Abstract
Human epilepsy is a common and heterogeneous condition in which genetics play an important etiological role. We begin by reviewing the past history of epilepsy genetics, a field that has traditionally included studies of pedigrees with epilepsy caused by defects in ion channels and neurotransmitters. We highlight important recent discoveries that have expanded the field beyond the realm of channels and neurotransmitters and that have challenged the notion that single genes produce single disorders. Finally, we project toward an exciting future for epilepsy genetics as large-scale collaborative phenotyping studies come face to face with new technologies in genomic medicine.
Introduction
Epilepsy is a common neurological condition defined by recurrent, unprovoked seizures that affects one percent of the population, including one in 200 children [1]. Epilepsy genetics encompasses two broad categories—(1) genes and loci discovered in association with primary epilepsy syndromes, in which the epilepsy is a primary presenting feature, and (2) genes discovered in association with disorders of brain development that are associated with epilepsy. In both cases, the genes identified provide an opportunity for the study of mechanisms of brain development and epileptogenesis in the context of the developing brain.
Though the causes of epilepsy are diverse and heterogeneous, epilepsy is considered a highly genetic and in many cases heritable condition [2,3]. An inherited predisposition to seizures (called an “epileptic diathesis” by Lennox and Lennox in 1960), combined with some trigger or additional factor(s), has long been suspected as a cause of many types of epilepsy [4]. Epidemiological studies of families and twins provide compelling evidence for the heritability of epilepsy [5-9]. For example, the risk of epilepsy among first-degree relatives of individuals with idiopathic generalized epilepsy is eight to 12 percent; this is well above the risk (approximately 0.5 percent) in the general population, illustrating a strong genetic component but a complex one that does not always show a pattern consistent with Mendelian inheritance [5].
The clinical classification of epilepsy includes the categories of symptomatic, presumed symptomatic, and so-called idiopathic epilepsy [10,11]. Over time, idiopathic epilepsy has been understood to mean epilepsy caused at least in part by genetics, and the majority of cases in this category are likely to be influenced by genetic susceptibility. Past studies of genetic epilepsies have validated the notion that genetics play a major role in epilepsy, largely by identifying channels and neurotransmitters important in epileptogenesis. More recent studies have moved the field of epilepsy genetics beyond the channelopathies, and, with the beginning of the era of whole genome exploration, we are now at the threshold of understanding more complex genetic mechanisms that underlie many forms of epilepsy, both common and rare.
The Past: Setting the stage of epilepsy genetics
The approach to epilepsy genetics has, until relatively recently, been based on Mendelian genetics, relying on the ascertainment of large pedigrees, linkage analysis of polymorphic markers to established disease-associated loci, and positional cloning within these loci to identify the pathogenic gene mutation. Inheritance is autosomal dominant in many of the familial epilepsy syndromes in which mutations have been identified, with many of the genes encoding subunits of ion channels or neurotransmitter receptors [2,3,12]. A prototypic epilepsy gene discovery is the example of Autosomal Dominant Partial Epilepsy with Auditory Features (ADPEAF), with mapping of the gene completed in 1995 and mutations in the gene LGI1 (leucine-rich glioma-inactivated 1) reported in 2002 [13,14]. The mechanisms by which LGI1 mutations produce epilepsy are not fully established but are postulated to involve a potassium channel mechanism, a glutamatergic mechanism, or altered binding of the secreted neuronal protein to a transmembrane receptor (ADAM22) [15,16].
While the early discovery of genes in families with Mendelian inheritance was exciting proof of principle that epilepsy may be genetically mediated, the genes discovered thus far (listed in Table 1 and shown in the Figure) collectively do not account for the majority of idiopathic epilepsy. This is illustrated by the example of Genetic Epilepsy with Febrile Seizures Plus (GEFS+), a familial syndrome in which family members are affected with a range of phenotypes ranging from simple febrile seizures to severe myoclonic epilepsy with infancy. Between 1998 and 2004, mutations were identified in families with GEFS+ in three sodium channel subunit genes (SCN1B, SCN1A, and SCN2A) and two GABA receptor subunit genes (GABRG2 and GABRD) [17-21], but these five GEFS+ genes explain only 10 percent of GEFS+ [22,23]. Linkage studies have identified seven additional regions associated with GEFS+ and five regions associated with familial febrile seizures [24-35]. The causative mutations in genes in these regions have not yet been uncovered, likely because of constraints of time, labor, and cost of Sanger sequencing methods, or because the mutations lie outside of the exonic regions typically targeted by the same methods. The variable penetrance and variable expressivity seen in these syndromes suggest that in addition to a primary genetic mutation required for disease, there may be other genes involved and potentially other non-genetic factors that influence phenotype.
Table 1.
