Abstract
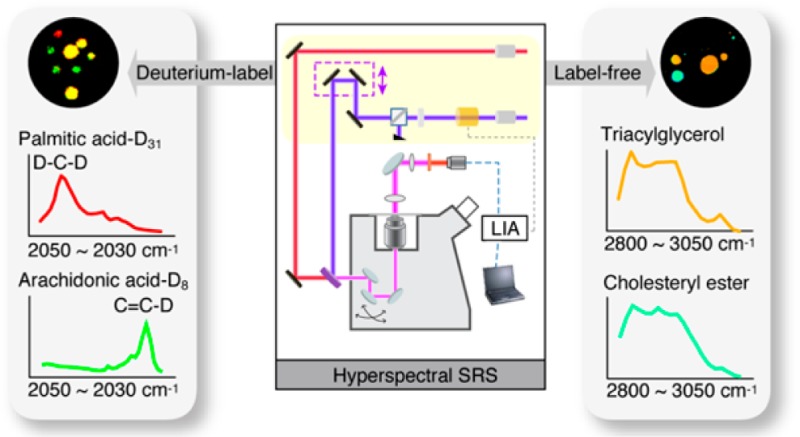
Metabolic fingerprinting provides valuable information on the physiopathological states of cells and tissues. Traditional imaging mass spectrometry and magnetic resonance imaging are unable to probe the spatial-temporal dynamics of metabolites at the subcellular level due to either lack of spatial resolution or inability to perform live cell imaging. Here we report a complementary metabolic imaging technique that is based on hyperspectral stimulated Raman scattering (hsSRS). We demonstrated the use of hsSRS imaging in quantifying two major neutral lipids: cholesteryl ester and triacylglycerol in cells and tissues. Our imaging results revealed previously unknown changes of lipid composition associated with obesity and steatohepatitis. We further used stable-isotope labeling to trace the metabolic dynamics of fatty acids in live cells and live Caenorhabditis elegans with hsSRS imaging. We found that unsaturated fatty acid has preferential uptake into lipid storage while saturated fatty acid exhibits toxicity in hepatic cells. Simultaneous metabolic fingerprinting of deuterium-labeled saturated and unsaturated fatty acids in living C. elegans revealed that there is a lack of interaction between the two, unlike previously hypothesized. Our findings provide new approaches for metabolic tracing of neutral lipids and their precursors in living cells and organisms, and could potentially serve as a general approach for metabolic fingerprinting of other metabolites.
Introduction
Understanding the complex metabolic processes that occur within living organisms provides important pathways to tackle major healthcare challenges such as diabetes and cancer. In the human body, each type of cell or tissue has a unique “metabolic fingerprint” that characterizes its specific function. The emerging field of metabolomics aims to uncover metabolic fingerprints of tissues at different physiopathological states. Its progress relies heavily on technological renovations with evolving capability of detecting and quantifying the thousands of metabolites (also known as metabolome) to be found in a biological sample.1 A wide range of mass spectrometry (MS) methods (usually coupled to gas chromatography or liquid chromatography) have been used to characterize the metabolome owing to their high sensitivity and specificity. To date, MS remains the key platform that is used to compare relative metabolite profile differences between biological samples. However, it only provides a snapshot of metabolite profile at a particular time point, and typically without any spatial context. Recent developments in imaging mass spectrometry (IMS) provide the much-needed spatial information for understanding disease mechanisms and their progresses. When combined with isotope labeling, IMS is extremely powerful in characterizing metabolic fate of small molecules at high resolution.2,3 Magnetic resonance imaging (MRI) is another technique that has been shown to be applicable for mapping the spatial distribution of metabolites, especially when combined with stable-isotope labeling (13C, 15N, 17O) and/or hyperpolarization.4,5 Even though its sensitivity and specificity is rather limited compared with IMS, MRI has a significant advantage of being able to trace the spatial-temporal dynamics of metabolites in live animals or human subjects.
Complementary to IMS or MRI, vibrational spectroscopy is another analytical technique that is capable of distinguishing different metabolites in intact biological samples. It has the advantages of minimal sample preparation, low cost, and high spatial resolution in imaging mode. Two different spectroscopic modalities are generally used for metabolic fingerprinting: Fourier-transform infrared (FTIR) and Raman.6 Raman imaging is advantageous over FTIR in that it is free from water interference and provides three-dimensional subcellular resolution. The major limitation of Raman imaging, however, is its slow speed due to the inefficiency of Raman scattering process. Coherent anti-Stokes Ramam scattering (CARS) and Stimulated Raman scattering (SRS) largely overcome this problem by improving the imaging sensitivity by 4–5 orders of magnitude.7,8 It has been used to image subcellular distribution of lipids, protein, nucleic acids, vitamins, drug molecules, etc., both in vitro and in vivo. However, the resolvability achieved with conventional CARS and SRS microscopy is limited. Molecular identification relies on unique sharp Raman peak features, which is not always available for metabolites. Few single-peak imaging of deuterated molecular species using CARS and SRS provides little information on the changing chemical state of metabolites in the tissue. Furthermore, a large number of metabolites in the tissue present overlapping Raman spectra, thus making separate quantification difficult. Recent technical advances in hyperspectral SRS (hsSRS) have enabled chemical mapping of molecules that have similar yet distinguishable Raman spectra,9−13 thus providing hope for metabolic fingerprinting of multiple molecular species.
In this report, we demonstrate the utility of hsSRS imaging in metabolic profiling of lipids. Lipids are important metabolites in the human body with vital effects on cell and organism physiology, ranging from membrane trafficking and cell maintenance to inflammation, metabolism and brain health. Dysfunction of lipid-related processes leads to the development of various human pathologies, such as metabolic disorders, cancer, and neurodegenerative diseases.14−16 However, tracking the spatiotemporal dynamics of structurally divergent lipid molecules in vivo has been technically challenging: lipid molecules are intrinsically nonfluorescent and labeling with fluorescent tag often alters the chemical activities of lipid molecules. Consequently, it has been difficult to differentially image various lipid molecules by the most commonly used fluorescence microscopic techniques. Although CARS and SRS have previously proved to be powerful tools for imaging lipid distribution in unfixed and unstained biological samples,17,18 metabolic profiling of lipids requires further differentiation of lipids with different chemical compositions. Here we demonstrate metabolic fingerprinting of neutral lipids at the individual lipid droplet level with hsSRS. Furthermore, we apply stable-isotope labeling with hsSRS to simultaneously track different fatty acid molecules in living cells and organisms. Our studies present a straightforward and powerful workflow to directly trace the metabolic dynamics of specific lipid molecules in various organisms under both physiological and pathological conditions. We believe that it opens up new avenues of investigation into spatial-temporal dynamics of many metabolites in live cells and animals, which will provide insights into aberrant metabolic processes in multiple diseases such as diabetes, steatohepatitis and cancer.
