Abstract
Accurate and consistent labeling of longitudinal cortical surfaces is essential to understand the early dynamic development of cortical structure and function in both normal and abnormal infant brains. In this paper, we propose a novel method for simultaneous, consistent, and unbiased labeling of longitudinal dynamic cortical surfaces in the infant brain MR images. The proposed method is formulated as minimization of an energy function, which includes the data fitting, spatial smoothness and temporal consistency terms. Specifically, in the spirit of multi-atlas based label fusion, the data fitting term is designed to integrate adaptive contributions from multi-atlas surfaces, according to the similarity of their local cortical folding with that of the subject surface. The spatial smoothness term is designed to adaptively encourage label smoothness based on the local folding geometries, i.e., also allowing label discontinuity at sulcal bottoms, where the cytoarchitecturally and functionally distinct cortical regions are often divided. The temporal consistency term is further designed to encourage the label consistency between temporal corresponding vertices with similar local cortical folding. Finally, the entire energy function is efficiently minimized by a graph cuts method. The proposed method has been successfully applied to the labeling of longitudinal cortical surfaces of 13 infants, each with 6 serial images scanned from birth to 2 years of age. Both qualitative and quantitative evaluation results demonstrate the validity of the proposed method.
Keywords: Infant cortical surface, longitudinal cortical surface labeling
1 Introduction
The human cerebral cortex develops dynamically in the first 2 years of life [1], with all primary and secondary cortical folding being well established at term birth [2]. Accurate and consistent labeling of longitudinal dynamic infant cortical surfaces into regions of interest (ROIs) is essential to understand postnatal development of cortical structure and function in both normal and abnormal infant brains. Many methods have been developed for the labeling of a single cortical surface. However, applying these methods to each longitudinal cortical surface independently is likely to generate longitudinally-inconsistent labeling results, especially in the ambiguous cortical regions, thus leading to inaccurate cortex development measurements. One strategy to ensure the longitudinal consistent labeling is to first label the cortical surface of a selected time point (usually the first or the last time point), and then propagate the labeling result to other time points. However, the surface labeling results by this strategy could be biased by the selected time point, in addition to the potential propagation of labeling errors. Accordingly, efforts have been made towards unbiased and consistent labeling of longitudinal cortical surfaces. For example, in the longitudinal pipeline of FreeSurfer, a within-subject template is first built by rigidly aligning all longitudinal images of a subject to a median image, and then the cortical surfaces of the within-subject template are reconstructed and labeled. These labeled cortical surfaces will be rigidly transformed back to the space of each longitudinal image as initialization and further deformed independently to refine the labeling results [3]. Although this independent refinement may be suitable for the adults with small longitudinal changes, it becomes problematic when applied to the infants with dynamic longitudinal changes.
In this paper, we propose a novel method for simultaneous, consistent, and unbiased labeling of longitudinal dynamic cortical surfaces in serial infant brain MR images. The proposed method is formulated as minimization of an energy function, which includes the data fitting, spatial smoothness, and temporal consistency terms. The data fitting term is designed to integrate adaptive contributions from multiple atlas surfaces, according to the similarities of their local cortical folding with the subject surface. The spatial smoothness term is also designed to adaptively encourage label smoothness based on the local folding geometries. The temporal consistency term is further designed to adaptively encourage longitudinal label consistency based on the temporal similarities of local cortical folding. The energy function is efficiently minimized by the alpha-expansion graph cuts method [4]. The proposed method has been successfully applied to labeling of longitudinal cortical surfaces of 13 infants, each with 6 serial images in the first 2 years of life. Both qualitative and quantitative evaluation results demonstrate the accuracy and consistency of the proposed method.