| GENE | ASSOCIATED EPILEPSY SYNDROME(S) |
|---|---|
| ARX | Infantile spasms Early infantile epileptic encephalopathy |
| ATP1A2 | Benign familial infantile convulsions Familial hemiplegic migraine and epilepsy |
| CACNA1A | Absence epilepsy and episodic ataxia |
| CACNB4 | Juvenile myoclonic epilepsy |
| CDKL5 (STK9) | Infantile spasms |
| CHRNA4 | Autosomal dominant nocturnal frontal lobe epilepsy |
| CHRNB2 | Autosomal dominant nocturnal frontal lobe epilepsy |
| CHRNA7 | Juvenile myoclonic epilepsy |
| CLCN2 | Childhood absence epilepsy Juvenile absence epilepsy Juvenile myoclonic epilepsy |
| EFHC1 | Juvenile myoclonic epilepsy |
| GABRD | Genetic epilepsy with febrile seizures plus |
| GABRA1 | Juvenile myoclonic epilepsy |
| GABRG2 | Childhood absence epilepsy Genetic epilepsy with febrile seizures plus |
| KCNQ2 | Benign familial neonatal convulsions |
| KCNQ3 | Benign familial neonatal convulsions |
| KCNMA1 | Generalized epilepsy with paroxysmal dyskinesia |
| LGI1 | Autosomal dominant partial epilepsy with auditory features |
| PCDH19 | Epilepsy in females with mental retardation |
| SCN1A | Genetic epilepsy with febrile seizures plus Severe myoclonic epilepsy of infancy (Dravet syndrome) |
| SCN1B | Genetic epilepsy with febrile seizures plus |
| SCN2A | Benign familial neonatal/infantile convulsions Genetic epilepsy with febrile seizures plus |
| SLC2A1 | Early-onset absence epilepsy Epilepsy with paroxysmal exercise-induced dyskinesia |
| STXBP1 | Early infantile epileptic encephalopathy Partial onset epilepsy with intellectual disability |
| TBC1D24 | Familial infantile myoclonic epilepsy Focal epilepsy with developmental disability |
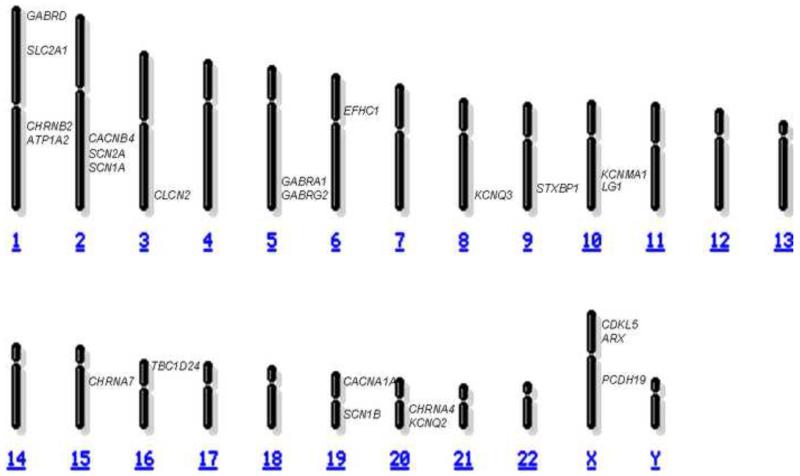
Figure.
The major epilepsy genes, including those highlighted in this review, are shown to depict their relative chromosomal locations on this schematic karyotype (adapted from the National Center for Biotechnology Information website www.ncbi.nlm.nih.gov).
When the status of the genetics of epilepsy was reviewed in 1980, the strongest evidence for a genetic role in the causation of epilepsy was the existence of pedigrees with epilepsy and the presence of animal models with epilepsy [36]. It is notable that among genetic mouse models for epilepsy, only one-quarter are caused by defects in channel-related genes [37], suggesting that there are multiple classes of epilepsy genes yet to be discovered. In parallel with the discovery of “epilepsy genes” in pedigrees with epilepsy as the primary phenotype, epilepsy genetics includes the discovery of genes associated with syndromes and cerebral cortical malformations highly associated with epilepsy. These genes include LIS1, DCX, ARX, FLNA, GPR56, and MECP2 [38-44]. The subject of brain malformations will be addressed in depth elsewhere in this issue, but there is a strong causal link between brain malformations and epilepsy. We have already seen some genes discovered in the setting of dramatic, radiographically evident brain malformations that are also responsible for epilepsy syndromes without brain malformations. A notable example is ARX, associated initially with X-linked lissencephaly but subsequently found to be associated with isolated infantile spasms without lissencephaly [45].
The Present: Recent Advances in the Genetics of Epilepsy
There have been several advances in epilepsy genetics in the past three years, including additional Mendelian genetics studies, further studies of previously discovered genes, and identification of genes encoding proteins in novel pathways not previously demonstrated to be relevant to epilepsy. Major advances in the field also include the recognition of the importance of copy number variation.
Genetically determined “interneuronopathies” associated with epilepsy
Our understanding of developmental “interneuronopathy” associated with severe epilepsy has been advanced by several recent studies of the role of ARX in the developing brain [46]. Refinement of the mechanisms involved in tangential neuronal migration has included the recognition that ARX has a cell autonomous role in proliferation as well as GABAergic interneuron migration [47,48]. In an early study of a human case of ARX-associated lissencephaly, the neocortical subventricular region showed cells positive for glutamic acid decarboxylase and calretinin as well as Mash-1 and nestin, suggesting a role for ARX in radial migration in addition to its traditional role in tangential GABAergic interneuron migration from the ganglionic eminence [49].
The translation of ARX genetics to a mouse model with real relevance to human epilepsy came in the form of a conditional knockout mouse in which Arx was deleted from neurons derived from the ganglionic eminence derived neurons; the result was a mouse with early life seizures resembling the human infantile spasms phenotype [50]. The body of work related to ARX provides a paradigm for epilepsy-related research with broad applicability. Stemming from the discovery of a gene for a severe form of infantile epilepsy, the ARX field now includes animal models that can be used to further study infantile spasms and to develop rational treatment strategies.
New epilepsy genes revealing novel mechanisms of epileptogenesis STXBP1
Early infantile epileptic encephalopathy with suppression-burst (EIEE) is a severe form of early onset epilepsy recently associated with mutations in the gene STXBP1 (also known as MUNC18-1) [51]. Importantly, the discovery of EIEE-associated sporadic heterozygous STXBP1 mutations arose from the initial identification of a microdeletion at 9q33.3-34.11 in a single case by comparative genomic hybridization (CGH), leading to candidate gene sequencing in other index cases of the genes in that region. In contrast to the previously mentioned epilepsy genes that encode subunits of ion channels or neurotransmitter receptors, the protein encoded by this gene, synaptic binding protein 1, is involved in synaptic vesicle release. The discovery of this gene has opened a new arena for targeted rational drug design, not only for the rare syndrome of EIEE but for epilepsy generally.