Materials and Methods
Hyperspectral SRS Imaging
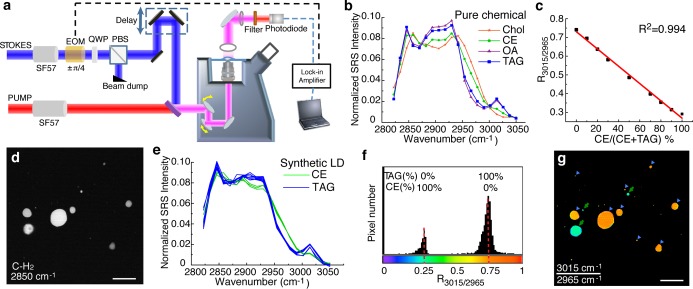
The spectral focusing hsSRS imaging method and experimental setup (Figure 1a) were described in details in our recent paper.9 In brief, two synchronized femtosecond lasers (with center wavelengths at 1040 and 795 nm, respectively) were chirped to about 2 ps using SF57 glass rods. The temporal delay between the two pulsed lasers was controlled by a motorized stage. The combined beam was sent into an inverted laser-scanning microscope (Olympus IX71, with Fluoview 300 scanning-head). A 60X water immersion objective (Olympus UPLSAPO 60XW/IR) was used to focus the beams onto the sample, which was typically sandwiched between a glass slide and a coverslip. On the detection side, the Stokes beam was filtered out by a bandpass filter (Chroma CARS 890/220 nm) and the pump beam was detected by an amplified Si photodiode (Advanced Photonix) biased at 50 V. The SRS signal was detected with a home-built lock-in amplifier.19 Each frame was scanned in 1.12 s, providing 512×512 pixels. hsSRS imaging of 50 mM Rhodamine 6G dye (R6G) solution was performed in each imaging session for laser power calibration. The spectrum of R6G was divided by all the sample spectra to obtain intensity-normalized hsSRS spectra shown in all figures.
Figure 1.
Quantitative analysis of different lipid molecules using hsSRS. (a) Schematic diagram of hsSRS setup. SF57, glass rod for pulse chirping; EOM, electro-optical modulator; QWP, quarter wave plate; PBS, polarizing beam splitter. (b) Four different lipid molecules—cholesterol (Chol), cholesteryl oleate (representing CE), oleic acid (OA), and triolein (representing TAG)—in CDCl3 solutions exhibit distinguishable hsSRS spectra. (c) Linear association between R3015/2965 and the percentage of CE in the CE/TAG mixture. R3015/2965, the ratio of the SRS signal at 3015 cm–1 over that at 2965 cm–1. (d) SRS image at 2850 cm–1 of a mixture of synthetic LDs containing either CE or TAG. (e) CE or TAG-containing LDs were classified into two groups by their distinct hsSRS spectra. (f) In the R3015/2965 histogram, pixels derived from LDs are distributed into two distinct classes. (g) The R3015/2965 image reveals the separation of CE-containing LDs (green arrow) and TAG-containing LDs (blue arrowheads). Scale bar = 20 μm.
Combined hsSRS Imaging with Deuterium Labeling
A picosecond one-box laser system (picoEmerald, APE) was used for hsSRS imaging in the C–D region. This system is chosen due to its high spectral resolution and low non-Raman background, which is advantageous for low signal imaging in the C–D region; the disadvantage is that it has poorer repeatability and slower tuning, compared with the previous system. The pump wavelength of the picoEmerald was sequentially tuned by using a home-built Labview program to control the Lyot filter inside the laser (while fixing the OPO crystal temperature), and a series of SRS images at evenly stepped pump wavelength was then acquired. The pump wavelength scan range was 855.6–870.3 nm, resulting in SRS spectra covering 2083–2280 cm–1. The scan range was limited by the tuning range of the Lyot filter. We typically used 32 steps, with an average step size of 6.35 cm–1. This method allows C–H imaging and C–D imaging on the same platform. For weak C–D SRS intensity at a low incorporation level, we removed non-SRS originated background signals that have a flat spectral response in the spectral range we scanned. To quantify the C–D signal, the signal intensity at 2110 cm–1 (for D–C–D) or 2250 cm–1 (for C=C–D) was subtracted with the off-resonance SRS signal at 2040 and 2280 cm–1, respectively.
Preparation of Synthetic Lipid Droplets (LDs)
To prepare CE-containing synthetic LDs, 300 μL of cholesteryl oleate (100 mg/mL, dissolved in hexane) and 200 μL of phosphatidylcholine solutions (5 mg/mL, in 1:1 hexane/chloroform) were mixed in a round-bottom Corex glass tube. After evaporating the solvent under dry nitrogen, 15 mL of PBS was added to the tube. The tube was then placed in a boiling water bath to melt the lipids. The mixture was immediately sonicated for 30 min. TAG-containing LDs were prepared with the same procedure by replacing cholesteryl oleate with triolein.
Yeast Strains and Culture
FYS252 strain: are1Δ::Kan are2Δ::His (no SE). FYS242 strain: dga1Δ::His lro1Δ::Kan (almost no TAG). Wild-type strain: BY4741 (MATa his3Δ0 leu2Δ0 met15Δ0 ura3Δ0).
Strains were created by routine lithium acetate transformation of PCR products amplified from deletion cassettes.20 FYS252 was made by transformation of are1Δ::Kan obtained from the Saccharomyces Genome Deletion Project collection with PCR product amplified from pFA6a-His3MX6 using forward primer ATGGACAAGAAGAAGGATCTACTGGAGAACGAACAATTTCCGGATCCCCGGGTTAATTAA and reverse primer AAAATTTACTATAAAGATTTAATAGCTCCACAGAACAGTTGCAGGATGCCGAATTCGAGCTCGTTTAAAC. FYS242 was similarly transformed using forward primer TAAGGAAACGCAGAGGCATACAGTTTGAACAGTCACATAACGGATCCCCGGGTTAATTAA and reverse primer TTTATTCTAACATATTTTGTGTTTTCCAATGAATTCATTAGAATTCGAGCTCGTTTAAAC in lro1Δ::Kan.
Yeast cells were cultured in YPD medium supplemented with 0.5 mM oleic acid. Oleic acid/BSA solution was first prepared with the following procedure: 5 mg of oleic acid was dissolved in hexane and neutralized with 1 M NaOH; after evaporating the solvent under dry nitrogen, oleic acid salt was dissolved in 1 mL of water and then added dropwise to 4 mL of warm bovine serum albumin (BSA) solution (10% w/v). The filter-sterilized oleic acid/BSA solution was then mixed with 50 mL of YPD medium. A single colony of yeast was cultured in oleic acid-supplemented YPD medium at 30 °C for 16 h (stationary phase).
Liver and Macrophage Cell Line culture
Mouse monocyte-macrophage RAW 264.7 cells and rat hepatic McA-RH7777 cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with penicillin (100 U/mL), streptomycin (100 μg/mL), 10% heat-inactivated fetal bovine serum, 100 μM oleic acid, and 50 μM cholesterol at 37 °C in a humidified incubator with 5% CO2 and 95% air.21
Mouse Culture, Dissection, and Frozen Section
C57BL/6J wild-type and ob/ob mice were purchased from Jackson Lab (Bar Harbor, Maine). All experimental procedures were carried out under a protocol approved by the Institutional Animal Care and Use Committee at Baylor College of Medicine and were in accordance with the National Institutes of Health guidelines for the care and the use of laboratory animals. Mice were maintained in a temperature-controlled facility with 12 h light/dark cycles and free access to regular chow and water. Male mice, 8–12 weeks old, were used for this study. To induce fatty liver, mice were injected with tunicamycin (0.5 mg/kg body weight). After tunicamycin injection, liver tissues were collected at 0, 24, and 48 h, flash-frozen in embedding medium containing a 3:1 mixture of Tissue Freezing Medium (Triangle Biomedical Sciences) and gum tragacanth (Sigma-Aldrich) at −80 °C, and sectioned to 10 μm-thick using Cryostat (Leica CM 3000).