2 Methods
2.1 Dataset and Image Preprocessing
Serial T1, T2, and diffusion-weighted MR images of 13 healthy infants (9 males/4 females) were acquired at every 3 months from 2 weeks to 1.5 years of age, using a Siemens 3T head-only MR scanner. T1 images (160 axial slices) were acquired with the following imaging parameters: TR/TE = 1900/4.38ms, flip angle = 7, resolution = 1 × 1 × 1 mm3. T2 images (70 axial slices) were acquired with the imaging parameters: TR/TE = 7380/119ms, flip angle = 150, resolution = 1.25 × 1.25 × 1.95 mm3. Diffusion-weighted images (DWI) (60 axial slices) were acquired with the parameters: TR/TE = 7680/82 ms, resolution = 2 × 2 × 2 mm3, 42 non-collinear diffusion gradients, and diffusion weighting b =1000s/mm 2. Distortion correction of DWI was also performed. T2 images and fractional anisotropy (FA) images, derived from DWI, were rigidly aligned onto their T1 images and further resampled to 1 × 1 × 1 mm3. For each set of aligned T1, T2, and FA images, non-cerebral tissues were removed. Then, all longitudinal images of the same infant were rigidly aligned. Brain tissue was segmented by a 4D level-set method by integration of the complementary information of T1, T2 and FA images [5]. After tissue segmentation, non-cortical structures were masked and filled, and each brain was separated into left and right hemispheres.
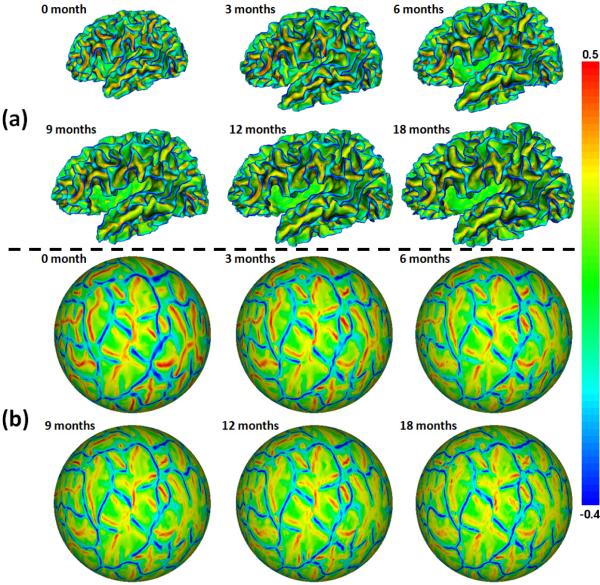
Then, the inner cortical surface (the interface between white matter (WM) and gray matter (GM)) of each hemisphere was reconstructed by correcting topological defects and tessellating WM as a triangular mesh [1]. The inner cortical surface was further inflated and mapped to a standard sphere [6]. All longitudinal cortical surfaces of the same infant were group-wisely aligned to establish within-subject correspondences using Spherical Demons [7]. Fig. 1(a) shows the longitudinal inner surfaces of a representative infant, color-coded by the mean curvatures. Fig. 1(b) shows group-wisely aligned longitudinal spherical surfaces of the infant, again color-coded by the mean curvatures. As can be seen, all primary and secondary cortical folding are well established at term birth. Moreover, the longitudinal cortical folding are quite stable during postnatal development and thus are well aligned by group-wise surface registration.
Fig. 1.
(a) Longitudinal inner cortical surfaces of a representative infant, color-coded by the mean curvatures. (b) Group-wisely aligned longitudinal spherical surfaces of the infant, color-coded again by the mean curvatures. Red colors indicate sulci and blue colors indicate gyri.
The publically available 39 cortical surfaces with manual parcellation based on sulcal bottoms by experts [7, 8] were adopted as multi-atlas surfaces. Information on image acquisition and demographics can be found in [8]. To warp atlas surfaces to subject surfaces, all atlas surfaces were first group-wisely aligned using Spherical Demons [7]. Then each longitudinal surface of the subject was aligned onto the group-wisely aligned atlas surfaces [7]. Finally, the deformation field from each atlas surface to the subject surface was computed by concatenating the deformation field from this atlas surface to the group-wisely aligned atlas surfaces and the deformation field from the group-wisely aligned atlas surfaces to the subject surface. Accordingly, each atlas surface can be warped to each subject surface. Note that the group-wise alignment is only required to perform one time, and can be used for all subjects, thus this strategy is computationally much more efficient than the conventional way of pair-wise registration between each atlas surface and each longitudinal surface of the subject. On the other hand, due to the inter-individual variability of the cortical folding patterns and also the regularization constraints in the registration, a subject surface and each warped atlas surface might not have the maximum similarity of their local cortical folding. This issue will be taken care for surface labeling in the section 2.2.
2.2 Simultaneous Labeling of Longitudinal Cortical Surfaces
The proposed method for simultaneous, consistent, and unbiased labeling of longitudinal cortical surfaces in the infant is formulated as an energy minimization problem:
| (1) |
where Ed is the data fitting term, Es is the spatial smoothness term, and is Et is temporal consistency term. αs and αt are the weighting parameters.
