linker with an arbitrary sequence, are complementary to the displaced strand of the DNA target. The PD-loop formation is extremely sequence-specific because nothing but the target site is accessible. After hybridization to the target, both ends of the linear ODN appear in juxtaposition and are covalently joined by DNA ligase (Fig. 1c,(2)). The resulting circle forms about two turns of a double helix with the displaced DNA strand, creating a truly topological link. To produce a strong signal from individual PD-loop probes, the RCA reaction was performed using a single primer and the phi29 DNA polymerase. Isothermal linear RCA of the circular probes yields a long single-stranded DNA concatemeric amplicon that contains thousands of copies of the target sequence37. These single-stranded amplicons were concurrently hybridized with linear oligonucleotides with CY-3-fluorophores at their termini - "decorator probe" (Table 1). This process yields a product with multiple fluorescent labels (Fig. 1c,(3)). The RCA product remains bound to its target site due to the naturally formed secondary structure and generates a strong fluorescent signal in every cell.
Targeting of specific oncoviral signature sites
To validate the method, we performed experiments on well-characterized EBV-positive cells, the BC-1 human cell line. In all experiments, the EBV-negative CCRF-CEM cell line was used as a negative control. We chose to detect three EBV signature sites as follows (Fig. 1b): LMP1 is a site within the gene encoding major transforming protein of EBV; EBNA-3 is a site within the gene for viral transcription factor that induces expression of LMP1 and LMP2; and EBNA-2IR is a multiple copy repeat (7–13 copies) within the gene for the nuclear antigen of latent infection that affects viral and cellular gene expression in EBV-infected cells29. Corresponding PNA-oligomers have been ordered to target LMP-1, EBNA-3 and EBNA-2IR sites. The recognition sequences within EBV genome targeted by PNA are shown in Fig. 1b.
DNA within fixed cells was treated with bis-PNA probes, hybridized with the padlock probe that was circularized by ligation. This was followed by the linear RCA reaction. When examined by fluorescent microscopy, the cells revealed multiple fluorescent spots in every nucleus of the BC-1 cell line (Fig. 1a). This result confirmed earlier data suggesting that the EBV virus can be present in 2–30 copies per cell21.
Next, the entire procedure was implemented in a cells-in-flow format. Specifically, we performed the experiments using the EBV-specific probes with both the BC-1 (EBV+) and the CCRL-CEM (EBV−) human cell lines. As can be seen in Fig. 2a, the BC-1 cell line, carrying the corresponding LMP-1 target site within the integrated provirus genome produced a fluorescent signal well above the level observed in the negative control CCRF-CEM cells. Additional negative controls were carried out in a cells-in-flow format as follows: 1) without PNA probes in the solution, and 2) substituting the EBV PNA probes with an irrelevant PNA probe cocktail consisting of four different PNA probes (Supplementary Fig. 4). In these experiments the PNA and DNA probes were at the same concentration as the specific PNA probes and DNA oligomers. The signal in both control experiments did not exceed that of the background, suggesting that the flow-based format of PNA-RCA assay allowed specific EBV-detection.
