Abstract
The elaboration of an effective immune response against pathogenic microbes such as viruses, intracellular bacteria or protozoan parasites relies on the recognition of microbial products called pathogen-associated molecular patterns (PAMPs) by pattern recognition receptors (PRRs) such as Toll-like receptors (TLRs). Ligation of the PRRs leads to synthesis and secretion of pro-inflammatory cytokines and chemokines. Infected cells and other stressed cells also release host-cell derived molecules, called damage-associated molecular patterns (DAMPs, danger signals, or alarmins), which are generic markers for damage. DAMPs are recognized by specific receptors on both immune and nonimmune cells, which, depending on the target cell and the cellular context, can lead to cell differentiation or cell death, and either inflammation or inhibition of inflammation. Recent research has revealed that DAMPs and PAMPs synergize to permit secretion of pro-inflammatory cytokines such as interleukin-1β (IL-1β): PAMPs stimulate synthesis ot pro-IL-1β, but not its secretion; while DAMPs can stimulate assembly ot an inflammasome containing, usually, a Nod-like receptor (NLR) member, and activation of the protease caspase-1, which cleaves pro-IL-1β into IL-1β, allowing its secretion. Other NLR members do not participate in formation of inflammasomes but play other essential roles in regulation of the innate immune response.
Keywords: cytokines, chemokines, danger signals, innate immunity, inflammasome, Nod-like receptors, Toll-like receptors
Pattern recognition receptors in innate immunity
The eradication of pathogenic microorganisms which are themselves constantly evolving relies on the development of an adequate immune response to ensure survival of the host. Key to successful elimination of infection is the ability to initiate a robust innate immune response that coordinates removal of pathogens, toxins, or allergenic structures, while avoiding excessive damage to self-tissues. Upon microbial sensing, the innate immune system initiates the inflammatory response by secreting cytokines and chemokines responsible for recruiting immune cells to the site of infection and inducing the establishment of adaptive immunity.
Over the past decade the field of immunology has witnessed tremendous advances in our understanding of microbial clearance via the discovery of germ-line-encoded pattern recognition receptors (PRRs) and their downstream signaling pathways. PRRs are evolutionarily conserved receptors that detect relatively invariant molecular patterns found in most microorganisms of a given class called pathogen-associated molecular patterns (PAMPs).[1] The best characterized PRRs are the Toll-like receptor (TLR) family members, which constitute at least 12 proteins expressed either on the plasma membrane or in endosomal/lysosomal organelles.[2]
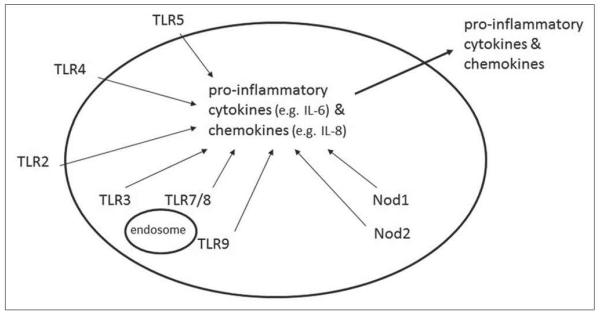
TLRs are composed of a leucine-rich repeat (LRR) domain that is usually involved in ligand binding and microbial sensing, and a cytoplasmic Toll/interleukin-1 (TL-1) receptor (TIR) domain that interacts with TIR domain-containing adaptor molecules such as myeloid differentiation-primary response gene 88 (MyD88).[3] The expression of TLRs is cell specific, assuring an adequate response depending on each cell type and the microbial challenges that they encounter.[4] Signaling through TLRs induces the activation of nuclear factor-κB (NF-κB), which subsequently translocates from the cytoplasm into the nucleus and drives the transcription of specific genes involved in inflammatory, cell survival, or cell proliferation responses.[5] Similarly, TLR3, through its adaptor protein TRIF, induces the activation of the transcription factor, interferon regulatory factor (IRF)-3, required for the production of type I interferon (IFN) responses during viral infection.[6] Ligation of most TLRs and the related cytosolic receptors, the Nod-like receptor (NLR) members Nod1 and Nod2, leads to synthesis of pro-inflammatory cytokines and chemokines, and their secretion [Figure 1].
Figure 1.
TLRs and the cytosolic proteins, Nod1 and Nod2, are “pattern recognition receptors” (PRRs) that recognize microbial products called “pathogen-associated molecular patterns” (PAMPs). As a result of PRR ligation by PAMPs, proinflammatory cytokines and chemokines are synthesized and secreted.