As is likely to be the case for many epilepsy-associated conditions, there are broader implications for STXBP1 that extend well beyond EIEE. There is already a report of heterozygous mutations in STXBP1 in patients with intellectual disability and nonsyndromic focal onset epilepsy, a phenotype much milder and less specific than EIEE [52]. The overall impact of STXBP1 in patients with epilepsy, with or without intellectual disability, is yet to be determined.
PCDH19
Epilepsy and mental retardation limited to females (EFMR) is an X-linked condition that is unusual in that it is transmitted through males and only carrier females show classic symptoms. The recognition of this unique mode of inheritance led to a study of seven families with EFMR that identified mutations in protocadherin 19 (PCDH19), a cadherin that is expressed in the developing brain [53]. This discovery is of particular interest on several counts: (a) the authors recognized this very unusual pattern of expression as a defining feature of an epilepsy syndrome with linkage to the X chromosome, (b) gene identification was possible due to increasing efficiency of resequencing methods and was accomplished by resequencing 737 genes on the X chromosome, and (c) the discovery of a link between cadherins and epilepsy expands the field to include still another class of proteins beyond ion channels and neurotransmitter receptors.
The implications of PCDH19 on epilepsy have already been shown to be broader than the familial EFMR syndrome. PCDH19 mutations have been identified by multiple groups in sporadic cases of epilepsy in females with intellectual disability as well as infantile onset epilepsy, even that resembling the severe myoclonic epilepsy of infancy phenotype of Dravet syndrome [54-57].
TBC1D24
Mutations in TBC1D24 have recently been identified in association with focal onset epilepsy with associated intellectual disability in a consanguineous pedigree [58]. Again, a novel category of epilepsy genes has emerged as this gene has been shown to have a role in axon specification in developing neurons [58]. While the brain imaging described in the original report includes evidence of a subtle anterior pachygyria pattern, it appears that this pattern is not a required feature of the phenotype associated with mutations in TBC1D24: simultaneous with the report of TBC1D24 mutations with the fairly non-specific phenotype of focal epilepsy and intellectual disability was a report of compound heterozygous mutations in a non-consanguineous pedigree with a distinct phenotype of familial infantile myoclonic epilepsy [59]. It would not be surprising to see the discovery of other effects of mutations in this gene in the months and years to come, perhaps with still different forms of phenotypic expression.
Cortical malformation genes with implications for epilepsy
Three microcephaly genes are particularly relevant to epilepsy genetics. ASPM, an established primary microcephaly gene, is associated with a microcephaly phenotype that now includes epilepsy (microcephaly with seizures) [60]. CDKL5, a gene traditionally associated with a Rett syndrome-like phenotype, is now considered on the differential diagnosis for individuals with microcephaly and infantile spasms [61,62]. Mutations in a novel microcephaly gene involved in DNA repair, PNKP, are responsible for microcephaly with early-onset epilepsy and developmental delay [63].
Another category of cortical malformation genes with implications for the field of epilepsy are the “tubulinopathies.” Mutations in TUBA1A have been associated not only with lissencephaly but also with polymicrogyria [64,65], both of which have a high prevalence of associated epilepsy. The role of TUBA1A as well as TUBB2B in brain development is reviewed elsewhere [66].
Structural genomic variations and epilepsy
Chromosomal deletions and duplications have been found in patients with epilepsy but are frequently in the context of a recognizable genetic syndrome and have been detectable by clinical methods such as karyotyping (e.g., trisomy 21, ring chromosome 20, fragile X syndrome) [67-69]. More recently, oligonucleotide arrays have allowed for evaluation of smaller duplications and deletion across the genome. This approach has revealed associations between copy number variations and neurodevelopmental conditions such as autism and intellectual disability, as well as a number of syndromes in which children have dysmorphic features [70, 71].
In the field of epilepsy genetics, large-scale studies of copy number variation have yielded results that are somewhat surprising in that the patients studied have idiopathic epilepsies without autism, intellectual disabilities, or dysmorphic features. One group studied 1234 Northern European subjects with idiopathic generalized epilepsy by high-density single nucleotide polymorphism arrays and identified microdeletions in almost 2 percent, most notably at 15q11.2 and 16p13.11 [72]. In another report, 517 subjects with idiopathic focal or generalized onset epilepsy were analyzed by whole-genome oligonucleotide array CGH, and nearly 3 percent were found to have deletions at 15q11.2, 15q13.3, or 16p13.11, loci previously associated with autism and intellectual disability [73]. Finally, a study of 3812 cases representing a wide range of epilepsy phenotypes found 23 cases to have 16p13.11 deletions 100 kb or larger [74]. These studies have identified structural genomic variations associated with epilepsy in the absence of dysmorphic features, representing a change from the conventional wisdom that chromosome microarray analysis is useful only for patients with intellectual disability or autism and abnormal physical features. As such, they may have important implications in the clinical arena.
Genome-wide association studies of epilepsy
Advances in techniques allow for genome-wide linkage analysis for familial disorders and large-scale association studies to establish allelic variants that may have broad impact in a population. These techniques present an opportunity to study a common yet heterogeneous condition such as epilepsy but present a challenge in that well-powered association studies need large numbers of affected individuals who are carefully phenotyped. A genome-wide association study (GWAS) of 3445 partial-onset epilepsy cases (and nearly 7000 controls) has been conducted but did not reveal any strong candidate genes [75].
The Future: Applying Advancing Genetic Technologies to Epilepsy Genetics
Though the results of the first large GWAS for epilepsy were disappointing in that no additional epilepsy genes or loci were identified, the study did show that a large consortium approach combined with powerful bioinformatics platforms can be successful in accruing enough samples for well-powered genetic studies in epilepsy and performing analyses on such large groups of cases. The large groups of samples already collected for this and other large studies, such as the Epilepsy Phenome/Genome Project [76], may provide a future reservoir in which to search for mutations in established and newly discovered genes.
