Abstract
The blood-brain barrier (BBB) represents an obstacle in targeting and delivering therapeutics to the central nervous system. In order to discover new BBB targeting molecules, we panned a phage-displayed nonimmune human single-chain antibody fragment (scFv) library against a representative BBB model comprised of hydrocortisone-treated primary rat brain endothelial cells. Parallel screens were performed with or without pre-subtraction against primary rat heart and lung endothelial cells in an effort to identify antibodies that may have binding selectivity towards brain endothelial cells. After three rounds of screening, three unique scFvs, scFv15, scFv38, and scFv29, were identified that maintained binding to primary rat brain endothelial cells, both in phage and soluble scFv format. While scFv29 and to a lesser extent, scFv15, exhibited some brain endothelial cell specificity in tissue culture, scFv29 did not appear to bind a BBB antigen in vivo. In contrast, both scFv15 and scFv38 were capable of immunolabeling rat brain vessels in vivo and displayed brain vascular selectivity with respect to all peripheral organs tested other than heart. Taken together, scFv15 and scFv38 represent two new antibodies that are capable of binding antigens that are expressed at the BBB in vivo.
Keywords: blood-brain barrier, phage display, brain targeting, combinatorial screening, antibody
INTRODUCTION
The vascular network of the brain forms a biological barrier known as the blood-brain barrier (BBB). The BBB possesses a unique vascular phenotype that is induced by neighboring cells such as pericytes, astrocytes and neurons that together form the neurovascular unit [1]. This phenotype is best characterized as a combination of properties designed to maintain brain homeostasis, including tight paracellular junctions, a significant transporter repertoire, and a low basal level of pinocytosis, thereby rendering the BBB selectively permeable to required ions, nutrients and cells [2]. While the BBB helps the brain to maintain the specific environment necessary for neuron function, it also prevents most small and large molecule therapeutics from gaining access to the brain [3]. The BBB is therefore a major impediment to the treatment of central nervous system disease, and effective delivery strategies remain scarce. One promising delivery method involves targeting known receptor-mediated transport systems with antibodies to mediate non-invasive drug delivery past the BBB. Two prominent examples of this approach are antibodies that target the transferrin and insulin receptors [4, 5]. While these systems allow for therapeutic amounts of drug to penetrate the BBB, they are inherently non-specific and relatively inefficient. The current lack of brain drug delivery systems with ideal specificity and efficiency has motivated the search for new antibodies capable of targeting and/or transporting therapeutic payloads into the brain [2].
To address this problem, one useful approach that has been employed is antibody-based screening. Such screens have been used to identify BBB cell surface proteins that can mediate brain targeting and, in some cases, transport. For instance, large combinatorial antibody libraries have been screened against brain endothelial cells in various formats, in vitro or in vivo, to discover both antibody targeting molecules and cognate brain endothelial cell proteins [6–9]. While, in some cases, the identified antibody-BBB antigen pairs look promising for circumventing the BBB [10], only a handful of new antibodies have been isolated [2]. However, multiple genomic and proteomic studies support substantial differences in gene expression between the brain microvascular endothelium and the peripheral microvasculature, particularly in areas of transport and signaling between the brain and bloodstream [8, 11–13],
Thus, in order to access the unexplored BBB proteome and expand the repertoire of BBB targeting antibodies, we panned a large phage-displayed library of nonimmune human scFv on an in vitro BBB model based on primary rat brain endothelial cells that are capable of mimicking key BBB characteristics such as elevated trans-endothelial electrical resistance, improved tight junction integrity, and a molecular signature that moves towards the in vivo BBB [6, 14]. Because of the aforementioned interest in antibodies that may be selective towards the BBB and/or mediate BBB internalization, our screen employed phage subtraction and internalization approaches. Three particularly interesting scFvs were isolated from the screen, with two that were subsequently shown to preferentially bind to the rat brain microvasculature in vivo.
MATERIALS AND METHODS
Cell isolation and culture
The brain microvascular endothelial cell (BMEC) isolation was performed as previously described [14]. The purified BMECs were plated on collagen type IV and fibronectin (Sigma-Aldrich, # C5533 and # F1141) coated tissue culture plates and cultured in endothelial cell culture medium consisting of DMEM supplemented with 20% platelet-poor bovine plasma derived serum (PDS, from Biomedical Technologies, # BT-214), heparin at 1 μg/mL (Sigma-Aldrich, # H3393), L-glutamine at 2 mM (Sigma-Aldrich, # G8540), 100× Antibiotic-Antimycotic (Life Technologies, # 15240-062), and basic fibroblast growth factor at 1 ng/mL (bFGF, R&D Systems, # 233-FB). For the first two days of culture, the medium also included 4 μg/mL of puromycin (Sigma-Aldrich, # P8833) for BMEC purification purposes. Upon reaching confluence, BBB properties were induced by changing to serum-free medium consisting of 50% DMEM and 50% Ham’s F-12 (Life Technologies, #11765-054) with L-glutamine at 2 mM and Antibiotic-Antimycotic supplemented with 550 nM hydrocortisone for 24 hours before use.
The primary rat heart and lung microvascular endothelial cells (HEC/LEC) were obtained from VEC Technologies (Rensselaer, NY), and cultured per manufacturer’s instructions on fibronectin coated tissue culture plates in MCDB-131 complete media (VEC technologies).
Screening on BMECs
All of the screening methods are based on protocols outlined in Zhou and Marks [15]. Before screening for phage internalization on BMECs, the human scFv displaying Fd-tet library [16, 17] was first pre-subtracted for common endothelial antigens by serially applying the library to T-75 flasks of HEC and LEC. Culture media for all cells was replaced with 3 mL of MCDB-131 complete media for HEC and LEC, and 20 mL of serum-free medium with hydrocortisone for BMECs, 1 hour before use. For the first round, 2×1011 colony forming units (cfu) of the scFv library were added to a flask of LECs containing 3 mL of MCDB-131 complete media and incubated for 1 hour at room temperature. MCDB-131 complete media from the HECs was removed and replaced with the phage containing MCDB-131 complete media from the LEC flask and incubated at room temperature for another hour. The serum free media with hydrocortisone from the BMEC T-75 flask was then removed and replaced with the medium from the HEC flask containing the now pre-subtracted library. In parallel, 2×1011 cfu of the Fd-tet library were added to a separate T-75 flask of BMECs containing 3 mL of serum free media for the non-subtracted screen. The BMEC flasks were kept at 4°C for 1 hour and rocked for 5 minutes every half-hour to encourage phage binding. The flasks were then washed quickly with 1 mL of phosphate buffered saline (PBS, 137 mM sodium chloride, 2.7 mM potassium chloride, 10 mM dibasic sodium phosphate, 1.8 mM monobasic potassium phosphate, pH 7.4) five times. Three mL of pre-warmed serum free media was added to the BMECs and the flasks were moved to an incubator at 37°C and 5% CO2 for 40 minutes and gently rocked several times to promote phage internalization. For rounds 2 and 3, the procedure described above was repeated except the BMEC flasks were washed 3 times with 25 mL of PBS, followed by addition of 20 mL of pre-warmed serum free media supplemented with hydrocortisone, prior to incubation at 37°C for 15 minutes.
