Abstract
RNA-protein interactions have critical roles in gene regulation. However, high-throughput methods to quantitatively analyze these interactions are lacking. We adapted an Illumina GAIIx sequencer to make several million such measurements with a High-Throughput Sequencing – RNA Affinity Profiling (HiTS-RAP) assay. Millions of cDNAs are sequenced, bound by the E. coli replication terminator protein Tus, and transcribed in situ, whereupon Tus halts transcription leaving RNA stably attached to its template DNA. The binding of fluorescently-labeled protein is then quantified in the sequencer. We used HiTS-RAP to measure the affinity of mutagenized libraries of GFP-binding and NELF-E binding aptamers to their respective targets and thereby identified regions in both aptamers that are critical for their RNA-protein interaction. We show that mutations additively affect binding affinity of the NELF-E binding aptamer, whose interaction depends mainly on a single-stranded RNA motif, but not that of the GFP aptamer, whose interaction depends primarily on secondary structure.
Introduction
RNA-protein interactions are ubiquitous in biology and critical at many regulatory steps of gene expression and various stages of development1.. Therefore, quantitative analysis of interactions between RNA and RNA Binding Proteins is necessary for a more profound understanding of basic biological mechanisms2. Several methods exist for determining RNA-protein affinities, though the quantitative methods are low-throughput and laborious3–7. Some high-throughput techniques can be used to determine binding motifs, but are either not quantitative or are difficult to scale8,9. High-throughput assays, such as RIP-Seq10, CLIP11, and PAR-CLIP12 can identify RNA molecules that interact with a protein, but have significant limitations: they depend on antibodies, cannot quantitatively measure the affinities of the interaction, and generally depend on crosslinking, which can introduce biases.
The Illumina platform allows simultaneous sequencing of hundreds of millions of DNA clusters, which are each derived from amplification of a single molecule with primers that are covalently linked to the glass flowcell13. These sequencers can also be used to automatically image and analyze the binding of fluorescently labeled proteins to these DNA clusters as demonstrated with the HiTS-FLIP protocol14. A corresponding assay for RNA binding to protein is conceivable, but has not been realized due to the difficulty of converting the DNA of clusters into RNA that is retained at the cluster15.
Here, we developed a method called High Throughput Sequencing- RNA Affinity Profiling (HiTS-RAP) that transcribes DNA at each cluster on an Illumina flowcell, and we used it to measure the affinity of mutants of two RNA aptamers at an unprecedented scale. We measured dissociation constants for 1,875 mutants of the GFP aptamer (GFPapt)16, and 9,832 mutants of NELFapt17, an RNA aptamer that binds Drosophila NELF-E (Negative Elongation Factor E), an RNA binding subunit of a protein complex involved in maintaining promoter proximally paused RNA Polymerase II18–21. We analyzed these data to determine sequence and structural features of these two aptamers that are most important for their protein interactions.
Results
Tus Stably Halts Transcription
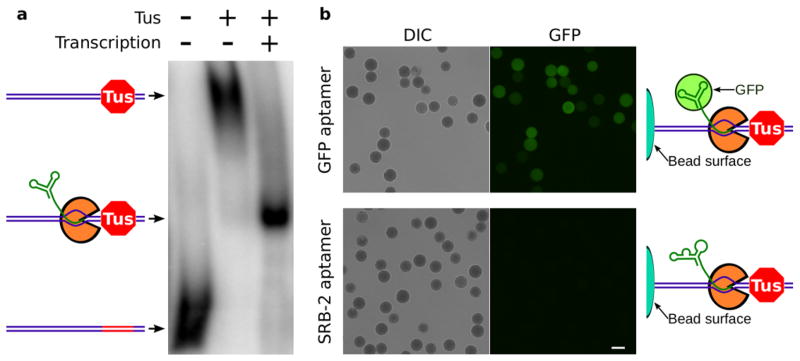
A critical feature of HiTS-RAP is to transcribe DNA on the Illumina flowcell with the RNA transcript stably retained at each DNA cluster. We used the E. coli replication terminator protein Tus as a roadblock to transcription to achieve this. Tus prevents DNA replication through sites of replication termination in an orientation-specific manner by binding to a 32 bp sequence element (ter) with high affinity, specificity, and stability22–25. In addition, Tus stops RNA polymerases from transcribing through Tus-bound ter sites in the non-permissive orientation, resulting in either terminated or halted transcription22,26. T7 RNAP produces only truncated transcripts, many of which remain anchored to the DNA, only when Tus is bound to DNA in the non-permissive orientation (Supplementary Fig. 1a). Electrophoretic Mobility Shift Assay (EMSA) with radiolabeled DNA shows that after transcription halting, nearly every DNA is engaged in a complex of intermediate mobility containing an RNA transcript (Fig. 1a, Supplementary Fig. 1b). Thus, T7 RNAP transcribing into a non-permissive Tus-ter complex halts upstream of the ter site with an RNA transcript bound to the DNA through the polymerase.
Figure 1. T7 RNA polymerase halting with Tus gives stable complexes containing DNA and functional RNA.
(a.) EMSA was carried out with radiolabeled DNA that encodes the GFP aptamer and has a binding site for Tus. Naked DNA runs with high mobility. Binding Tus to the ter DNA element (red segment) retards DNA mobility. After transcription, nearly every DNA participates in a complex of intermediate mobility, containing Tus, T7 RNA polymerase (orange), and RNA (green). This band is very sharp, indicating that each DNA-Tus-T7RNAP-RNA complex is of homogeneous composition. b. 454 Life Sciences polystyrene beads covered in covalently linked DNA templates for transcription. After transcription halting, beads were incubated with EGFP and then washed. EGFP bound to beads presenting halted GFP aptamer RNA, but not to beads presenting SRB-2 aptamer RNA. Scale bar is 20 μm.
We used an RNA aptamer that has high affinity and specificity for GFP and its derivatives (i.e. EGFP)16 to develop HiTS-RAP. As an initial test, we attached aptamer template DNAs to beads and generated halted transcription complexes. EGFP binds beads with halted GFPapt transcription complexes but not negative control SRB-2 aptamer27 (Fig. 1b). A similar experiment with single-round transcription showed that this interaction is due to the full length GFPapt RNA (Supplementary Fig. 2).