Massively parallel sequencing strategies now allow whole exome and whole genome sequencing experiments to be performed at increasingly affordable costs and with analysis tools that allow comparison to a growing number of reference cases, such as presented in the 1000 Genomes Project [77]. In the immediate future, we should expect efforts to resolve the unanswered questions of which genes are responsible for epilepsies for which excellent linkage data are available, such as the GEFS+ and familial febrile seizures loci mentioned above. Moving forward, gene discovery will continue in pedigrees identified with epilepsy. We now face the exciting prospect of identifying mutations among large groups of individuals with specific epilepsy syndromes and then applying these discoveries to the broader epilepsy field and beyond.
We present a summary of the past, present, and future of the actively evolving field of epilepsy genetics in Table 2. Along with the ability to identify vast numbers of genetic variants potentially associated with epilepsy will come the challenges of validating genetic associations and analyzing their significance. Indeed, ongoing successes in gene discovery—whether in families with Mendelian inheritance or in large cohorts of patients with epilepsy—will unearth questions regarding the functional significance of the mutations discovered thus far. The true measure of our successes will be the ability to translate these genetic discoveries into a deeper mechanistic understanding of epilepsy that can be translated into effective therapies.
Table 2. The Evolution of Epilepsy Genetics.
Past
|
Present
|
Future
|
Acknowledgments
Annapurna Poduri was supported by the NINDS (K23 NS069784). Daniel Lowenstein was supported by the NINDS (U01 NS053998, Epilepsy Phenome Genome Project).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
Papers of particular interest have been highlighted with an asterisk (*).
Papers of outstanding interest have been highlighted with two asterisks (**).
- [1].Cowan LD. The epidemiology of the epilepsies in children. Ment Retard Dev Disabil Res Rev. 2002;8:171–181. doi: 10.1002/mrdd.10035. [DOI] [PubMed] [Google Scholar]
- [2]*.Prasad AN, Prasad C. Genetic Influences on the Risk for Epilepsy. In: Pellock JM, Bourgeois BFD, Dodson WE, et al., editors. Pediatric Epilepsy. Demos; New York: 2008. pp. 117–134. The authors present a comprehensive review of the evidence for a genetic influence on epilepsy.
- [3].Sigurdardottir L, Poduri A. Inherited Epilepsies. In: Lynch DR, editor. Neurogenetics: Scientific and Clinical Advances. Taylor and Francis; New York: 2006. pp. 427–467. [Google Scholar]
- [4].Lennox WG, Lennox MA. Epilepsy and related disorders. Little, Brown; Boston: 1960. The genetics of epilepsy. [Google Scholar]
- [5].Steinlein OK. Genes and mutations in human idiopathic epilepsy. Brain Dev. 2004;26:213–218. doi: 10.1016/S0387-7604(03)00149-9. [DOI] [PubMed] [Google Scholar]
- [6].Berkovic SF, Howell RA, Hay DA, Hopper JL. Epilepsies in twins: genetics of the major epilepsy syndromes. Ann Neurol. 1998;43:435–445. doi: 10.1002/ana.410430405. [DOI] [PubMed] [Google Scholar]
- [7].Kjeldsen MJ, Kyvik KO, Friis ML, Christensen K. Genetic and environmental factors in febrile seizures: a Danish population-based twin study. Epilepsy Res. 2002;51:167–177. doi: 10.1016/s0920-1211(02)00121-3. [DOI] [PubMed] [Google Scholar]
- [8].Kjeldsen MJ, Corey LA, Christensen K, Friis ML. Epileptic seizures and syndromes in twins: the importance of genetic factors. Epilepsy Res. 2003;55:137–146. doi: 10.1016/s0920-1211(03)00117-7. [DOI] [PubMed] [Google Scholar]
- [9].Kjeldsen MJ, Corey LA, Solaas MH, Friis ML, Harris JR, Kyvik KO, Christensen K, Pellock JM. Genetic factors in seizures: a population-based study of 47,626 US, Norwegian and Danish twin pairs. Twin Res Hum Genet. 2005;8:138–147. doi: 10.1375/1832427053738836. [DOI] [PubMed] [Google Scholar]
- [10].Proposal for classification of epilepsies and epileptic syndromes. Commission on Classification and Terminology of the International League Against Epilepsy. Epilepsia. 1985;26:268–278. [PubMed] [Google Scholar]
- [11].Proposal for revised classification of epilepsies and epileptic syndromes. Commission on Classification and Terminology of the International League Against Epilepsy. Epilepsia. 1989;30:389–399. doi: 10.1111/j.1528-1157.1989.tb05316.x. [DOI] [PubMed] [Google Scholar]
- [12]**.Mulley JC, Scheffer IE, Petrou S, Berkovic SF. Channelopathies as a genetic cause of epilepsy. Curr Opin Neurol. 2003;16:171–176. doi: 10.1097/01.wco.0000063767.15877.c7. This review provides an excellent summary of epilepsy-related channelopathies, which account for the majority of genetic epilepsies described to date.
