Abstract
We conducted imputation to the 1000 Genomes Project of four genome-wide association studies of lung cancer in populations of European ancestry (11,348 cases and 15,861 controls) and genotyped an additional 10,246 cases and 38,295 controls for follow-up. We identified large-effect genome-wide associations for squamous lung cancer with the rare variants of BRCA2-K3326X (rs11571833; odds ratio [OR]=2.47, P=4.74×10−20) and of CHEK2-I157T (rs17879961; OR=0.38 P=1.27×10−13). We also showed an association between common variation at 3q28 (TP63; rs13314271; OR=1.13, P=7.22×10−10) and lung adenocarcinoma previously only reported in Asians. These findings provide further evidence for inherited genetic susceptibility to lung cancer and its biological basis. Additionally, our analysis demonstrates that imputation can identify rare disease-causing variants having substantive effects on cancer risk from pre-existing GWAS data.
Lung cancer causes over 1 million deaths each year worldwide1. While primarily caused by tobacco smoking, studies have also implicated inherited genetic factors in its etiology; notably genome-wide association studies (GWAS) in Europeans have consistently identified polymorphic variation at 15q25.1 (CHRNA5-CHRNA3-CHRNB4), 5p15.33 (TERT-CLPTM1) and 6p21.33 (BAT3-MSH5) as determinants of lung cancer risk2-6. Additionally, susceptibility loci for lung cancer at 3q28, 6q22.2, 13q12.12, 10q25.2 and 22q12.2 in Asians have been identified through GWAS7-9.
Non-small cell lung cancer (NSCLC) is the commonest lung cancer histology, comprised primarily of adenocarcinoma (AD) and squamous cell carcinoma (SQ). These lung cancer histologies have different molecular characteristics reflective of differences in etiology and carcinogenesis10. Perhaps not surprisingly there is variability in genetic effects on lung cancer risk by histology with subtype-specific associations at 5p15.33 (TERT-CLPTM1) for AD11,12 and at 9p21 (CDKN2A/CDKN2B)13 and 12q13.33 (RAD52)14 for SQ. In addition the 6p21.33 associations are stronger for SQ than AD13.
To identify additional lung cancer susceptibility loci we conducted a meta-analysis of four lung cancer GWAS in populations of European ancestry, the MD Anderson Cancer Center (MDACC) GWAS; the Institute of Cancer Research (ICR) GWAS; the National Cancer Institute (NCI) GWAS and the International Agency for Research on Cancer (IARC) GWAS (Online Methods) that were genotyped using either Illumina HumanHap 317, 317+240S, 370Duo, 550, 610 or 1M arrays (Supplementary Table 1). After filtering the studies provided genotypes on 11,348 cases and 15,861 controls (Supplementary Table 1). Prior to undertaking meta-analysis of the GWAS, we searched for potential errors and biases in the datasets. Quantile-quantile (Q-Q) plots of genomewide association test statistics showed minimal inflation rendering substantial cryptic population substructure or differential genotype calling between cases and controls unlikely (λ=1.01 to 1.05; Supplementary Figure 1). To bring genotype data obtained from different arrays into a common platform and recover untyped genotypes, we imputed >10 million SNPs using 1000 Genomes Project data as reference. Q-Q plots for all SNPs and restricted to rare SNPs (minor allele frequency (MAF) <1%) post imputation did not show evidence of substantive over-dispersion introduced by imputation (λ=0.99-1.06 and 0.82-1.05 respectively; Supplementary Figure 1).
Pooling data from each GWAS, we derived joint odds ratios (ORs) and 95% confidence intervals (CIs) under a fixed effects model for each SNP and associated per allele P-values. To explore the variability in associations according to tumour histology we derived ORs for all lung cancer, AD and SQ.
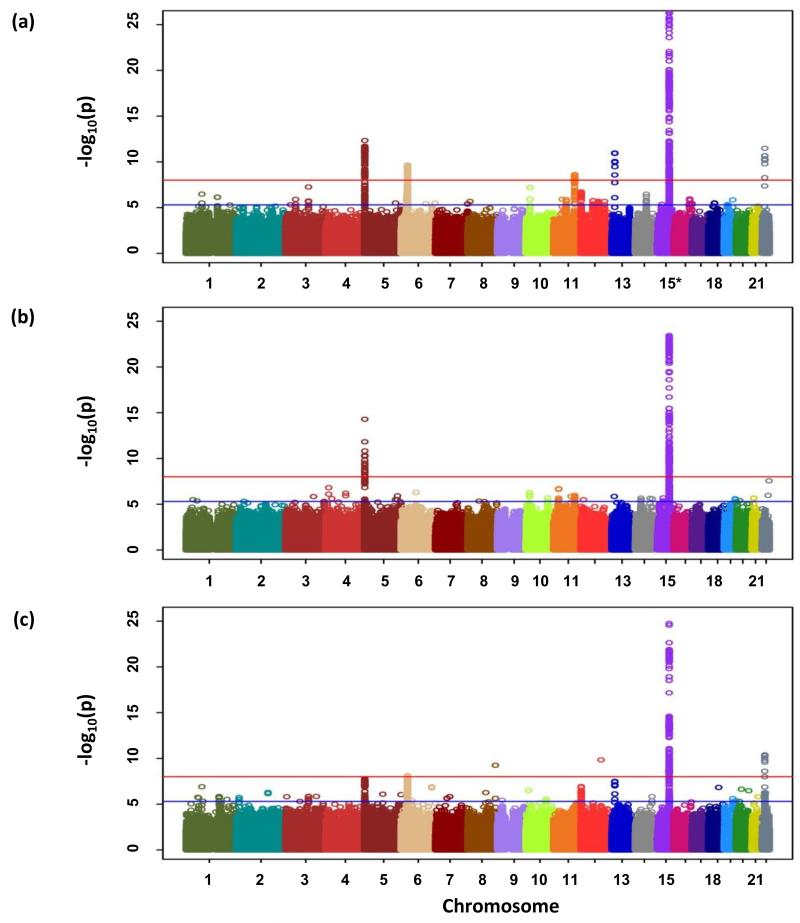
Meta-analysis identified 50 SNPs that showed evidence of an association with either lung cancer, AD or SQ (P<5.0×10−6; Figure 1) at loci not previously reported in Europeans (Figure 1). 1Mb regions encompassing these 50 SNPs were evaluated for association through in silico replication in the Harvard15 and deCODE16 series. Nine of the SNPs within these 50 regions showed support for an association (combined P-value <5.0×10−7). Genotyping of these nine SNPs was attempted in four additional series, Heidelberg-EPIC replication, ICR replication, IARC replication and Toronto replication (Supplementary Table 3 (b), Online Methods). rs185577307 could not be genotyped due to repetitive sequence. Collectively genotypes are available from 21,594 cases and 54,156 controls, providing 80% power to detect a variant with MAF of 0.01 conferring a relative risk of ≥1.5. In the combined analysis of all GWAS plus replication series, SNPs mapping to 13q13.1 (rs11571833, rs56084662), 22q12.1 (rs17879961) and 3q28 (rs13314271) showed evidence for an association, which was statistically significant after adjustment for multiple testing (i.e. P<3.0×10−9; Figure 2, Supplementary Table 2). We confirmed the high fidelity of imputation by genotyping rs11571833, rs17879961 and rs13314271 in subsets of ICR-GWAS, IARC-GWAS, NCI-GWAS and MDACC-GWAS (Supplementary Table 3, Online Methods). The NCI-GWAS comprised samples from Finland, Italy and the US. The IARC-GWAS comprised samples from 10 series from Western and Eastern Europe, and the US. While adjustment of test statistics for principle components generated on common SNPs had been applied to these GWAS, confounding of rare variants in spatially structured populations is not necessarily corrected by such methods17. We therefore investigated if country of origin had an impact on the associations at 13q13.1 and 22q12.1; the associations remained statistically highly significant (Supplementary Table 4).