Data Fitting Term
To define the data fitting term, we take advantage of multi-atlas based methods, which account for atlas-subject variability [9]. Given K atlas surfaces with each having L labels, the data fitting term is defined as:
| (2) |
where Px(lx) indicates the probability of assigning a label lx ∈ {1, ... L} to a vertex x in a subject surface. The label probability at a vertex is computed based on the shape information of labels in the atlas surfaces, as well as the differences of local cortical folding between the subject surface and atlas surfaces. The latter is defined based on the average absolute difference of mean curvatures in local surface patches:
| (3) |
where D(·,·) is the coritcal folding difference between two surface patches, and S(·) is a local surface patch, defined as a circular region ΩS on the spherical surface with the radius of 2.5mm (set experimentally) surrounding a center vertex. |ΩS| is the number of vertices in the surface patch. xk is the corresponding point in atlas surface k, for the vertex x in a subject surface. y is a vertex in the subject surface patch, and yk is its corresponding point in the atlas surface k. H(·) is the mean curvature.
To use the shape information of labels in atlas surfaces, we adopt the logarithm of odds model [9] based on the signed geodesic distance map on the original cortical surface, computed by the fast marching method on triangular meshes. Denoting dk,lx (·) as the signed geodesic distance map of label lk in the atlas surface k that also setting the inside of the label being positive values, the label probability of the vertex x is finally defined as:
| (4) |
where is the partition function for atlas surface k. The first term in Eq. (4) is the weight of the atlas surface k, and the second term in Eq. (4) is the probability of observing label lx at subject vertex x based on the atlas surface k. Positive parameters β and γ are experimentally set as 1.0 and 2.0, respectively. could be the corresponding point xk in the atlas surface k for the subject vertex x determined by surface registration. However, it might not achieve the maximum similarity of local cortical folding due to registration errors. Therefore, after surface registration, a better corresponding point in the atlas surface k for the subject vertex x can be further determined by local searching for the most similar surface patch: , . N(·) is the search range, defined as circular region on the spherical surface with the radius 2.5mm surrounding the vertex x.
Spatial Smoothness Term
The spatial smoothness term represents the sum of the costs of labeling a pair of spatial neighboring vertices in a subject surface:
| (5) |
where NS indicates the cost of labeling a pair of spatial neighboring vertices pairs in a cortical surface. indicates the cost of labeling a pair of spatial neighboring vertices x and y as lx and ly, respectively. The costs of discontinuous labeling are set as small values at highly bended cortical regions, e.g. sulcal bottoms, where the cytoarchitecturally and functionally distinct cortical regions are often divided, similarly as done by the manual labels in atlas surfaces by experts [8]. The costs of discontinuous labeling are set as large values at other regions, i.e. flat cortical regions. is thus defined as:
| (6) |
where n is the normal direction and δ is the Dirac delta function. If lx = ly , δ(|lx – ly|) = 1; otherwise, δ(|lx – ly|) = 0. Therefore, is 0, if lx = ly. At highly bended cortical regions, e.g. sulcal bottoms, x and y belonging to different regions generally have quite different normal directions and also large magnitudes of mean curvatures, therefore, both the first and second terms inEq. (6) are small values; while x and y in the same region generally have the similar normal direction but large magnitudes of mean curvatures, only the second term in Eq. (6) is a small value. If x and y are at other cortical regions, i.e. flat cortical regions, their normal directions will be quite similar and their magnitudes of mean curvatures are close to 0, thereby, both the first and second terms in Eq. (6) are close to 1.
Temporal Consistency Term
The temporal consistency term represents the sum of the costs of labeling a pair of temporal corresponding vertices between a pair of longitudinal cortical surfaces:
| (7) |
where Nt is the set of temporal corresponding vertex pairs, and defined in any two longitudinal surfaces in a subject. indicates the cost of labeling a pair of temporal corresponding vertices x and y as lx and ly, respectively. The cost of discontinuous labeling of a pair of temporal corresponding vertices is set based on their local cortical folding similarity. To avoid bias and reduce computational cost in coventional pair-wise registration, temporal correspondences are determined by group-wise registration of all longitudinal surfaces of the same infant as mentioned in section 2.1.