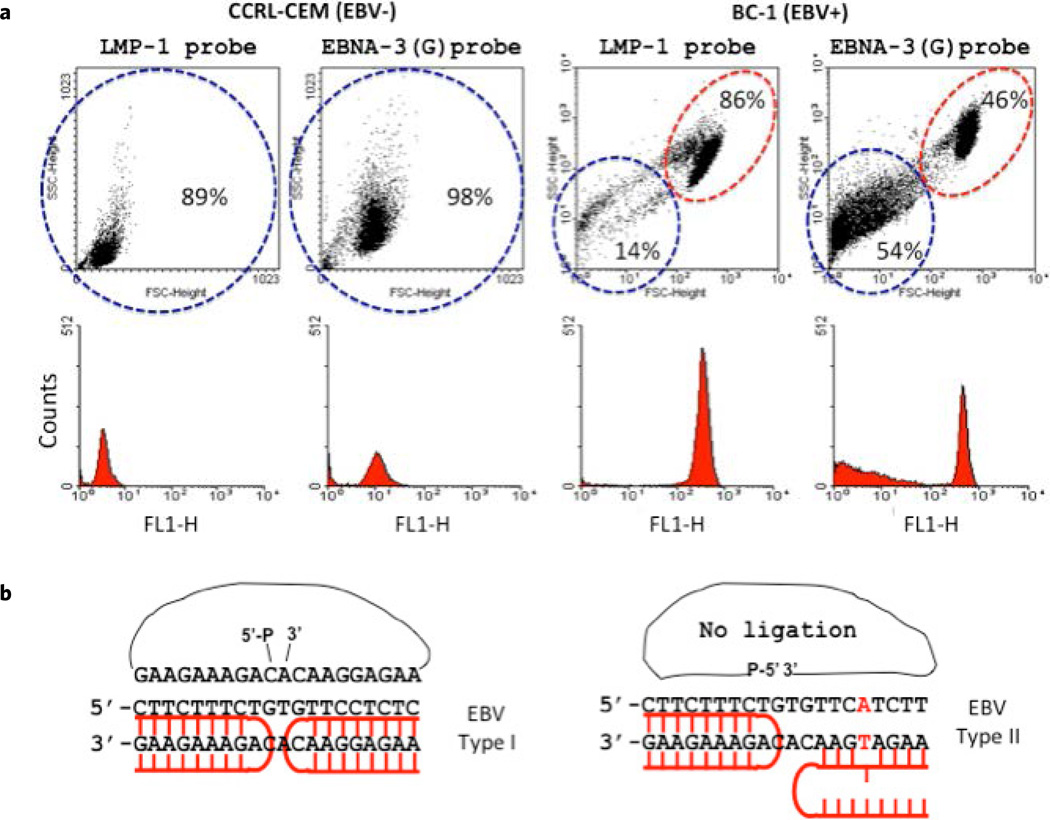
Figure 2.
FACS analysis discriminates single nucleotide polymorphism in types 1 and 2 EBV. (a) The BC-1 (EBV-positive) and the CCRF-CEM (EBV-negative) cell lines were subjected to PNA-RCA assay and analyzed by FACS. (b). Scheme of EBV typing based on known polymorphisms in the latent genes EBNA-3(G) EBNA-3(T) signature sites within EBNA-3 gene. The ligation of the padlock probe was accomplished only for the EBV-type 1 BC-1 cells while SNP at EBNA-3(T) prohibits the ligation and consequently the RCA detection of the EBV-type 2 BC-1 cells.
Typing of viral DNA with single nucleotide resolution
Different types of the EBV display different levels of transforming efficiency and prevalence pattern; therefore it is important to distinguish between various types. Even though types 1 and 2 EBV are highly homologous, the viral genomes can be typed based on known polymorphisms in the latent genes EBNA2, 3A, 3B, and 3C29–31. In order to test sequence specificity of our method and its feasibility for EBV classification, we selected EBNA-3(G)-EBV type 1 signature site within the EBNA-3 gene that differs by a single nucleotide (SNP) from EBNA-3(T)-EBV type 2 in the PNA binding sequence (Fig. 2b). It was shown previously that the BC-1 cell line demonstrated the presence of both potential intertypic recombinants of the EBV types 1 and 227,38–40. We therefore applied the LMP-1 probe, which is common for both types of EBV in the BC-1 cell line, and probe EBNA-3(G) EBV that is specific for EBV type 1 to determine type specificity of the EBV. Figure 2b shows the structure of the PNA-DNA complex that allows ligation of the padlock probe followed by RCA and thus detection of the EBV type 1 signature sequence. Due to the stringent strand-matching requirement for PD-loop formation and the ligation of the padlock probe, the mismatch in type 2 at EBNA-3(T) prohibits the RCA amplification of the type 2 EBNA-3 gene (Fig. 2b). Figure 2a shows the results obtained by FACS analysis on the BC-1 (EBV+) cell line which was tested for EBNA-3(G) probe. The fluorescent signal with the EBNA-3(G) probe was detected in approximately half of the BC-1 (EBV+) cells, while LMP-1 probe (common for both EBV types) gave a fluorescent signal in virtually all cells. No signal was observed with both probes in the EBV-negative control (CCRL-CEM) cell line. Collectively, these results suggest successful SNP discrimination within the EBV EBNA-3A gene in BC-1 cells. Such high sequence specificity has been demonstrated earlier in PNA-based detection assays confirming the zero mismatch tolerance of the PNA-RCA approach19,41.