Figure 1. Genome-wide P-values (−log10P, y axis) plotted against their respective chromosomal positions (x axis).
(a) All lung cancer, (b) AD and (c) SQ. Shown are the genomewide P-values (two-sided) obtained using the Cochran-Armitage trend test from analysis of 8.9 million successfully imputed autosomal SNPs in 11,348 cases and 15,861 controls from discovery phase. The red and blue horizontal lines represent the significance threshold of P=5.0×10−8 and P=5.0×10−6 respectively. Any region contains at least one association signal better than P=5.0×10−6 were selected for the in silico replication.
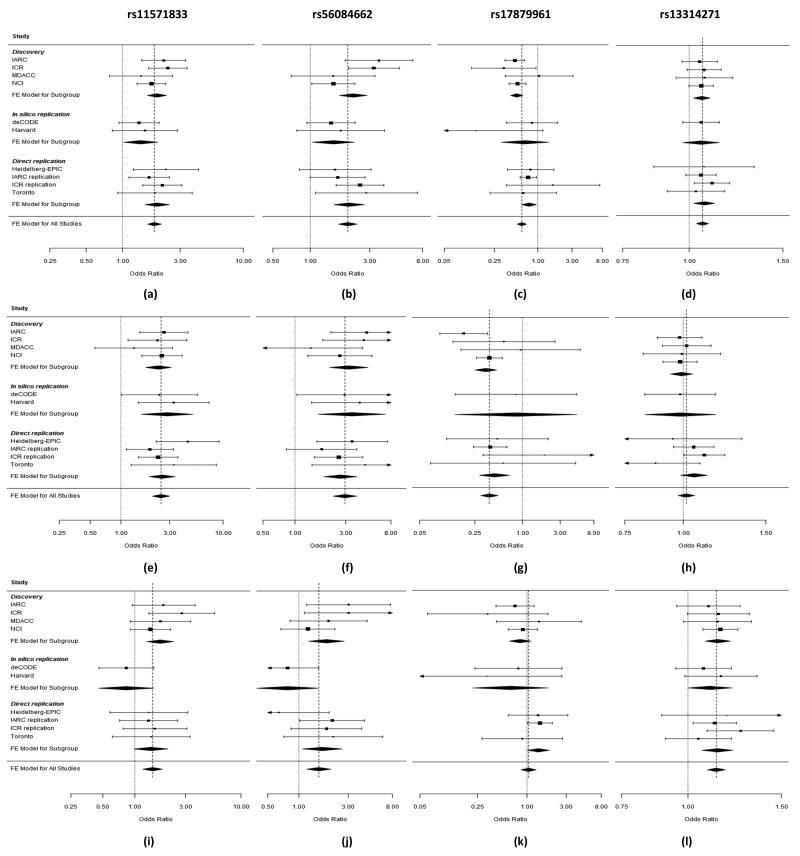
Figure 2. Plot of the odds ratios of lung cancer associated with 13q13.1 (rs11571833 and rs56084662), 22q12.1 (rs17879961) and 3q28 (rs13314271) risk loci (a-l).
All lung cancer based on 21,594 lung cancer cases and 54,156 controls (a-d), SQ based on 6,477 SQ and 53,333 controls (e-h) and AD based on 7,031 AD and 53,189 controls (i-l). Studies are weighted according to the inverse of the variance of the log of the OR calculated by unconditional logistic regression. Horizontal lines: 95% confidence intervals (95% CI). Box: OR point estimate; its area is proportional to the weight of the study. Diamond (and broken line): overall summary estimate, with confidence interval given by its width. Unbroken vertical line: at the null value (OR = 1.0).
Both rs11571833 and rs56084662 localizing to 13q13.1, near or within BRCA2, are rare SNPs (MAF<0.01), map 103kb apart (32,972,376bps, 32,869,614bps) and are moderately correlated (r2=0.45, D′=0.82, based on genotypes from Heidelberg-EPIC, IARC replication, ICR-replication and Toronto-replication series; Figure 3). rs11571833 (c.9976A>T) is responsible for BRCA2-K3326X whereas rs56084662 is located in the 3′UTR of FRY. While the association provided by rs11571833 was substantially stronger than rs56084662 in the combined analysis (OR=1.83, P=2.11×10−19 and P=1.88×10−15) conditional analysis based on directly genotyped samples in the replication series was consistent with the two SNPs tagging the same haplotype. The rs11571833 association is primarily driven by a relationship with SQ rather than AD histology (OR=2.47, P=4.74×10−20 and OR=1.47, P=4.66×10−4 respectively; Figure 2, Supplementary Table 2). A more significant role for BRCA2 in SQ etiology than in AD is reflected in the higher observed mutational frequency in respective lung cancers (~6% and 1%18,19). c.9976T has recently been shown to confer a 1.26-fold increased breast cancer risk20 and previously suggested as a risk factor for esophageal and pancreatic cancers21,22. We found no evidence for an association between c.9976T and lung cancer risk in non-smokers using directly genotyped samples (Supplementary Table 3), however these cases comprised <10% of each cohort hence our power to demonstrate a relationship was limited. Previous analysis of families carrying highly penetrant BRCA2 mutations have either found no evidence for any excess or a reduced lung cancer risk in carriers23,24. A possible explanation for these observations is that members of studied breast-ovarian cancer families tend to smoke less than the general population24.
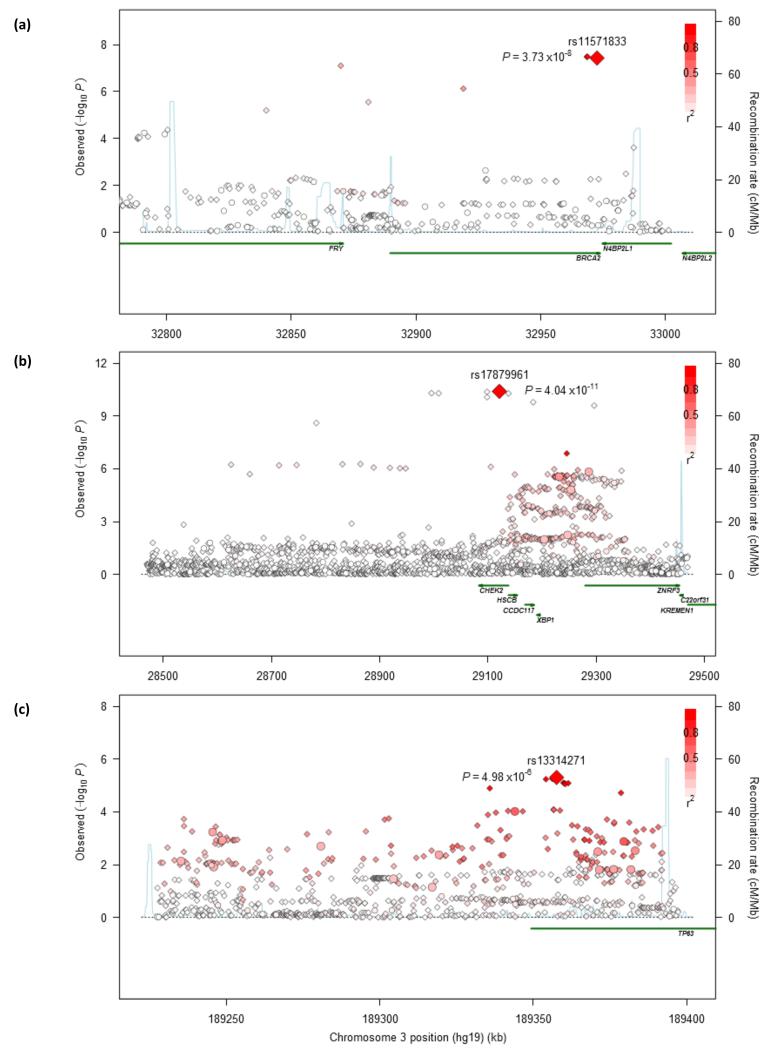
Figure 3. Regional plots of association results and recombination rates for the 13q13.1 in SQ (a), 22q12.1 in SQ (b) and 3q28 susceptibility loci in AD (c).