Finally, the multi-label alpha-expansion graph cuts method [4] is adopted to minimize the above defined energy function. Specifically, longitudinal surfaces are represented as an undirected weighted graph , where is the set of noes, including all vertices on the longitudinal surface and the terminals represented by labels. is the collection of edges, where is edges formed by spatial neighboring and temporal corresponding vertices, called n-links, and is the edges formed by vertices to terminals. called t-links. In this graph, Dx describes the edge weight of t-links, and and describe the edge weights of n-links. For more details of the graph cuts method, please refer to [4].
3 Results
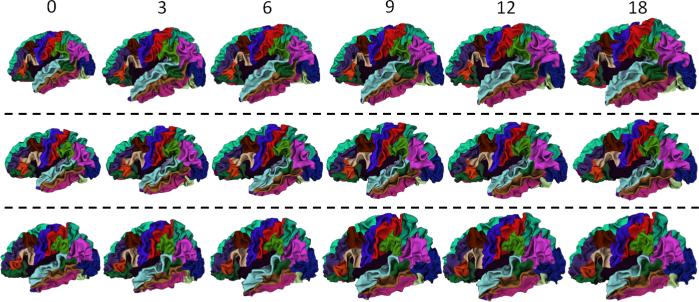
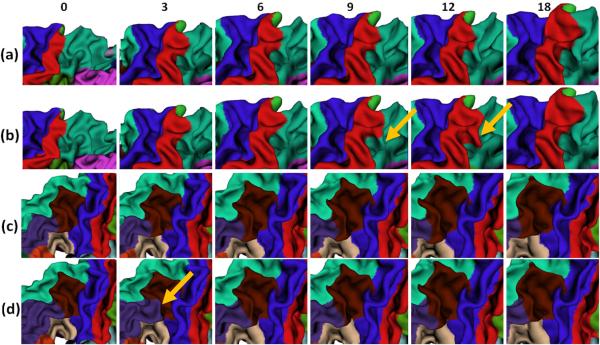
The proposed method has been applied to the labeling of longitudinal cortical surface in 13 healthy infants, each with serial images acquired at 0, 3, 6, 9, 12 and 18 months. Parameters αs and αt in Eq. (1) are both set as 0.15 in all experiments. Our method takes around 10 minutes (for energy computation and minimization) to label all longitudinal cortical surfaces for each subject on a PC with Intel Xenon 2.26GHz CPU and 4GB memory. Fig. 2 shows longitudinal surface labeling results on left hemispheres of 3 typical subjects by the proposed method. As can be seen, the labeling results are visually quite reasonable and consistent. Fig. 3 shows close-up views of representative longitudinal surface labeling results by the proposed method, and also FreeSurfer [3,8] where the same atlases were used to train the classifiers. As can be observed, the proposed method achieves longitudinally more consistent results than FreeSurfer.
Fig. 2.
Longitudinal cortical surface labeling results in 3 representative infant subjects, each with 6 longitudinal cortical surfaces at 0, 3, 6, 9, 12 and 18 months
Fig. 3.
Close-up views of representative longitudinal cortical surface labeling results. (a) and (c) Results by the proposed method. (b) and (d) Results by FreeSurfer. Yellow arrows indicate several regions with longitudinally-inconsistent labels by FreeSurfer.
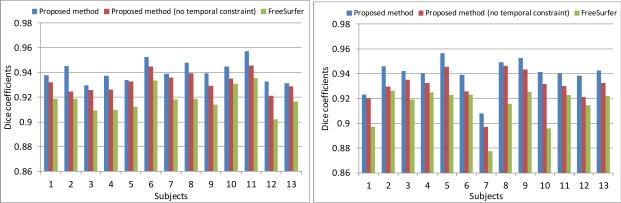
To quantitatively evaluate the accuracy of the longitudinal cortical surface labeling results, we have an expert manually annonate the precentral gyrus (PreCG) and superior temporal gyrus (STG) in the first and last time-point cortical surfaces of the left hemisphere in each of the 13 subjects, according to the mean-curvature based cortical surface labeling protocol in [8]. We calculate the Dice coefficients between automatic and manual labeling regions. Fig. 4 shows the Dice coefficients of PreCG and STG on the 13 subjects by 3 different methods. The average Dice coefficients for PreCG/STG are 0.941/0.939 (proposed method), 0.932/0.930 (proposed method with out temporal constraint (by setting αt as 0)), and 0.918/0.914 (FreeSurfer), respectively. As can be seen, the proposed method achieves the highest Dice coefficient.