C. elegans Strains and Culture
Wild-type N2 strain from the Caenorhabditis Genetics Center were grown on standard NGM plates (containing 5 mg/L cholesterol) with E. coli OP50 at 20 °C using standard protocols.22
Deuterated Fatty Acid Supplementation
McA-RH7777 liver cells were supplemented with 0.4 mM palmitic acid-D31 (PA-D31) or oleic acid-D34 (OA-D34) for 7 h. The fatty acid/BSA solution was prepared as described before, and then supplemented to culture medium.
For C. elegans fatty acid supplementation, OP50 bacterial culture was mixed well with 4 mM PA-D31, or OA-D34, or 2 mM PA-D31 + 2 mM arachidonic acid (AA-D8), and then seeded onto NGM plates. After 1-day-old adult worms were fed for the desired time, they were mounted onto 2% agarose pads with 0.5% NaN3 as anesthetic on glass microscope slides.
Thin Layer Chromatography (TLC)
For yeast TLC, 10 OD units of yeast cells were broken with glass beads and lipids were extracted with chloroform:methanol (1:1 v/v). Lipids were dried under nitrogen gas, loaded onto TLC plates (Whatman), developed in hexane:ethyl ether:acetic acid (80:20:1 v/v), and charred with cupric sulfate until lipid bands were visible.
For C. elegans TLC, about 5000 worms for each sample were collected and homogenized. Total lipids were extracted and loaded onto silica gel G TLC plates (Analtech), developed in hexane:diethyl ethyl:acetic acid (75:25:2 v/v). The TLC plate was stained with 0.05% Primuline (Sigma-Aldrich, in acetone:H2O, 8:2 v/v). Lipids were visualized using a Gel Imaging System (Bio-Rad) with fluorescence (Ex 488 nm/Em 555 nm).
For mouse liver TLC, 200 mg liver tissues for each sample were homogenized in PBS and used for lipid extraction. TLC plates were developed in petroleum ether:ether:glacial acetic acid (85:25:1 v/v) and visualized in saturated iodine chamber.
Results
Quantitatively Differentiating Two Classes of Neutral Lipid Using hsSRS Imaging
To test the capability of hsSRS in imaging different lipid molecules, we first used this technique to acquire the spectra of cholesteryl oleate, triolein, cholesterol and oleic acid in deuterated chloroform solutions. We acquired a total of 20 images that were evenly spaced between 2825 and 3050 cm–1 for each sample, which covers the C–H stretching region of most biological molecules and provides spectra at a wavenumber spacing of 12.5 cm–1 (Figure 1b). The four lipid molecules lack characteristic chemical bonds needed to separate them when using conventional SRS, but they generate distinguishable spectra when imaged with hsSRS (Figure 1b). Interestingly, we found that cholesteryl oleate and triolein exhibit significant differences in the hsSRS spectra ranging from 2950 to 3025 cm–1 (Figure 1b). Cholesteryl esters (CEs) and triacylglycerols (TAGs) are two major classes of neutral lipid molecules. The significant spectral differences between cholesteryl oleate and triolein suggest that hsSRS can be utilized as a quantitative method to study spatiotemporal distribution differences between CEs and TAGs in vivo. We verified the quantification ability of hsSRS by imaging a series of mixtures containing cholesteryl oleate and triolein. We calculated the ratio of the signal intensity between 3015 and 2965 cm–1 (R3015/2965) and plotted it against the percentage of cholesteryl oleate. As expected, the intensity ratio exhibited a linear relationship with the percentage of cholesteryl oleate in the mixture (Figure 1c; R2 = 0.994).
Inside a cell, CE and TAG are predominantly located in specialized cellular organelles called lipid droplets (LDs). It is now widely accepted that LDs are not inert inclusions of lipids, but rather are highly dynamic organelles that are heterogeneous in size, in localization, and in their associated proteins.23 To test the ability of hsSRS in differentiating CE and TAG within LDs, we first generated artificial LDs that contained either cholesteryl oleate or triolein, and then imaged their mixture using hsSRS. Although both classes of LDs varied in size by more than an order of magnitude (Figure 1d), we were able to separate them based on their normalized spectra (Figure 1e). We then calculated R3015/2965 for each pixel in the image and found that the pixels derived from the LDs were separated into two groups in the histogram (Figure 1f). The R3015/2965 peaks of these two groups—0.29 and 0.75—are correlated with 100% CE and 100% TAG, respectively. In the R3015/2965 image, CE- or TAG-containing LDs in the mixture are now clearly distinguishable (Figure 1g), which were not possible with conventional SRS (Figure 1d). Importantly, the spectra and R3015/2965 that were obtained with artificial LDs (Figure 1e,f) exhibit remarkable consistency with those of the pure chemical solutions (Figure 1b,c), thereby indicating a high degree of spectral repeatability achieved with hsSRS. We note that the acquired hsSRS spectra is system dependent, and thus comparison of sample spectra to standard chemical spectra measured with the same system is necessary.
Metabolic Fingerprinting of Neutral Lipids in Vivo
Next, we validated that hsSRS is efficacious for metabolic fingerprinting of neutral lipids in vivo. In Saccharomyces cerevisiae, Are1 and Are2 encode acyl-coenzyme A (CoA):cholesterol acyltransferase-related enzymes, which are essential for the synthesis of steryl esters (SE, similar to CE in animal cells).24 On the other hand, Dga1 and Lro1 encode acyl-CoA:diacylglycerol acyltransferase and lecithin-cholesterol acyltransferase, respectively, which are the major contributors to TAG synthesis.25 The yeast mutant strains—FYS252 (lacking Are1 and Are2) and FYS242 (lacking Dga1 and Lro1)—have defective SE and TAG synthesis, respectively. With hsSRS imaging of these yeast strains, we found that the FYS252 mutants contain LDs that exhibit TAG-like R3015/2965 and hsSRS spectra and the LDs of the FYS242 mutants exhibit CE-like R3015/2965 and hsSRS spectra (Figure 2a–g and Figure S1a,b). For the LDs in wild-type yeast cells, both the spectra and R3015/2965 fell mostly in between those of the FYS252 and the FYS242 mutants and exhibit no apparent clustering (Figure 2f,g and Figure S1c). Biochemical analyses of lipid composition using thin-layer chromatography (TLC) confirmed our findings (Figure 2h).
Figure 2.