SQ related panels (a, b) were based on 3,275 SQ and 15,038 controls from discovery phase; and AD related panel (c) was based on 3,442 AD and 14,894 controls from discovery phase. Association results of both genotyped (circles) and imputed (diamonds) SNPs in the GWAS samples and recombination rates for each locus: For each plot, −log10P values (y axis) of the SNPs are shown according to their chromosomal positions (x axis). The top genotyped SNP in each combined analysis is a large diamond and is labeled by its rsID. The color intensity of each symbol reflects the extent of LD with the top genotyped SNP: white (r2=0) through to dark red (r2=1.0). Genetic recombination rates (cM/Mb), estimated using HapMap CEU samples, are shown with a light blue line. Physical positions are based on NCBI build 37 of the human genome. Also shown are the relative positions of genes and transcripts mapping to each region of association. Genes have been redrawn to show the relative positions; therefore, maps are not to physical scale.
The rad51-brca2 interaction is pivotal for brca2-mediated double stranded break repair (DSBR) and exon 27 of BRCA2 encodes one of the highly conserved rad51 binding domains; homozygous deletion of exon 27 in mice confers susceptibility to tumours including lung cancer25. c.9976T leads to the loss of the C-terminal domain of brca2 inviting speculation that the SNP is functional. While the deleted region is distal to the rad51 binding domain and an impact on nuclear localisation is debated26,27 the nearby mutation at BRCA2 T3387A interrupts chk2-phosphorylation and abrogates BRCA2-Chk2-Rad51 mediated recombination repair28. Alternatively, the association might be a consequence of linkage disequilibrium (LD) with another BRCA2 mutation. Studies of breast cancer families with northern European ancestry show the BRCA2 c.6275delTT and c.4889C>G mutations which are highly penetrant for breast and ovarian cancer originated on a K3326X haplotype29. To gain further insight into a probable genetic basis of the 13q13.1 lung cancer association we sequenced germline DNA from 70 lung cancer cases which carried c.9976A>T from the UK Genetic Lung Cancer Predisposition Study for c.6275delTT and c.4889C>G mutations. In none were c.6275delTT and c.4889C>G mutations identified. Similarly sequencing the coding region of BRCA2 identified no clearly pathogenic mutations amongst 13 individuals from 1958BC, 11 IARC lung cancer cases or 24 TCGA lung cancer cases carrying c.9976T. In Iceland c.9976T is not correlated with the founder BRCA2 mutation p.256_257del (999del5) which greatly increases breast and ovarian cancer risk. Paradoxically while c.9976T is a risk factor for lung cancer in this population the SNP is not associated with breast or ovarian cancer risk cancer (Supplementary Table 5). Although in vitro studies have failed to demonstrate K3326X affects DNA repair30 our findings raise the possibility K3326X may have a direct effect on lung cancer risk. Since somatic mutation of BRCA2 is not associated with K3326X carrier status 19 (Supplementary Table 6 (a)) it suggests that any impact the SNP has on lung cancer risk is mediated through alternative mechanisms.
The relationship at 22q12.1 between the SNP rs17879961 (c.470T>C) and SQ in the combined series (OR=0.38, P=1.27×10−13) validates an association previously reported31,32 (Figure 2, Supplementary Table 2, Supplementary Table 4). The frequency of rs17879961 varies significantly between populations with the MAF being ~5% in Eastern Europeans (e.g. IARC series) but almost monomorphic in most Northern Europeans. This is likely to account for a failure to demonstrate a significant relationship in the ICR, MDACC, Toronto and deCODE series which comprise largely Western European populations (Figure 2, Supplementary Table 2). rs17879961 is responsible for the I157T missense mutation in CHEK2, a cell cycle control gene encoding a pluripotent kinase that can cause arrest or apoptosis in response to DNA damage. Acquired mutation of CHEK2 is rarely seen in lung cancer and CHEK2-I57T genotype does not appear to correlate with mutation (Supplementary Table 6 (a)) raising the possibility that carrier status per se influences cancer risk. I157T lies in a functionally important domain of chek2 causing reduced or abolished binding of principal substrates. While c.470C increases breast cancer risk33 here c.470C was associated with reduced lung cancer risk. A mechanism for the paradoxical associations is not immediately apparent. CHEK2 can however have opposite effects on damaged stem cells retarding stem cell division until DNA damage is repaired, or activating apoptosis if damage cannot be repaired. Although speculative, in the presence of continued DNA damage to squamous epithelia by tobacco smoke the normal stem cell defences involving chek2 might be attenuated by a reduction in chek2 activity as a result of I151T31. Concordant with such a model is that a paradoxically increased lung cancer risk was seen in non-smokers (P=0.05), and correlated subgroups of AD and women, albeit based on small numbers (Supplementary Table 3).
The association between variation at 3q28 marked by rs13314271 and lung cancer risk was restricted to AD (OR=1.13, P=7.22×10−10Figure 2, Supplementary Table 2). rs13314271 maps within intron 1 of TP63 (Figure 3). Variation at TP63 defined by the intron 1 SNP rs4488809, which is in complete LD with rs13314271 (r2=1.00, D′=1.00) is associated with AD in Asians8. Our findings provide robust evidence for the generalisability of a relationship between 3q28 variation and AD. A weak association between rs13314271 and lung cancer risk was shown in non-smokers (P=0.03; Supplementary Table 3 (b)). TP63 is a member of the tumor suppressor TP53 gene family, which is pivotal to cellular differentiation and responsiveness to cellular stress34,35. Exposure of cells to DNA damage leads to induction of TP63 and both isoforms have the ability to transactivate TP53 target genes, hence impacting on cellular responsiveness to DNA damage36. While rs13314271 does not map to an evolutionary conserved region (ECR), rs7636839 which is correlated with rs13314271 and rs4488809 (r2=1.0) maps to an ECR and has predicted enhancer activity (Supplementary Table 6 (b)). Moreover, rs4488809 has been shown to be an eQTL for p63 in lung tissue37. Although the mechanism by which 3q28 variation affects AD development is unknown, accumulation of DNA damage and lack of response to genotoxic stress is recognized to contribute to lung carcinogenesis; hence loss of fidelity of repair as a consequence of differential TP63 expression is likely to be deleterious.
There was no association between rs11571833, rs17879961 and rs13314271 genotypes and cigarette consumption using smoking information on 43,693 Icelandic subjects (Supplementary Table 7); in contrast to the 15q25 association and risk of lung cancer.
While there is overlap distinct DNA lesions are ostensibly repaired by different DNA repair pathways and the histology specific relationships seen implicate the brca2-chek2-rad52 DSBR–homologous recombination pathways as a determinant of SQ and defective tp53/tert apoptosis-telomerase regulation as a basis of AD risk.