Transcription Halting on Illumina GAIIx Sequencer
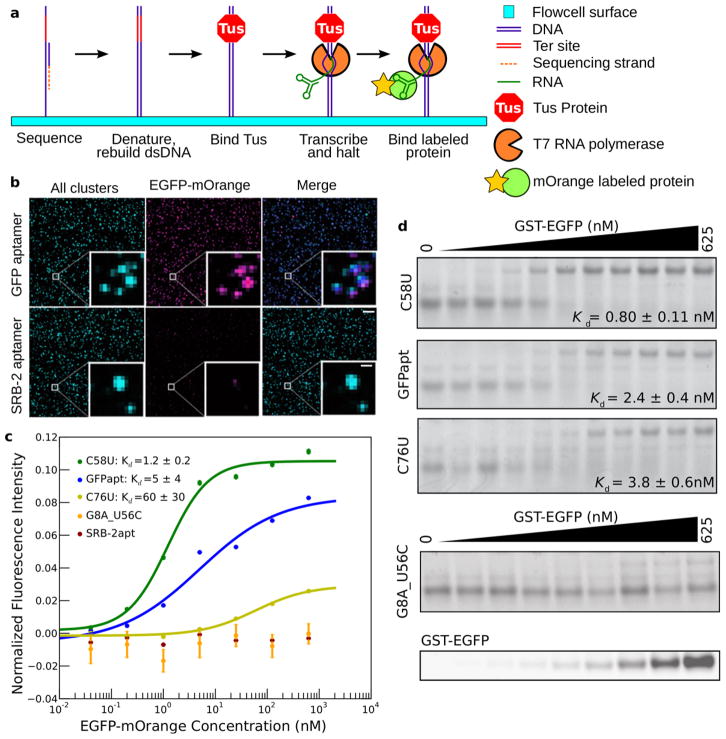
To couple Tus-dependent halting of T7 RNA polymerase with sequencing on an Illumina GAIIx, we constructed DNA libraries containing a template to be transcribed flanked by a T7 promoter upstream and the ter sequence downstream (Supplementary Fig. 3). All steps of HiTS-RAP are carried out automatically (Fig 2a). After sequencing, a new second DNA strand is generated at all clusters in the flowcell. Transcription halting is then carried out, presenting the RNAs encoded by the millions of DNA clusters. The binding properties of these RNAs of known sequence is then probed by allowing fluorescently-labeled protein to interact with the halted RNAs13,28. Illumina’s software is used to image protein binding at equilibrium and measure fluorescence intensity of bound mOrange fusion protein at each cluster. The TIRF microscopy of the sequencer enables equilibrium measurements, as excess protein in solution does not interfere with imaging of protein bound at clusters13,14.
Figure 2. RNA-protein interactions can be assayed by HiTS-RAP on an Illumina GAIIx instrument.
(a) HiTS-RAP schematic. Sequencing is done following the standard Illumina workflow. The strand synthesized during sequencing is then stripped away, a primer is annealed and the second strand is regenerated with Klenow enzyme. Tus is then bound to the ter site, and DNAs on the flowcell are transcribed. T7 RNAP initiates at its promoter, transcribes through the sequence of interest, and halts just upstream of the Tus bound ter site. The RNA transcript is stably linked to its DNA template through the polymerase. Fluorescently labeled protein is then bound to the RNA and imaged. (b) Images from a HiTS-RAP run with GFP and SRB-2 aptamers. ‘All clusters’ (left panels) are labeled during sequencing and shown as a maximum intensity projection of the four channels. After transcription halting and EGFP-mOrange binding, the flowcell is imaged at 625 nM EGFP-mOrange. GFPapt clusters are labeled by mOrange while SRB-2 aptamer clusters are not. Scale bars: 6.75 μM, 1.125 μM in inset (c) Binding curves for the GFP aptamer (n =2,665,064), and mutants C58U (n =3,833), C76U (n =4,758), and G8U_U56C (n =29), and the SRB-2 aptamer (n =1,588,404). G8U_U56C and SRB-2 aptamer are scored as not binding. Data are from one lane of the sequencer (SRB-2 aptamer is from a separate lane). Intensities are the average of all clusters of each sequence in the lane, normalized by dividing by their average sequencing intensity and subtracting their average intensity at no EGFP-mOrange. Error bars represent standard error. Error of fitted Kds are the square root of variances returned by the fitting algorithm. (d) EMSA of RNAs in part c. Kds are determined from a single fit to two replicate EMSA experiments for each sequence, ± standard deviation fitted by IGOR. The G8U_U56C gel was also scanned to visualize EGFP.
The GFP aptamer, a population of point mutants, and control RNA were assayed by HiTS-RAP for their affinity to EGFP-mOrange fusion protein. EGFP-mOrange was used because EGFP is not detectable with the optics of the sequencer13,28. The vast majority of GFPapt DNA clusters produce halted RNA capable of binding to EGFP-mOrange, while those in a lane where all clusters encode the SRB-2 aptamer, a negative control, do not (Fig. 2b). We repeatedly measured the intensity of GFPapt clusters at a high protein concentration in the sequencer to estimate that the assay is sensitive to measurements above background for the first 48 cycles, or 72 hours, given an approximate cycle time of 1.5 hours (Online Methods, Supplementary Fig. 4a). Thus, the halted transcription complexes are sufficiently stable to carry out the several sequential measurements necessary to determine dissociation constants (Kds).
Measuring Kds for the GFP Aptamer and Its Mutants
HiTS-RAP was carried out using a flowcell with three lanes containing GFPapt template that was subjected to many rounds of PCR and thereby accumulated a population of mutants in addition to the canonical GFP aptamer. Seven protein concentrations increasing in five-fold increments from 0.04 to 625 nM were used to measure the Kds of interactions between RNAs and EGFP-mOrange. We identified 1,875 mutant GFP aptamer sequences that had at least 10 copies in at least one of the three lanes with quality scores greater than 25 at the mutated residue (Supplementary Table 1). We measured a Kd of 4.27 ×/1.11 nM (geometric mean ×/(times/divide) geometric standard deviation29) for the EGFP-GFP aptamer interaction, in agreement with its published affinity of 5–15 nM Kd 16, while the SRB-2 aptamer negative control shows no appreciable binding in HiTS-RAP (Fig. 2c).
Most GFPapt mutants do not differ substantially in sequence from the canonical aptamer, and therefore bind EGFP with similar affinity. However, some showed altered affinity. To verify HiTS-RAP measured Kds, we picked eight such mutants and measured their binding affinity by EMSA4 (Fig. 2d, Supplementary Fig. 5a). HiTS-RAP and EMSA measured affinities are correlated (Supplementary Fig. 5b, Supplementary Table 2). Some of the GFPapt mutants that we identified were represented by only a small fraction of the total clusters on the flowcell, demonstrating that HiTS-RAP can measure Kds of even low copy number sequences in a library (Supplementary Table 1).