- [13].Ottman R, Risch N, Hauser WA, Pedley TA, Lee JH, Barker-Cummings C, Lustenberger A, Nagle KJ, Lee KS, Scheuer ML, et al. Localization of a gene for partial epilepsy to chromosome 10q. Nat Genet. 1995;10:56–60. doi: 10.1038/ng0595-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kalachikov S, Evgrafov O, Ross B, Winawer M, Barker-Cummings C, Martinelli Boneschi F, Choi C, Morozov P, Das K, et al. Mutations in LGI1 cause autosomal-dominant partial epilepsy with auditory features. Nat Genet. 2002;30:335–341. doi: 10.1038/ng832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Fukata Y, Adesnik H, Iwanaga T, Bredt DS, Nicoll RA, Fukata M. Epilepsy-related ligand/receptor complex LGI1 and ADAM22 regulate synaptic transmission. Science. 2006;313:1792–1795. doi: 10.1126/science.1129947. [DOI] [PubMed] [Google Scholar]
- [16].Ottman R, Rosenberger L, Bagic A, Kamberakis K, Ritzl EK, Wohlschlager AM, Shamim S, Sato S, Liew C, Gaillard WD, et al. Altered language processing in autosomal dominant partial epilepsy with auditory features. Neurology. 2008;71:1973–1980. doi: 10.1212/01.wnl.0000336923.29538.5b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wallace RH, Wang DW, Singh R, Scheffer IE, George AL, Jr., Phillips HA, Saar K, Reis A, Johnson EW, Sutherland GR, Berkovic SF, Mulley JC. Febrile seizures and generalized epilepsy associated with a mutation in the Na+-channel beta1 subunit gene SCN1B. Nat Genet. 1998;19:366–370. doi: 10.1038/1252. [DOI] [PubMed] [Google Scholar]
- [18].Escayg A, MacDonald BT, Meisler MH, Baulac S, Huberfeld G, An-Gourfinkel I, Brice A, LeGuern E, Moulard B, Chaigne D, et al. Mutations of SCN1A, encoding a neuronal sodium channel, in two families with GEFS+ Nat Genet. 2000;24:343–345. doi: 10.1038/74159. [DOI] [PubMed] [Google Scholar]
- [19].Sugawara T, Tsurubuchi Y, Agarwala KL, Ito M, Fukuma G, Mazaki-Miyazaki E, Nagafuji H, Noda M, Imoto K, Wada K, et al. A missense mutation of the Na+ channel alpha II subunit gene Na(v)1.2 in a patient with febrile and afebrile seizures causes channel dysfunction. Proc Natl Acad Sci U S A. 2001;98:6384–6389. doi: 10.1073/pnas.111065098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Baulac S, Huberfeld G, Gourfinkel-An I, Mitropoulou G, Beranger A, Prud’homme JF, Baulac M, Brice A, Bruzzone R, LeGuern E. First genetic evidence of GABA(A) receptor dysfunction in epilepsy: a mutation in the gamma2-subunit gene. Nat Genet. 2001;28:46–48. doi: 10.1038/ng0501-46. [DOI] [PubMed] [Google Scholar]
- [21].Dibbens LM, Feng HJ, Richards MC, Harkin LA, Hodgson BL, Scott D, Jenkins M, Petrou S, Sutherland GR, Scheffer IE, et al. GABRD encoding a protein for extra- or peri-synaptic GABAA receptors is a susceptibility locus for generalized epilepsies. Hum Mol Genet. 2004;13:1315–1319. doi: 10.1093/hmg/ddh146. [DOI] [PubMed] [Google Scholar]
- [22].Nakayama J, Arinami T. Molecular genetics of febrile seizures. Epilepsy Res. 2006;70:S190–S198. doi: 10.1016/j.eplepsyres.2005.11.023. [DOI] [PubMed] [Google Scholar]
- [23].Marini C, Mei D, Temudo T, Ferrari AR, Buti D, Dravet C, Dias AI, Moreira A, Calado E, Seri S, et al. Idiopathic epilepsies with seizures precipitated by fever and SCN1A abnormalities. Epilepsia. 2007;48:1678–1685. doi: 10.1111/j.1528-1167.2007.01122.x. [DOI] [PubMed] [Google Scholar]
- [24].Audenaert D, Claes L, Claeys KG, Deprez L, Van Dyck T, Goossens D, Del-Favero J, Van Paesschen W, Van Broeckhoven C, De Jonghe P. A novel susceptibility locus at 2p24 for generalised epilepsy with febrile seizures plus. J Med Genet. 2005;42:947–952. doi: 10.1136/jmg.2005.031393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hedera P, Ma S, Blair MA, Taylor KA, Hamati A, Bradford Y, Abou-Khalil B, Haines JL. Identification of a novel locus for febrile seizures and epilepsy on chromosome 21q22. Epilepsia. 2006;47:1622–1628. doi: 10.1111/j.1528-1167.2006.00637.x. [DOI] [PubMed] [Google Scholar]
- [26].Nabbout R, Baulac S, Desguerre I, Bahi-Buisson N, Chiron C, Ruberg M, Dulac O, LeGuern E. New locus for febrile seizures with absence epilepsy on 3p and a possible modifier gene on 18p. Neurology. 2007;68:1374–1381. doi: 10.1212/01.wnl.0000260062.02829.e3. [DOI] [PubMed] [Google Scholar]
- [27].Baulac S, Gourfinkel-An I, Couarch P, Depienne C, Kaminska A, Dulac O, Baulac M, LeGuern E, Nabbout R. A novel locus for generalized epilepsy with febrile seizures plus in French families. Arch Neurol. 2008;65:943–951. doi: 10.1001/archneur.65.7.943. [DOI] [PubMed] [Google Scholar]
- [28].Peiffer A, Thompson J, Charlier C, Otterud B, Varvil T, Pappas C, Barnitz C, Gruenthal K, Kuhn R, Leppert M. A locus for febrile seizures (FEB3) maps to chromosome 2q23-24. Ann Neurol. 1999;46:671–678. doi: 10.1002/1531-8249(199910)46:4<671::aid-ana20>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- [29].Deprez L, Claes LR, Claeys KG, Audenaert D, Van Dyck T, Goossens D, Van Paesschen W, Del-Favero J, Van Broeckhoven C, De Jonghe P. Genome-wide linkage of febrile seizures and epilepsy to the FEB4 locus at 5q14.3-q23.1 and no MASS1 mutation. Hum Genet. 2006;118:618–625. doi: 10.1007/s00439-005-0077-x. [DOI] [PubMed] [Google Scholar]
- [30].Johnson EW, Dubovsky J, Rich SS, O’Donovan CA, Orr HT, Anderson VE, Gil-Nagel A, Ahmann P, Dokken CG, Schneider DT, Weber JL. Evidence for a novel gene for familial febrile convulsions, FEB2, linked to chromosome 19p in an extended family from the Midwest. Hum Mol Genet. 1998;7:63–67. doi: 10.1093/hmg/7.1.63. [DOI] [PubMed] [Google Scholar]
- [31].Nabbout R, Prud’homme JF, Herman A, Feingold J, Brice A, Dulac O, LeGuern E. A locus for simple pure febrile seizures maps to chromosome 6q22-q24. Brain. 2002;125:2668–2680. doi: 10.1093/brain/awf281. [DOI] [PubMed] [Google Scholar]