After round 1, BMECs were washed three times with 1 mL ice cold PBS. Subsequent rounds were washed three times with 10 mL of PBS. The loosely bound phage were stripped from the surface of the cells by adding of 4 mL stripping buffer I (500 mM sodium chloride, 0.2 M urea, 2 mg/mL polyvinylpyrrolidone in 50 mM glycine, pH 2.8) three times at room temperature for 5 minutes. The stripping buffer fractions were recovered and neutralized with 2 mL of 1 M Tris-HCL (pH 7.4), then placed on ice, and reserved for titering and storage. The still adherent cells were washed two times in 10 mL of PBS at room temperature and then once with 2 mL of 0.25% trypsin/EDTA (Life Technologies, # 25200-056). Then, 2 mL of fresh trypsin/EDTA was added to the BMECs and incubated at 37°C for no longer than 10 minutes to detach the cells and further remove phage bound to the outside of the BMEC. The detached cells were moved to a conical tube and centrifuged at 300g at 4°C for 5 minutes. The cell pellet was then washed twice with 10 mL ice cold hydrocortisone supplemented serum free media and then resuspended in 1 mL of ice cold lysis Buffer (100 mM triethanolamine in ddH2O), triturated, and incubated on ice for 10 minutes. The lysate containing the “cell-associated fraction” was neutralized by triturating with 0.5 mL of 1 M Tris-HCL (pH 7.4).
The phage were recovered from the cell-associated fraction by incubation with an excess of log phase TG1 Escherichia coli cells from Agilent Technologies (Santa Clara, CA). Briefly, 0.75 mL of phage-containing fractions were added to 10 mL of log phase TG1 E. coli and incubated at 37°C for 30 minutes, followed by another 30 minute incubation at 37°C while shaking. A volume of 300 μL of the phage bearing TG1 was used for titer determination. The rest was plated on two 150 mm 2xYT (16 g/L Bacto Tryptone, 10 g/L Bacto Yeast Extract, and 5 g/L sodium chloride, pH 7.0) agar plates with 15 μg/mL tetracycline and incubated at 37°C overnight. The phage harboring bacteria were subsequently scraped off the plates using 2xYT media, expanded in 200 mL culture and phage in the culture supernatant was recovered by standard polyethylene glycol (PEG) precipitation [15]. For subsequent screening rounds, 1×1011 cfu of phage from the cell-associated fraction were used, except for round 2 of the pre-subtracted library screen. The recovery of phage from round 1 in this pool was lower than expected, so round 2 of the pre-subtracted screen was treated the same way as round 1 (using less stringent conditions than round 2 for the non-subtracted pool) except phage was applied in a ratio of 5:1 of cell-associated fraction to third stripping fraction, and stripping buffer II (150 nM sodium chloride, 100 mM glycine, pH 2.5) was used in place of stripping buffer I.
DNA fingerprinting by BstN1 digestion
Estimates of post-screen pool diversity were determined by BstN1 digestion of scFv-encoding inserts. Briefly, bacteria infected from phages isolated from the post-screen pools were spread on TYE (10 g/L bacto tryptone, 15 g/L bacto yeast extract, and 8 g/L sodium chloride, pH to 7.0) agar plates with 15 μg/mL tetracycline and allowed to grow overnight at 37°C into isolated colonies. Ninety-six of these were picked off the plate, expanded, and stored in 15% glycerol at −80°C until experiment was performed. Whole cell PCR was performed using Platinum® Taq (Life Technologies, # 10966-034) per manufacturer’s instructions on each of the wells described above using primers that flank the scFv gene in the phage DNA. The primer sequences were 5′-TTTTTGGAGATTTTCAACGTGA-3′, and 5′-GAATTTTCTGTATGAGGTTTTGCTAAA-3′ for the forward and reverse primers, respectively. The PCR product was then digested with the restriction enzyme BstN1 (New England Biolabs # R0168L) per manufacturer’s instructions. The digestion products were run on a 3% agarose gel, stained with ethidium bromide and imaged using the Molecular Imager Gel Doc XR System from Bio-Rad (Hercules, CA). The resulting images were analyzed for distinct patterns in each lane and categorized accordingly.
Enzyme Linked Immunosorbent Assay (ELISA) for phage binding
BMECs were cultured in 96 well tissue culture plates as described above. The day of the assay, each well of BMECs was blocked with 250 μL of PBSCM (PBS with 1 mM of calcium chloride and 0.5 mM of magnesium sulfate) supplemented with 40% goat serum (PBSCMG) (Sigma-Aldrich, #G6767). The wells were washed three times with 250 μL of PBSCM. Overnight cultures of phage harboring bacteria were centrifuged, and 50 μL of the phage containing supernatant from each sample was incubated directly on the BMECs in the presence of 100 μL of fresh PBSCMG. The plate was incubated for one hour at 4°C and then washed once. An anti-M13-HRP antibody (GE Healthcare, # 27942101) diluted 1:500 in PBSCMG was incubated in each well for one hour at 4°C. Following this, the cells were washed three times in PBSCM and a colorimetric substrate was added to each well and incubated for 30 minutes (ABTS (Sigma-Aldrich, # A9941) prepared by manufacturer’s instructions). The plate was then read at 405 nm using an EL800 Universal Microplate Reader from BioTek (Winooski, VT).