Comprehensive mutagenesis of the GFP Aptamer
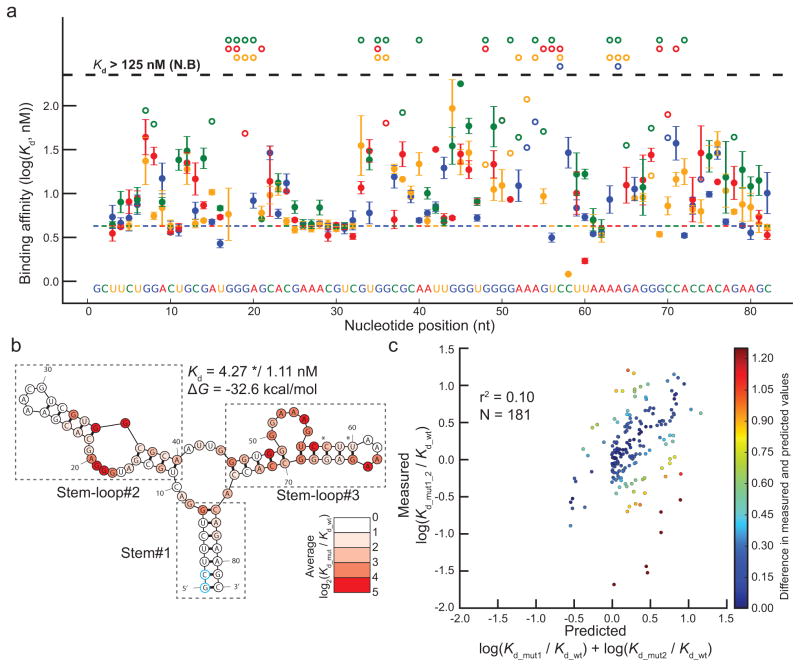
We used our library of mutants to carry out an in-depth analysis of the 82 nt of the GFP aptamer. Of the 246 possible single base substitutions, 236 are present in our data set (Supplementary Table 3), and have either a measured affinity, or an assigned Kd of >125 nM if no binding was observed (Supplementary Fig. 4b–d). Most GFPapt mutations have a negative effect on binding affinity (log2(Kd_mut/Kd_wt) > 0), consistent with its highly optimized nature (Fig 3a). However, C58U (Kd = 1.21 ×/1.03 nM) and U60A (Kd = 1.71 ×/1.13 nM) are two notable exceptions. Both do not alter the predicted secondary structure of GFPapt, but reduce the free energy of folding (Supplementary Fig. 6a), so they likely make the overall structure more flexible. The Kds of both of these mutants were verified by EMSA (Supplementary Fig. 5). In contrast, two single point mutants which have Kds >125 nM, G68C and G46C, cause gross alterations in the predicted structure of GFPapt, consistent with their substantial negative effects on binding affinity (Supplementary Fig. 6b).
Figure 3. Analysis of GFPapt by HiTS-RAP.
(a) GFP binding affinities of all 236 measured single-point mutants of GFPapt. Binding affinities in nM are plotted in logarithmic scale. Mutations at each position are color-coded. Wild-type GFPapt binding affinity is indicated by the colored dashed line and the sequence is shown at the bottom of the graph. Error bars represent the standard deviations in log(Kd). 175 mutants qualify as binding in all three lanes. Single point mutants that qualify as not binding and are thus assigned an affinity of 125 nM in at least one lane are plotted with open circles, with no error bars. Those that do not bind in all lanes are at the top of the plot. (b) Predicted secondary structure of GFPapt. GFPapt is predicted to fold into a three stem-loop structure connected by a central 3-way junction. Each position is colored by the average absolute effect |log2(Kd_mut/Kd_wt)| of all its measured mutants. Mutations that qualify as not binding were assigned an affinity of 125 nM for this plot. Positions where the average effects are greater than 4 (>16-fold effect in affinity) are colored red. Most mutations have a negative effect or less than a 2-fold positive effect on binding affinity, except C58U and U60A (indicated by asterisk). (c) Correlation between measured and predicted effects of GFPapt double mutants. Measured log10(Kd_mut1_2/Kd_wt) is plotted against the value predicted based on single-point mutants (log10 (Kd_mut1/Kd_wt) + log10 (Kd_mut2/Kd_wt)). Points are colored based on the difference between the measured and the predicted effects. There is a positive but small correlation (r2 = 0.10) between the measured and predicted effects, and they differ as much as 1.98, or ~100-fold.
To estimate the contribution of each nucleotide in GFPapt to its interaction with EGFP, we calculated the average effect of all measured mutations at each position (Fig. 3b). In agreement with the original study16, the majority of the high impact positions are located in either stem-loop #2 or #3.
To gain further insight into the relationship between the binding affinity and sequence of the GFPapt, the relationship between the affinity of double mutants and their two corresponding single point mutants was analyzed. We identified 181 double mutants where the double mutant and two corresponding single mutants bound EGFP above the threshold for signal increase and had high confidence EGFP-binding affinities (Supplementary Table 4). If two single point mutations affect binding independently of each other, the binding of a double mutant would be predicted by the additive effect of the two individual mutations. However, if they affect binding in either a cooperative or antagonistic manner, the measured effect of the double mutant would differ from the additive effect of the two individual mutations. We used a metric analogous to ΔΔG (the difference in Gibbs free energy change) to make this comparison: the effect of a mutation is represented by the log ratio of the mutant and canonical aptamers’ Kds (log(Kd_mut/Kd_wt)). There is only a weak correlation (r2 = 0.10) between the measured and the predicted effect of the GFPapt double mutants (Fig 3c).
Double mutants with substantially higher or lower affinity than predicted can highlight features important for the interaction with EGFP (Supplementary Table 4). For example, A23G_U34C reconstitutes an AU base pair as a GC base pair in the predicted secondary structure and binds with higher affinity than predicted, while the same U34C mutation in combination with C58U fails to rectify the structural perturbation caused by U34C and has lower affinity than predicted (Supplementary Fig. 7). Overall, mutational analysis and structural predictions indicate that the interaction between the GFP aptamer and its target protein EGFP is complex in nature, depending upon an intricate structure dictated by its sequence.
High-Throughput Affinity Profiling of Drosophila NELF-E
We also used HiTS-RAP to examine an interaction between NELF-E, an RNA binding protein with a highly conserved RNA recognition motif (RRM)17, and NELFapt, an RNA aptamer which binds NELF-E17. Within NELFapt, NELF-E recognizes a 7 nucleotide motif (CUGAGGA), called the NELF-E Binding Element (NBE), which is located within a putative k-turn motif that forms a sharp bend between two stems to present the NBE on a single stranded loop region17. To characterize this interaction, we mutated NELFapt through error prone PCR (epPCR), and performed HiTS-RAP to probe NELF-E binding using a single lane of an Illumina GAIIx flowcell. We measured high-confidence binding affinities for 9,832 mutants (Supplementary Table 5). The Kd for full length NELFapt was measured to be 5.2 nM, in good agreement with EMSA (Supplementary Fig. 8)
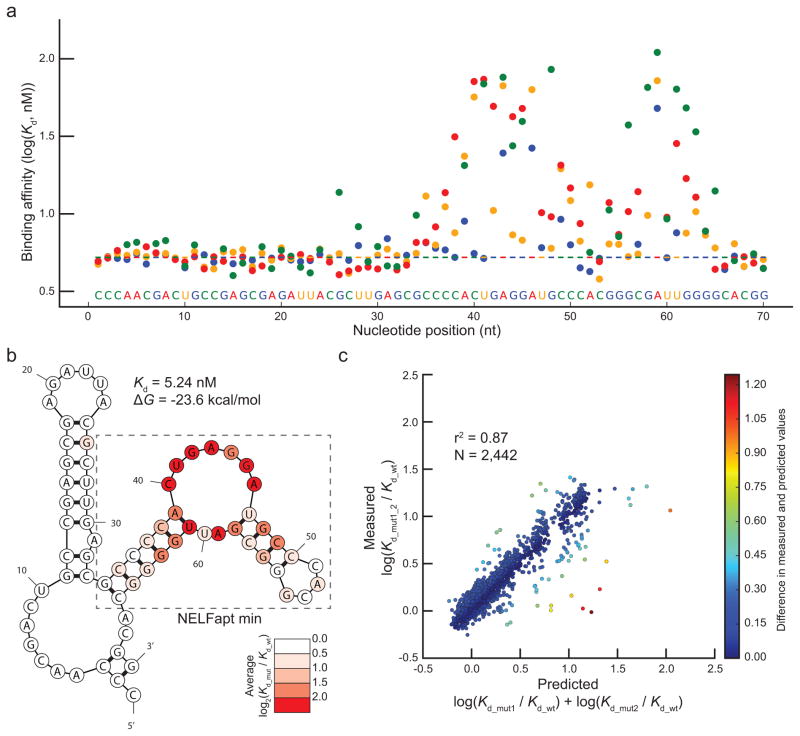
All of the 210 possible single base substitution mutants within the 70 nt NELFapt were identified (Supplementary Table 6), 206 with high-confidence fits for Kds (Fig. 4a). The majority of single point mutations did not have a notable effect on binding affinity, and none had substantially higher affinity than the canonical aptamer. Mutations within the NBE were among the most disruptive to the interaction between the aptamer and NELF-E (Fig 4b), with measured affinities as low as 76 nM for A43C. Interestingly, bases in the loop region opposite of the NBE in the predicted secondary structure (GAUU, nucleotides 58–61) were also found to be important for binding (Fig. 4b), with the most critical residues outside of the NBE located at positions G58 and A59. The deleterious effects of these two mutations were confirmed by EMSA (Supplementary Fig. 8). These residues likely interact with G45 and A46 through the non-Watson-Crick base pairing characteristic of k-turn structures30. The observed effects of the single point mutations strongly support a k-turn structure of this aptamer with the presentation of the NBE as a single stranded loop.