The immune response relies also on the contribution of non-TLR PRR family members such as C-type lectin receptors that recognize complex carbohydrates expressed on the cell surface of multiple pathogens, especially fungi, whose activation triggers endocytosis and phagocytosis of the pathogen as well as cell signaling pathways regulating the innate and adaptive immune system.[7] Some of these receptors, such as Dectin-1, have been shown to cooperate with TLRs, particularly TLR2, to increase the production of pro-inflammatory cytokines, indicating that different PRRs can synergize for better microbial control and effective host resistance.[8]
Recently, the characterization of a novel class of intracellular PRRs has shed light on our understanding of host innate immunity as well as pathogenesis due to inflammation. These are the nucleotide binding and oligomerization domain NLRs, and retinoid acid inducible gene-I (RIG)-like receptors (RLRs), which are intracellular proteins that survey the cytoplasm for signs revealing the presence of not only pathogen-encoded molecules, but also pathogen-encoded activities termed “patterns of pathogenesis”.[9,10] Studies focusing on RIG-I have revealed its function in detecting viral PAMPs and subsequent production of antiviral cytokines such as type I IFNs.[11] Until recently, TLR3 was the main receptor known to be involved in recognition of viral double stranded (ds) RNA and anti-viral signaling via NF-κB and IRF activation. The RIG-I structure consists of two N-terminal caspase recruitment domains (CARDs) followed by an RNA helicase domain. Upon activation by dsRNA, RIG-1 interacts through its CARD domain with a CARD-containing adaptor called CARD adaptor inducing IFN-β (Cardif). As a result, this platform recruits appropriate signaling intermediates, such as IκB kinases (IKKs), to activate NF-κB and IRF transcription factors in order to modulate anti-viral responses.[10,12]
The NLR family members are emerging as regulators of immunity in response to a surprisingly broad variety of pathogens. Members of this family share a similar pattern of domain organization, which consists of an N -terminal effector region such as CARD or a CARD-related ‘X’ domain, pyrin (PYD) or baculovirus inhibitor repeat (BIR) involved in protein-protein interactions, a central nucleotide-binding domain (NBD) required for self oligomerization, and finally an array of C-terminal LRR motifs which are believed to be involved in sensing microbial patterns and modulating NLR activity by folding back onto the NOD domain, thereby inhibiting spontaneous oligomerization.[13,14]
Yet the precise mechanisms whereby NLR LRRs sense their ligands are still unknown. Bioinformatic studies pointed out the presence of 23 NLR proteins in the human genome; however, their functions as well as their respective activators are still under investigation.[15]
Under most circumstances, when the host cell is confronted with a pathogen that expresses a battery of different PAMPs, activation of different families of PRRs governs the combined immune response in order to successfully overcome the microbial challenge with minimal side effects to the host. The tissue distribution and subcellular localization of the PRRs is a good example of the potential strategy that the host has developed in order to discriminate not only between pathogens and commensals, but also between a virulent organism and one that has lower disease-causing potential.
DAMPs and PAMPs
Previous studies have suggested that innate immunity can discriminate between pathogenic microorganisms and commensals. But this raises the question of how the immune system interprets the microbial environment in order to discriminate between a virulent microorganism from one that has lower infectious potential or none.[16] Interestingly, some PAMPs are sensed by the immune system only when they are present at specific locations, such as flagellin, dsRNA, and DNA, which have different effects depending on whether they are present extracellularly or within an infected cell, indicating that the immune system initiates responses based not only on the identity of the PAMPs, but on where and under what cellular context they are presented. Besides the recognition of microbial PAMPs, the immune system senses other signals associated with infection, such as host components released from infected or necrotic cells, which activate and amplify the immune response.[17] These observations show that the immune system provokes a robust response not only against foreign antigens but also against any sign that the host is encountering a dangerous situation, which is reflected by the release of host molecules from the cell.
During trauma or damage-induced responses, many endogenous molecules such as DNA, ATP, uric acid, DNA binding proteins, and reactive oxygen species (ROS) can stimulate the immune response when released to the extracellular milieu.[17,18] These molecules had been variously named damage-associated molecular pattern (DAMPs), alarmins, danger signals, or endogenous adjuvants in order to identify these molecules as host-derived molecules that signal tissue and cell damage [Table 1], Cells of the innate immune system respond to these warning signals via specific receptors and appropriate signaling pathways and thus directly or indirectly promote adaptive immune responses.
Table 1.