Preparation of soluble hexahistidine-tagged scFv
The following method for secreting the soluble scFv-His6 fusion is based on a protocol described in Zhou and Marks [15]. An overnight bacterial culture harboring the scFv secretion plasmid was used to inoculate 2xYT medium containing 100 μg/mL ampicillin and 0.1% glucose, which was then grown at 37°C until an OD600nm of 0.9 was reached. Expression was induced by adding 1mM isopropyl-β-D-thiogalactopyranoside (IPTG, Fisher Scientific, # 50213380) and allowed grow for 4 hours at 30°C. The bacteria was harvested and the scFv recovered by serial incubation with a periplasmic extraction buffer (PPB, 200 g/L sucrose, 1 mM EDTA, 30 mM tris-HCl, pH 8.0) supplemented with DNAse I (Roche Applied Sciences, # 10104159001) to 100 μg/mL, and cOmplete protease inhibitor cocktail tablets, mini (Roche Applied Sciences, # 11836153001) and an osmotic shock buffer (OSB, 5 mM magnesium sulfate in ddH2O) supplemented with DNAse I and cOmplete, Mini. The result was syringe filter sterilized, and dialyzed against PBS + 10mM imidazole. The scFv were purified from the crude extract with HisPur Ni-NTA spin columns (Thermo Scientific, # 88224) using manufacturer recommended protocol for purification by gravity flow. The purified scFvs were eluted and subsequently dialyzed against PBS, and the purity of the scFv was verified by SDS polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining. Finally, the elution fractions were sterile filtered using Ultrafree centrifugal filtration units (Millipore # UFC30GV25), and quantified using UV 280 absorbance and extinction coefficients generated by ExPASy (http://web.expasy.org/protparam/).
Immunocytochemical labeling of cultured cells with soluble scFv
BMEC, LEC, or HEC were cultured as described above. The cells were washed once with 300 μL ice cold PBSCM, blocked for 30 minutes on ice with 300 μL of PBSCMG. The cells were incubated with 300 μL of 100 μg/mL (~3 μM) scFv monomer in PBSCMG for 2.5 hours on ice. The cells were washed 2 times with ice cold PBSCM, and then the surface was labeled with 250 μL of a mouse anti-c-myc antibody (Covance # MMS-150P) at a 1:250 dilution in PBSCMG for 30 minutes on ice. This was followed by 2 washes in ice cold PBSCM and a 30 minute incubation with 250 μL of goat anti-mouse Alexa Fluor® 594 (Life Technologies # A11032) diluted 1:400 in PBSCMG. The nuclear stain, DAPI (Life Technologies, # D1306) was applied for 4 minutes at room temperature at a concentration of 300 nM in 300 μL of PBS, and then the cells were washed three times with ice cold PBSCM and post fixed for 8 minutes at room temperature with paraformaldehyde (4% w/v in PBS). Finally, the cells were washed three times in PBSCM, with 0.5 mL of wash buffer and visualized using an Olympus fluorescence microscope connected to a Diagnostic Instruments camera run by MetaVue.
ScFv internalization was also assayed using a procedure similar to above except the following: The scFv were pre-dimerized with the mouse anti-c-myc antibody at a molecular ratio of 4 scFv to 1 anti-c-myc antibody in PBSCMG. The pre-dimerized scFv was incubated with BMECs for 1 hour on ice and then transferred to an incubator at 37°C and 5% CO2 for 30 minutes to allow for internalization. The cells were next incubated with Alexa Fluor® 594 as described previously. The cells were then fixed in paraformaldehyde for 8 minutes and washed 2 times in ice cold PBSCM. The cells were permeabilized using 0.6% Triton™ X-100 (Sigma-Aldrich, # X100) in PBSCMG for 30 minutes. Next, the cells were washed 2 times in ice cold PBSCM and incubated with goat anti-mouse Alexa Fluor® 488 (Life Technologies, # A11029) diluted 1:400 in PBSCMG. The cells were then labeled with DAPI, fixed again, and viewed under the fluorescent microscope as previously described. A positive control antibody against the rat transferrin receptor (AbD Serotec, # MCA155G) was used at a dilution of 1:200.
Flow cytometric analysis of cultured cells with soluble scFv
BMEC, LEC, or HEC were cultured as described above. The cells (~2×106 cells/T-25 flask) were washed in PBS and detached from the T-25 culture flasks using 1 mL of accutase (Life Technologies, # A11105-01). The cells were transferred to a conical tube, centrifuged at 800g for 10 minutes, and resuspended in PBSG (PBS with 40% goat serum) and blocked on wet ice for 30 minutes. The cells from each flask were separated into 5 equal samples containing ~4×105 cells which were centrifuged and resuspended in 300 μL scFv monomer at 100 μg/mL (~3 μM) in PBSG and incubated for 60 minutes on ice. The cells were centrifuged as above and washed once in 1 mL of ice cold wash buffer (PBS with 5% goat serum) and centrifuged again. The samples were resuspended in mouse-anti-c-myc in 300 μL of PBSG at a 1:250 dilution and incubated on ice for 30 minutes. The cells were washed once in 1 mL of ice cold wash buffer, centrifuged and resuspended in a goat anti-mouse antibody conjugated to allophycocyanin (APC, Life Technologies, # A865) in 300 μL of PBSG and incubated for 30 minutes. The cells were washed two times and resuspended in flow buffer (PBS + 0.1% BSA + 5 mM EDTA) supplemented with Sytox® (Life Technologies, # S7020)) diluted 1:10,000 and analyzed on a flow cytometer (Becton Dickinson FACSCalibur).
Immunohistochemical labeling of rat tissue sections with soluble scFv
Rat brain, heart, lung, liver, and kidney were dissected from a male Sprague-Dawley rat, snap frozen, and 7 μM cryosections were prepared. Prior to use, the sections were removed from the freezer and allowed to thaw and air dry for approximately 20 minutes. The sections were wetted in sterile PBS at room temperature. Next, tissue sections were blocked in PBSG for 30 minutes. ScFv monomer was diluted to 100 μg/mL (~3 μM) in 300 μL of PBSG and incubated on the tissue sections on ice for 2.5 hours (kidney used 50 μg/mL scFv). The sections were washed twice with ice cold PBS and the mouse anti-c-myc antibody diluted 1:250 in 300 μL of PBSG was incubated on the sections for 30 minutes on ice. The sections were next washed twice and incubated with an Alexa Flour® 594 conjugated anti-mouse antibody diluted 1:400 and isolectin B4 conjugated FITC (IB4-FITC, Sigma-Aldrich # L2895) diluted 1:100 in PBSG for 30 minutes. The samples were washed three times and fixed in 4% paraformaldehyde for 10 minutes at room temperature. Kidney sections serial to the scFv labeled sections were labeled in a similar manner but with an anti-rat CD31 antibody (Thermo Scientific, # MA1-81051) to visualize endothelial cells in place of the IB4-FITC. The sections were viewed using an Olympus fluorescence microscope connected to a Diagnostic Instruments camera run by MetaVue.