Figure 4. Analysis of NELFapt by HiTS-RAP.
(a) NELF-E binding affinities of 206 single-point mutants of NELFapt. Binding affinities in nM are plotted in logarithmic scale. Mutations and the canonical aptamer sequence shown at the bottom of the graph are colored as in Fig. 3a. (b) Predicted secondary structure of NELFapt. NELFapt is predicted to have 2 stem-loops connected via a loop-stem-loop structure in between. Predicted secondary structure is drawn and colored as in Fig 3b. Most mutations show a less than 2-fold effect (log2(Kd_mut/Kd_wt) ≤ 1) on binding affinity. (c) Correlation between measured and predicted effects of NELFapt double mutants (n = 2,442). Measured fold effect of double mutants is plotted against the fold effect predicted by single mutations as in Figure 3c. In this case, there is a strong positive correlation (r2 = 0.87) between measured and predicted fold effect.
To assess the interplay of different nucleotides, we compared affinities of mutant NELF-E aptamers with single and double point mutations. We identified 2,442 double mutants with high-confidence Kds (Supplementary Table 7). In contrast to GFPapt, the affinities of NELFapt double mutants were predicted well by the affinities of the individual single mutants (r2 = 0.87) (Fig. 4c). Most double mutants have small effects on affinity, and are well predicted by an additive model. Many that do not follow this trend are notable. Some are compensatory: for example, both A39G_U61C and A39U_U61A bind NELF-E better than predicted by their corresponding single mutants, presumably because the double mutants reconstitute a predicted AU base-pair as GC and UA base-pairs, respectively, on a critical stem near the NBE (Supplementary Fig. 9a). Others, such as A39G_G63A bind with lower affinity than predicted by individual mutations because they may result in sequestration of the NBE within a double stranded region (Supplementary Fig. 9b), which was shown to be deleterious for NELF-E interaction17. Altogether, this analysis shows that most mutations within NELFapt affect its interaction with NELF-E in an additive way with only a few notable and informative exceptions.
Discussion
Here, we developed a new method that combines binding affinity measurements for RNA and sequencing in one high-throughput assay, which we named HiTS-RAP. This method satisfies a need in the field of RNA protein interactions for a quantitative, high throughput assay, which was lacking despite considerable interest. In fact, while this work was in review, Buenrostro et al published a similar technique using halting of E. coli RNA polymerase by a biotin/streptavidin roadblock after single-round transcription to perform an assay very similar to ours31. After a normal sequencing run, all additional manipulations for HiTS-RAP are carried out automatically by incorporating these steps into the .xml recipe used for the run, so that it adds very little to cost and hands-on time, but generates a large and useful data set. Because the sequencing chemistry used is identical across all of Illumina’s instruments, this would be possible with HiSeq and MiSeq instruments as well, for increased data output or faster small-scale readout, respectively. Using HiTS-RAP, we have accurately measured the affinities of two aptamers that bind GFP and NELF-E. In addition to the canonical aptamers, affinities of thousands of mutants of each aptamer were accurately measured. Careful filtering minimized the contribution of sequencing errors to these real mutants (Online Methods): the close correspondence of Kds measured by HiTS-RAP and by EMSA of synthesized mutants suggests that the vast majority of sequence variations are due to mutation. The effects of these mutations on affinity provided insight into the structure of the aptamers and identified regions that are critical for their RNA-protein interactions.
The interaction between GFPapt and EGFP is based upon the complex three dimensional structure of the RNA aptamer. Analysis of single base substitutions showed that most positions critical for EGFP binding reside within stem-loops #2 and #3, while the rest of the aptamer is still required for proper folding (Fig. 3b). Of particular interest, two GFPapt mutants with higher affinities than the canonical aptamer were identified, highlighting the potential impact of HiTS-RAP for in-depth analysis and optimization of a pre-selected aptamer in a cost-effective high throughput manner.
In contrast, the interaction between NELFapt and NELF-E is primarily due to a short consensus motif presented on a single-stranded region. This analysis can also be used to identify a minimal aptamer: in fact, HiTS-RAP shows that the sequence outside of the 35 nt minimal aptamer defined in the original publication17 does not influence binding appreciably (Fig. 4b). The strong effect of mutations in bases opposite the NBE sequence motif in the predicted secondary structure shows that the presentation of the NBE within NELFapt, in the single stranded region of a k-turn, is just as important as its presence for recognition by this RNA binding protein. Thus, while the sequence specified recognition of RNA by the RRM of NELF-E occurs within a single stranded loop, the structural context of that loop is also important for the interaction.
The behavior of double mutants shows that the interaction between EGFP and GFPapt is fundamentally different from that of NELF-E with NELFapt. Multiple mutations within the intricate structure of GFPapt confound each other, so that the affinity of a double mutant is poorly predicted by its two individual mutations. In contrast, most double mutants of NELFapt are well predicted as the additive effect of their individual mutations, as we would expect for an interaction where only a small fraction of the total RNA is indispensable for binding. Moreover, many of the double mutants that deviate from this trend occur within the NBE, suggesting that mutations within this short consensus motif are less likely to affect the interaction additively. The large number of quantitative binding constants determined by HiTS-RAP enable such insights. With a larger library of mutants, it may be possible to derive a comprehensive model of an RNA-protein interaction.