In conclusion, our findings provide further evidence for inherited genetic susceptibility to lung cancer and underscore the importance of searching for histology-specific risk variants. Our data also provide an important proof of principle that 1000 Genomes imputation can be used to detect novel, low frequency-large effect associations, thereby extending the utility of pre-existing GWAS data. Notably this has facilated the identification of BRCA2 c.9976T which represents by far the stongest genetic association in lung cancer reported so far. For a smoker carrying this variant (2% of the population) the risk of developing lung cancer is approximately doubled, which may have implications for identifying high risk ever-smoking subjects for lung cancer screening. Additionally, study of the effect of PARP inhibition for smokers with lung cancer carrying BRCA2 c.9976T may be warranted.
ONLINE METHODS
The study was conducted under the auspices of the Transdisciplinary Research In Cancer of the Lung (TRICL) Research Team, which is a part of the Genetic Associations and MEchanisms in ONcology (GAME-ON) consortium, and associated with the International Lung Cancer Consortium (ILCCO). Tumours from patients were classified as adenocarcinomas (AD), squamous carcinomas (SQ), large-cell carcinomas (LCC), mixed adenosquamous carcinomas (MADSQ) and other non-small cell lung cancer (NSCLC) histologies following either the International Classification of Diseases for Oncology (ICD-O) or World Health Organisation (WHO) coding. Tumours with overlapping histologies were classified as mixed.
Ethics
All participants provided informed written consent. All studies were reviewed and approved by institutional ethics review committees at the involved institutions.
Genome-wide association studies
The meta-analysis was based on data from four previously reported lung cancer GWAS of European populations: the MD Anderson Cancer Center lung cancer study (MDACC-GWAS)3; the UK lung cancer GWAS from the Institute for Cancer Research (ICR-GWAS)6; the NCI lung cancer GWAS (NCI-GWAS)13 and the IARC lung cancer GWAS (IARC-GWAS)2. In each of the studies, SNP genotyping had been performed using Illumina HumanHap 317, 317+240S, 370, 550, 610 or 1M arrays (Supplementary Table 1).
IARC-GWAS
The IARC-GWAS2 comprised 3,062 lung cancer cases and 4,455 controls derived from five case-control studies: (i) Carotene and Retinol Efficacy Trial (CARET) cohort38; (ii) The Central Europe multicenter hospital-based case-control39,40; (iii) The hospital-based case-control study from France40; (iv) The hospital based case-control lung cancer study from Estonia41,42; and (v) The population-based HUNT2/Tromsø IV lung cancer studies43. Patient and control DNAs were derived from EDTA-venous blood samples. The lung cancer patients were classified according to ICD-O-3; SQ: 8070/3, 8071/3, 8072/3, 8074/3; AD: 8140/3, 8250/3, 8260/3, 8310/3, 8480/3, 8560/3, 8251/3, 8490/3, 8570/3, 8574/3; with tumours with overlapping histologies classified as mixed. After applying standardized quality control procedures 2,533 cases and 3,791 controls were included in the current analysis (Supplementary Table 1).
NCI-GWAS
Details of the NCI-GWAS have been previously reported. Briefly, the study comprised samples from four series: (i) The Environment and Genetics in Lung cancer Etiology (EAGLE), a population-based case-control study of 2,100 lung cancer cases and 2,120 healthy controls enrolled in Italy between 2002 and 200544. Cancers were classified according to the ICD-O coding for histology and grading. Histology of ~10% of tumours was confirmed by an independent pathologist from NCI. (ii) The Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study (ATBC), a randomized primary prevention trial of 29,133 male smokers enrolled in Finland between 1985 and 199345; ICD-O-2 and ICD-O-3 was used to classify tumours. Cases diagnosed between 1985 and 1999 had histology reviewed by at least one pathologist. After 1999, histological coding (ICD-O-2 and ICD-O-3) was derived from the Finnish Cancer Registry. (iii) The Prostate, Lung, Colon, Ovary Screening Trial (PLCO), a randomized trial of 150,000 individuals enrolled in ten U.S. study centers between 1992 and 200146; ICD-O-2 was used to classify tumors and quality assurance measures included reabstraction of 50 lung cancer diagnoses per year; (iv) The Cancer Prevention Study II Nutrition Cohort (CPS-II), a cohort study of approximately 184,000 individuals enrolled by the American Cancer Society between 1992 and 1993 in 21 U.S. states of which 109,379 provided a blood (36%) or buccal (64%) sample between 1998 and 200312,47. Tumour histology was abstracted from Certified Tumor Registrars and coded using WHO ICD-O-2 and ICD-O-3. Quality assurance was done by re-abstracting 10% of all cancer diagnoses per year. After initial data control, the NCI-GWAS included 5,739 cases and 5,848 controls; however, an additional 26 cases and 112 controls were excluded due to changes in case status and further quality control filtering. The current meta-analysis included 5,713 lung cancer cases and 5,736 controls from the NCI-GWAS (Supplementary Table 1).
ICR-GWAS
This comprised 1,952 cases (1,166 male; mean age at diagnosis 57 years, SD 6) with pathologically confirmed lung cancer ascertained through the Genetic Lung Cancer Predisposition Study (GELCAPS) conducted between March 1999 and July 200448. All cases were British residents and self-reported to be of European Ancestry. To ensure that data and samples were collected from bona fide lung cancer cases and avoid issues of bias from survivorship only incident cases with histologically or cytologically (only if not AD) confirmed primary disease were ascertained. Tumours from patients were classified according to ICD-O3; Specifically, SQ: 8070/3, 8071/3, 8072/3, 8074/3; AD: 8140/3, 8250/3, 8260/3, 8310/3, 8480/3, 8560/3, 8251/3, 8490/3, 8570/3, 8574/3; with tumours with overlapping histologies classified as mixed. Patient DNA was derived from EDTA-venous blood samples using conventional methodologies. Genotype frequencies were compared with publicly accessible data generated by the UK Wellcome Trust Case-Control Consortium 2 (WTCCC2) study49 of individuals from the 1958 British Birth Cohort (58BC) and blood service typed using Illumina Human1.2M-Duo Custom_v1 Array BeadChips.
MDACC-GWAS
Cases and controls were ascertained from a case-control study at the U.T. M.D. Anderson Cancer Center conducted between 1997 and 20073. Cases were newly diagnosed, patients with histologically-confirmed lung cancer presenting at M.D. Anderson Cancer and who had not previously received treatment other than surgery. Clinical and pathological data were abstracted from patient medical records and lung cancer histology was coded according to the major histological groups. Specifically, as per ICD-O-2 these groups were, SQ: 8070/3, AD: 8140/3, 8250/3, 8260/3, 8310/3, 8480/3, 8251/3 and 8490/3. Only patients with predominantly or wholly AD or SQ cancers were included; those with mixed histology or unspecified lung cancers, were excluded from the study. Controls were healthy individuals seen for routine care at Kelsey-Seybold Clinics, in the Houston Metropolitan area. Controls were frequency matched to cases according to smoking behaviour, age in 5-year categories, ethnicity, and sex. Former smoking controls were further frequency matched to former smoking cases according to the number of years since smoking cessation (in 5-year categories). After applying quality control data were available on 1,150 cases and 1,134 controls.
Quality control of GWAS datasets
Standard quality control was performed on all scans excluding individuals with low call rate (<90%) and extremely high or low heterozygosity (i.e. P<1.0×10−4), as well as all individuals evaluated to be of non-European ancestry (using the HapMap version 2 CEU, JPT/CHB and YRI populations as a reference; Supplementary Table 1). For apparent first-degree relative pairs, we removed the control from a case-control pair; otherwise, we excluded the individual with the lower call rate.
Replication series
To validate promising associations from meta-analysis were made use of in silico data and imputed genotypes from Harvard and deCODE GWAS datasets together with data from direct genotyping Heidelberg-EPIC, ICR, IARC and Toronto replication series.