RESULTS
Screening of phage display scFv library on BMECs
The screen was performed using a rat in vitro BBB model described previously [14]. The model employs primary rat brain microvascular endothelial cells (BMECs) that are purified by puromycin treatment, and after confluence, BBB properties are induced with hydrocortisone [14]. This model was chosen as the cellular screening platform because hydrocortisone induction enhances the in vivo relevance of the model by leading to increases in trans-endothelial electrical resistance, improvements in tight junction morphology, and molecular changes that move the model more towards the in vivo situation [6, 14].
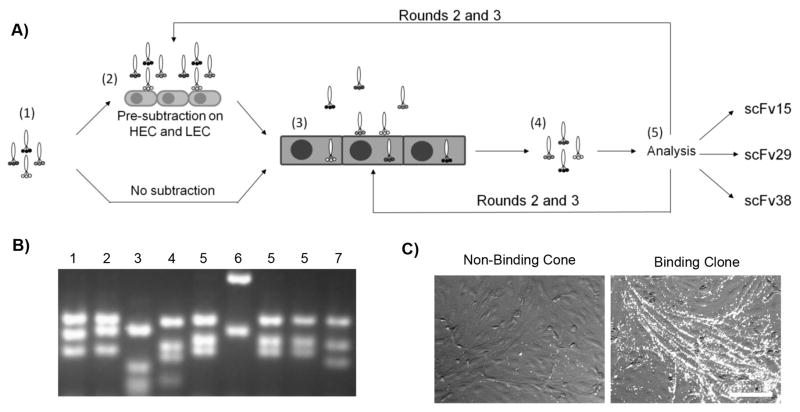
A library of 5×108 human-derived scFvs displayed on the surface of Fd-tet phage [16, 17] was panned against the BMECs. This library was chosen primarily for its multivalent display of scFv (3–5 copies per phage) to help bias the screen towards antibodies capable of internalization [18]. To this end, the screen was designed to enrich the recovered pools for internalized phages; but, as the data below indicate, the screen will also recover those antibody-bearing phage that bind well to the BMEC cell surface (Figure 1A). Within this functional screen, one path incorporated a pre-subtraction step prior to each round of screening in which the phage libraries were first incubated successively on primary rat heart (HEC) and rat lung (LEC) endothelial cell lines prior to using the unbound phage for screening on BMECs (Figure 1A, see Materials and Methods for details). The rationale was to attempt to remove phages that bind to common endothelial cell antigens and help promote brain selectivity. The parallel screening path did not employ subtraction in order to access the diversity of the entire antibody library and corresponding BMEC antigens. The pre-subtracted or non-subtracted phage pools were then incubated on hydrocortisone-induced BMECs grown in a tissue culture flask, first on ice as a binding step and then at 37°C to allow for possible phage internalization. The surface of the BMECs was subsequently washed and stripped with a low pH buffer. After trypsinization and lysis, the phages that had either internalized or were incompletely stripped were recovered in TG1 E. Coli, titered and this cell-associated fraction was used for the next round of screening (Table 1). As one exception to this strategy, it was noted that the diversity of the cell-associated fraction resulting from round 1 of the pre-subtraction screen was quite low and thus for round 2 phage recovered in the last stripping wash were combined with the cell-associated fraction (Table 1). The screen progress was followed both by phage titer and by BstN1 restriction-based DNA fingerprinting (Table 1 and Figure 1B). While the titer in the pre-subtracted screens continued to increase throughout three rounds of enrichment, by round 3 the diversity had been reduced to just two unique digestion patterns (Table 1). By contrast, the amount of phage recovered from the non-subtracted library increased from round 1 to round 2, but plateaued in round 3, and the diversity remained comparatively high. At this point, the scFv-bearing phages were analyzed on a clonal basis.
Figure 1.
Antibody library screening on in vitro BBB model. A) Step 1: Display of a library of human scFv on fd-tet phage. Step 2: One path employed pre-subtraction techniques using rat HEC and LEC in an attempt to promote brain specificity. The parallel screening path did not employ library subtraction. Step 3: Library incubation with BMECs to allow for binding and internalization of the antibody bearing phage. Following incubation, the BMECs were washed and stripped. Step 4: The BMECs were lysed to recover the phage stilal associated with the cells because of internalization or stripping-resistant binding. Step 5: Library pools were analyzed on a clonal basis by phage titer, BstNI restriction mapping, and in the final rounds, phage immunocytochemistry. B) Sample BstN1 digestion pattern on an agarose gel. Patterns are categorized by number to indicate the clustering used to compile the statistics reported in Table 1. C) Sample images from clonal phage immunocytochemistry using BMECs to determine which clones displayed a binding phenotype. Image on left is a non-binding clone, and image on right is scFv38 in phage format. Scale bar is 50 μm
Table 1.
Assessment of screen progress.
| Pre-subtracted | Non-subtracted | |||
|---|---|---|---|---|
| Titer (cfu)a | Digestion Patternsb | Titer (cfu)a | Digestion Patternsb | |
| Round 1 | 9.45×103 | 8.98×104 | ||
| Round 2 | 3.01×105 | 33/92 | 1.29×106 | 28/71 |
| Round 3 | 1.51×106 | 2/94 | 3.44×105 | 25/94 |
Phage titers from the internalized fraction of screens
The fraction of unique BstN1 digestion patterns
Clonal assessment of recovered scFvs
Antibody-bearing phage clones were individually assessed in a BMEC cell-based phage ELISA to determine those clones capable of binding BMECs. Out of 395 individual clones sampled from rounds 2 and 3 of the pre-subtracted and non-subtracted pools, 39 clones yielded an elevated ELISA signal. Evaluation of these clones based on BstN1 digestion patterns, reduced the number of potentially unique binding antibody-bearing phage clones to 22. Further evaluation of these 22 clones by phage-based immunocytochemistry revealed 10 phage clones with definitive binding to cultured BMECs (Figure 1C). Sequencing of the 10 clones yielded three unique scFv sequences, one designated scFv38 appeared in both pre-subtracted and non-subtracted pools resulting from round 3 of screening, scFv29 was found in round 3 of the pre-subtracted pool, and scFv15 was found only in round 2 of the pre-subtracted pool.