HiTS-RAP has as its foundation a technique of halting transcription by sequence-specific binding of Tus to ter sites so that the RNA transcript emanating from the polymerase (i.e. T7 RNA polymerase) remains stably halted on DNA template and competent for interaction with fluorescently-labeled molecules for many hours. This could effectively be used to convert assays deemed to be restricted to DNA, such as microarrays, to measure properties of RNA as well. Additionally, although we have used HiTS-RAP to measure affinities of RNAs to a protein, its utility extends to any entity that can be fluorescently labeled (e.g., small molecules and peptides). Lastly, DNA libraries tailored for many applications, including random sequence libraries, aptamer libraries generated by SELEX, random genomic fragments, or targeted genomic libraries (such as nascent RNA, mRNA, ncRNA, enhancer regions, or pre-mRNA) can be easily adapted or constructed de novo for HiTS-RAP. Thus, HiTS-RAP could facilitate genome-wide, direct, quantitative measurement of RNA affinity for regulatory proteins.
Online Methods
Purification of proteins
The gene encoding Tus protein was PCR amplified from a vector provided by B. Mohanty (Medical University of South Carolina), and then inserted between BamHI and XhoI restriction sites in the expression vector pGST-parallel 32. GST-Tus was then overexpressed in BL21 (DE3) RIPL bacteria for 4 hours after induction with 1mM IPTG at 37°C. The protein was purified using glutathione coupled agarose resin (Pierce), dialyzed into 40 mM Tris-HCl pH 7.5, 40 mM NaCl, 2 mM EDTA, 10 mM β-mercaptoethanol22, mixed with an equal volume of 80% glycerol, and stored at −80°C.
An EGFP-mOrange construct with a 4x GGGS flexible linker was produced by overlap extension PCR, and inserted between the BamHI and XhoI restriction sites of the expression vector pHis-parallel32. 6xHis-EGFP-mOrange was overexpressed in as for GST-Tus. The protein was purified using Ni-NTA coupled agarose resin (Pierce), dialyzed into 10 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 5 mM β-mercaptoethanol, allowed to mature at 4°C for one month, mixed with an equal volume of 80% glycerol, and stored at −80°C.
NELF-E mOrange was made following a protocol similar to EGFP-mOrange. mOrange was first PCR amplified with BglII and BamHI sites at either end, digested with those enzymes, and inserted into the BamHI site of pHis-parallel, conserving its entire multiple cloning site. NELF-E was then subcloned from pHis-parallel into the mOrange containing pHis-parallel using EcoRI and SpeI restriction sites. Expression and purification were carried out exactly as for EGFP-mOrange.
Library preparation
Supplementary Figure 3 shows a schematic of a HiTS-RAP template. For GFPapt, mutations were introduced by PCR amplification of the template over a total of ~100 cycles. Realizing that PCR induced mutations were limited and the throughput of the HiTS-RAP assay could accommodate far greater number of mutants, NELFapt was subjected to error-prone PCR (epPCR) as described elsewhere33 in order to generate more mutants. DNA templates were prepared for sequencing and transcription halting by sequential addition of primers by PCR. First, a T7 promoter was added to the 5′ ends of the GFP, Sulforhodamine B (SRB), and NELF-E binding aptamers, and the Illumina sequencing primer site to the 3′ ends. Then, adaptors complementary to oligos on the Illumina flowcell were added to either end along with a ter site immediately 3′ of the Illumina sequencing primer site. The ter site was positioned 3′ of the Illumina sequencing primer site to ensure that the target RNA is fully emerged from the polymerase upon halting and so that sequencing begins with the target RNA. A third PCR step was used to add adaptors for the 454 Life Sciences sequencing platform for templates used to make polystyrene beads with covalently linked halting templates. Sequences of primers used in this work are listed in Supplementary Table 8.
Final halting templates for HiTS-RAP (T7 RNAP promoter in italics, aptamer sequence underlined, ter sequence in bold):
GFPapt halting template used for HiTS-RAP:
5′- CAAGCAGAAGACGGCATACGAGATCGGTGATAATACGACTCACTATAGGGAATGGATCCACATC TACGAATTCAGCTTCTGGACTGCGATGGGAGCACGAAACGTCGTGGCGCAATTGGGTGGGGAAA GTCCTTAAAAGAGGGCCACCACAGAAGCTAGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGTAA ATTAGTATGTTGTAACTAAAGTCACGTCATGAGATCTCGGTGGTCGCCGTATCATT-3′ SRB-2 aptamer halting template used for HiTS-RAP: 5′- CAAGCAGAAGACGGCATACGAGATCGGTGATAATACGACTCACTATAGGGAATGGATCCACATC TACGAATTCGGAACCTCGCTTCGGCGATGATGGAGAGGCGCAAGGTTAACCGCCTCAGGTTCCA GATCGGAAGAGCGTCGTGTAGGGAAAGAGTGTAAATTAGTATGTTGTAACTAAAGTCACGTCAT GAGATCTCGGTGGTCGCCGTATCATT-3′ NELFapt template used for epPCR: 5′- GATAATACGACTCACTATAGGGAATGGATCCACATCTACGAATTCCCAACGACTGCCGAGCGAG ATTACGCTTGAGCGCCCCACTGAGGATGCCCACGGGCGATTGGGGCACGGCTTCACTGCAGACTTGACGAAGCTT-3′ NELFapt halting template used for HiTS-RAP: 5′- CAAGCAGAAGACGGCATACGAGATCGGTGATAATACGACTCACTATAGGGAATGGATCCACATC TACGAATTCCCAACGACTGCCGAGCGAGATTACGCTTGAGCGCCCCACTGAGGATGCCCACGGG CGATTGGGGCACGGCTTCACTGCAGACTTGACGAAGCTTATGGCTAGATCGGAAGAGCGTCGTG TAGGGAAAGAGTGTAAATTAGTATGTTGTAACTAAAGTCACGTCATGAGATCTCGGTGGTCGCC GTATCATT -3′
EMSA of halted transcription complex
An electrophoretic mobility shift assay (EMSA) was performed to resolve DNAs engaged in different complexes with Tus, T7 RNA polymerase, and RNA. The DNA template contains the GFP aptamer template flanked by a T7 RNA polymerase promoter at the 5′ end and a ter element at the 3′ end. This was then end labeled with 32P using T4 Polynucleotide Kinase and γ 32P- ATP, and purified using a P30 size exclusion column (BioRad). Radiolabeled DNA was mixed with either 1x T7 transcription buffer or GST-Tus at ~100 times the concentration of DNA in 1x T7 transcription buffer (30 mM HEPES pH 7.8, 80mM Potassium Glutamate, 15mM MgAc, 0.25 mM EDTA, 5 mM DTT, 0.05% Tween-20, 2 mM Spermidine), and incubated at 37°C for 30 minutes. Then, an equal volume of 1x T7 transcription buffer was added to DNA alone and DNA + Tus protein samples, and an equal volume of 2x transcription reaction was added to another DNA + Tus sample to make the Tus + DNA + transcription sample, and incubated at 37°C for another 30 minutes. The final transcription reaction consists of 1x T7 transcription buffer, 0.5 mM NTPs, 3 ng/μL T7 RNAP, 0.001 unit/μL YIPP (New England Biolabs), 0.2 units/μL SUPERase In (Ambion), 0.5 μM GST-Tus). Glycerol was then added to a final concentration of 10%, and equal volumes of the resulting reactions were run on a 4% native polyacrylamide gel. The gel was dried, exposed to a PhosphorImager screen, scanned by a Typhoon 9400 Imager, and analyzed by ImageQuant software.