Examples of well-characterized DAMPs (danger signals or alarmins)
| DAMPs | Receptors | Outcome of receptor ligation |
|---|---|---|
| Extracellular nucleotides (ATP, ADP, adenosine) |
PI, P2X and P2Y receptors (ATP, ADP); Al, A2A, A2B and A3 receptors (adenosine) |
Dendritic cell (DC) maturation, chemotaxis, secretion of cytokines (IL-1β, IL-18), inflammation |
| Extracellular heat shock proteins |
CD14, CD91, scavenger receptors, TLR4, TLR2, CD40 |
DC maturation, cytokine induction, DC, migration to lymph nodes |
| Extracellular HMGB1 | RAGE, TLR2, TLR4 | Chemotaxis, cytokine induction, DC activation, neutrophil recruitment, inflammation, activation of immune cells |
| Uric acid crystals | CD14, TLR2, TLR4 | DC activation, cytokine induction, neutrophil recruitment, gout induction |
| Oxidative stress | Intracellular redox-sensitive proteins | Cell death, release of endogenous DAMPs, inflammation |
| Laminin | Integrins | Neutrophil recruitment, chemotaxis |
| S100 proteins or calgranulins |
RAGE | Neutrophil recruitment, chemotaxis, cytokine secretion, apoptosis |
| Hyaluronan | TLR2, TLR4, CD44 | DC maturation, cytokine production, adjuvant activity |
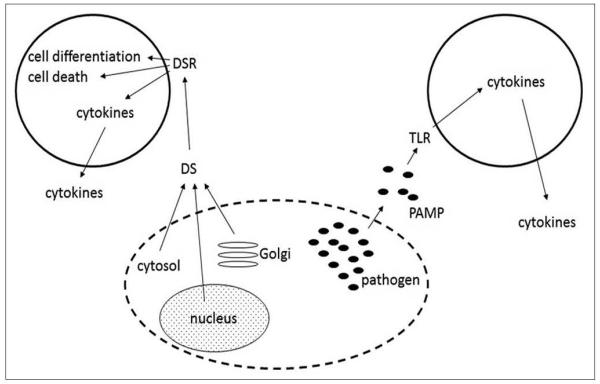
During an infection in vivo, both PAMPs and DAMPs would likely synergize to stimulate a coordinated immune response to infection. PAMPs produced by the pathogen and released into the extracellular space should bind to TLRs and initiate secretion of cytokines. DAMPs released from infected or stressed host cells bind to specific DAMP receptors (DAMPRs), which, depending on the cell expressing the DAMPR and the cellular context, can stimulate cell differentiation, cell death, or secretion of inflammatory or anti-inflammatory mediators [Figure 2]. For example, ATP stimulates inflammation, while adenosine can inhibit inflammation following ligation of the receptor A2a.[19-22]
Figure 2.
DAMPs (danger signals or alarmins) are released from stressed or infected cells. The extracellular DAMPs bind to specific danger signal receptors (DAMPRs), whose ligation can lead to cell differentiation, cell death, or secretion of proinflammatory cytokines. Depending on the DAMP and the target cell, ligation of DAMPRs such as the adenosine receptor A2a can result in inhibition of inflammation.
Some of the DAMPs that have been extensively studied are discussed below.
High-mobility group protein B1 (HMGB1)
This DAMP behaves as both an intracellular transcriptional regulator and an extracellular cytokine/inflammatory mediator. HMGB1 is a nuclear protein that binds to nucleosomes and promotes DNA bending.[23] HMGB1 can be released into the extracellular space primarily by nonapoptotic dying cells, or after translocation into the cytoplasm when acetylated or ADP ribosylated. Once released into the extracellular space, HMGB 1 mediates endothelial cell activation, angiogenesis, stem cell migration, and recruitment and activation of innate immune effectors through TLR2, TLR4 and RAGE (a multi-ligand receptor binding advanced glycation end product.[24-30] Since it has potent extracellular functions, extracellular HMGB 1 is neutralized by soluble RAGE and thrombomodulin. Furthermore, anti-HMGB 1 -neutralizing antibodies had been observed to occur physiologically.[31,32] Several articles have described a role for HMGB1 as a pro-inflammatory agent whose secretion is regulated by NLRP3 and its adaptor, an apoptosis associated speck-like protein containing a CARD domain (ASC).[33,34]
Reactive oxygen species (ROS)
ROS are highly reactive molecules originating from molecular oxygen that are involved in a broad variety of pathologies and aging. A major pool of ROS is produced during oxidative phosphorylation within mitochondria, where electrons are passed through a series of proteins until they reach their final destination on an oxygen molecule.[35] In phagocytic cells, microbial ingestion leads to NADPH oxidase activation, which generates ROS for the main purpose of killing invading microbes. Besides the damaging properties of these oxidant molecules, several studies have highlighted their importance at lower concentrations as cell signaling molecules mediating tyrosine kinase receptor signaling in specific cellular subdomains.[36] The role of ROS in secondary signaling, cross-linking of the cell wall, DNA laddering, protein modification, and cell death is well known.[37] A more recent role for ROS has been described in activation of the NLRP3 inflammasome (see below). Most NLRP3 triggers that have been examined, including ATP, bacterial pore-forming toxins, asbestos and silica, stimulate the generation of short-lived ROS, and treatment with various ROS scavengers blocks NLRP3 activation.[38-39] The molecular mechanisms whereby ROS induce the activation of the NALP3 inflammasome are not fully understood but may involve a thioredoxin-interacting protein (TXNIP; also known as VDuPl) which associates with NALP3 in a ROS-dependent manner.[40] Furthermore, an interesting study suggested that inhibition of autophagy, an evolutionarily conserved cellular process which promotes the turnover of damaged proteins and organelles, disturbs mitochondrial integrity, leading to an increase in mitochondrial ROS production and the release of mitochondrial DNA, which is responsible for activation NLRP3-dependent easpase-1 activation.[41]
Adenosine triphosphate (ATP)
ATP is an important signaling molecule belonging to the purine family. It is produced by cellular respiration and is an indispensable factor for the proper function of a huge variety of enzymes and structural proteins.[42] Extracellular release of ATP takes place in healthy tissue under different circumstances, and is involved in triggering a variety of cellular responses such as neurotransmission, vasodilatation, muscle contraction, and cell growth.[43,44] ATP mediates its effects through ligation of distinct cell-surface purinergic receptors called P2Y (G protein-coupled receptors) and P2X (ligand-gated cation channels), which have distinct effector functions, and pharmacological and tissue distribution profiles.[43]
A substantial body of literature demonstrates the role of ATP as a proinfammatory immunomediator affecting different immune cell functions. Thus, in healthy tissues, release of ATP is tightly controlled and its extracellular concentration is kept low by ubiquitous ecto-ATP/ADPases (CD39). [45] During tissue injury, trauma or cellular stress, the levels of extracellular ATP become elevated and are sensed as a potential threat to surrounding tissues. A growing body of evidence has shown the involvement of extracellular ATP in the recruitment and activation of neutrophils, macrophages and dendritic cells; for instance, ATP acts as a chemotactic agent for dendritic cells (DCs), promotes their maturation, and modulates their production of cytokines.[46,47] ATP can also stimulate monocyte and microglial cell migration, and has a profound impact on ROS and nitric oxide production and nitric oxide synthase expression.[48,49] Evidence based on in vitro and in vivo studies with P2X-deficient mice conclusively identified extracellular ATP as a potent activator of NLRP3-dependent IL-1β release from macrophages. [50-52] The same studies identified the purinergic receptor, P2X7, as the specific receptor responsible for NLRP3 activation. Thus, extracellular ATP triggers K+ efflux, inducing gradual recruitment of the pannexin-1 pore and allowing extracellular NLRP3 agonists to access the cytosol.[53] As many agonists and antagonists exist for P2X7 and new ones are being developed, further elucidation of the link between purinergic signaling and the inflammasome pathway should lead to the discovery of novel therapeutic drugs that could suppress inflammation-related pathogenesis.