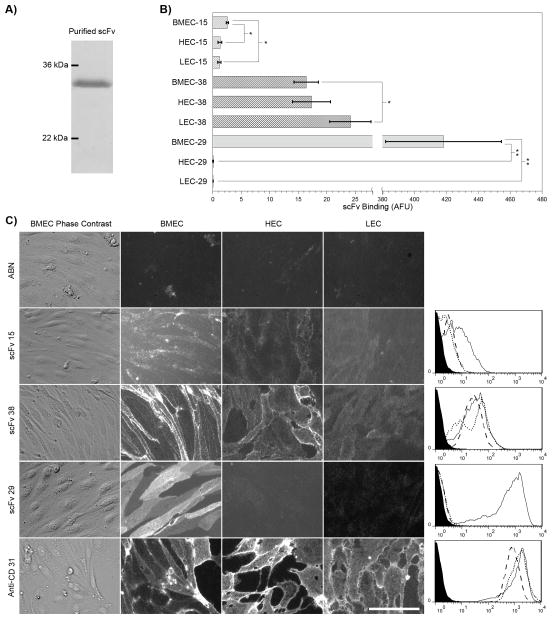
These three scFv were subcloned, expressed in bacteria and purified via a hexahistidine tag (Figure 2A). Purified scFv were used to immunolabel BMECs, HECs, and LECs. Fluorescent microscopy and flow cytometry were used to assess their qualitative and quantitative BMEC binding specificity, respectively. ScFv29 originated from the pre-subtracted screens and exhibited very clear preferential binding to BMEC’s without detectable binding to either the HECs or LECs (Figure 2C), and this finding was corroborated by flow cytometry (Figure 2B and 2C). In contrast, scFv15, which also originated in the pre-subtracted pool, exhibited more limited binding to BMECs and also yielded detectable binding to HECs and LECs (Figure 2B). While there was certainly less BMEC specificity for scFv15 compared with scFv29, flow cytometric quantification indicated that scFv15 binding to BMECs was increased compared with binding to either HECs or LECs (Figure 2C, p<.05). ScFv38 was found in both non-subtracted and pre-subtracted pools, and accordingly bound all three cell types with roughly the same intensity (Figure 2B and C).
Figure 2.
Production and assessment of soluble scFv binding profiles in cultured cells. A) Purified scFv electrophoresed on an SDS-PAGE gel stained with Coomassie blue. scFv38 is shown. B) Quantitative antibody binding resulting from flow cytometric analyses of scFv15, scFv38, and scFv29 on BMEC, HEC, and LEC. Experiments were performed on cells cultured in separate triplicate wells. Expressed are the specific signals after subtraction of the irrelevant, anti-botulinum (ABN) scFv background as mean ± S.D., n=3. *p<0.05, **p<0.005 computed using a student’s two-tailed t-test assuming unequal variances. Bar graph is representative of 3 independent experiments. C) Purified ABN scFv (negative control), scFv15, scFv38, and scFv29 scFv as well as anti-CD31 antibody (positive control) were incubated on BMECs, HECs, and LECs. The scFv immunolabeling concentration was 100 μg/ml (~3 μM) and is most likely saturating the targeted receptors. Scale bar is 50 μm and images are representative of 5 independent experiments. The histograms at the right of each row of images are flow cytometry histograms for each respective antibody representative of the triplicate samples quantified in Panel B. The x-axis of the histograms represents antibody binding intensity in arbitrary fluorescent units. The filled line represents the ABN scFv negative control, the solid line is binding signal arising from BMEC, the dotted line is binding signal from HEC, and the dashed line is binding signal from LEC.
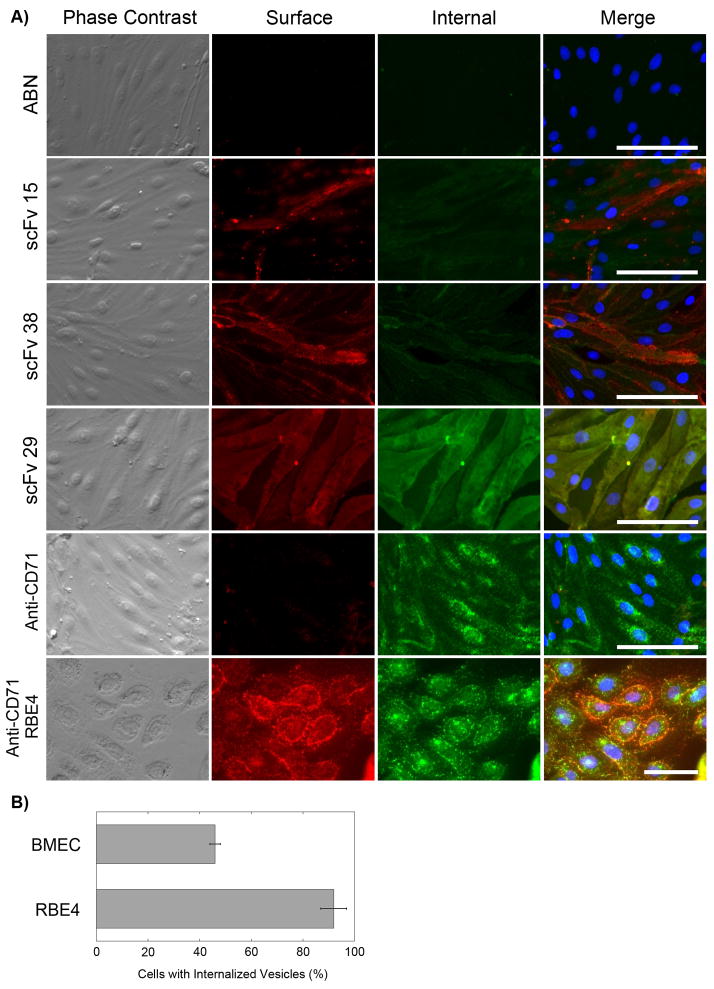
Since one of the objectives of the screen design was to bias towards internalizing antibodies, the endocytosis capability of the three antibodies was assessed. Briefly, purified scFv38, scFv29, and scFv15 were pre-dimerized with an anti-c-myc antibody since scFv multimerization has been shown to help promote internalization and the antibodies were selected using the multivalent Fd phage display system [19, 20]. The pre-dimerized scFvs were incubated with BMECs on ice for 30 minutes and then placed at 37°C for 30 minutes to allow for internalization. The antibody distribution was then visualized by a dual fluorophore detection to assay for both external and internal antibody localization (Figure 3A). ScFv38, scFv29, and scFv15 did not appear to internalize appreciably, although with limited overall in vitro labeling of scFv15, internalization could not be completely ruled out. In contrast, a positive control antibody, OX-26, that recognizes the rat transferrin receptor, which is known to internalize [18], did demonstrate internalization in this assay as evidenced by punctate vesicles in the perinuclear and cytoplasmic regions of the cell (Figures 3A and 3B).