Quantitation of gene copy number in microfluidic droplet format
To demonstrate that the entire process could be performed in a one-step, droplet-based microfluidic system, we integrated the reagents for the PNA-RCA reaction within monodisperse aqueous-emulsion nanoliter droplets containing single cells. In order to test the quantitative capability of our approach, we focused on detection of single copy LMP1 and EBNA-3 and multiple copy EBNA-2IR signature sites. Specifically, after formation of the PD-loops, cells were provided with the circular probes and then encapsulated in distinct nanoliter-sized drops of RCA reagents for screening with single-cell resolution. The PDMS microfluidic system for single cell encapsulation was fabricated using standard soft lithography methods, as previously described32,34,35. The system contained the droplet generation chip and an in-channel incubation chamber. The generation of monodisperse droplets in a microchannel through shearing flow at a flow-focusing zone is illustrated in Fig. 3a (see methods for details). After cell encapsulation in the nanoliter reaction volume, the individual droplets were delivered to the incubation channel and maintained for 2 hours at 37°C for the RCA amplification reaction. Since RCA is isothermal, there is no need for thermal cycling that would require sophisticated and expensive instrumentation, as required for other methods33,42. After incubation, we evaluated the number of specific signature sites per genome of each encapsulated cell by measuring the intensity of the fluorescent signal in individual cells.
Quantification of target sites is possible due to the unbiased, linear nature of the RCA process. This capability is essential for mechanistic studies and diagnostic applications. This system allowed us to distinguish between single and multiple copies of the target sites based on fluorescent intensity of single cells. Assuming a replication rate of phi29 DNA polymerase of 1400–1500 bases min−1,43 and a circle size of 80 nucleotides, each rolling-circle product (RCA amplicon) increases by ~80,000 nucleotides/h, with total circle amplification as high as 1000-fold per hour, resulting in accumulation of fluorescent product directly within the cell. In our experiments, signal was recorded and quantified for encapsulated cells and compared to empty droplets, which represented a background signal. An average of at least ten droplets, each containing a single cell, for every EBV target site was calculated (Fig. 3b). Figure 3c shows a surface plot of the fluorescence intensity distribution (z axis) of the cells within each droplet at the end of the amplification process. For the single copy genes LMP-1 and EBNA-3, the fluorescent intensity was demonstrated to be about seven times lower than for the EBNA-2 IR repeat inserts. This number correlates well with the known copy number (7–13) of the EBNA-2 IR repeat, indicating the potential for quantitative analysis. Control experiments were conducted to establish whether nonspecific fluorescence background was observed in the droplet format. No false positives were detected in the EBV-negative control (CCRF-CEM) cell line, once again confirming that the method is highly specific (Fig. 3c and Supplementary Fig. 5). Collectively, these results clearly demonstrate that the fluorescent intensity of the cells reflects the copy number of the target sites and confirms that the method is useful for quantitative studies.
DISCUSSION
Variations in the human genome range from chromosomal alterations that involve millions of nucleotides to single nucleotide polymorphisms (SNPs). Therefore, it is not surprising that tremendous efforts have been made to develop molecular technologies that would enhance our understanding of allelic variations. However, most of the available cytogenetic techniques are geared toward detecting DNA targets that are at least 1–2 kilobases long, which means that they cannot resolve SNPs, the most common source of genetic variation. In situ detection of short-sequence elements in genomic DNA requires short probes with high molecular resolution as well as powerful and specific signal amplification processes. Unlike simple hybridization probes, padlock probes can differentiate single base variations. Several attempts have been made to detect short DNA sequences in the human genome based on padlock probe design.
In this report, we demonstrate the feasibility of the PNA-RCA method for sensitive and sequence-specific detection of oncoviral DNA inserts within human cell lines in a convenient cells-in-flow format. The high sequence specificity of the PNA approach and signal amplification provided by RCA allowed us to detect genetic variations with single nucleotide resolution, and thus to potentially classify EBV type infection. It should be emphasized that the PNA-based DNA circle formation keeps the rest of the genomic DNA in its duplex form and therefore inaccessible for non-specific interactions. Another significant advantage of the PD-loop design consists in the fact that its assembly is possible only as a result of a multiple coincidence event: the local opening of dsDNA by two PNA openers, ODN probe hybridization, and closure. Finally, the circular probe is threaded between two strands of dsDNA and is truly topologically linked to the target site. These important traits make our approach background-insensitive and mutation-immune in contrast to PCR or any assay based on probe circularization on single-stranded DNA. Although PNA openers pose certain sequence limitations on their binding sites (the binding site must consist of purines in one strand and pyrimidines in the other), these limitations are relatively minor since the binding sites can be as short as 7 nucleotides14. Statistically, one can expect on average one PD-loop site per about every 800 base pairs of a random DNA sequence15. Moreover, our preliminary data indicate that, to form the PD-loop, the PNA openers can be as short as hexamers. If they again can be separated by up to 10 bp of an arbitrary sequence, then we expect to have one such site per 200 bp, on average20. PNA openers have already found applications in several diagnostics based on fluorescence detection44,45.