Inflammasomes and signalosomes
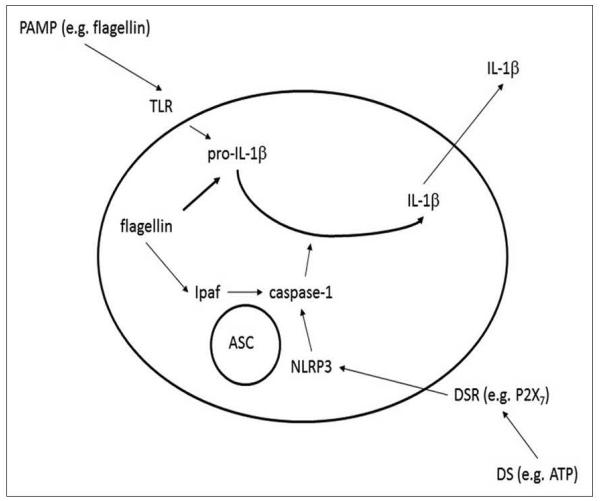
Keeping the body free from invading pathogens is an intricate task that requires cooperation between different PRRs. While responses to extracellular PAMPs are mediated by membrane bound receptors such as TLRs and C-type lectin receptors, many NLRs are specialized for detection of PAMPs that had reached the cytosol or subcellular organelles. Thus, several members of the NLR gene family are involved in the assembly of a macromolecular protein complexes termed ‘inflammasomes”that lead to the activation of the inflammatory cysteine protease, caspase-1 (also known as interleukin-1 converting enzyme or ICE). Caspase-1 in turn cleaves pro-IL-1β or pro-IL-18, resulting in secretion of the mature form of these cytokines [Figure 3].[54,55]
Figure 3.
Danger signals can activate an inflammasome containing an NLR, the adaptor protein ASC, and the protease caspase-1. An extracellular PAMP stimulates synthesis of pro-IL-1β, but not its secretion. Ligation of a DAMP receptor (DAMPR) with a DAMP such as extra-cellular ATP activates an inflammasome and caspase-1. Caspase-1 thus cleaves pro-IL-1β, allowing secretion of the mature cytokine. Different NLR inflammasomes detect different types of clanger: the NLRP3 inflammasome is sensitive to extracellular ATP or toxins that damage the plasma membrane, while NLRC4 is activated by cytosolic flagellin.
Secretion of IL-1β generates fever, induces leukocyte migration to the site of infection, and activates adaptive immunity. [56] On the other hand, IL-18 triggers T helper 1 (TH1) polarization in the presence of IL-12, and a TH2 response via the production of IL-4, IL-5 and IL-10 when IL-12 is lacking. Recent studies have shown that IL-18 can synergize with IL-23 in driving TH17 cell responses.[57-59]
The current widely accepted model of inflammasome assembly proposes that NLRs sense directly or indirectly the presence of pathogens via their C-terminal LRRs, causing the latter to unfold and exhibiting the NOD domains which become free to oligomerize. This exposes the N-terminal effector domain responsible for the recruitment of the adaptors, and leads to initiation of cell signaling.[60] The bipartite adaptor protein ASC probably plays a key role in inflammasome assembly and caspase-1 activation by bridging the NLRs proteins and caspase-1. In fact, ASC contains both a CARD and PYD domain and is responsible of bringing the CARD of the procaspase-1 to the PYD domain of the inflammasome-forming proteins such as NLRP3 and NLRP1 through homotypic interactions, leading to procaspase-1 autocleavage and formation of the active caspase-1 pl0/p20 tetramer.