Harvard For the Harvard Lung Cancer Susceptibility Study, details of participant recruitment have been described previously50. Replication was based on data derived from 1,000 cases and 1,000 controls genotyped using Illumina Humanhap610-Quad arrays. Cases were patients aged >18 years, with newly diagnosed, histologically confirmed primary NSCLC. Controls were healthy non-blood-related family members and friends of patients with cancer or with cardiothoracic conditions undergoing surgery. The histological classification of lung tumors was performed by two staff pulmonary pathologists at the Massachusetts General Hospital according to ICD-O-3; Specifically, AD: 8140/3, 8250/3, 8260/3, 8310/3, 8480/3 8560/3; LCC: 8012/3, 8031/3; SQ: 8070/3, 8071/3, 8072/3, 8074/3; and other NSCLC: 8010/3, 8020/3, 8021/3, 8032/3, 8230/3. Unqualified samples were excluded if they fit the following QC criteria: (i) overall genotype completion rates <95%; (ii) gender discrepancies; (iii) unexpected duplicates or probable relatives (based on pairwise identity by state value, PI_HAT in PLINK>0.185); (iv) heterozygosity rates >6 times the standard deviation from the mean; or (v) individuals evaluated to be of non-Caucasians (using the HapMap release 23 including JPT, CEPH, CEU and YRI populations as a reference). Unqualified SNPs were excluded when they fit the following QC criteria: (i) SNPs were not mapped on autosomes; (ii) SNPs had a call rate <95% in all GWAS samples; (iii) SNPs had MAF <0.01; or (iv) the genotype distributions of SNPs deviated from those expected by Hardy-Weinberg equilibrium (P<1.0×10−6). After applying these pre-specified quality controls genotype data were available for 984 cases and 970 controls.
deCODE The Icelandic lung cancer study has been described previously4. The primary source of information on the Icelandic lung cancer cases is the Icelandic Cancer Registry (ICaR) which covers the entire population of Iceland (http://www.cancerregistry.is). The sources of data in the ICaR are all pathology and hematology laboratories, all hospital departments and health care facilities in the country. ICaR registration is based on the ICD system and included information on histology (Systemized Nomenclature of Medicine, SNOMED). The ICaR registration also uses the ICD-O system which takes histology diagnosis into account. Over 94% of diagnoses in the ICaR have histological confirmation. According to the ICaR, Briefly, according to the ICaR a total of 4,252 lung cancer patients were diagnosed from January 1, 1955, to December 31, 2010. Recruitment of both prevalent and incident cases was initiated in 1998, the recruitment is ongoing and DNA samples from lung cancer cases are subjected to whole-genome genotyping as they are collected. The controls used in this study consisted of individuals from other GWASs, age and sex-matched to cases with no individual disease group accounting for >10% of all controls. Samples were assayed with the Illumina HumanHap300, HumanCNV370, HumanHap610, HumanHap1M, HumanHap660, Omni-1, Omni 2.5 or Omni Express bead chips at deCODE genetics. SNPs were excluded if they had (i) a yield <95%, (ii) MAF <1% in the population, (iii) deviation from Hardy-Weinberg equilibrium (HWE; P<10−6), (iv) inheritance error rate (>0.001) or (v) if there was a substantial difference in allele frequency between chip types (in which case the SNP was removed from a single chip type if that resolved the difference, but if it did not then the SNP was removed from all chip types). All samples with a call rate of <97% were removed from the analysis. The Icelandic sample set is drawn from the Icelandic population, a small homogeneous founder population with almost no detectable population substructure. Thus there was no need adjust for such substructure in the association analysis. In addition, the comprehensive Icelandic genealogy database allowed us to exclude individuals not of Icelandic origin from the analysis. SNP genotypes were phased using the method of long range phasing51; for the HumanHap series of chips, 304,937 SNPs were used for long range phasing, whereas for the Omni series of chips 564,196 SNPs were used. An initial imputation step was carried out on each chip series separately to create a single harmonized, long-range phased genotype dataset consisting of 707,525 SNPs for 95,085 Icelandic individuals. Two sets of genotypes were imputed into this dataset with methods previously described 52: (i) genotypes for about 38 million variants using the 1000 genomes phase I integrated variant set (v3) as training set, and (ii) genotypes for about 34 million variants identified in 2,230 whole genome sequenced Icelanders. The first set of imputed genotypes was used for replicating the association with variants in the 5p15.33, 9p21 and 12q13.33 regions, using IMPUTE (v2.1.1)53 to perform the cases-control analysis. The second set was used when testing the relationship between K3326X, 999del5 genotypes and risk of different cancer types in the Icelandic population using a method that allowed including individuals that had not been chip typed, but for which genotype probabilities were imputed using methods of familial imputation51.
Heidelberg-EPIC comprised 1,253 EPIC-Heidelberg controls and 1,362 lung cancer cases from the Heidelberg lung cancer study recruited between 1994-1998 and 1996-2007, respectively. Details of the EPIC-Heidelberg controls and the Heidelberg lung cancer study have been previously described 54,55. All subjects were aged 18 years or older and information on lifestyle risk factors, medical and family history was collected through interviews based on standardised questionnaires. The EPIC Lung and the Heidelberg-EPIC studies were performed independently with no sample overlap with those analysed as part of the IARC-replication series. Histological classification of tumours was obtained from pathology reports, where it was recorded by staff pulmonary pathologist according to WHO. Blood samples from patients with malignant lung disease categorized as follows were included: AD, SCLC, NSCLC, LCC, carcinoid, mixed lung tumors, mixed without SCLC. The above EPIC Lung and the Heidelberg-EPIC studies were performed independently with no sample overlap. Genotypes for SNPs showed no significant departure from Hardy-Weinberg equilibrium with the exception of rs13314271 in cases.
ICR-replication comprised 2,448 cases (1,664 male; mean age at diagnosis 71.8 years, SD 6.7) with pathologically confirmed lung cancer ascertained through GELCAPS48 and 2,989 controls (1,469 male, mean age at sampling 60.6 years, SD 12.0) collected through the National Study of Colorectal Cancer Genetics56 with no personal history of malignancy. Cases were sub-classified into histological subtypes based on ICD coding as described above (Study description: ICR-GWAS). Both cases and controls were British residents and had self-reported European Ancestry. The genotype distributions of genotypes for each of the SNPs typed in replication showed no significant departure from HWE.
IARC-replication comprised three studies: (i) EPIC Lung2,57, a nested case control study performed within the EPIC (European Prospective Investigation into Cancer and Nutrition) prospective cohort totalling 1,119 lung cancer cases and 2,546 controls (matched 1-2 to cases for age, sex, centre, and time of recruitment), selected from eight of the 10 countries participating in EPIC (Sweden, Netherlands, UK, France, Germany, Spain, Italy and Norway); (ii) Szczecin case-control study32 a consecutive series of 849 incident lung cancer cases ascertained from the outpatient oncology clinic in the regional hospital of Szczecin between 2004-2007. The 1,072 controls were individuals without a diagnosed cancer or family history of cancer matched to cases by sex, age and region recruited via general medical practitioners; (iii) Moscow L2, 1,081 newly diagnosed lung cancer cases and 2,119 controls recruited from three hospitals within the Moscow area of Russia between 2007 and 2011. Information on lifestyle risk factors, medical and family history was collected from subjects by interview using a standard questionnaire. Cases were sub-classified into histological subtypes based on ICD-O3 coding as described above (Study description: IARC-GWAS). The genotype distributions of genotypes for each of the SNPs typed in replication showed no departure from HWE in each country/study series.