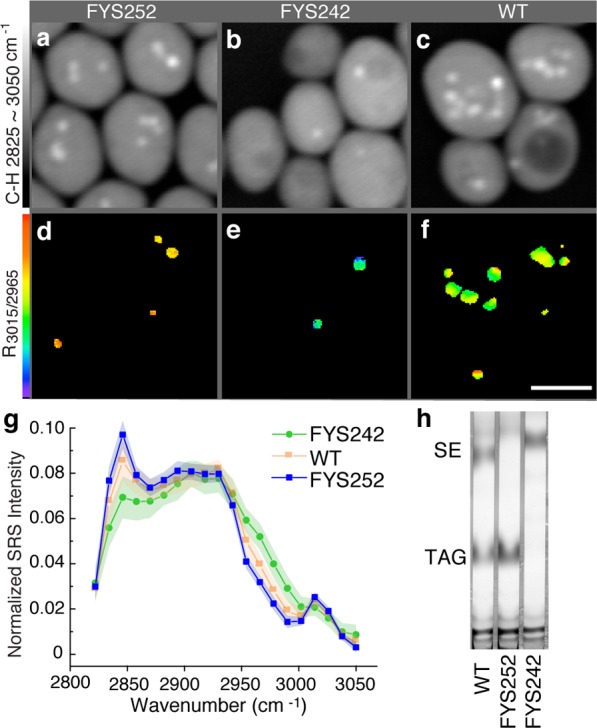
hsSRS imaging of different neutral lipid molecules in vivo in yeast cells. (a–c) Maximum intensity projection of 20 slices from 2825 to 3050 cm–1 shows the overall morphology of yeast cells. (d–f) R3015/2965 images show CE/TAG composition in these LDs. FYS252 mutants contain only TAG (d), whereas FYS242 mutants contain only CE (e). Wild-type (WT) LDs have approximately equal amounts of both CE and TAG (f). Scale bar = 5 μm. (g) The average hsSRS spectra of LDs in FYS252 and FYS242 yeast mutants are similar to TAG and CE, respectively, while WT LDs show an average spectrum that falls in between TAG and CE. Shading along the dotted lines represents the standard deviation, FYS252, n = 42; FYS242, n = 33; WT, n = 53. (h) Biochemical analysis using thin-layer chromatography shows that WT yeast cells contain both TAG and SE, but the FYS252 and the FYS242 mutants consist exclusively of TAG and SE, respectively.
Revealing Different Distributions of Neutral Lipids in Mammalian Cells and Tissues
We then used hsSRS to image neutral lipids in mammalian cells and tissues. In the murine macrophage cell line RAW 264.7 supplemented with oleic acid and cholesterol, we observed CE-like spectra and R3015/2965 images (Figure 3a,c,e), suggesting predominant storage of CE in macrophage cells. In contrast, the hepatic cell line McA-RH7777 with the same culture medium and supplements showed TAG-like spectra and R3015/2965 images (Figure 3b,d,e), suggesting predominant TAG storage in hepatic cells.
Figure 3.
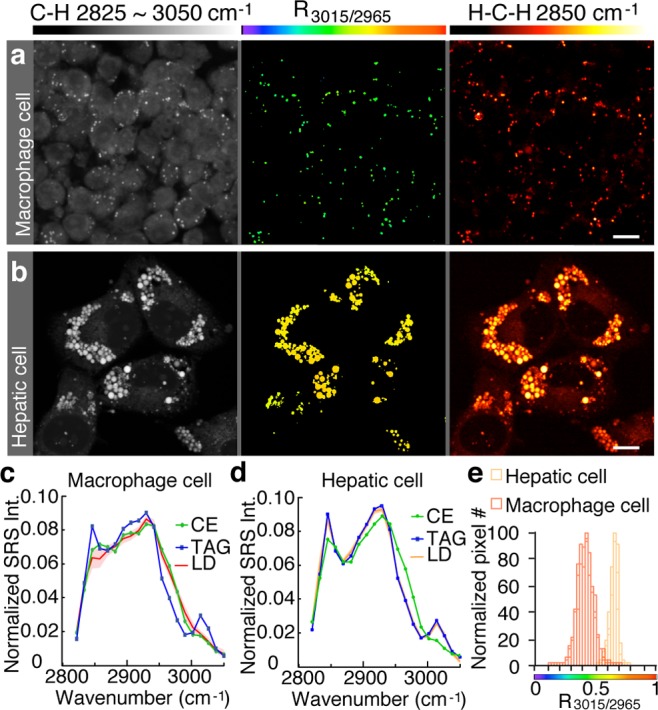
Different distribution of neutral lipids in mammalian cells visualized with hsSRS. (a,b) Images of cultured mouse macrophage cells RAW 264.7 (a) and rat hepatic cells McA-RH7777 (b). The maximum intensity projection of 20-frame SRS images from 2825 to ∼3050 cm–1 shows the overall cell morphology. R3015/2965 images reveal predominant storage of CE in macrophage cells and TAG in hepatic cells. Total lipid levels were visualized by targeting the H–C–H bonds in all fatty acyl chains at 2850 cm–1. Scale bar = 10 μm. (c,d) The average spectra of LDs in macrophage cells (c, n = 142) and hepatic cells (d, n = 115) closely resemble those of CE and TAG, respectively. Shading along the line represents the standard deviation. (e) Macrophage and hepatic cells contain two different classes of neutral lipid molecules with R3015/2965 peaks at 0.4 and 0.65, respectively.
In wild-type mice fed chow diet, we found that the adrenal gland, an important steroidogenic tissue, contains a small number of LDs (Figure S2a), and most of these LDs exhibit R3015/2965 and hsSRS spectra that are close to CE (Figure S2a,c). In contrast, LDs in the liver tissue show R3015/2965 and hsSRS spectra similar to pure TAG (Figure 4a,b, 0 h). These differences can be visualized simply by comparing the R3015/2965 images (Figure 4a and Figure S2a). Our analyses thus show that neutral lipid molecules are differentially distributed in different mammalian cells and tissues. In addition, we found that R3015/2965 of all the LDs in the cells and tissues are narrowly distributed in a small range (Figures 3e and 4a), suggesting a relatively homogeneous lipid composition among different LDs within the same type of cells and tissues.
Figure 4.
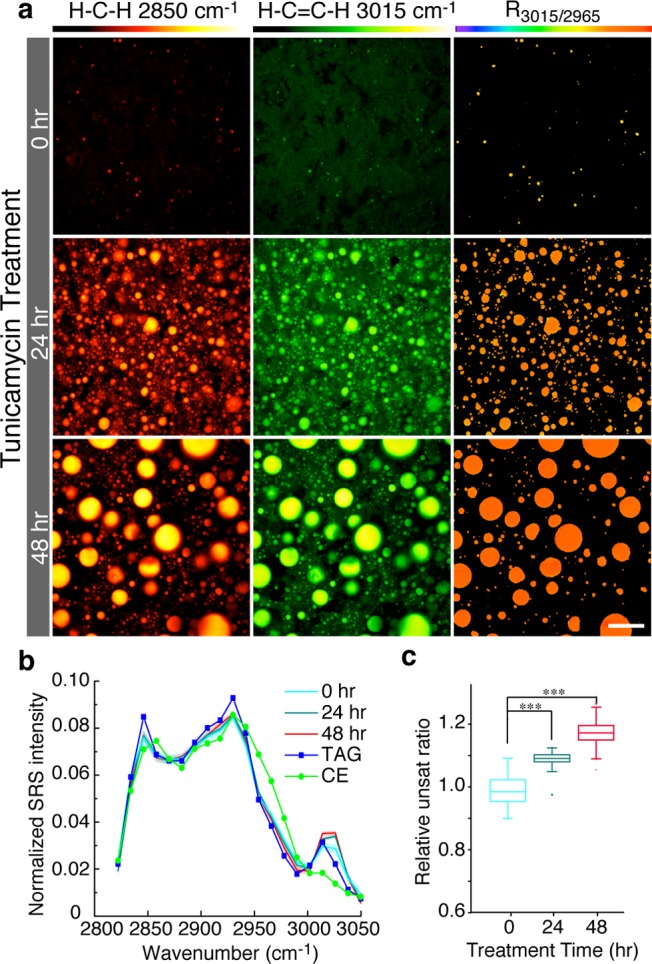
Lipid compositional changes associated with hepatic steatosis during endoplasmic reticulum (ER) stress. (a) Wild-type mice were injected with tunicamycin to induce ER stress in the liver. hsSRS images show rapid expansion of LDs after 24 and 48 h of the treatment, with increased storage of both total lipids and unsaturated lipids. Scale bar = 10 μm. (b) The average hsSRS spectra of hepatic LDs at different post-treatment time points closely resemble the TAG spectrum but with increased signals at 3015 cm–1. Shading along the lines represents the standard deviation, 0 h, n = 37; 24 h, n = 28; 48 h, n = 169. (c) The unsaturation ratio (signal intensity at 3015 to 2850 cm–1) of hepatic LDs is increased by 9% and 17% after 24 and 48 h of the treatment. *** p < 0.001.