All steps of the PNA-RCA approach — genomic DNA opening with PNA probes, circular probe ligation and RCA — require ambient temperatures, that eliminate genomic DNA denaturation needed for other DNA analysis. Moreover, physiologic reaction conditions maintain the interrogated cell integrity, and the entire procedure can be implemented in cells-in-flow format, thus omitting the DNA isolation step. Our current work integrates the PNA-RCA method with FACS to simplify DNA analysis using FISH, and with droplet microfluidics to enhance an ability of PNA-RCA method to visualize specific target genes in single cells. As a proof of concept, we used this new approach to detect Epstein-Barr Virus (EBV) infection in human B-lymphocytes. First, we showed the ability to discriminate SNPs using PNA-RCA method coupled with FACS analysis. Second, we were able, using microfluidic nanoliter droplets, to visualize the viral genomes in each infected cell as well as to quantify the gene copy number per cell. The advantages of the microfluidic droplet PNA-RCA method include very low reaction volumes (which minimize reagent costs), and its inherent capability for rapid and highly parallel statistical single-cell analysis. In the future, we intend to enhance the current assay by incorporating a custom droplet array device with integrated optics to provide automatic quantitative measurements of real-time PNA-RCA signals in a high-throughput format.
In summary, our results demonstrate the development of a microfluidic nanodroplet system with capability of single-nucleotide polymorphism (SNP) discrimination and detection of copy-number variations (CNV) under isothermal, non-denaturing conditions. This method is ideal for various applications exploiting the selective screening of single mammalian cells by analysing the genomic characteristics of individual cells.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. Natalia Broude for critical reading and valuable comments on the manuscript.
FUNDING
This work was supported by the National Institutes of Health [1R21AI100180-01 to I.S.]. Funding for open access charge: National Institutes of Health.
REFERENCES
- 1.Naidoo N, Pawitan Y, Soong R, Cooper DN, Ku CS. Human genetics and genomics a decade after the release of the draft sequence of the human genome. Hum. Genomics. 2011;5:577–622. doi: 10.1186/1479-7364-5-6-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pomerantz MM, Freedman ML. The genetics of cancer risk. Cancer J. 2011;17:416–422. doi: 10.1097/PPO.0b013e31823e5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kasarskis A, Yang X, Schadt E. Integrative genomics strategies to elucidate the complexity of drug response. Pharmacogenomics. 2011;12:1695–1715. doi: 10.2217/pgs.11.115. [DOI] [PubMed] [Google Scholar]
- 4.Seeman NC. Structural DNA nanotechnology: Growing along with Nano Letters. Nano Lett. 2010;10:1971–1978. doi: 10.1021/nl101262u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shu D, Moll WD, Deng Z, Mao C, Guo P. Bottom-up assembly of RNA arrays and superstructures as potential parts in nanotechnology. Nano Lett. 2004;4:1717–1723. doi: 10.1021/nl0494497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Novak R, et al. Single-cell multiplex gene detection and sequencing with microfluidically generated agarose emulsions. Angew. Chem. Int. Ed. Engl. 2011;50:390–395. doi: 10.1002/anie.201006089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huebner A, et al. Microdroplets: A sea of applications? Lab on a Chip. 2008;8:1244–1254. doi: 10.1039/b806405a. [DOI] [PubMed] [Google Scholar]
- 8.Clausell-Tormos J, et al. Droplet-based microfluidic platforms for the encapsulation and screening of Mammalian cells and multicellular organisms. Chem. Biol. 2008;15:427–437. doi: 10.1016/j.chembiol.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 9.Breitenstein M, Nielsen PE, Holzel R, Bier FF. DNA-nanostructure-assembly by sequential spotting. J. Nanobiotechnol. 2011;9:54. doi: 10.1186/1477-3155-9-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singer A, et al. Nanopore based sequence specific detection of duplex DNA for genomic profiling. Nano Lett. 2010;10:738–742. doi: 10.1021/nl100058y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Debaene F, Winssinger N. Self-assembly of PNA-encoded peptides into microarrays. Methods Mol. Biol. 2009;570:299–307. doi: 10.1007/978-1-60327-394-7_15. [DOI] [PubMed] [Google Scholar]