The Toronto study was conducted in the Great Toronto Area from 2008 to 2013. Lung cancer cases were recruited at the hospitals in the network of University of Toronto. Controls were randomly selected from individuals registered in the family medicine clinics databases, frequency matched with cases on age and sex. All subjects were interviewed and information on lifestyle risk factors, occupational history, medical and family history collected using a standard questionnaire. Tumours were centrally reviewed by the reference pathologist (a member of the IASLC committee) and a second pathologist in the University Health Network. If reviews conflicted, consensus was arrived at following discussion. Coding of histology was based on 2001 WHO/IASLC. After applying standardized quality control procedures and restricting to participants with self-reported European ancestry, data and samples were available on 1,084 cases and 966 controls. The genotype distributions of genotypes for each of the SNPs typed in replication showed no significant departure from HWE.
Replication genotyping
Genotyping of rs1519542, rs13314271, rs55731496, rs149423192, rs4592420, rs11571833, rs56084662 and rs17879961 was performed using either competitive allele-specific PCR KASPar chemistry (LGC, Hertfordshire, UK; UK replication series), Sequenom (Sequenom, Inc. San Diego, US; Toronto replication, Heidelberg-EPIC replication [rs1519542, rs55731496, rs149423192, rs4592420, rs11571833, rs56084662, rs17879961],) or Taqman (Carlsbad, CA; IARC-replication series, Heidelberg-EPIC replication [rs13314271]). All primers, probes and conditions used are available on request. Call rates for SNP genotypes were >95% in each of the replication series.
To ensure quality of genotyping in all assays, at least two negative controls and 1-10% duplicates (showing a concordance >99%) were genotyped at each centre. To exclude technical artefact in genotyping, at the ICR and IARC we performed cross-platform validation of 96 samples and sequenced a set of 96 randomly selected samples from each case and control series to confirm genotyping accuracy. Assays were found to be performing robustly; concordance >99%.
Statistical and bioinformatic analysis
Data were imputed for all scans for over 10 million SNPs using data from the 1000 Genomes Project (Phase 1 integrated release 3, March 2012) as reference, using IMPUTE2 v2.1.153, MaCH58 v1.0 or minimac (version 2012.10.3)59 software (Supplementary Table 1). Genotypes were aligned to the positive strand in both imputation and genotyping. Imputation was conducted separately for each scan in which prior to imputation each GWAS dataset was pruned to a common set of SNPs between cases and controls. As previously advocated we set thresholds for imputation quality to retain both potential common and rare variants for validation13,60. Specifically, poorly imputed SNPs defined by an RSQR<0.30 with MaCH or an information measure Is<0.40 with IMPUTE2 were excluded from the analyses. Tests of association between imputed SNPs and lung cancer was performed under a probabilistic dosage model in SNPTEST v2.561, ProbABEL62, MaCH2dat v.12458 or glm function in R. Principle components generated using common SNPs were included in the analysis in order to limit the effects of cryptic population stratification that might cause inflation of test statistics. The association between each SNP and lung cancer risk was assessed by the Cochran-Armitage trend test. The adequacy of the case-control matching and possibility of differential genotyping of cases and controls were formally evaluated using quantile-quantile (Q-Q) plots of test statistics. Meta-analysis was undertaken using inverse-variance approaches. The inflation factor λ was based on the 90% least significant directly typed SNPs63. Odds ratios (ORs) and associated 95% confidence intervals (CIs) were calculated by unconditional logistic regression using R (v2.6), Stata v.10 (State College, Texas, US) and PLINK64 (v1.06) software. Cochran’s Q-statistic to test for heterogeneity and the I2 statistic to quantify the proportion of the total variation due to heterogeneity were calculated65. I2 values ≥75% are considered characteristic of large heterogeneity65. Additionally analyses stratified by histology, sex, age and smoking status (current, former, never) were performed. All statistical tests are two-sided.
The fidelity of imputation as assessed by the concordance between imputed and directly genotyped SNPs was examined in a subset of samples from the UK-GWAS, MDACC-GWAS, IARC-GWAS and NCI-GWAS discovery series (Supplementary Table 3).
LD metrics were calculated in PLINK using 1000 genomes data and plotted using SNAP66. LD blocks were defined on the basis of HapMap recombination rate (cM/Mb) as defined using the Oxford recombination hotspots and on the basis of distribution of confidence intervals defined by Gabriel et al.67
Relationship between genotypes and smoking
To examine the relationship between rs11571833 (BRCA2 K3326X), rs17879961 (CHEK2 I157T) and rs13314271 (TP63) genotype and cigarette consumption (cigarette per day)68 we made use of data on using 43,693 Icelandic subjects (including 34,850 chip typed individuals).
Sequence analysis of BRCA2 in constitutional DNA
At the ICR targeted sequencing for c.6275delTT and c.4889C>G BRCA2 mutations was performed by Sanger implemented on an ABI3700 analyzer (Applied Biosystems; primer sequences and conditions available on request). Mutational analysis of the complete coding region of BRCA2 was based on exome sequencing data generated using Illumina TruSeq capture technology (Illumina, Inc, San Diego, CA 92122 USA). Analysis of Illumina HiSeq2000 (Illumina, Inc, San Diego, USA) sequence data from was performed using an in-house pipeline based on the GATK tool kit.
At IARC Qiagen Generead (SABiosciences/Qiagen Hilde, Germany) was used to amplify the coding region of BRCA2 in rs11571833 heterozygotes. Following library preparation (New England Biolabs, Ipswich, MA, USA) sequencing was performed using an IonTorrent PGM desktop sequencer (Life Technologies, Guilford, San Francisco, CA). Genotypes were called using Ionsuite software. Sequence changes were referenced to Leiden Open Variation Database (LOVD2) and BReast CAncer IARC databases.
Analysis of TCGA data
The exomes of 243 LUSC and 338 LUAD TCGA individuals (Project Number #3230) were analyzed at IARC using an in-house pipeline based on the GATK tool set. Variant calls were annotated using ANNOVAR making use of use the NHLBI Exome Sequencing Project and 1000 Genomes data.