Fig. 4.
Dice coefficients of precentral (left) and superior temporal (right) gyri by the proposed method, the proposed method without temporal constraint, and FreeSurfer on 13 subjects
To further demonstrate the consistency of the longitudinal surface labeling results, Fig. 5(a) shows label boundaries of the aligned longitudinal spherical surfaces of a typical subject by the proposed method and FreeSurfer. As can be seen, the proposed method achieves temporally more consistent labeling boundaries than FreeSurfer. To quantitatively evaluate the consistency, we compute the average value of mean symmetric distance of boundaries [8] for labeled regions between each pair of aligned longitudinal surfaces in each of 13 infants, as shown in Fig. 5(b). The average boundary distance by 3 methods are 0.52±0.002mm (proposed method), 0.75±0.03mm (proposed method without temporal constraint), and 0.90±0.03mm (FreeSurfer), respectively. The proposed method achieves the lowest boundary distance.
Fig. 5.
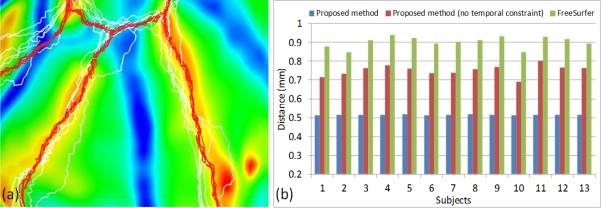
(a) Label boundaries of all aligned longitudinal surfaces of a typical subject, overlaid on the mean spherical surface that is color-coded by the average mean curvatures. Red curves are the results by the proposed method, and white curves are the results by FreeSurfer. (b) Average boundary distance of all labels between every pair of aligned longitudinal surfaces in each of 13 infants by the proposed method (with and without temporal constraint) and FreeSurfer.
4 Discussion and Conclusion
This paper presented a novel method for consistent labeling of longitudinal dynamic infant cortical surface using multi-atlas surfaces. The preliminary results on 13 infants demonstrated its promising performance. Our main contributions are: first, we proposed a data fitting term based on the shape information adaptively derived from atlas surfaces; second, we proposed a spatial smoothness term adaptive to the cortical folding geometries and also a temporal consistency term adaptive to the temporal similarities of the cortical folding. Our future work includes parameter optimization, more validation, and application to a large-scale dataset for early brain development study.
References
- 1.Li G, Nie J, Wang L, et al. Mapping Region-Specific Longitudinal Cortical Surface Expansion from Birth to 2 Years of Age. Cereb. Cortex. 2012 doi: 10.1093/cercor/bhs265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hill J, Dierker D, Neil J, et al. A surface-based analysis of hemispheric asymmetries and folding of cerebral cortex in term-born human infants. J. Neurosci. 2010;30:2268–2276. doi: 10.1523/JNEUROSCI.4682-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reuter M, Schmansky NJ, Rosas HD, Fischl B. Within-subject template estimation for unbiased longitudinal image analysis. Neuroimage. 2012;61:1402–1418. doi: 10.1016/j.neuroimage.2012.02.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boykov Y, Kolmogorov V. An experimental comparison of min-cut/max-flow algorithms for energy minimization in vision. IEEE Trans. PAMI. 2004;26:1124–1137. doi: 10.1109/TPAMI.2004.60. [DOI] [PubMed] [Google Scholar]
- 5.Dai Y, Shi F, Wang L, Wu G, Shen D. iBEAT: A Toolbox for Infant Brain Magnetic Resonance Image Processing. Neuroinformatics. 2012 doi: 10.1007/s12021-012-9164-z. [DOI] [PubMed] [Google Scholar]
- 6.Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999;9:195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- 7.Yeo BT, Sabuncu MR, Vercauteren T, et al. Spherical demons: fast diffeomorphic landmark-free surface registration. IEEE Trans. Med. Imaging. 2010;29:650–668. doi: 10.1109/TMI.2009.2030797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desikan RS, Segonne F, Fischl B, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 9.Sabuncu MR, Yeo BT, Van Leemput K, et al. A generative model for image segmentation based on label fusion. IEEE Trans. Med. Imaging. 2010;29:1714–1729. doi: 10.1109/TMI.2010.2050897. [DOI] [PMC free article] [PubMed] [Google Scholar]