Transcription halting on 454 beads
Halting DNA templates compatible with the 454 sequencing were used to coat 454 polystyrene beads with either SRB-2 aptamer or GFP aptamer templates by PCR (without an emulsion) using the bead-bound 454 Primer A and in solution 454 Primer B. An aliquot of these beads was washed with 1x T7 transcription buffer, incubated with 1 μM GST-Tus in 1x T7 transcription buffer for 30 minutes at room temperature, washed in transcription buffer, and resuspended in a transcription reaction. After transcription (30 minutes at 37°C), beads were washed with GFP aptamer binding buffer (1x PBS, 5 mM MgCl2, 0.01% Tween-20), and incubated with 1 μM 6xHis-EGFP. After 20 minutes of binding at room temperature, beads were washed with GFP aptamer binding buffer and imaged with a Zeiss Axioplan II epifluorescence microscope using a FITC/Fluo filter set (Chroma Technology Corp. Cat # 41001). Both DIC and fluorescence images were taken.
Transcription halting on glutathione beads
DNA templates were prepared with the GFP aptamer template flanked on the 5′ end by a T7 promoter and an 11 nt C-less cassette, and two ter sites on its 3′ end. First, these templates were bound to GST-Tus in 10x molar excess in 1x T7 transcription buffer for 30 minutes at 37°C. The resulting Tus-DNA complexes were then incubated with glutathione coupled agarose beads (Pierce) in 1x T7 transcription buffer for 30 minutes at 37°C. An equal volume of 2x transcription reaction lacking CTP was then added to the resulting bead slurry, and transcription was allowed to proceed for 30 minutes at 37°C. At this time, the one aliquot of beads was washed three times in 1x T7 transcription buffer and then mixed with an equal volume of 2x transcription reaction mix lacking polymerase but containing all four NTPs. Transcription was allowed to proceed for another 30 minutes at 37°C. Both bead treatments were then washed with GFP aptamer binding buffer, incubated with 1 μM 6xHis-EGFP for 30 minutes at RT, washed again, and imaged as described before for 454 beads.
HiTS-RAP
A standard sequencing run was performed on an Illumina GAIIx using an 82 cycle read length on a paired end flowcell. The same .xml recipe used for the sequencing run included all subsequent steps to effect transcription halting and binding of mOrange labeled protein to the sequenced DNA clusters, so that fresh solutions are added at once for all steps through transcription halting, and then once for the binding curve. Thus, reagents for HiTS-RAP are loaded onto the Paired End Module (PEM) twice. The recipe program used for the GFP run is included as Supplementary Software 1; it details the exact reagent delivery used for HiTS-RAP.
2.25 mL of each solution was loaded onto its own position on the PEM of the GAIIx. The .xml recipe delivered reagents in the proper sequence, and set the flowcell temperature using the Peltier heater in the instrument. During each reaction or binding step, 75 μL of solution is flowed through each lane, and then the flowcell is incubated for 30 minutes to equilibrate. During the incubation, 15 μL of fresh solution is delivered to each lane every 5 minutes. The second strand generated during sequencing was stripped away and 1 μM primer (IllumFORAdapt_T1_IllumFORSeq, Supplementary Table 8) for double stranded DNA regeneration annealed as per the standard Illumina protocol. Excess primer was then washed away with 1x NEB buffer 4 with 0.01 % Tween-20 (New England Biolabs). DNA was then made double stranded by flowing in a Klenow exo- enzyme reaction mix (1x NEB Buffer 4, 0.01% Tween-20, 0.2 mM dNTPs, Klenow exo-(New England Biolabs)) and incubating for 30 minutes at 37°C. The flowcell containing double stranded DNA clusters was then equilibrated with 1x T7 transcription buffer. Tus was allowed to bind the DNA templates’ ter elements by equilibrating with 1 μM GST-Tus in 1x T7 transcription buffer for 30 minutes at 37°C. The flowcell was then equilibrated with a transcription reaction (1x T7 transcription buffer, 0.5 mM NTPs, T7 RNAP, YIPP, Superase In, ~0.5 μM GST-Tus). Transcription and halting was allowed to proceed for 30 minutes at 37°C.
After transcription halting, the flowcell was equilibrated with GFP or NELF (10 mM HEPES, pH 7.5, 100 mM NaCl, 25 mM KCl, 5 mM MgCl2, 0.02% Tween-20) aptamer binding buffer at room temperature. It was imaged immediately, just as during sequencing, to measure the background intensity at every cluster. It was then equilibrated successively with increasing concentrations of mOrange labeled protein in binding buffer and imaged. Imaging at each concentration was carried out in equilibrium binding after 30 minutes equilibration at room temperature. Concentrations of EGFP-mOrange varied from 0.04 nM to 625 nM, increasing in fivefold increments. The images in Figure 2b are from .tif files collected by the Illumina SCS during this binding. HiTS-RAP for NELF-E mOrange was identical to EGFP-mOrange, except that concentrations of protein varied from 0.064 to 1000 nM.
Sequencing and data extraction
Sequencing, transcription, and protein binding are executed as a single run on a GAIIx. In the case of NELF-E, transcription and protein binding were carried out twice. The first binding curve was used for all analyses. Intensity data were collected and basecalling done using the Illumina SCS version 2.9. During the run, .cif and .bcl files were saved, along with 5% of the raw .tif images taken. We used scripts generously provided by R. Friedman (Institut Pasteur) to extract intensities of mOrange fluorescence for every cluster from .cif file. This gives the coordinates of each cluster, together with its protein binding intensities (in the T channel) and average intensity during sequencing. These data were then matched by cluster coordinates to their sequence from .qseq.txt files generated by the Illumina OLB version 1.9.4. Only clusters that passed filter were included. If a cluster contained a mutation (deviating from NELFapt or GFPapt), it was only included if the mutated base had a quality score greater than 25 (<0.003 probability of an incorrect base call). A threshold of quality score >20 is common practice for SNP calling algorithms34.
Double mutant analysis was restricted to base substitutions and to the region in the center of the aptamer (76 nt for GFPapt, 64 for NELFapt) so that the 3 bp at the ends could be used to ensure against insertions and deletions.