- 12.Egholm M, et al. Efficient pH-independent sequence-specific DNA binding by pseudoisocytosine-containing bis-PNA. Nucleic Acids Res. 1995;23:217–222. doi: 10.1093/nar/23.2.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuhn H, Demidov VV, Frank-Kamenetskii MD, Nielsen PE. Kinetic sequence discrimination of cationic bis-PNAs upon targeting of double-stranded DNA. Nucleic Acids Res. 1998;26:582–587. doi: 10.1093/nar/26.2.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bukanov NO, Demidov VV, Nielsen PE, Frank-Kamenetskii MD. PD-loop: A complex of duplex DNA with an oligonucleotide. Proc. Natl. Acad. Sci. U.S.A. 1998;95:5516–5520. doi: 10.1073/pnas.95.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Demidov VV, Frank-Kamenetskii MD. Two sides of the coin: Affinity and specificity of nucleic acid interactions. Trends Biochem. Sci. 2004;29:62–71. doi: 10.1016/j.tibs.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 16.Kuhn H, Demidov VV, Frank-Kamenetskii MD. Topological links between duplex DNA and a circular DNA single strand. Angew. Chem. Int. Ed. 1999;38:1446–1449. doi: 10.1002/(SICI)1521-3773(19990517)38:10<1446::AID-ANIE1446>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 17.Kuhn H, Demidov VV, Frank-Kamenetskii MD. An earring for the double helix: Assembly of topological links comprising duplex DNA and a circular oligodeoxynucleotide. J. Biomol. Struct. Dyn. 2000;17:221–225. doi: 10.1080/07391102.2000.10506625. [DOI] [PubMed] [Google Scholar]
- 18.Smolina I, Lee C, Frank-Kamenetskii M. Detection of low-copy-number genomic DNA sequences in individual bacterial cells by using peptide nucleic acid-assisted rolling-circle amplification and fluorescence in situ hybridization. Appl. Environ. Microbiol. 2007;73:2324–2328. doi: 10.1128/AEM.02038-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smolina I, Miller NS, Frank-Kamenetskii MD. PNA-based microbial pathogen identification and resistance marker detection: An accurate, isothermal rapid assay based on genome-specific features. Artif. DNA PNA XNA. 2010;1:76–82. doi: 10.4161/adna.1.2.13256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yaroslavsky AI, Smolina IV. Fluorescence imaging of single-copy DNA sequences within the human genome using PNA-directed padlock probe assembly. Chem. Biol. 2013;20:445–453. doi: 10.1016/j.chembiol.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mazouni C, et al. Epstein-Barr virus as a marker of biological aggressiveness in breast cancer. Br. J. Cancer. 2011;104:332–337. doi: 10.1038/sj.bjc.6606048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller G, et al. Selective switch between latency and lytic replication of Kaposi's sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J. Virol. 1997;71:314–324. doi: 10.1128/jvi.71.1.314-324.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arvanitakis L, et al. Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus. Blood. 1996;88:2648–2654. [PubMed] [Google Scholar]
- 24.Khanim F, et al. Analysis of Epstein-Barr virus gene polymorphisms in normal donors and in virus-associated tumors from different geographic locations. Blood. 1996;88:3491–3501. [PubMed] [Google Scholar]
- 25.Hoshino Y, et al. Long-term administration of valacyclovir reduces the number of Epstein-Barr virus (EBV)-infected B cells but not the number of EBV DNA copies per B cell in healthy volunteers. J. Virol. 2009;83:11857–11861. doi: 10.1128/JVI.01005-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cesarman E, et al. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood. 1995;86:2708–2714. [PubMed] [Google Scholar]