Copy number variation
This was assessed from Human SNP Array 6.0 data. We retrieved level 3 TCGA data comprising normalized log2 ratios of the fluorescence intensities between the target sample and a reference sample. We included in our analysis only tumour-normal paired data. We considered a log2 ratio <−0.5 as reflecting loss, and a log2 ratio >0.5 reflecting gain. Annotation was performed adding the genes contained in each of the remaining segments using EnsEMBL databases.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank all individuals who participated in this study. Additionally we are grateful to patients’ clinicians and allied health care professions. We thank Zhuo Chen and Kevin Boyle for sample handling and data management of the Toronto study; additionally, Laura Admas and Li Rita Zhang for the field recruitment. We thank Li Su, Yang Zhao, Geoffrey Liu, John Wain, Rebecca Heist and Kofi Asomaning. We thank G. Thomas and Synergy Lyon Cancer (Lyon France) for High Performance Computing support and Dr J. Olivier and A. Chabrier for IARC’s PGM Ion torrent sequencing optimisation and Taqman genotyping, respectively. We thank David Goldgar for sharing information from CIMBA on sequence variation in BRCA2 from familial breast cancer analysis. We gratefully acknowledge the Icelandic Cancer Registry (www.krabbameinsskra.is) for assistance in the ascertainment of the Icelandic lung cancer patients. The ICR study made use of genotyping data from the WTCCC2; a full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. We acknowledge The Cancer Genome Atlas (TCGA) for their contribution of lung cancer genomic data to this study (TCGA Project Number 3230). Finally we acknowledge support from the NIHR biomedical research centre at the Royal Marsden Hospital. This study was supported by The National Institute of Health (NIH) (U19CA148127, R01CA055769, 5R01CA127219, 5R01CA133996, 5R01CA121197). At ICR: Cancer Research UK (C1298/A8780, C1298/A8362) NCRN, HEAL and Sanofi-Aventis, National Health Service funding to the Royal Marsden/Institute of Cancer Research; National Institute for Health Research Biomedical Research Centre. Sir John Fisher Foundation PhD studenship to BK. NIH GM103534 and the Institute for Quantitative Biomedical Sciences at Dartmouth to CA. At Toronto: The Canadian Cancer Society Research Institute (020214), Ontario Institute of Cancer and Cancer Care Ontario Chair Award to R.H and G.L, and the Alan Brown Chair and Lusi Wong Programs at the Princess Margaret Hospital Foundation. At Heidelberg: Deutsche Krebshilfe (70-2387; 70-2919) and German Federal Ministry of Education and Research (EPIC-Heidelberg). At IARC: Institut National du Cancer, France, European Community (LSHG-CT-2005-512113), Norwegian Cancer Association, Functional Genomics Programme of Research Council of Norway, the European Regional Development Fund and State Budget of the Czech Republic (RECAMO, CZ.1.05/2.1.00/03.0101), NIH (R01-CA111703 and UO1-CA63673) Fred Hutchinson Cancer Research Center, US National Cancer Institute (NCI) (R01 CA092039), FP7 grant (REGPOT 245536), the Estonian Government (SF0180142s08), RDF in the frame of Centre of Excellence in Genomics and Estoinian Research Infrastructure’s Roadmap and by University of Tartu (SP1GVARENG) and an IARC Postdoctoral Fellowship (MNT). NCI: Intramural Research Program of NIH, NCI, U.S. Public Health Service contracts NCI (N01-CN-45165, N01-RC-45035, N01-RC-37004, NO1-CN-25514, NO1-CN-25515, NO1-CN-25512, NO1-CN-25513, NO1-CN-25516, NO1-CN-25511, NO1-CN-25524, NO1-CN-25518, NO1-CN-75022, NO1-CN-25476, NO1-CN-25404), The American Cancer Society, The NIH Genes, Environment and Health Initiative partly by HG-06-033-NCI-01 and RO1HL091172-01, genotyping at the Johns Hopkins University Center for Inherited Disease Research (U01HG004438, NIH HHSN268200782096C) and study coordination at the GENEVA Coordination Center (U01 HG004446). NIH grants (P50 CA70907, R01CA121197, RO1 CA127219, U19 CA148127, RO1 CA55769) and CPRIT grant (RP100443). Genotyping was provided by the Center for Inherited Disease Research (CIDR). At Harvard: NIH (CA074386, CA092824, CA090578). The Icelandic study: partly by NIH DA17932.
URLs
URLs
The R suite can be found at http://www.r-project.org/
1000Genomes: http://www.1000genomes.org/
SNAP: http://www.broadinstitute.org/mpg/snap/
IMPUTE2: http://mathgen.stats.ox.ac.uk/impute/impute_v2.html
MACH: http://www.sph.umich.edu/csg/abecasis/MACH/
Minimac: http://genome.sph.umich.edu/wiki/Minimac
SNPTEST: https://mathgen.stats.ox.ac.uk/genetics_software/snptest/snptest.html
ProbABEL: http://www.genabel.org/packages/ProbABEL
mach2dat: http://genome.sph.umich.edu/wiki/Mach2dat:_Association_with_MACH_output
Wellcome Trust Case Control Consortium: www.wtccc.org.uk
RegulomeDB: http://regulome.stanford.edu
HaploReg v2: http://www.broadinstitute.org/mammals/haploreg/haploreg.php
Transdisciplinary Research In Cancer of the Lung (TRICL): http://u19tricl.org/
Genetic Associations and MEchanisms in ONcology (GAME-ON) consortium: http://epi.grants.cancer.gov/gameon/
International Lung cancer Consortium (ILCO): http://ilcco.iarc.fr
Icelandic Cancer Registry: www.krabbameinsskra.is
Genome Analysis Toolkit (GATK): http://www.broadinstitute.org/gatk/
The Cancer Genome Atlas (TCGA): http://cancergenome.nih.gov/
Leiden Open Variation Databasehttp (LOVD): //chromium.liacs.nl/LOVD2/
BReast CAncer IARC database: http://brca.iarc.fr/
Footnotes
COMPETING INTERESTS STATEMENT: The authors declare no competing issues.
REFERENCES
- 1.Ferlay J, et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Hung RJ, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–7. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 3.Amos CI, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–22. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thorgeirsson TE, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–42. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKay JD, et al. Lung cancer susceptibility locus at 5p15.33. Nat Genet. 2008;40:1404–6. doi: 10.1038/ng.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet. 2008;40:1407–9. doi: 10.1038/ng.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Z, et al. A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12.2 in Han Chinese. Nat Genet. 2011;43:792–6. doi: 10.1038/ng.875. [DOI] [PubMed] [Google Scholar]
- 8.Miki D, et al. Variation in TP63 is associated with lung adenocarcinoma susceptibility in Japanese and Korean populations. Nat Genet. 2010;42:893–6. doi: 10.1038/ng.667. [DOI] [PubMed] [Google Scholar]
- 9.Lan Q, et al. Genome-wide association analysis identifies new lung cancer susceptibility loci in never-smoking women in Asia. Nat Genet. 2012;44:1330–5. doi: 10.1038/ng.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Travis WD, et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society: international multidisciplinary classification of lung adenocarcinoma: executive summary. Proc Am Thorac Soc. 2011;8:381–5. doi: 10.1513/pats.201107-042ST. [DOI] [PubMed] [Google Scholar]
- 11.Broderick P, et al. Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer Res. 2009;69:6633–41. doi: 10.1158/0008-5472.CAN-09-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Landi MT, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85:679–91. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Timofeeva MN, et al. Influence of common genetic variation on lung cancer risk: meta-analysis of 14 900 cases and 29 485 controls. Hum Mol Genet. 2012;21:4980–95. doi: 10.1093/hmg/dds334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi J, et al. Inherited variation at chromosome 12p13.33, including RAD52, influences the risk of squamous cell lung carcinoma. Cancer Discov. 2012;2:131–9. doi: 10.1158/2159-8290.CD-11-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang YT, et al. Cigarette smoking increases copy number alterations in nonsmall-cell lung cancer. Proc Natl Acad Sci U S A. 2011;108:16345–50. doi: 10.1073/pnas.1102769108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rafnar T, et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat Genet. 2009;41:221–7. doi: 10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathieson I, McVean G. Differential confounding of rare and common variants in spatially structured populations. Nat Genet. 2012;44:243–6. doi: 10.1038/ng.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imielinski M, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–20. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–25. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michailidou K, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45:353–61. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akbari MR, et al. Germline BRCA2 mutations and the risk of esophageal squamous cell carcinoma. Oncogene. 2008;27:1290–6. doi: 10.1038/sj.onc.1210739. [DOI] [PubMed] [Google Scholar]