- [32].Nakayama J, Yamamoto N, Hamano K, Iwasaki N, Ohta M, Nakahara S, Matsui A, Noguchi E, Arinami T. Linkage and association of febrile seizures to the IMPA2 gene on human chromosome 18. Neurology. 2004;63:1803–1807. doi: 10.1212/01.wnl.0000144499.34164.e0. [DOI] [PubMed] [Google Scholar]
- [33].Wallace RH, Berkovic SF, Howell RA, Sutherland GR, Mulley JC. Suggestion of a major gene for familial febrile convulsions mapping to 8q13-21. J Med Genet. 1996;33:308–312. doi: 10.1136/jmg.33.4.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Dai XH, Chen WW, Wang X, Zhu QH, Li C, Li L, Liu MG, Wang QK, Liu JY. A novel genetic locus for familial febrile seizures and epilepsy on chromosome 3q26.2-q26.33. Hum Genet. 2008;124:423–429. doi: 10.1007/s00439-008-0566-9. [DOI] [PubMed] [Google Scholar]
- [35].Poduri A, Wang Y, Gordon D, Barral-Rodriguez S, Barker-Cummings C, Ulgen A, Chitsazzadeh V, Hill RS, Risch N, Hauser WA, Pedley TA, et al. Novel susceptibility locus at chromosome 6q16.3-22.31 in a family with GEFS+ Neurology. 2009;73:1264–1272. doi: 10.1212/WNL.0b013e3181bd10d3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Newmark ME, Penry JK. Genetics of Epilepsy: A Review. Raven Press; New York: 1980. [Google Scholar]
- [37].Frankel WN. Genetics of complex neurological disease: challenges and opportunities for modeling epilepsy in mice and rats. Trends Genet. 2009;5:361–367. doi: 10.1016/j.tig.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Reiner O, Carrozzo R, Shen Y, Wehnert M, Faustinella F, Dobyns WB, Caskey CT, Ledbetter DH. Isolation of a Miller-Dieker lissencephaly gene containing G protein β-subunit-like repeats. Nature. 1993;364:717–721. doi: 10.1038/364717a0. [DOI] [PubMed] [Google Scholar]
- [39].Gleeson JG, Allen KM, Fox JW, Lamperti ED, Berkovic S, Scheffer I, Cooper EC, Dobyns WB, Minnerath SR, Ross ME, Walsh CA. Doublecortin, a brain-specific gene mutated in human X-linked lissencephaly and double cortex syndrome, encodes a putative signaling protein. Cell. 1998;92:63–72. doi: 10.1016/s0092-8674(00)80899-5. [DOI] [PubMed] [Google Scholar]
- [40].des Portes V, Pinard JM, Billuart P, Vinet MC, Koulakoff A, Carrié A, Gelot A, Dupuis E, Motte J, Berwald-Netter Y, et al. A novel CNS gene required for neuronal migration and involved in X-linked subcortical laminar heterotopia and lissencephaly syndrome. Cell. 1998;92:51–61. doi: 10.1016/s0092-8674(00)80898-3. [DOI] [PubMed] [Google Scholar]
- [41].Kitamura K, Yanazawa M, Sugiyama N, Miura H, Iizuka-Kogo A, Kusaka M, Omichi K, Suzuki R, Kato-Fukui Y, Kamiirisa K, et al. Mutation of ARX causes abnormal development of forebrain and testes in mice and X-linked lissencephaly with abnormal genitalia in humans. Nat Genet. 2002;32:359–369. doi: 10.1038/ng1009. [DOI] [PubMed] [Google Scholar]
- [42].Fox JW, Lamperti ED, Ekşioğlu YZ, Hong SE, Feng Y, Graham DA, Scheffer IE, Dobyns WB, Hirsch BA, Radtke RA, et al. Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron. 1998;21:1315–1325. doi: 10.1016/s0896-6273(00)80651-0. [DOI] [PubMed] [Google Scholar]
- [43].Piao X, Hill RS, Bodell A, Chang BS, Basel-Vanagaite L, Straussberg R, Dobyns WB, Qasrawi B, Winter RM, Innes AM, Voit T, et al. G protein-coupled receptor-dependent development of human frontal cortex. Science. 2004;303:2033–2036. doi: 10.1126/science.1092780. [DOI] [PubMed] [Google Scholar]
- [44].Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- [45].Sherr EH. The ARX story (epilepsy, mental retardation, autism, and cerebral malformations): one gene leads to many phenotypes. Curr Opin Pediatr. 2003;15:567–571. doi: 10.1097/00008480-200312000-00004. [DOI] [PubMed] [Google Scholar]
- [46].Kato M, Dobyns WB. X-linked lissencephaly with abnormal genitalia as a tangential migration disorder causing intractable epilepsy: proposal for a new term, “interneuronopathy.”. J Child Neurol. 2005;20:392–397. doi: 10.1177/08830738050200042001. [DOI] [PubMed] [Google Scholar]
- [47].Colasante G, Collombat P, Raimondi V, Bonanomi D, Ferrai C, Maira M, Yoshikawa K, Mansouri A, Valtorta F, Rubenstein JL, Broccoli V. Arx is a direct target of Dlx2 and thereby contributes to the tangential migration of GABAergic interneurons. J Neurosci. 2008;28:10674–10686. doi: 10.1523/JNEUROSCI.1283-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Friocourt G, Kanatani S, Tabata H, Yozu M, Takahashi T, Antypa M, Raguénès O, Chelly J, Férec C, Nakajima K, Parnavelas JG. Cell-autonomous roles of ARX in cell proliferation and neuronal migration during corticogenesis. J Neurosci. 2008;28:5794–5805. doi: 10.1523/JNEUROSCI.1067-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Okazaki S, Ohsawa M, Kuki I, Kawawaki H, Koriyama T, Ri S, Ichiba H, Hai E, Inoue T, Nakamura H, et al. Aristaless-related homeobox gene disruption leads to abnormal distribution of GABAergic interneurons in human neocortex: evidence based on a case of X-linked lissencephaly with abnormal genitalia (XLAG) Acta Neuropathol. 2008;116:453–462. doi: 10.1007/s00401-008-0382-2. [DOI] [PubMed] [Google Scholar]
- [50]*.Marsh E, Fulp C, Gomez E, Nasrallah I, Minarcik J, Sudi J, Christian SL, Mancini G, Labosky P, Dobyns W, Brooks-Kayal A, Golden JA. Targeted loss of Arx results in a developmental epilepsy mouse model and recapitulates the human phenotype in heterozygous females. Brain. 2009;132:1563–1576. doi: 10.1093/brain/awp107. This reported Arx mouse model approximates the human ARX phenotype. As such, this model can be used not only for future studies of interneuron development but also studies of potential therapeutic interventions.