- 27.Yao QY, et al. Epidemiology of infection with Epstein-Barr virus types 1 and 2: Lessons from the study of a T-cell-immunocompromised hemophilic cohort. J. Virol. 1998;72:4352–4363. doi: 10.1128/jvi.72.5.4352-4363.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rowe M, et al. Epstein-Barr virus (EBV)-associated lymphoproliferative disease in the SCID mouse model: Implications for the pathogenesis of EBV-positive lymphomas in man. J. Exp. Med. 1991;173:147–158. doi: 10.1084/jem.173.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen A, Zhao B, Kieff E, Aster JC, Wang F. EBNA-3B-and EBNA-3C-regulated cellular genes in Epstein-Barr virus-immortalized lymphoblastoid cell lines. J. Virol. 2006;80:10139–10150. doi: 10.1128/JVI.00854-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dambaugh T, Hennessy K, Chamnankit L, Kieff E. U2 region of Epstein-Barr virus DNA may encode Epstein-Barr nuclear antigen 2. Proc. Natl. Acad. Sci. U.S.A. 1984;81:7632–7636. doi: 10.1073/pnas.81.23.7632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sample J, et al. Epstein-Barr virus types 1 and 2 differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes. J. Virol. 1990;64:4084–4092. doi: 10.1128/jvi.64.9.4084-4092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agresti JJ, et al. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc. Natl. Acad. Sci. U.S.A. 2010;107:4004–4009. doi: 10.1073/pnas.0910781107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiss MM, et al. High-throughput quantitative polymerase chain reaction in picoliter droplets. Anal. Chem. 2008;80:8975–8981. doi: 10.1021/ac801276c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koster S, et al. Drop-based microfluidic devices for encapsulation of single cells. Lab on a Chip. 2008;8:1110–1115. doi: 10.1039/b802941e. [DOI] [PubMed] [Google Scholar]
- 35.Konry T, Smolina I, Yarmush JM, Irimia D, Yarmush ML. Ultrasensitive detection of low-abundance surface-marker protein using isothermal rolling circle amplification in a microfluidic nanoliter platform. Small. 2011;7:395–400. doi: 10.1002/smll.201001620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Konry T, Dominguez-Villar M, Baecher-Allan C, Hafler DA, Yarmush ML. Droplet-based microfluidic platforms for single T cell secretion analysis of IL-10 cytokine. Biosens. Bioelect. 2011;26:2707–2710. doi: 10.1016/j.bios.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lizardi PM, et al. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat. Genet. 1998;19:225–232. doi: 10.1038/898. [DOI] [PubMed] [Google Scholar]
- 38.Aguirre AJ, Robertson ES. Characterization of intertypic recombinants of the Epstein-Barr virus from the body-cavity-based lymphomas cell lines BC-1 and BC-2. Virology. 1999;264:359–369. doi: 10.1006/viro.1999.0015. [DOI] [PubMed] [Google Scholar]
- 39.Aguirre AJ, Robertson ES. Epstein-Barr virus recombinants from BC-1 and BC-2 can immortalize human primary B lymphocytes with different levels of efficiency and in the absence of coinfection by Kaposi's sarcoma-associated herpesvirus. Journal of Virology. 2000;74:735–743. doi: 10.1128/jvi.74.2.735-743.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horenstein MG, et al. Epstein-Barr virus latent gene expression in primary effusion lymphomas containing Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8. Blood. 1997;90:1186–1191. [PubMed] [Google Scholar]
- 41.Demidov VV, Kuhn H, Lavrentieva-Smolina IV, Frank-Kamenetskii MD. Peptide nucleic acid-assisted topological labeling of duplex DNA. Methods. 2001;23:123–131. doi: 10.1006/meth.2000.1113. [DOI] [PubMed] [Google Scholar]
- 42.Joensson HN, et al. Detection and analysis of low-abundance cell-surface biomarkers using enzymatic amplification in microfluidic droplets. Angew. Chem. Int. Ed. Engl. 2009;48:2518–2521. doi: 10.1002/anie.200804326. [DOI] [PubMed] [Google Scholar]
- 43.Melin J, Jarvius J, Goransson J, Nilsson M. Homogeneous amplified single-molecule detection: Characterization of key parameters. Anal. Biochem. 2007;368:230–238. doi: 10.1016/j.ab.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 44.Phillips KM, et al. Application of single molecule technology to rapidly map long DNA and study the conformation of stretched DNA. Nucleic Acids Res. 2005;33:5829–5837. doi: 10.1093/nar/gki895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brind'Amour J, Lansdorp PM. Analysis of repetitive DNA in chromosomes by flow cytometry. Nat. Methods. 2011;8:484–486. doi: 10.1038/nmeth.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.