However, it should be noted that an involvement for murine caspase-11 (also known as caspase-4) has recently been described for caspase-1 activation and IL-1β in macrophages infected with Escherichia coli, Citrobacter rodentium or Vibrio cholcni.” As the caspase-1-deficient mouse lines used by all laboratories until now also lack caspase-11, further studies will be needed to better define the respective roles of caspase-1 and caspase-11 in immune responses to different microbial infections.
Other NLR proteins such as NLRP1 and NLRC4 contain a caspase recruitment domain (CARD) at their carboxyl and amino termini and can therefore interact directly with caspase-1, forming an inflammasome and sometimes bypassing the requirement for the adaptor ASC.[62,63]
Several studies showed that potent caspase-1 activation and the production of IL-1β and IL-18 are markedly decreased in ASC-deficient macrophages infected with different pathogens, or exposed to a variety of DAMPs and crystalline substances.[54,64,65] The exact mechanisms directing NLRC4 inflammasome activation and the participation of ASC remain to be determined. It appears that maximal caspase-1 activation by the NLRC4 inflammasome requires ASC, but NLRC4-de-pendent cell death is independent of ASC upon macrophage infection with S. flexneri[66,67] and P. aeruginosa.[65]
Listed below are the different inflammasomes and signalosomes that have been described until now, as well as the mechanisms leading to inflammasome activation and the role played by inflammasomes in regulating the inflammatory response.
The NLRP3 inflammasome
The NLRP3 (Nalp3) inflammasome is currently the best characterized inflammasome. It is composed of NLRP3, a CARD-containing adaptor ( ASC), and caspase-1. Several bacterial species including Staphylococcus aureus and Listeria monocytogenes can activate the NLRP3 inflammasome by triggering K+ efflux due to their toxins, listeriolysin O, and α-toxin, β-toxin or γ-toxin.[52] Mycobacterium tuberculosis and Mycobacterium marinum use their ESX-1 secretion system to regulate secretion of IL-1β and IL-18 in a process that requires the NLRP3 inflammasome.[68] On the other hand, Klebsiella pneumoniae requires the NLRP3 inflammasome not only for caspase-1 dependent IL-1β secretion, but also for inducing macrophage cell death. Similarly, Neisseria gonorrhoeae infection of monocytes causes IL-1β production, pyronecrosis (a form of cell death involving caspase-1), and HMGB1 release in an NLRP3-dependent manner through a process triggered by the cysteine protease cathepsin B.[69] In contrast, Porphyromonas gingivalis infection of primary epithelial cells fails to activate an inflammasome on its own, but induces synthesis of pro-IL-1β subsequent stimulation with a second signal such as extracellular ATP then allows NLRP3-dependent cleavage and secretion of IL-1β.[70] Conversely, M. tuberculosis subverts the innate immune response by inhibiting NLRP3 inflammasome activation.[71] Chlamydia trachomatis stimulates NLRP3-inflammasome dependent caspase-1 activation through its type III secretion system, by causing K+ efflux-dependent and ROS production in epithelial cells. Caspase-1 activation was shown to be crucial for chlamydial growth and survival,[72] and was required for IL-1β secretion from monocytes or macrophages infected with C. trachomatis or C. pneumoniae,[73-75] The NLRP3 inflammasome can also be activated by viruses such as the Sendai virus, adenovirus, and influenza A virus, whose infections trigger caspase-1-dependent IL-1β secretion.[76,77] Recently, modified vaccinia virus Ankara (MVA), an attenuated poxvirus used as a vector for AIDS vaccines, was shown to activate the NLRP3 inflammasome, leading to secretion of IL-1β.[78] Furthermore, the fungus Candida albicans, signaling through the tyrosine kinase Syk, causes NLRP3 inflammasome-mediated caspase-1 activation and IL-1β secretion in a process triggered by K+ efflux and ROS production.[79,80] The transition of Candida from the yeast to the filamentous form (hyphae formation) is important for activation of the NLRP3 inflammasome.[81] Heat-killed Saccharomyces cerevisiae can also activate the NLRP3 inflammasome.[82] In contrast, the fungal pathogen, Aspergillus fumigatus, activates the NLRP3 inflammasome only after gemination into a mycelium, which is a more threatening form to the host than the conidial stage of this fungus.[83] Moreover, both abiotic and biological crystals such as asbestos, silica and amyloid-β fibrils can activate the NLRP3 inflammasome through “frustrated phagocytosis” of the large components or lysosomal leakage.[84-86] Destabilization of lysosomes by silica crystals, amyloid-β fibrils and alum salts leads to release of the lysosomal protease, cathepsin B, which is somehow sensed by the NLRP3 inflammasome, inducing inflammasome assembly and activation.[85,86] More recently, hemozoin, a heme crystal produced by the malaria-causing parasite, Plasmodium falciparum, was shown to activate the NLRP3 inflammasome through the Lyn/Syk kinase pathway.[39,87] Long-term exposure to these crystals and other environmental irritants can induce aberrant NLRP3 activation, thus inducing inflammation.[65,88,89] Crystals can in fact cause autoinflammatory diseases such as gout, pseudogout, silicosis, and asbestosis. The NLRP3 inflammasome can also be activated by host-derived molecules, demonstrating that our immune system is not only capable of recognizing foreign particles but can also react to endogenous indicators of cellular danger such as extracellular ATP,[52] metabolic stress,[90] fibrillar amyloid-(3 peptide in Alzheimer’s patients,[86] and hyaluronan, which is one of the chief components of the extracellular matrix that is released during tissue injury.[91]
Even though the NLRP3 inflammasome may give the impression of being promiscuous since it can be activated by a wide variety of stimuli, recent studies have shown that all NLRP3 inflammasome activators engage a small number of shared pathways such as K+ efflux through P2X7-gated ion channel,[49,53] production of ROS from the plasma membrane (NADPH oxidase) or mitochondria,[38,92,94] or lysosomal damage or mitochondrial dysfunction, resulting in cytosolic release of lysosomal or mitochondrial contents.[74,85,86,94,95] Interestingly, release of oxidized DNA from mitochondria was recently identified as a trigger for NLRP3 inflammasome activation in cells undergoing apoptosis.[96] Since K+ efflux, ROS production, and perturbation of lysosomal stability can all lead to apoptosis, it is possible that many of the ligands that were previously identified as inducers of NLRP3 inflammasome activation may in fact exert their effect via apoptosis.