- [51]**.Saitsu H, Kato M, Mizuguchi T, Hamada K, Osaka H, Tohyama J, Uruno K, Kumada S, Nishiyama K, Nishimura A, et al. De novo mutations in the gene encoding STXBP1 (MUNC18-1) cause early infantile epileptic encephalopathy. Nat Genet. 2008;40:782–788. doi: 10.1038/ng.150. The authors used comparative genomic hybridization to identify a critical region for Ohtahara syndrome and identified mutations in STXBP1, one of the first non-channel, non-neurotransmitter-related genes to be associated with epilepsy. This represents one of the few identifiable etiologies associated with a severe neonatal epilepsy.
- [52].Hamdan FF, Piton A, Gauthier J, Lortie A, Dubeau F, Dobrzeniecka S, Spiegelman D, Noreau A, Pellerin S, Côté M, et al. De novo STXBP1 mutations in mental retardation and nonsyndromic epilepsy. Ann Neurol. 2009;65:748–753. doi: 10.1002/ana.21625. [DOI] [PubMed] [Google Scholar]
- [53]**.Dibbens LM, Tarpey PS, Hynes K, Bayly MA, Scheffer IE, Smith R, Bomar J, Sutton E, Vandeleur L, Shoubridge C, et al. X-linked protocadherin 19 mutations cause female-limited epilepsy and cognitive impairment. Nat Genet. 2008;40:776–781. doi: 10.1038/ng.149. The identification of the PCDH19 mutations in females with epilepsy and cognitive impairment provides a novel class of epilepsy genes.
- [54].Depienne C, Bouteiller D, Keren B, Cheuret E, Poirier K, Trouillard O, Benyahia B, Quelin C, Carpentier W, Julia S, et al. Sporadic infantile epileptic encephalopathy caused by mutations in PCDH19 resembles Dravet syndrome but mainly affects females. PLoS Genet. 2009;5:e1000381. doi: 10.1371/journal.pgen.1000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Hynes K, Tarpey P, Dibbens LM, Bayly MA, Berkovic SF, Smith R, Raisi ZA, Turner SJ, Brown NJ, Desai TD, et al. Epilepsy and mental retardation limited to females with PCDH19 mutations can present de novo or in single generation families. J Med Genet. 2010;47:211–216. doi: 10.1136/jmg.2009.068817. [DOI] [PubMed] [Google Scholar]
- [56].Marini C, Mei D, Parmeggiani L, Norci V, Calado E, Ferrari A, Moreira A, Pisano T, Specchio N, Vigevano F, Battaglia D, Guerrini R. Protocadherin 19 mutations in girls with infantile-onset epilepsy. Neurology. 2010;75:646–653. doi: 10.1212/WNL.0b013e3181ed9e67. [DOI] [PubMed] [Google Scholar]
- [57].Jamal SM, Basran RK, Newton S, Wang Z, Milunsky JM. Novel de novo PCDH19 mutations in three unrelated females with epilepsy female restricted mental retardation syndrome. Am J Med Genet A. 2010;152:2475–2481. doi: 10.1002/ajmg.a.33611. [DOI] [PubMed] [Google Scholar]
- [58].Corbett MA, Bahlo M, Jolly L, Afawi Z, Gardner AE, Oliver KL, Tan S, Coffey A, Mulley JC, Dibbens LM, et al. A focal epilepsy and intellectual disability syndrome is due to a mutation in TBC1D24. Am J Hum Genet A. 2010;87:371–375. doi: 10.1016/j.ajhg.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Falace A, Filipello F, La Padula V, Vanni N, Madia F, De Pietri Tonelli D, de Falco FA, Striano P, Dagna Bricarelli F, Minetti C, et al. TBC1D24, an ARF6-interacting protein, is mutated in familial infantile myoclonic epilepsy. Am J Hum Genet. 2010;87:365–370. doi: 10.1016/j.ajhg.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Shen J, Eyaid W, Mochida GH, Al-Moayyad F, Bodell A, Woods CG, Walsh CA. ASPM mutations identified in patients with primary microcephaly and seizures. J Med Genet. 2005;42:725–729. doi: 10.1136/jmg.2004.027706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Archer HL, Evans J, Edwards S, Colley J, Newbury-Ecob R, O’Callaghan F, Huyton M, O’Regan M, Tolmie J, Sampson J, et al. CDKL5 mutations cause infantile spasms, early onset seizures, and severe mental retardation in female patients. J Med Genet. 2006;43:729–734. doi: 10.1136/jmg.2006.041467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Bahi-Buisson N, Nectoux J, Rosas-Vargas H, Milh M, Boddaert N, Girard B, Cances C, Ville D, Afenjar A, Rio M, et al. Key clinical features to identify girls with CDKL5 mutations. Brain. 2008;131:2647–2661. doi: 10.1093/brain/awn197. [DOI] [PubMed] [Google Scholar]
- [63]*.Shen J, Gilmore EC, Marshall CA, Haddadin M, Reynolds JJ, Eyaid W, Bodell A, Barry B, Gleason D, Allen K, et al. Mutations in PNKP cause microcephaly, seizures and defects in DNA repair. Nat Genet. 2010;42:245–249. doi: 10.1038/ng.526. The syndrome of microcephaly with seizures caused by PNKP highlights the importance of recognizing epilepsy in the context of microcephaly.