Tracking Metabolic Changes of Neutral Lipids Associated with Obesity and Fatty Liver
Next, we applied hsSRS based metabolic fingerprinting of neutral lipids to study lipid composition changes associated with metabolic diseases. Ectopic lipid accumulation in non-adipose tissues is a key pathological consequence of obesity, causing various metabolic dysfunctions.26 The ob/ob mouse with leptin deficiency is a widely used genetic animal model of obesity, in which excess lipids are deposited in both adipose and non-adipose tissues.27 Normal steroidogenic tissues showed predominant CE distribution (Figure S2a). In the ob/ob mouse, the LD number and size of the adrenal gland increased dramatically (Figure S2b). More importantly, their hsSRS spectra also exhibit significant changes (Figure S2c): although the average spectrum is still close to that of CE, the intensity of the 3015 cm–1 peak (originate from C=C–H stretching8) is significantly elevated in the ob/ob mouse, suggesting an increased deposition of unsaturated lipid molecules into LDs in the obese mouse.
In the liver, excess lipid accumulation is not only associated with obesity, but also seen in patients with alcoholic and nonalcoholic fatty liver diseases, leading to insulin resistance and liver cancer.28 Recent studies implicated endoplasmic reticulum (ER) stress in the development and progression of nonalcoholic steatohepatitis.29 Injecting mice with the ER stress-inducing drug tunicamycin results in rapid lipid accumulation in the liver.29,30 Using hsSRS, we confirmed that both the number and size of hepatic LDs are dramatically increased in wild-type mice injected with tunicamycin (Figure 4a). The expansion of LDs is predominantly due to increased deposition of TAG, which is reflected by their TAG-resembling spectra (Figure 4b). This result is also corroborated by TLC analysis of extracted lipids (Figure S3). Furthermore, we noticed that, after tunicamycin treatment, the intensity of the peak at 3015 cm–1 is significantly elevated (Figure 4b), the unsaturated ratio (signal intensity at 3015 to 2850 cm–1) is significantly elevated by 9% and 17% after 24 and 48 h of treatment, respectively (Figure 4c). These results suggest that ER stress induces rapid deposition of TAG into hepatic LDs and that these quickly expanded LDs preferentially utilize unsaturated fatty acids for TAG synthesis.
Metabolic Tracing of Stable-Isotope Labeled Fatty Acids in Living Cells with hsSRS
Stable-isotope labeling is a commonly used strategy to improve the specificity of molecular detection. In particular, stable-isotope labeling is extremely useful in quantifying the dynamics of molecular changes with pulse-chase experiments. Stable isotope labeling by amino acids in cell culture (SILAC) is a widely adopted approach for in vivo incorporation of a label into proteins for MS-based quantitative proteomics.31 MS can also measure metabolic flux with 13C labeled precursors.32 Alternatively, hyperpolarized 13C NMR/MRI is powerful for mapping metabolites in the glycolytic pathway and the Krebs cycle.5
A similar strategy can be applied to Raman-based imaging. In fact, a number of Raman imaging studies have been carried out to characterize the metabolic incorporation of fatty acids, amino acids, and carbohydrates into cells.33−35 C–D imaging with nonlinear Raman techniques including CARS and SRS provides better sensitivity and high imaging speed.36−38 When the C–H bonds of fatty acid molecules are replaced with C–D, their stretching Raman signals are shifted into a “silence window” (Figure 5a), while the biological properties of the fatty acid molecules remain mostly unmodified. However, so far CARS and SRS imaging both rely on the total C–D signal without distinguishing different species. By combining hsSRS imaging with stable-isotope labeling, it is possible to differentiate labeled lipid metabolites and detect changes in their chemical states. Here we demonstrate hsSRS imaging of the metabolic dynamics of pulse labeled deuterated fatty acids in living cells as well as in living animals.
Figure 5.
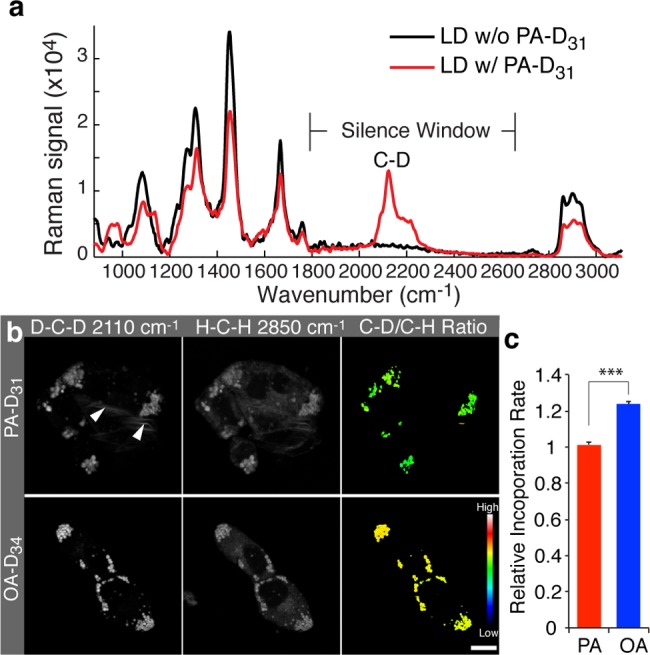
Tracking incorporation dynamics of different fatty acid molecules in hepatic cells with deuterium-labeling-coupled hsSRS. (a) Spontaneous Raman spectra of LDs in wild-type C. elegans fed with or without deuterated palmitic acid (PA-D31). The Raman signal peak of C–D bonds at 2110 cm–1 is located in the “silence window” that contains no signals from unlabeled samples. (b) hsSRS images of hepatic cells (McA-RH7777) labeled with either PA-D31 or OA-D34 for 7 h. The incorporation of deuterated fatty acids was imaged at 2110 cm–1, and the total lipid level was imaged at 2850 cm–1. The ratio between the C–D and the C–H signal intensities was used to measure the level of fatty acid incorporation into LDs. Arrowheads indicate abnormal membrane-like structures caused by PA feeding. Scale bar = 10 μm. (c) The incorporation rate of OA-D34 is 24% faster than PA-D31 in hepatic cells. n = 93 for PA-D31; n = 244 for OA-D34. *** p < 0.001.