The NLRP1 inflammasome
Although less studied than NLRP3, the NLRP1 (Nalpl) inflammasome was the first inflammasome described.[97] It contains a CARD domain that can interact directly with procaspase-1 and bypass the requirement for ASC. However, ASC inclusion in the complex has been described and shown to increase human NLRP1 inflammasome activity.[13] The human NLRP1 inflammasome is stimulated by the presence of cytosolic muramyl dipeptide (MDP, derived from peptidogly-can), which results in activation of caspase-1.[97] In a cell-free system, activation of caspase-1 was shown to occur through a two-step mechanism whereby MDP triggers a conformational change in NLRP1, allowing it to oligomerize into the inflammasome after binding nucleotides.[13] Separate studies on the NLRP1 inflammasome were carried out in mice, which possess three NLRP1 paralogues (NLRPla-c) compared to the sole NLRP1 gene in humans, but they lack functional PYD domains and are therefore incapable of interacting with ASC.[63,98] Susceptibility of mouse macrophages to lethal toxin (LT), a metalloproteinase crucial for Bacillus anthracis pathogenesis, is mediated by theNLRPlb inflammasome.[98] Transfection of fibroblasts with NLRPlb and caspase-1 conferred susceptibility of these cells to anthrax LT.[99]
The NLRC4 inflammasome
The NLRC4 (Ipaf) inflammasome is activated by some Gram-negative bacteria possessing type IE or type IV secretion systems. These bacteria include Salmonella typhimurium, Shigella flexneri, Pseudomonas aeruginosa and Legionella pneumophila. [64-66,100-102] An initial study showed that caspase-1 activation and subsequent IL-1β and IL-18 maturation in response to S. typhimurium infection requires the assembly of the NLRC4 inflammasome,[64] and later work demonstrated that NLRC4 senses the presence of flagellin in the cytosol.[103,104] Similarly, L. pneumophila utilizes its Dot-Icm type IV secretion system to activate the NLRC4 inflammasome and caspase-1, which then restricts growth of Legionella in macrophages.[102] Flagellin from Legionella was shown to be the trigger for assembly of the NLRC4 inflammasome.[105] Unlike Salmonella and Legionella, S. flexneri does not possess flagellin, yet is still capable of activating the NLRC4 inflammasome through a process that requires an intact type III secretion system.[66] Although initial studies showed thatNLRC4 inflammasome activation following P. aeruginosa infection requires flagellin and an intact type HI secretion system,[100,101] it was also observed that either the nonflagellated strain PA103AU or the flagellin-deficient mutant strain PAKAfliC can also activate the NLRC4 inflammasome, leading to IL-1β secretion.[65] These results suggested that NLRC4 could be activated via flagellin-independent pathways. [66,106] In fact, it was recently shown that NLRC4 could also be activated by the type EI secretion apparatus, some of whose components are analogous to flagellin.[107,108] Unlike NLRP3, NLRC4 is not affected by K+ efflux and does not contain a PYD domain, but rather has a CARD domain capable of direct interaction with procaspase-1, making ASC dispensable for NLRC4-dependent caspase-1 activation. [39,109]
The AIM2 inflammasome
AIM2 was identified as a PYHIN (pyrin and HIN domain-containing protein) family member “absent in melanoma 2” that assembles into an inflammasome and controls maturation and secretion of IL-1β and IL-18, whose proinflammatory activities direct host responses against cytosolic double-stranded DNA (dsDNA) from virus, bacteria, or the host itself.[110-113] AIM2 is the first non-NLR discovered so far that can interact with ASC via homotypic PYD-PYD interactions, allowing the ASC CARD domain to recruit procas-pase-1 to the complex whereas its C-terminal HIN domain recognizes and binds to dsDNA. Further characterization of the AIM2 inflammasome could contribute to design of new therapies since the aberrant presence of host DNA has been shown to induce pathological autoimmunity.[114]
The NLRP6 inflammasome
NLRP6 (Nalp6) possesses the structural motifs of molecular sensors and is recruited to “specks” formed in the cytosol by ASC oligomerization.[115,116] It has been known to activate caspase-1,[115] but a role in immunity was not demonstrated until recently, when NLRP6 was found to regulate microbial ecology.[117] Thus, NLRP6-deficient mice revealed an altered gut microbiota, with prominent changes in the representation of members of several bacterial phyla and reduced levels of IL-18.[117] In addition, NLRP6 plays an important role in suppressing inflammation and carcinogenesis by regulating tissue repair and preserving the integrity of the epithelial barrier.[118]
NOD signalosomes