Loss of signal correction
After the GFP binding curve, the flowcell was imaged 9 successive times after reequilibrating with 625 nM EGFP mOrange. After three of these cycles, imaging was not carried out, but rather the flowcell was allowed to sit in 625 nM EGFP-mOrange for the time that it would take for one equilibration and binding cycle. Thus, the 9 imaging steps span the time that it would take for 12 equilibration and imaging steps. A similar rate of decay was observed between successive and staggered imaging cycles, indicating that most loss of signal is due to time rather than photobleaching caused by the number of times that the flowcell was imaged. We find that in the regime where our imaging takes place (i.e. where time is much less than the characteristic lifetime), the observed decay is described well by a linear approximation to exponential decay. Thus, we took the average intensities of clusters with canonical GFP aptamer sequence though the nine cycles imaged and used scipy.optimize.curve_fit to perform a weighted linear least squares regression, using weights determined from the standard deviation of the intensities at each time point. The equation used was:
where Iobserved is the measured mOrange intensity, I0 is the initial intensity, t is the time in units of cycles, and τ is the characteristic lifetime in cycles. I0 and τ were solved for the GFP aptamer clusters in each of the three lanes (each containing ~2.7 million clusters with canonical GFP aptamer sequence); the final characteristic lifetime is the average of these three fitted lifetimes. In this regime, halted complexes are decaying at a rate corresponding to a characteristic lifetime of 137 ± 7 cycles, or 206 ± 11 hours (at an average cycle time of ~1.5 hours). This lifetime simply describes the rate at which the halted transcription complexes are decaying, independent of the actual intensity. Given that the background intensity in these lanes is approximately 85, this means that fluorescence signal will be above background for 48 cycles, or 72 hours.
The characteristic lifetime was used to apply a correction to each measured fluorescence intensity in binding curves, using the equation:
where Iactual is the fluoresce intensity used in the fit for Kd, Iobserved is the measured intensity, 0.00730 is the decay rate in cycle−1 (the inverse of τ), and t is the time since transcription, in cycles. Supplementary Figure 4a shows intensity data from three lanes, together with the same intensities after applying the correction factor.
Kd calculation
Sequential measurements of the corrected mOrange intensities at increasing concentrations give a binding curve for each cluster. Intensities were normalized for cluster size and position in the tile by dividing each intensity by the average sequencing intensity14. This correction is applied so that all intensities from a lane can be averaged, to be representative of each sequence. Clusters with the same sequence in each lane were matched, and average binding curves were generated by taking the mean of their normalized intensities for each protein concentration. If more than 10 clusters had a given sequence in a lane, this binding curve was fit to a Hill equation, solving the equilibrium dissociation constant (Kd) of the interaction35. We have used the following Hill equation:
Where b is the background fluorescence intensity at the cluster, m is the maximum fluorescence intensity, Kd is the dissociation constant, n is the Hill coefficient, and C is the concentration of target protein. The intensities measured by imaging at several different concentrations are then used to solve m, b, Kd, and n in a weighted non-linear least squares regression. We have found that letting these four parameters vary gives reliable fits; this also means that the Kd that we solve is independent of the intensity values, so that it shows the inflection point in the binding curve. Only fits for which the scipy.optimize.curve_fit algorithm of the NumPy Python package returned a variance of less than 1,000,000 were considered to be high confidence and used in these analyses.
The GFPapt run contained three lanes of GFPapt. To generate a single data set from all three lanes, Kds for every unique sequence in each lane were determined by fitting to average binding curves. The Kds for all unique sequences in the flowcell were then determined by geometrically averaging the fitted Kds across the three lanes: therefore the Kd values are reported as the average Kd ×/(multiply or divide by) standard deviation.
The GFPapt data have the added complication of very low background binding. Thus, there are sequences in this data set which do not bind GFP measurably, while for NELF-E, background binding for this RNA binding protein is high enough that every sequence is expected to bind to some extent. To mitigate this problem, only sequences which show a 3% increase (determined in Supplementary Fig. 4b–d) between the first and last two measured intensities were considered as binding: all others were called as not binding, meaning that they effectively have a Kd greater than 125 nM. We set the limit at 125 nM because this was the second highest concentration probed, so we do not expect to be able to measure affinities greater than this. For analysis of single mutants, any mutant scored as not binding based on its intensity increase was assigned an affinity of 125 nM; this value was averaged with other real measurements from other lanes if it was called as binding there. This results in a Kd measurement of greater than that average. Sequences for which any lane was called as not binding are indicated in all tables. Such mutants were excluded from analysis of double mutants.
EMSA of GFPapt and NELFapt and mutants
EMSA was used to verify the HiTS-RAP measured binding affinities of wild-type and mutant GFP binding aptamers to EGFP. To this end, in vitro transcribed aptamers were 3′ end-labeled with AlexaFluor647 Hydrazide (Invitrogen) as described elsewhere5 and quantified by Qubit Fluorometer (Invitrogen). Fluorescently labeled aptamers were mixed with recombinant GST-EGFP protein at 25°C for 45 min. The GST-EGFP concentration in the binding reaction was varied as a 2/3 dilution series starting from 500 nM, and a no protein control. The final 20 μl binding reaction was composed of 1X PBS, 5 mM MgCl2, 0.4 U of Superase In, 1 μg of yeast tRNA, 0.005% NP-40, and 5 nM fluorescently-labeled aptamer. After addition of Bromocresol Green containing 30% glycerol to 6% final glycerol concentration, binding reactions were loaded on a 6% polyacrylamide gel (0.5X TBE, 5 mM MgCl2) that was pre-equilibrated to 4°C and pre-run at 120V for 10 min at 4°C. Loaded gels were run at 120V for 90 min at 4°C, and then imaged with a Typhoon 9400 scanner using Cy5 settings. Images were quantified by ImageQuant5.2 software, and data were fitted to Hill Equation to determine the Kd values using Igor software. EMSA was carried out with fluorescein labeled minimal NELFapt as described elsewhere17.
RNA secondary structure predictions
The average absolute effect of all three mutations at each position was calculated using the following equation: .
We used Kinefold36 to predict the folded structures and the associated folding free energies. Kinefold, unlike other programs which predict secondary structures based on minimal free energy, predicts the structure of RNA as it is being synthesized, and thus recapitulates what is happening in HiTS-RAP, where the RNA folds co-transcriptionally. This predicted structure is consistent with the published secondary structure of the GFP aptamer which was supported by extensive mutational analysis16.
Predicted secondary structures and the Gibbs Free Energy (ΔG) of the folded structures are obtained from Kinefold, unless indicated otherwise, however the actual drawings are obtained from mFold37, because they are easier to manipulate.
Supplementary Material
Acknowledgments
The authors would like to thank R. Friedman (Institut Pasteur) for providing scripts to extract intensity data, B. Mohanty (Medical University of South Carolina) for providing vectors containing the Tus gene, K. Szeto and D. Shalloway (Cornell University) for advice on data analysis, W. Zipfel and A. Singh (Cornell University) for help in understanding the optics of the GAIIx, C.T. Waters and H. Kwak (Cornell University) for bioinformatic advice, the Cornell sequencing core facility for help in learning the GAIIx, and H. Craighead (Cornell University) and the members of the Lis lab for helpful discussions on experimental design and the manuscript. This work was supported by NIH grants GM090320 and DA030329 to J.T.L.