Fatty acids, a key class of small metabolites, are crucial precursors of lipid molecules. We first examined the incorporation dynamics of different deuterated fatty acid molecules in McA-RH7777 hepatic cells with SRS imaging at 2110 cm–1. We chose PA-D31 to represent saturated fatty acids and oleic acid-D34 (OA-D34) to represent unsaturated fatty acids. The ratio between their signal intensities at 2110 cm–1 directly measures the ratio of their concentration. We incubated the cells with either PA-D31 or OA-D34 for 7 h and found that the signal intensity at 2110 cm–1 from PA-D31 is significantly lower than that from OA-D34 (Figure 5b). To compare the incorporation rate, we normalized the intensity of the C–D signals to that of the C–H signals, and showed that the incorporation of OA-D34 is 24% faster than that of PA-D31 (Figure 5c).
Surprisingly, we also observed abnormal membrane-like structures with strong C–D signals in the cytosol of PA-D31-labeled cells, but not in OA-D34-labeled cells (Figure 5b and Figure S4). These structures were detected in all the images that we captured and in about 61% of PA-D31-labeled cells (Figure S4). We analyzed the spectra of those membrane-like structures and found that they are identical to those from LDs, indicating that those structures are rich in PA-D31. We also found that the morphology of PA-D31-labeled cells appears less healthy than that of OA-D34-labeled cells (Figure S4). Those phenotypic changes in PA-labeled cells may be due to alterations in membrane fluidity and/or structure as a result of increased saturation of membrane lipids, which is likely associated with the cytotoxicity of saturated fatty acids.39−41
Metabolic Tracing of Stable-Isotope Labeled Fatty Acids in Living Animals
Next, we tracked the dynamics of deuterated fatty acid molecules at the whole organism level in C. elegans. C. elegans is an excellent model system for metabolic studies based on Raman imaging due to its whole-body transparency.42 We imaged deuterated fatty acids using hsSRS in live C. elegans and traced their uptake, transportation and incorporation over time. These fatty acids exhibit distinct spectra and therefore allow profiling with hsSRS (Figure 6a). Within 5 h of supplementation with OA-D34, we detected its distribution in several different tissues of adult C. elegans, including the intestine, the hypodermis, oocytes, and embryos (Figure S5). These results suggest that absorbed fatty acid molecules can be rapidly transported from the intestine to peripheral tissues.
Figure 6.
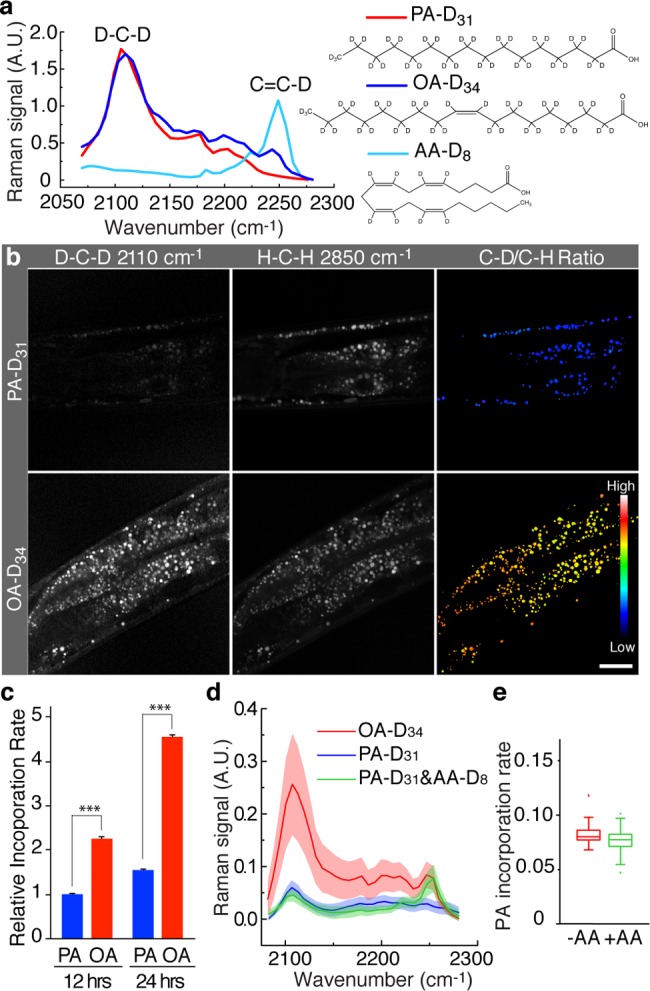
Tracking transportation and incorporation dynamics of different fatty acid molecules in C. elegans. (a) hsSRS spectra and chemical structures of PA-D31, OA-D34, and AA-D8. Both OA-D34 and AA-D8 exhibit a C=C–D peak at 2250 cm–1, which are clearly separated from the D–C–D peak at 2110 cm–1. (b) hsSRS images of wild-type worms labeled with either PA-D31 or OA-D34. The incorporation of deuterated fatty acids and the total lipid level were imaged at 2110 and 2850 cm–1, respectively. The ratio between the C–D and the C–H signal intensities was used to determine the level of fatty acid incorporation into LDs. Scale bar = 20 μm. (c) Compared with PA-D31, the incorporation rate of OA-D34 is 2.3 and 2.9 times higher at 12 and 24 h of supplementation, respectively. For 12 h, PA-D31, n = 503; OA-D34, n = 512. For 24 h, PA-D31, n = 429; OA-D34, n = 516. *** p < 0.001. (d) The average spectra of intestinal LDs in wild-type worms labeled with OA-D34, PA-D31, and PA-D31 + AA-D8 for 24 h. The shaded curve represents standard deviation, PA-D31, n = 159; OA-D34, n = 147; PA-D31 + AA-D8, n = 226. (e) The incorporation rate of PA-D31 is not affected by the presence of AA-D8. PA-D31 alone, n = 127; PA-D31 + AA-D8, n = 159. p > 0.5.
In C. elegans, the intestine not only is a digestive organ but also combines the functions of liver and adipose tissue to store lipids and regulate metabolism.43 Using hsSRS, we found that C. elegans intestinal LDs predominantly contain TAG, similar to mouse hepatic LDs (Figure S6). Consistent with the results in mammalian hepatic cells, we also found that the signal intensity at 2110 cm–1 from PA-D31 is lower than that from OA-D34 in the intestinal cells after 12 and 24 h of supplementation (Figure 6b). After normalized to the C–H signals, the incorporation rate of OA-D34 is about 2 times faster than that of PA-D31 within 12 h of supplementation and 3 times faster within 24 h (Figure 6c). Together our results show that unsaturated fatty acids are utilized preferentially as substrates for TAG synthesis and then are incorporated into LDs.