Figure 3.
Assay for antibody internalization. A) BMECs or RBE4 (last row only) cells were incubated with 100 μg/mL (~3 μM) of scFv pre-dimerized with 9E10 (anti-c-myc) first on ice for 30 minutes and then at 37°C for 30 minutes. As an internalization positive control an anti-CD71 (anti-transferrin receptor IgG, OX-26) antibody was employed. The BMEC or RBE4 cell surface was subsequently labeled with Alexafluor594 (red) for 30 minutes. The cells were fixed with 4% paraformaldehyde, permeabilized with triton X100 and the interior of the cell labeled with Alexafluor488 (green). Subsequently the cells were labeled with the nuclear stain, DAPI (merged column only). Note the accumulated, punctate intracellular green fluorescence in the anti-CD71 samples indicative of internalized antibody, and the enhanced surface and internalized fluorescence visible in the RBE4 rat brain endothelial cell line. Scale bar is 50 μm and the images are representative of 4 independent experiments. B) Quantitation of anti-CD71 antibody internalization. The percentage of cells with multiple internalized vesicles is depicted as mean ± S.D., n=3 microscope fields (RBE4 cells 92±5% versus primary BMECs 46±2%, p<.001 computed using a student’s two-tailed t-test assuming unequal variances).
In vivo organ binding distribution of scFv38, scFv29, and scFv15
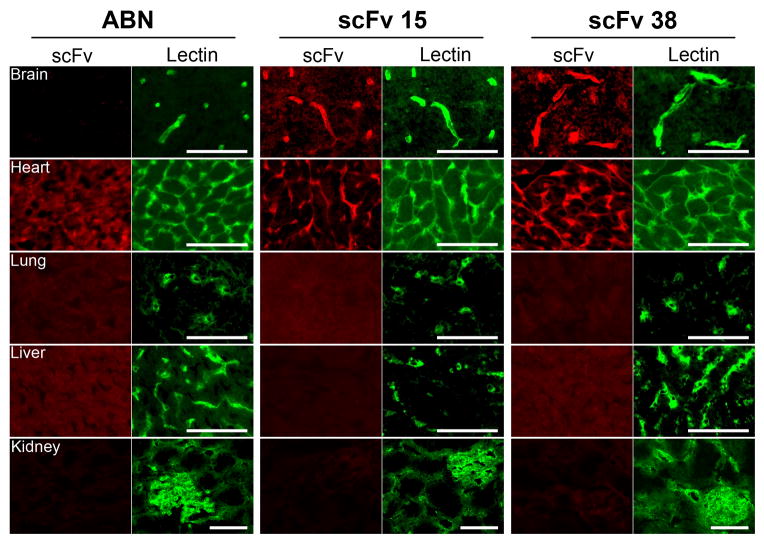
Since the screen was performed in vitro and some interesting cellular binding selectivity was exhibited for clones scFv29, and scFv15, the in vivo organ binding distribution was assessed using rat tissue cryosections. First, purified scFv38, scFv29, and scFv15 were used to immunolabel rat brain sections (Figure 4). Interestingly, scFv clones scFv38 and scFv15 clearly bind to brain capillaries having excellent co-localization with the vascular marker, IB4 lectin, but no brain capillary labeling was detected for scFv29 although this scFv produced the highest and most selective binding signal on cultured BMECs.
Figure 4.
Tissue distribution of antibody binding. Purified ABN scFv (negative control), scFv15, and scFv38 were incubated on rat brain, heart, liver, lung or kidney tissue sections at a concentration of 100 μg/mL (~3 μM) except for kidney labeling which was done at 50 μg/mL (~1.5 μM) to limit nonspecific background (labeling of brain vasculature was still detected using the 1.5 μM concentration). The sections were co-labeled with FITC-conjugated IB4 lectin as an endothelial cell marker, except for kidney in which an anti-rat CD31 antibody was used to label kidney sections serial to the sections labeled with scFv. The scale bar is 50 μm and the images are representative of 5 independent experiments for the brain panel and 3 independent experiments for peripheral tissue panels.
To assess the organ specificity of the scFv that bind to brain endothelial cells in vivo, scFv15 and scFv38 were also used to immunolabel rat heart, lung, liver, and kidney cryosections (Figure 4). As predicted by the binding patterns in cultured cells in vitro (figure 2b and 2c), scFv15 and scFv38 immunolabeled the heart vasculature. However, in contrast to the in vitro-based immunocytochemistry results, neither scFv15 or scFv38 appeared to bind the in vivo lung vasculature, nor did they immunolabel the in vivo kidney or liver vasculatures. Thus, the antigens targeted by these two antibodies are at the very least differentially expressed in the brain compared with these peripheral vascular beds. Taken together, scFv38 and scFv15 both target a BBB resident antigen in vivo and do so with vascular selectivity with respect to all peripheral organs tested other than heart.
DISCUSSION
This study demonstrates that scFvs having the capability to selectively bind the BBB in vivo can be identified using the hydrocortisone-induced primary BMEC model as a phage display-based screening substrate. While other in vitro BBB models based on primary BMECs or immortalized BMECs have been used as screening substrates [10, 19], it was expected that the hydrocortisone-induced model might provide a more robust screening platform given the advantages in BBB phenotype previously demonstrated [14, 21, 22]. After evaluating roughly 400 phage clones by applying phage ELISA, phage immunocytochemistry and DNA sequence filters, the overall diversity was reduced to three scFvs capable of binding BMECs as soluble proteins. The imposition of these various evaluation filters, along with pre-subtraction and internalization screening pressures, likely contributed to the low numbers of recovered scFvs. Moreover, as a result of antibody affinity, antigen abundance and clonal expression bias, it is relatively commonplace for such screens to result in a limited diversity of targeting antibodies [10, 18, 23]. While the resultant scFv diversity was lower than desired, it may be possible to further expand the diversity by performing pure binding screens rather than including internalization selection pressure. For example, employing a binding screen on the immortalized RBE4 brain endothelial cell line using a yeast display library yielded 34 different binding antibodies [19].