The NLR members, Nod1 and Nod2, are cytosolic sensors of breakdown products of bacterial cell walls (mesodiaminopimelic acid and MDP, respectively), whose recognition leads to activation of the NF-κB pathway through homophilic interactions with the CARD domain containing adaptor serine/threonine kinase, Rip2.[119,120] Previous studies had shown that overexpression of Nod1 and Nod2 leads to their auto-oligomerization, resulting in exposure of their CARD domains, which recruit Rip2, triggering NF-κB activation.[121,122] Recently, the CARD-containing protein, CARD9, has been found to interact with Nod2 and Rip2, and thus to be involved in the activation of JNK and p38.[123] Importantly, mutations in the genes encoding Nod1 and Nod2 are associated with autoinflammatory diseases: mutations in the LRR domains of Nod2 result in loss of bacterial sensing, leading to decreased tolerance toward commensal bacteria seen in Crohn’s disease;[124] and gain of function mutations in the NACHT domain of Nod2 are responsible for Blau syndrome, a rare autoinflammatory disorder affecting the eyes and joints.[125] A recent study suggested that Nod1 also plays a role in the activation of the type IIFN pathway, which plays a key role in pathogen clearance during Helicobacter pylori infection.[126] On the other hand, Nod2 is activated in response to infection with the protozoan parasite Toxoplasma gondii,[121] and Nod1 and Nod2 had been previously reported to sense infection by a large number of bacteria.[128-134] Another study revealed a key role for Nod2 in viral infection, since Nod2-deficient mice are more susceptible to challenge with respiratory syncytial virus (RSV) than wild type mice, and show weakened IFN responses resulting in increased viral burden and death of the infected Nod2-deficient animals.[135]
The function of the different inflammasomes and signalosomes in triggering inflammation in response to a large variety of pathogens is now well established. Indeed, the identification of the ligands and triggers for the inflammasome [Table 2] and the role of the inflammasome in several diseases that had been previously linked to caspase-1 represent a tremendous ad vance in our understanding of the critical importance of NLR family members and their mutations in these disorders.
Table 2.
Main inflammasomes and triggers for their activation
| Inflammasome | Ligands or twriggers |
|---|---|
| NLRP1 | Muramyldipeptide (MDP) from peptidoglycan, for human NLRP1; anthrax lethal toxin (LT), for the murine NLRP1b isoform |
| NLRP3 | Bacteria or bacterial toxins that damage plasma membrane, pathogens that induce cytosolic ROS, some crystals, alum adjuvant |
| NLRC4 | Flagellin, type III secretion apparatus |
| AIM2 | dsDNA |
Immune function of other NLR family members
Possibly the best-characterized member of the NLR family is class II, major histocompatibility complex (MHC), transactivator (CIITA), which regulates expression of MHC class II molecules.[136] However, the functions of other recently-described NLRs that are not known predominantly as inflammasome components have also been described.
NLRC5
NLRC5 (Nalp5) is an NLR protein whose N-terminal domain is most closely related to Nod1, Nod2, and the transcription regulator for MHC class n genes, CIITA.[137,138] Recent publications have described conflicting roles for NLRC5. According to one view, NLRC5 is a negative regulator of type I IFN and NF-κB pathways through interaction, respectively, with RIG-I/MDA5 and IKKα/β,[137,139] and knock-down of NLRC5 enhances innate and antiviral immunity. However, two other studies reported contradictory results: NLRC5 was reported to be a positive regulator of the IFN pathway in epithelial cells and monocytes, and to be required for robust levels of IFN secretion, which potentiates the immune viral response.[140,141] A different role for NLRC5 was reported with regards to MHC class I gene expression.[138] Thus NLRC5 could directly bind to the promoter region of MHC class I and associated genes and induce expression of MHC class I genes,[142] whereas another study described an opposite effect in other cell lines.[139] A separate study revealed a role for NLRC5 in inflammasome formation upon challenge with pathogens, PAMPs and DAMPs, by interacting directly with NLRP3 in order to regulate caspase-1 activity.[143] Moreover, macrophages from NLRC5-deficient mice produced normal levels of IFNγ, IL-6, and TNFα after incubation with RNA viruses, DNA viruses, and bacteria,[144] implying thatNLRC5 was not necessary for cytokine induction during infection. Further understanding of the role of NLRC5 in innate and adaptive immunity awaits a deeper analysis of the immune response of NLRC5-deficient mice.