- [64].Poirier K, Keays DA, Francis F, Saillour Y, Bahi N, Manouvrier S, Fallet-Bianco C, Pasquier L, Toutain A, Tuy FP, et al. Large spectrum of lissencephaly and pachygyria phenotypes resulting from de novo missense mutations in tubulin alpha 1A (TUBA1A) Hum Mutat. 2007;28:1055–1064. doi: 10.1002/humu.20572. [DOI] [PubMed] [Google Scholar]
- [65].Bahi-Buisson N, Poirier K, Boddaert N, Saillour Y, Castelnau L, Philip N, Buyse G, Villard L, Joriot S, Marret S, et al. Refinement of cortical dysgeneses spectrum associated with TUBA1A mutations. J Med Genet. 2008;45:647–653. doi: 10.1136/jmg.2008.058073. [DOI] [PubMed] [Google Scholar]
- [66].Jaglin XH, Chelly J. Tubulin-related cortical dysgeneses: microtubule dysfunction underlying neuronal migration defects. Trends Genet. 2009;25:555–66. doi: 10.1016/j.tig.2009.10.003. [DOI] [PubMed] [Google Scholar]
- [67].Smigielska-Kuzia J, Sobaniec W, Kulak W, Boćkowski L. Clinical and EEG features of epilepsy in children and adolescents in Down syndrome. J Child Neurol. 2009;24:416–420. doi: 10.1177/0883073808324542. [DOI] [PubMed] [Google Scholar]
- [68].Conlin LK, Kramer W, Hutchinson AL, Li X, Riethman H, Hakonarson H, Mulley JC, Scheffer IE, Berkovic SF, Hosain SA, Spinner NB. Molecular analysis of ring chromosome 20 syndrome reveals two distinct groups of patients. J Med Genet. 2010 Oct 23; doi: 10.1136/jmg.2010.080382. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- [69].Musumeci SA, Hagerman RJ, Ferri R, Bosco P, Dalla Bernardina B, Tassinari CA, De Sarro GB, Elia M. Epilepsy and EEG findings in males with fragile X syndrome. Epilepsia. 1999;40:1092–1099. doi: 10.1111/j.1528-1157.1999.tb00824.x. [DOI] [PubMed] [Google Scholar]
- [70].Weiss LA, Shen Y, Korn JM, Arking DE, Miller DT, Fossdal R, Saemundsen E, Stefansson H, Ferreira MA, Green T, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- [71].Manning M, Hudgins L, Professional Practice and Guidelines Committee Array-based technology and recommendations for utilization in medical genetics practice for detection of chromosomal abnormalities. Genet Med. 2010 Oct 18; doi: 10.1097/GIM.0b013e3181f8baad. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72]*.de Kovel CG, Trucks H, Helbig I, Mefford HC, Baker C, Leu C, Kluck C, Muhle H, von Spiczak S, Ostertag P, et al. Recurrent microdeletions at 15q11.2 and 16p13.11 predispose to idiopathic generalized epilepsies. Brain. 2010;133:23–32. doi: 10.1093/brain/awp262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73]**.Mefford HC, Muhle H, Ostertag P, von Spiczak S, Buysse K, Baker C, Franke A, Malafosse A, Genton P, Thomas P, et al. Genome-wide copy number variation in epilepsy: novel susceptibility loci in idiopathic generalized and focal epilepsies. PLoS Genet. 2010;6:e1000962. doi: 10.1371/journal.pgen.1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74]*.Heinzen EL, Radtke RA, Urban TJ, Cavalleri GL, Depondt C, Need AC, Walley NM, Nicoletti P, Ge D, Catarino CB, et al. Rare deletions at 16p13.11 predispose to a diverse spectrum of sporadic epilepsy syndromes. Am J Hum Genet. 2010;86:707–718. doi: 10.1016/j.ajhg.2010.03.018. The above three studies provide evidence that a small but notable subset of patients with idiopathic epilepsy have associated copy number abnormalities. Given that such copy number abnormalities have traditionally been associated with dysmorphic features, these findings are important.
- [75]*.Kasperaviciūte D, Catarino CB, Heinzen EL, Depondt C, Cavalleri GL, Caboclo LO, Tate SK, Jamnadas-Khoda J, Chinthapalli K, Clayton LM, et al. Common genetic variation and susceptibility to partial epilepsies: a genome-wide association study. Brain. 2010;133:2136–2147. doi: 10.1093/brain/awq130. This study represents the largest epilepsy GWAS study. Though the study was “negative” (with no genes reaching genome-wide significance), the report is instructive in that it illustrates the strength of multicenter collaboration to achieve sufficient power to conduct such a large analysis.
- [76].Kelley MS, Jacobs MP, Lowenstein DH. NINDS Epilepsy Benchmark Stewards. The NINDS epilepsy research benchmarks. Epilepsia. 2009;50:579–582. doi: 10.1111/j.1528-1167.2008.01813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].1000 Genomes Project Consortium. Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]