The screen was designed in an attempt to bias towards identification of endocytosing antibodies [10, 18, 24, 25], although the recovered scFvs did not appear to be capable of internalization, at least using the in vitro assay employed in Figure 3. For the most part, internalization screens have been performed using immortalized cell lines or cancer cell lines [18, 24] that endocytose avidly given their enhanced proliferative status. By comparison, primary rat BMECs were used as the screening platform in this study and these cells are fairly non-proliferative, as is the in vivo BBB [14]. Moreover, one of the prevailing phenotypes of the in vivo BBB is a substantially reduced amount of vesicle-based trafficking [26], a property that in our experience also appears to manifest itself in primary in vitro BBB models (Figure 3B). This phenomenon can be visualized in Figure 3, where both primary endothelial cells and the RBE4 immortalized endothelial cell line were assayed for anti-transferrin receptor antibody internalization. It can be seen that the amount of transferrin receptor labeling on the cell surface (red) and the number of cells possessing multiple internalized vesicles (green) is decreased in primary BMECs when compared to the highly proliferative immortalized RBE4 cell line (Figure 3). Thus, the beneficial signal-to-noise often afforded by an internalization selection pressure was likely muted in the BMEC screen, leading to the identification of antibodies that either do not internalize, or do so sparingly in primary cultured BMECs. Interestingly, in an attempt to further explore the internalization capacity of scFv15 and scFv38 in the RBE4 cell line because of its enhanced endocytosis phenotype, it was found that scFv15 and scFv38 did not cross-react with the RBE4 cells. Therefore, while the primary culture based BMEC model used in the screen was apparently non-ideal for internalization screening, the model offered benefits in that scFv15 and scFv38 which interact with the BBB in vivo would not have been identified using the RBE4 cell line.
Pre-subtraction has been used to predispose screens toward cell-type specificity [10, 24, 27], and this approach was effective in this study as well. ScFv29 originated from the pre-subtraction screen and appeared to display BMEC specificity on the in vitro cultured cell screening substrates. ScFv15 also originated from the pre-subtraction screen and while it still bound to HECs and LECs, scFv15 still exhibited elevated binding to BMECs. When moving toward the in vivo environment in terms of binding the vasculature in rat brain tissue sections, scFv29 proved to bind an antigen that is expressed at detectable levels only in the cultured cells. This in vitro artifact is a well-known challenge of performing screens using in vitro cell-based platforms because, while a primary BMEC expression profile is more in vivo-like than an immortalized cell line, it is still substantially different from the true in vivo situation [6, 28]. In particular, although the addition of hydrocortisone to the rat BMEC model does beneficially move many transcripts towards the in vivo BBB expression profile, some transcripts are also artificially upregulated as well [6]. This phenomenon, along with typical aberrant cellular regulation associated with in vitro culture can help explain the identification of scFvs like scFv29 that only bind in vitro. In contrast to scFv29, scFv15 and scFv38 bound brain capillaries in vivo. In addition, these two antibodies demonstrated binding to an antigen expressed in brain and heart vasculature that was not detected by scFv immunolabeling of the lung, liver, or kidney. Despite the heart cross-reactivity, there are very few if any potential brain targeting antibodies or peptides that have been demonstrated to have the BBB selectivity that is demonstrated by scFv15 and scFv38, as oftentimes the targeted receptors are ubiquitously expressed like the transferrin or insulin receptors [2, 3]. While it remains to be determined whether or not scFv15 or scFv38 will transport into the brain parenchyma, it should be noted that simply targeting the BBB and providing a local drug reservoir, or internalizing into the BBB and using the endothelial cells as reservoirs for trophic molecules can also be beneficial for therapeutic efficacy [29, 30]. Thus, given their tissue selectivity, scFv15 and scFv38 offer promise as BBB targeting reagents, pending further in vivo evaluation of their targeting and transport attributes.
Acknowledgments
The authors would like to acknowledge Dr. Shelly M. Cook and the Department of Pathology and Laboratory Medicine histology laboratory for assistance with immunohistochemistry experiments. This work was supported by the Michael J. Fox Foundation for Parkinson’s Research and the National Institutes of Health (1 R01 NS071513-01). A.R.J. was supported by a National Science Foundation Graduate Research Fellowship.
ABBREVIATIONS
- BBB
blood-brain barrier
- scFv
single-chain variable fragment
- BMEC
brain microvascular endothelial cell
- DMEM
dulbecco’s modified eagle medium
- PDS
platelet depleted serum
- HEC
heart microvascular endothelial cell
- LEC
Lung microvascular endothelial cell
- PBS
phosphate buffered saline
- PEG
polyethylene glycol
- ELISA
enzyme linked immunosorbent assay
- PBSCM
PBS supplemented with calcium and magnesium
- PBSCMG
PBSCM supplemented with 40% goat serum
- APC
allophycocyanin
- SDS
sodium dodecyl sulfate
- PAGE
polyacrylamide gel electrophoresis
- DAPI
4′,6-diamidino-2-phenylindole
- FITC
fluorescein isothiocyanate
Footnotes
The authors have no conflicts of interest to declare.[30]
References
- 1.Abbott NJ, Friedman A. Overview and introduction: The blood-brain barrier in health and disease. Epilepsia. 2012;53:1–6. doi: 10.1111/j.1528-1167.2012.03696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stutz C, Zhang X, Shusta E. Combinatorial Approaches for the Identificiation of Brain Drug Delivery Targets. Curr Pharm Des. 2013 doi: 10.2174/13816128113199990459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones AR, Shusta EV. Blood-brain barrier transport of therapeutics via receptor-mediation. Pharm Res. 2007;24:1759–1771. doi: 10.1007/s11095-007-9379-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pardridge WM, Kang YS, Buciak JL. Transport of human recombinant brain-derived neurotrophic factor (BDNF) through the rat blood-brain barrier in vivo using vector mediated pepted drug delivery. Pharm Res. 1994;11:738–746. doi: 10.1023/a:1018940732550. [DOI] [PubMed] [Google Scholar]