- 22.Martin ST, et al. Increased prevalence of the BRCA2 polymorphic stop codon K3326X among individuals with familial pancreatic cancer. Oncogene. 2005;24:3652–6. doi: 10.1038/sj.onc.1208411. [DOI] [PubMed] [Google Scholar]
- 23.Cancer risks in BRCA2 mutation carriers. The Breast Cancer Linkage Consortium. J Natl Cancer Inst. 1999;91:1310–6. doi: 10.1093/jnci/91.15.1310. [DOI] [PubMed] [Google Scholar]
- 24.van Asperen CJ, et al. Cancer risks in BRCA2 families: estimates for sites other than breast and ovary. J Med Genet. 2005;42:711–9. doi: 10.1136/jmg.2004.028829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McAllister KA, et al. Cancer susceptibility of mice with a homozygous deletion in the COOH-terminal domain of the Brca2 gene. Cancer Res. 2002;62:990–4. [PubMed] [Google Scholar]
- 26.Spain BH, Larson CJ, Shihabuddin LS, Gage FH, Verma IM. Truncated BRCA2 is cytoplasmic: implications for cancer-linked mutations. Proc Natl Acad Sci U S A. 1999;96:13920–5. doi: 10.1073/pnas.96.24.13920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yano K, et al. Nuclear localization signals of the BRCA2 protein. Biochem Biophys Res Commun. 2000;270:171–5. doi: 10.1006/bbrc.2000.2392. [DOI] [PubMed] [Google Scholar]
- 28.Bahassi EM, et al. The checkpoint kinases Chk1 and Chk2 regulate the functional associations between hBRCA2 and Rad51 in response to DNA damage. Oncogene. 2008;27:3977–85. doi: 10.1038/onc.2008.17. [DOI] [PubMed] [Google Scholar]
- 29.Mazoyer S, et al. A polymorphic stop codon in BRCA2. Nat Genet. 1996;14:253–4. doi: 10.1038/ng1196-253. [DOI] [PubMed] [Google Scholar]
- 30.Wu K, et al. Functional evaluation and cancer risk assessment of BRCA2 unclassified variants. Cancer Res. 2005;65:417–26. [PubMed] [Google Scholar]
- 31.Brennan P, et al. Uncommon CHEK2 mis-sense variant and reduced risk of tobacco-related cancers: case control study. Hum Mol Genet. 2007;16:1794–801. doi: 10.1093/hmg/ddm127. [DOI] [PubMed] [Google Scholar]
- 32.Cybulski C, et al. Constitutional CHEK2 mutations are associated with a decreased risk of lung and laryngeal cancers. Carcinogenesis. 2008;29:762–5. doi: 10.1093/carcin/bgn044. [DOI] [PubMed] [Google Scholar]
- 33.Han FF, Guo CL, Liu LH. The effect of CHEK2 variant I157T on cancer susceptibility: evidence from a meta-analysis. DNA Cell Biol. 2013;32:329–35. doi: 10.1089/dna.2013.1970. [DOI] [PubMed] [Google Scholar]
- 34.Flores ER. The roles of p63 in cancer. Cell Cycle. 2007;6:300–4. doi: 10.4161/cc.6.3.3793. [DOI] [PubMed] [Google Scholar]
- 35.Katoh I, Aisaki KI, Kurata SI, Ikawa S, Ikawa Y. p51A (TAp63gamma), a p53 homolog, accumulates in response to DNA damage for cell regulation. Oncogene. 2000;19:3126–30. doi: 10.1038/sj.onc.1203644. [DOI] [PubMed] [Google Scholar]
- 36.Petitjean A, et al. Properties of the six isoforms of p63: p53-like regulation in response to genotoxic stress and cross talk with DeltaNp73. Carcinogenesis. 2008;29:273–81. doi: 10.1093/carcin/bgm258. [DOI] [PubMed] [Google Scholar]
- 37.Hao K, et al. Lung eQTLs to help reveal the molecular underpinnings of asthma. PLoS Genet. 2012;8:e1003029. doi: 10.1371/journal.pgen.1003029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Omenn GS, et al. The beta-carotene and retinol efficacy trial (CARET) for chemoprevention of lung cancer in high risk populations: smokers and asbestos-exposed workers. Cancer Res. 1994;54:2038s–2043s. [PubMed] [Google Scholar]
- 39.Scelo G, et al. Occupational exposure to vinyl chloride, acrylonitrile and styrene and lung cancer risk (europe) Cancer Causes Control. 2004;15:445–52. doi: 10.1023/B:CACO.0000036444.11655.be. [DOI] [PubMed] [Google Scholar]
- 40.Feyler A, et al. Point: myeloperoxidase −463G --> a polymorphism and lung cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:1550–4. [PubMed] [Google Scholar]
- 41.Nelis M, et al. Genetic structure of Europeans: a view from the North-East. PLoS One. 2009;4:e5472. doi: 10.1371/journal.pone.0005472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valk K, et al. Gene expression profiles of non-small cell lung cancer: survival prediction and new biomarkers. Oncology. 2010;79:283–92. doi: 10.1159/000322116. [DOI] [PubMed] [Google Scholar]
- 43.Holmen J, et al. The nord-Trondelag Health Study 1995-97 (HUNT2): objectives, contents, methods and participation. Norsk Epidemiologi. 2003;13:1932. [Google Scholar]
- 44.Landi MT, et al. Environment And Genetics in Lung cancer Etiology (EAGLE) study: an integrative population-based case-control study of lung cancer. BMC Public Health. 2008;8:203. doi: 10.1186/1471-2458-8-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.The ATBC Cancer Prevention Study Group The alpha-tocopherol, beta-carotene lung cancer prevention study: design, methods, participant characteristics, and compliance. Ann Epidemiol. 1994;4:1–10. doi: 10.1016/1047-2797(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 46.Hayes RB, et al. Methods for etiologic and early marker investigations in the PLCO trial. Mutat Res. 2005;592:147–54. doi: 10.1016/j.mrfmmm.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 47.Calle EE, et al. The American Cancer Society Cancer Prevention Study II Nutrition Cohort: rationale, study design, and baseline characteristics. Cancer. 2002;94:2490–501. doi: 10.1002/cncr.101970. [DOI] [PubMed] [Google Scholar]
- 48.Eisen T, Matakidou A, Houlston R. Identification of low penetrance alleles for lung cancer: the GEnetic Lung CAncer Predisposition Study (GELCAPS) BMC Cancer. 2008;8:244. doi: 10.1186/1471-2407-8-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Su L, et al. Genotypes and haplotypes of matrix metalloproteinase 1, 3 and 12 genes and the risk of lung cancer. Carcinogenesis. 2006;27:1024–9. doi: 10.1093/carcin/bgi283. [DOI] [PubMed] [Google Scholar]
- 51.Kong A, et al. Parental origin of sequence variants associated with complex diseases. Nature. 2009;462:868–74. doi: 10.1038/nature08625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Styrkarsdottir U, et al. Nonsense mutation in the LGR4 gene is associated with several human diseases and other traits. Nature. 2013;497:517–20. doi: 10.1038/nature12124. [DOI] [PubMed] [Google Scholar]
- 53.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boeing H, Wahrendorf J, Becker N. EPIC-Germany--A source for studies into diet and risk of chronic diseases. European Investigation into Cancer and Nutrition. Ann Nutr Metab. 1999;43:195–204. doi: 10.1159/000012786. [DOI] [PubMed] [Google Scholar]
- 55.Dally H, et al. The CYP3A4*1B allele increases risk for small cell lung cancer: effect of gender and smoking dose. Pharmacogenetics. 2003;13:607–18. doi: 10.1097/00008571-200310000-00004. [DOI] [PubMed] [Google Scholar]
- 56.Penegar S, et al. National study of colorectal cancer genetics. Br J Cancer. 2007;97:1305–9. doi: 10.1038/sj.bjc.6603997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Timofeeva MN, et al. Genetic polymorphisms in 15q25 and 19q13 loci, cotinine levels, and risk of lung cancer in EPIC. Cancer Epidemiol Biomarkers Prev. 2011;20:2250–61. doi: 10.1158/1055-9965.EPI-11-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–34. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–9. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zeggini E, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–45. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 62.Aulchenko YS, Struchalin MV, van Duijn CM. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Clayton DG, et al. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nat Genet. 2005;37:1243–6. doi: 10.1038/ng1653. [DOI] [PubMed] [Google Scholar]
- 64.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Johnson AD, et al. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–9. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gabriel SB, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–9. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 68.Thorgeirsson TE, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42:448–53. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.