We then utilized the spectral difference between deuterated unsaturated fatty acid and deuterated saturated fatty acid to track their states after incorporation into LDs. Different from the saturated ones, deuterated unsaturated fatty acids have an additional peak at 2250 cm–1 arising from C=C–D stretching (Figure 6a). The intensity of this peak is proportional to the number of C=C–D bonds. By comparing the hsSRS spectra of C. elegans LDs to that of pure solutions, we concluded that neither PA-D31 nor OA-D34 underwent any substantial modification such as desaturation. More importantly, the difference in the spectra between C=C–D and D–C–D offers a unique opportunity to simultaneously trace saturated and unsaturated fatty acids in vivo and to analyze their interactions during their incorporation into LDs. To demonstrate this new approach, we supplemented C. elegans with deuterated palmitic acid (PA-D31) and deuterated archidonic acid (AA-D8) and acquired hsSRS spectra of LDs in these dual-labeled animals. As expected, the hsSRS spectra exhibit two separate peaks at 2110 and 2250 cm–1 that are derived from PA-D31 and AA-D8, respectively (Figure 6d). Furthermore, the signal intensity at 2110 cm–1 in the dual-labeled sample is comparable to that of the sample labeled with PA-D31 alone (Figure 6d,e), revealing that the presence of unsaturated fatty acids does not accelerate the rate at which saturated fatty acids are incorporated, unlike previously proposed by Listenberger.41 These studies demonstrate that hsSRS, when coupled with deuterium labeling, provides a new method for simultaneously tracing multiple lipid molecules specifically in vivo.
Discussion
Metabolomics provides crucial readouts on the universal outcome of influencing factors on disease states, and therefore has great potentials in early diagnosis, therapy monitoring, and understanding the pathogenesis of diseases. To fully realize those potentials, innovative analytical technologies are needed. hsSRS imaging potentially offers a complementary approach to MS and NMR/MRI in monitoring metabolic states and the dynamics of metabolites in living biological systems. We demonstrated this possibility with lipid metabolite fingerprinting. Similar to proteins, the physiological activities of lipid molecules are tightly associated with their composition, spatial distribution, and temporal dynamics. In this study, we reported a general method based on hsSRS and deuterium labeling to quantitatively image different types of lipid molecules in vivo and to track their spatiotemporal dynamics in living cells and organisms. Based on this approach, we were able to distinguish two classes of neutral lipid molecules, TAG and CE at the single-LD level in yeast, C. elegans, cultured mammalian cells, and mouse tissues, and to elucidate the dynamics of different fatty acids molecules during their incorporation and transportation in vivo.
Our imaging data show that the distribution of neutral lipids is heterogeneous between different tissues and between different organisms. Yeast cells consist of a mixture of TAG and CE, while C. elegans predominantly contains TAG. In mammalian cells and tissues, TAG and CE are heterogeneously distributed depending on the type of the cell or the tissue. This lipid compositional heterogeneity is likely related to the physiological activities of different tissues and organisms. Recently, Hsieh and colleagues used fatty acid BODIPY 558/568 C12 and cholesteryl BODIPY 500/510 FL C12 to show that there are distinct TAG- and CE-containing LDs in McA-RH7777 hepatic cells, and different Perilipin families of proteins coat distinct LD subpopulation.21 However, using label-free hsSRS, we only detected TAG-containing LDs in the same hepatic cell line and found that lipid compositions of LDs are homogeneous inside the cell. The discrepancy could be due to the BODIPY labeling methods, which could change the molecular properties of lipid molecules and result in mischaracterization of LDs.
Furthermore, we found that lipid compositional changes are associated with obesity and steatohepatitis. In the ob/ob mouse or in the mouse challenged with ER-stress-induced hepatic steatosis, lipid accumulation in non-adipose tissues is accompanied by an increased level of unsaturation in neutral lipids. Due to the important role of fatty acid unsaturation play in biophysical properties of membrane, it is conceivable that this compositional change in neutral lipids will influence the cell physiology.44,45 With deuterated fatty acid tracing, we have already observed abnormal membrane structures built up when cells take up significant amount of saturated fatty acids. Recently, Ariyama and colleagues showed that a decrease in membrane phospholipid unsaturation induces the unfolded protein response in the ER.46 Thus, it will be very interesting for future studies to dissect whether and how the compositional changes of neutral lipids influence membrane phospholipid composition, and consequently affect lipotoxicity in non-adipose tissues.
Our results also show that different lipid molecules exhibit great heterogeneity in their incorporation and transportation dynamics. The incorporation rate of unsaturated fatty acids into existing LDs is much faster than saturated fatty acids. This might be related to the preference of lipid-synthesizing enzymes for their substrates; for example, in vitro studies suggest that acyl-coenzyme A:diacylglycerol acyltransferase 2 (DGAT2), which is localized at the LD membrane and catalyzes the last step of TAG synthesis, preferentially utilizes monounsaturated fatty acids.47 Through metabolic tracing of two different deuterium labeled fatty acids, we found that the presence of unsaturated fatty acids (AAs) does not accelerate the incorporation of saturated fatty acids (PAs), which challenges the previous hypothesis by Listenberger and colleagues that unsaturated fatty acids facilitate the incorporation of saturated fatty acids into TAG.41 Future studies based on the stable-isotope labeling hsSRS imaging approach are expected to uncover more previously unknown interactions between different fatty acid molecules.
Even though we only demonstrated fingerprinting of lipids in this work, it is straightforward to extend the approach to image other metabolites such as glucose and amino acids. A major limitation is the sensitivity of the imaging. Most recently, Wei demonstrated that by tagging an alkyne group to small molecules, the detection sensitivity can be increased to 200 μM.48 The sensitivity can be further improved with longer averaging and higher laser power, with a trade-off of potential photodamage to the sample. We believe hsSRS imaging provides a complementary approach to IMS and MRI for metabolic profiling of biological samples, which fills the niche of live cell and tissue imaging at subcellular resolution. Because no sample preparation is required for hsSRS imaging, it is also straightforward to combine different metabolic imaging modalities to characterize biological systems across different temporal and spatial scales.
Acknowledgments
We thank C. Freudiger, M. Ji, and F. Lu for helpful discussions. We thank W. Yang for providing the amplified photodiode for the hsSRS imaging experiments. We also thank J. Qi and W. Shih for helping with the Raman spectra measurements. This work was supported by an Ellison Medical Foundation New Scholar Award (M.C.W.) and National Institutes of Health grants RO0AG034988 (M.C.W.), RO1AG045183 (M.C.W.), RO1EB010244 (X.S.X.), and RO1GM099844 (R.V.F.).