- 5.Pardridge WM, Kang YS, Buciak JL, Yang J. Human insulin receptor monoclonal antibody undergoes high affinity binding to human brain capillaries in vitro and rapid transcytosis through the blood-brain barrier in vivo in the primate. Pharm Res. 1995;12:807–816. doi: 10.1023/a:1016244500596. [DOI] [PubMed] [Google Scholar]
- 6.Calabria AR, Shusta EV. A genomic comparison of in vivo and in vitro brain microvascular endothelial cells. J of Cereb Blood Flow Metab. 2008;28:135–148. doi: 10.1038/sj.jcbfm.9600518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shusta EV, Boado RJ, Mathern GW, Pardridge WM. Vascular genomics of the human brain. J of Cereb Blood Flow Metab. 2002;22:245–252. doi: 10.1097/00004647-200203000-00001. [DOI] [PubMed] [Google Scholar]
- 8.Li JY, Boado RJ, Pardridge WM. Blood-brain barrier genomics. J Cereb Blood Flow Metab. 2001;21:61–68. doi: 10.1097/00004647-200101000-00008. [DOI] [PubMed] [Google Scholar]
- 9.Li JY, Boado RJ, Pardridge WM. Rat blood-brain barrier genomics. II. J Cereb Blood Flow Metab. 2002;22:1319–1326. doi: 10.1097/01.WCB.0000040944.89393.0f. [DOI] [PubMed] [Google Scholar]
- 10.Muruganandam A, Tanha J, Narang S, Stanimirovic D. Selection of phage-displayed llama single-domain antibodies that transmigrate across human blood-brain barrier endothelium. FASEB J. 2001;15:240–242. doi: 10.1096/fj.01-0343fje. [DOI] [PubMed] [Google Scholar]
- 11.Calabria AR, Shusta EV. Blood-brain barrier genomics and proteomics: elucidating phenotype, identifying disease targets and enabling brain drug delivery. Drug Discov Today. 2006;11:792–799. doi: 10.1016/j.drudis.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 12.Agarwal N, Lippmann ES, Shusta EV. Identification and expression profiling of blood-brain barrier membrane proteins. J Neurochem. 2010;112:625–635. doi: 10.1111/j.1471-4159.2009.06481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daneman R, Zhou L, Agalliu D, Cahoy JD, et al. The Mouse Blood-Brain Barrier Transcriptome: A New Resource for Understanding the Development and Function of Brain Endothelial Cells. PLoS One. 2010:5. doi: 10.1371/journal.pone.0013741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calabria AR, Weidenfeller C, Jones AR, de Vries HE, et al. Puromycin-purified rat brain microvascular endothelial cell cultures exhibit improved barrier properties in response to glucocorticoid induction. J Neurochem. 2006;97:922–933. doi: 10.1111/j.1471-4159.2006.03793.x. [DOI] [PubMed] [Google Scholar]
- 15.Zhou Y, Marks JD. Identification of target and function specific antibodies for effective drug delivery. In: Dimitrov A, editor. Therapeutic Antibodies: Methods and Protocols. Humana Press; Totowa: 2009. pp. 145–160. [DOI] [PubMed] [Google Scholar]
- 16.Sheets MD, Amersdorfer P, Finnern R, Sargent P, et al. Efficient construction of a large nonimmune phage antibody library: The production of high-affinity human single-chain antibodies to protein antigens. Proc Natl Acad Sci USA. 1998;95:6157–6162. doi: 10.1073/pnas.95.11.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Connell D, Becerril B, Roy-Burman A, Daws M, et al. Phage versus phagemid libraries for generation of human monoclonal antibodies. J Mol Bio. 2002;321:49–56. doi: 10.1016/s0022-2836(02)00561-2. [DOI] [PubMed] [Google Scholar]
- 18.Poul MA, Becerril B, Nielsen UB, Morisson P, et al. Selection of tumor-specific internalizing human antibodies from phage libraries. J Mol Bio. 2000;301:1149–1161. doi: 10.1006/jmbi.2000.4026. [DOI] [PubMed] [Google Scholar]
- 19.Wang XX, Cho YK, Shusta EV. Mining a yeast library for brain endothelial cell-binding antibodies. Nat Methods. 2007;4:143–145. doi: 10.1038/nmeth993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adams GP, Tai MS, McCartney JE, Marks JD, et al. Avidity-mediated enhancement of in vivo tumor targeting by single-chain Fv dimers. Clin Can Res. 2006;12:1599–1605. doi: 10.1158/1078-0432.CCR-05-2217. [DOI] [PubMed] [Google Scholar]
- 21.Perriere N, Yousif S, Cazaubon S, Chaverot N, et al. A functional in vitro model of rat blood-brain barrier for molecular analysis of efflux transporters. Brain Res. 2007;1150:1–13. doi: 10.1016/j.brainres.2007.02.091. [DOI] [PubMed] [Google Scholar]
- 22.Perriere N, Demeuse PH, Garcia E, Regina A, et al. Puromycin-based purification of rat brain capillary endothelial cell cultures. Effect on the expression of blood-brain barrier-specific properties. J Neurochem. 2005;93:279–289. doi: 10.1111/j.1471-4159.2004.03020.x. [DOI] [PubMed] [Google Scholar]
- 23.Li JW, Feng L, Fan L, Zha Y, et al. Targeting the brain with PEG-PLGA nanoparticles modified with phage-displayed peptides. Biomaterials. 2011;32:4943–4950. doi: 10.1016/j.biomaterials.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heitner T, Moor A, Garrison JL, Marks C, et al. Selection of cell binding and internalizing epidermal growth factor receptor antibodies from a phage display library. J Immunol Methods. 2001;248:17–30. doi: 10.1016/s0022-1759(00)00340-9. [DOI] [PubMed] [Google Scholar]
- 25.Florea BI, Molenaar TJM, Bot I, Michon IN, et al. Identification of an internalising peptide in differentiated Calu-3 cells by phage display technology; application to gene delivery to the airways. J Drug Target. 2003;11:383–390. doi: 10.1080/10611860310001642389. [DOI] [PubMed] [Google Scholar]
- 26.Stewart PA. Endothelial vesicles in the blood-brain barrier: Are they related to permeability? Cell Mol Neurobiol. 2000;20:149–163. doi: 10.1023/A:1007026504843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huie MA, Cheung MC, Muench MO, Becerril B, et al. Antibodies to human fetal erythroid cells from a nonimmune phage antibody library. Proc Natl Acad Sci USA. 2001;98:2682–2687. doi: 10.1073/pnas.051631798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lyck R, Ruderisch N, Moll AG, Steiner O, et al. Culture-induced changes in blood-brain barrier transcriptome: implications for amino-acid transporters in vivo. J Cereb Blood Flow Metab. 2009;29:1491–1502. doi: 10.1038/jcbfm.2009.72. [DOI] [PubMed] [Google Scholar]
- 29.Chen YH, Chang M, Davidson BL. Molecular signatures of disease brain endothelia provide new sites for CNS-directed enzyme therapy. Nat Med. 2009;15:1215–U145. doi: 10.1038/nm.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bickel U, Yoshikawa T, Pardridge WM. Delivery of peptides and proteins through the blood-brain barrier. Adv Drug Deliv Rev. 2001;46:247–279. doi: 10.1016/s0169-409x(00)00139-3. [DOI] [PubMed] [Google Scholar]