Footnotes
Accession Codes:
Accession codes. Addgene: 53302 (EGFP-mOrange pHis-//), 53303 (NELFE-mOrange pHis-//), 53304 (mOrange pHis-//), and 53305 (Tus pGST-//).
Author contributions:
These authors contributed equally to this work:
Jacob M. Tome and Abdullah Ozer
Initial idea: AO and JTL. Experimental design: JMT, AO, DG, GPS, and JTL. Experimental implementation: JMT, AO, and JMP. HiTS-RAP experiments and bioinformatics analysis: JMT. Project coordination: JTL. Paper writing: JMT, AO, JMP, and JTL.
References
- 1.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–9. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 2.König J, Zarnack K, Luscombe NM, Ule J. Protein-RNA interactions: new genomic technologies and perspectives. Nat Rev Genet. 2011;13:77–83. doi: 10.1038/nrg3141. [DOI] [PubMed] [Google Scholar]
- 3.Wong I, Lohman TM. A double-filter method for nitrocellulose-filter binding: application to protein-nucleic acid interactions. Proc Natl Acad Sci U S A. 1993;90:5428–32. doi: 10.1073/pnas.90.12.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ryder SP, Recht MI, Williamson JR. Quantitative analysis of protein-RNA interactions by gel mobility shift. Methods Mol Biol. 2008;488:99–115. doi: 10.1007/978-1-60327-475-3_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pagano JM, Clingman CC, Ryder SP. Quantitative approaches to monitor protein-nucleic acid interactions using fluorescent probes. RNA. 2011;17:14–20. doi: 10.1261/rna.2428111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salim NN, Feig AL. Isothermal titration calorimetry of RNA. Methods. 2009;47:198–205. doi: 10.1016/j.ymeth.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katsamba PS, Park S, Laird-Offringa IA. Kinetic studies of RNA-protein interactions using surface plasmon resonance. Methods. 2002;26:95–104. doi: 10.1016/S1046-2023(02)00012-9. [DOI] [PubMed] [Google Scholar]
- 8.Campbell ZT, et al. Cooperativity in RNA-protein interactions: global analysis of RNA binding specificity. Cell Rep. 2012;1:570–81. doi: 10.1016/j.celrep.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin L, et al. Systematic reconstruction of RNA functional motifs with high-throughput microfluidics. Nat Methods. 2012;9:1192–4. doi: 10.1038/nmeth.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tenenbaum SA, Carson CC, Lager PJ, Keene JD. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc Natl Acad Sci U S A. 2000;97:14085–90. doi: 10.1073/pnas.97.26.14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Licatalosi DD, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–9. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–41. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–9. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nutiu R, et al. Direct measurement of DNA affinity landscapes on a high-throughput sequencing instrument. Nat Biotechnol. 2011;29:659–64. doi: 10.1038/nbt.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evanko D. Next-generation protein binding. Nat Methods. 2011;8:619–619. doi: 10.1038/nmeth0811-619. [DOI] [PubMed] [Google Scholar]
- 16.Shui B, et al. RNA aptamers that functionally interact with green fluorescent protein and its derivatives. Nucleic Acids Res. 2012;40:e39. doi: 10.1093/nar/gkr1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pagano JM, et al. Defining NELF-E RNA Binding in HIV-1 and Promoter-Proximal Pause Regions. PLoS Genet. 2014;10:e1004090. doi: 10.1371/journal.pgen.1004090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu CH, et al. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev. 2003;17:1402–14. doi: 10.1101/gad.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu CH, et al. Molecular characterization of Drosophila NELF. Nucleic Acids Res. 2005;33:1269–79. doi: 10.1093/nar/gki274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamaguchi Y, et al. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 21.Yamaguchi Y, Inukai N, Narita T, Wada T, Handa H. Evidence that Negative Elongation Factor Represses Transcription Elongation through Binding to a DRB Sensitivity-Inducing Factor/RNA Polymerase II Complex and RNA. 2002;22:2918–2927. doi: 10.1128/MCB.22.9.2918-2927.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohanty BK, Sahoo T, Bastia D. The relationship between sequence-specific termination of DNA replication and transcription. EMBO J. 1996;15:2530–9. [PMC free article] [PubMed] [Google Scholar]
- 23.Kamada K, Horiuchi T, Ohsumi K, Shimamoto N, Morikawa K. Structure of a replication-terminator protein complexed with DNA. Nature. 1996;383:598–603. doi: 10.1038/383598a0. [DOI] [PubMed] [Google Scholar]
- 24.Mulugu S, et al. Mechanism of termination of DNA replication of Escherichia coli involves helicase-contrahelicase interaction. Proc Natl Acad Sci U S A. 2001;98:9569–74. doi: 10.1073/pnas.171065898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mulcair MD, et al. A molecular mousetrap determines polarity of termination of DNA replication in E. coli. Cell. 2006;125:1309–19. doi: 10.1016/j.cell.2006.04.040. [DOI] [PubMed] [Google Scholar]
- 26.Guajardo R, Sousa R. Characterization of the effects of Escherichia coli replication terminator protein (Tus) on transcription reveals dynamic nature of the Tus block to transcription complex progression. Nucleic Acids Res. 1999;27:2814–2824. doi: 10.1093/nar/27.13.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holeman LA, Robinson SL, Szostak JW, Wilson C. Isolation and characterization of fluorophore-binding RNA aptamers. Fold Des. 1998;3:423–31. doi: 10.1016/S1359-0278(98)00059-5. [DOI] [PubMed] [Google Scholar]
- 28.Shaner NC, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp red fluorescent protein. Nat Biotechnol. 2004;22:1567–72. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 29.Limpert E, Stahel Wa. Problems with using the normal distribution--and ways to improve quality and efficiency of data analysis. PLoS One. 2011;6:e21403. doi: 10.1371/journal.pone.0021403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001;20:4214–21. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buenrostro JD, et al. Quantitative analysis of RNA-protein interactions on a massively parallel array reveals biophysical and evolutionary landscapes. Nat Biotechnol. 2014 doi: 10.1038/nbt.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheffield P, Garrard S, Derewenda Z. Overcoming Expression and Purification Problems of RhoGDI Using a Family of “ Parallel ” Expression Vectors. 1999;39:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- 33.Mccullum EO, Williams BAR, Zhang J, Chaput JC. In Vitro Mutagenesis Protocols. 2010;634:103–109. doi: 10.1007/978-1-60761-652-8_7. [DOI] [PubMed] [Google Scholar]
- 34.Nielsen R, Paul JS, Albrechtsen A, Song YS. Genotype and SNP calling from next-generation sequencing data. Nat Rev Genet. 2011;12:443–51. doi: 10.1038/nrg2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barlow R, Blake JF. Hill coefficients and the logistic equation. Trends Pharmacol Sci. 1989;10:440–1. doi: 10.1016/S0165-6147(89)80006-9. [DOI] [PubMed] [Google Scholar]
- 36.Xayaphoummine A, Bucher T, Isambert H. Kinefold web server for RNA/DNA folding path and structure prediction including pseudoknots and knots. Nucleic Acids Res. 2005;33:W605–10. doi: 10.1093/nar/gki447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.