Supporting Information Available
Supplementary figures as described in the text. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
# D.F. and Y.Y. contributed equally.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Zenobi R. Science 2013, 34261631243259 . [DOI] [PubMed] [Google Scholar]
- Steinhauser M. L.; Bailey A. P.; Senyo S. E.; Guillermier C.; Perlstein T. S.; Gould A. P.; Lee R. T.; Lechene C. P. Nature 2012, 4817382516–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura D.; Fujimura Y.; Wariishi H. J. Proteomics 2012, 75165052–5060. [DOI] [PubMed] [Google Scholar]
- Golman K.; in ‘t Zandt R.; Thaning M. Proc. Natl. Acad. Sci. U.S.A. 2006, 1033011270–11275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias N. M.; Chan H. R.; Sailasuta N.; Ross B. D.; Bhattacharya P. J. Am. Chem. Soc. 2011, 1342934–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis D. I.; Dunn W. B.; Griffin J. L.; Allwood J. W.; Goodacre R. Pharmacogenomics 2007, 891243–1266. [DOI] [PubMed] [Google Scholar]
- Evans C. L.; Xie X. S. Annu. Rev. Anal. Chem. 2008, 11883–909. [DOI] [PubMed] [Google Scholar]
- Freudiger C. W.; Min W.; Saar B. G.; Lu S.; Holtom G. R.; He C.; Tsai J. C.; Kang J. X.; Xie X. S. Science 2008, 32259091857–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D.; Holtom G.; Freudiger C.; Zhang X.; Xie X. S. J. Phys. Chem. B 2013, 117164634–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozeki Y.; Umemura W.; Otsuka Y.; Satoh S.; Hashimoto H.; Sumimura K.; Nishizawa N.; Fukui K.; Itoh K. Nat. Photonics 2012, 6127. [Google Scholar]
- Zhang D.; Wang P.; Slipchenko M. N.; Ben-Amotz D.; Weiner A. M.; Cheng J. X. Anal. Chem. 2013, 85198–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhalim J. L.; Chung C. Y.; Lilledahl M. B.; Lim R. S.; Levi M.; Tromberg B. J.; Potma E. O. Biophys. J. 2012, 10281988–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P.; Li J.; Wang P.; Hu C. R.; Zhang D.; Sturek M.; Cheng J. X. Angew. Chem. 2013, 524913042–13046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg A. S.; Coleman R. A.; Kraemer F. B.; McManaman J. L.; Obin M. S.; Puri V.; Yan Q. W.; Miyoshi H.; Mashek D. G. J. Clin. Invest. 2011, 12162102–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T. C.; Farese R. V. Jr. Annu. Rev. Biochem. 2012, 81, 687–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenk M. R. Nat. Rev. Drug Discovery 2005, 47594–610. [DOI] [PubMed] [Google Scholar]
- Folick A.; Min W.; Wang M. C. Curr. Opin. Genet. Dev. 2011, 215585–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y.; Ramachandran P. V.; Wang M. C. BBA-Mol. Cell Biol. Lipids 2014, 10.1016/j.bbalip.2014.02.003. [DOI] [Google Scholar]
- Saar B. G.; Freudiger C. W.; Reichman J.; Stanley C. M.; Holtom G. R.; Xie X. S. Science 2010, 33060091368–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S.; McKenzie A. III; Demarini D. J.; Shah N. G.; Wach A.; Brachat A.; Philippsen P.; Pringle J. R. Yeast 1998, 1410953–961. [DOI] [PubMed] [Google Scholar]
- Hsieh K.; Lee Y. K.; Londos C.; Raaka B. M.; Dalen K. T.; Kimmel A. R. J. Cell Sci. 2012, 125174067–4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle T. WormBook 2006, 10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T. C.; Farese R. V. Annu. Rev. Biochem. 2012, 811687–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen-Pergakes K.; Guo Z.; Giattina M.; Sturley S. L.; Bard M. J. Bacteriol. 2001, 183174950–4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger D.; Daum G. J. Bacteriol. 2002, 1842519–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger R. H.; Scherer P. E. Trends Endocrin. Metab. 2010, 216345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Proenca R.; Maffei M.; Barone M.; Leopold L.; Friedman J. M. Nature 1994, 3726505425–432. [DOI] [PubMed] [Google Scholar]
- Farrell G. C.; Larter C. Z. Hepatology 2006, 432 Suppl. 1S99–S112. [DOI] [PubMed] [Google Scholar]
- Pagliassotti M. J. Annu. Rev. Nutr. 2012, 32, 17–33. [DOI] [PubMed] [Google Scholar]
- Yamamoto K.; Takahara K.; Oyadomari S.; Okada T.; Sato T.; Harada A.; Mori K. Mol. Biol. Cell 2010, 21172975–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S.-E.; Blagoev B.; Kratchmarova I.; Kristensen D. B.; Steen H.; Pandey A.; Mann M. Mol. Cell. Proteomics 2002, 15376–386. [DOI] [PubMed] [Google Scholar]
- Zamboni N.; Fendt S.-M.; Ruhl M.; Sauer U. Nat. Protocols 2009, 46878–892. [DOI] [PubMed] [Google Scholar]
- Li M.; Huang W. E.; Gibson C. M.; Fowler P. W.; Jousset A. Anal. Chem. 2012, 8531642–1649. [DOI] [PubMed] [Google Scholar]
- Matthaus C.; Krafft C.; Dietzek B.; Brehm B. R.; Lorkowski S.; Popp J. Anal. Chem. 2012, 84208549–8556. [DOI] [PubMed] [Google Scholar]
- van Manen H.-J.; Lenferink A.; Otto C. Anal. Chem. 2008, 80249576–9582. [DOI] [PubMed] [Google Scholar]
- Xie X. S.; Yu J.; Yang W. Y. Science 2006, 3125771228–230. [DOI] [PubMed] [Google Scholar]
- Zhang D.; Slipchenko M. N.; Cheng J.-X. J. Phys. Chem. Lett. 2011, 2111248–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L.; Yu Y.; Shen Y.; Wang M. C.; Min W. Proc. Natl. Acad. Sci. U.S.A. 2013, 1102811226–11231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricchi M.; Odoardi M. R.; Carulli L.; Anzivino C.; Ballestri S.; Pinetti A.; Fantoni L. I.; Marra F.; Bertolotti M.; Banni S.; Lonardo A.; Carulli N.; Loria P. J. Gastroen. Hepatol. 2009, 245830–840. [DOI] [PubMed] [Google Scholar]
- Herms A.; Bosch M.; Ariotti N.; Reddy B. J.; Fajardo A.; Fernandez-Vidal A.; Alvarez-Guaita A.; Fernandez-Rojo M. A.; Rentero C.; Tebar F.; Enrich C.; Geli M. I.; Parton R. G.; Gross S. P.; Pol A. Curr. Biol. 2013, 23151489–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listenberger L. L.; Han X.; Lewis S. E.; Cases S.; Farese R. V. Jr.; Ory D. S.; Schaffer J. E. Proc. Natl. Acad. Sci. U.S.A. 2003, 10063077–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulme S. E.; Whitesides G. M. Angew. Chem., Int. Ed. 2011, 50214774–4807. [DOI] [PubMed] [Google Scholar]
- Mak H. Y. J. Lipid Res. 2012, 53128–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowhan W. Annu. Rev. Biochem. 1997, 66, 199–232. [DOI] [PubMed] [Google Scholar]
- Martin C. E.; Hiramitsu K.; Kitajima Y.; Nozawa Y.; Skriver L.; Thompson G. A. Biochemistry 1976, 15245218–5227. [DOI] [PubMed] [Google Scholar]
- Ariyama H.; Kono N.; Matsuda S.; Inoue T.; Arai H. J. Biol. Chem. 2010, 2852922027–22035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man W. C.; Miyazaki M.; Chu K.; Ntambi J. J. Lipid Res. 2006, 4791928–1939. [DOI] [PubMed] [Google Scholar]
- Wei L.; Hu F.; Shen Y.; Chen Z.; Yu Y.; Lin C.-C.; Wang M. C.; Min W. Nat. Methods 2014, 114410–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.