NLRX1
NLRX1 is a newly characterized NLR protein that contains a CARD-related ‘X’ domain rather than a typical CARD domain. Its function as well as its subcellular localization is still a matter of debate. Recent articles reported that NLRX1 is targeted to the mitochondria. In one case, NLRX1 was located within the mitochondrial matrix; but in the other, it was proposed that NLRX1 is attached to the outer mitochondrial membrane.[145-147] Two different functions have been assigned to NLRX1: either suppressing MAVS-dependent antiviral pathways, or promoting the generation of ROS through interactions with a molecule of the respiratory chain complex HI, called UQCRC2.[145-147] Additional roles have recently been described in cell signaling pathways in the eytosol[148-151] or efficiency of bacterial infection,[152] raising the possibility that the complete functions of this pleiotropic intracellular receptor have yet to be deciphered.
NLRP12
NLRP12 is expressed mainly in the myeloid lineage.[153] Several studies have revealed, that NLRP12 is a negative regulator of TLR-dependent NF-κB activation by preventing hyper-phosphorylation of the TLR adaptor IL-1 receptor associated kinase 1 (IRAK-1) and increasing degradation of theNF-κB-indueing kinase (NIK).[154,155] Activation of NLRP12 requires ATP binding to the NATCH domain, and mutations in this area have been associated with hereditary periodic fever syndromes resembling the autoinflammatory disorders caused by gain-of-function mutations in NLRP3.[156] In agreement with its anti-inflammatory effect, NLRP12 is downregulated following activation by TLR agonists, M. tuberculosis infection, or TNFoc or IFNy treatment.[155]
Concluding remarks
Despite substantial progress in understanding the activation, regulation and function of the different inflammasomes, many gaps remain in our understanding of this sophisticated danger and infection detection system. A full characterization is complicated by the abundance of NLRs and their adaptors and regulators, and the various pathogens and danger signals that trigger the innate immune system. Nonetheless, the production of mice that are deficient for d ifferent NLRs is already aiding in elucidating the roles and physiological functions of these receptors during infection and development of autoinflammatory diseases. As new research elucidates the role of the inflammasome in inflammation, it is not surprising that many studies have focused on identifying its regulators and the evaluation of their physiological importance. Based on sequence studies, regulators of inflammasomes are divided into two families: Those containing a CARD domain, and those with a PYD domain. The PYD domains should interfere with PYD-dependent interactions between the NLRs and their adaptors, and the CARD domains will affect the recruitment of caspase-1 to the inflammasome by modulating CARD-CARD interactions. PYD-containing regulators include Pyrin, POP1, POP2, and viral PYDs (vPYDs), whose mutations are correlated with excessive caspase-1 activation and IL-1 [3 secretion, suggesting that, by interacting with ASC, these regulators may decrease its recruitment to the appropriate inflamamsome. [157,158] Morec>ver9 several autoinflammatory disorders such as familial Mediterranean fever (FMF) and pyogenic arthritis, pyoderma gangrenosum, and acne (PAPA) syndromes have been described in patients who exhibit loss-of-function mutations in, respectively, pyrin or pyrin-interacting proteins, leading to inflammasome hyperactivation.[159,160] Overexpression or gene silencing of these regulators will help to clarify the synergism, feedback loops, and checkpoints that ultimately control inflammasome activation.
Taken together, these studies show that the host innate immune system, through the activation of NLR proteins, is able to respond to “patterns of pathogenesis”, i.e. signals related to the strategies that live pathogens use to invade host cells, replicate intracellularly, and spread through tissues. Regardless of the lessons that may be learned in the future, the discovery of the NLRs and elucidation of their function have already contributed critical information for understanding how an effective innate immune response is established.
Acknowledgments
Work on the inflammasome in our laboratory has been supported by National Institutes of Health grant R01 DE019444-01 and a University of California Presidential Chair.
Biography
Prof. David M. Ojcius

REFERENCES
- 1.Janeway CA., Jr. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;54(Pt 1):1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Takeda K, Akira S. Toll-like receptors in innate immunity. Int Immunol. 2005;17:1–14. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- 3.Takeda K, Kaisho T, Akira S. Toll-like receptors. Arum Rev Immunol. 2003;21:335–76. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 4.Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat Immunol. 2004;5:987–95. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- 5.Brasier AR. The NF-kappaB regulatory network. Cardiovasc Toxicol. 2006;6:111–30. doi: 10.1385/ct:6:2:111. [DOI] [PubMed] [Google Scholar]
- 6.Sato S, Sugiyama M, Yamamoto M, Watanabe Y, Kawai T, Takeda K, et al. Toll/IL-1 receptor domain-containing adaptor inducing IFN-beta (TRIF) associates with TNF receptor-associated factor 6 and TANK-binding kinase 1, and activates two distinct transcription factors, NF-kappa B and IFN-regulatory factor-3, in the Toll-like receptor signaling. J Immunol. 2003;171:4304–10. doi: 10.4049/jimmunol.171.8.4304. [DOI] [PubMed] [Google Scholar]
- 7.Apostolopoulos V, McKenzie IF. Role of the marmose receptor in the immune response. Curr Mol Med. 2001;1:469–74. doi: 10.2174/1566524013363645. [DOI] [PubMed] [Google Scholar]
- 8.Rogers NC, Slack EC, Edwards AD, Nolte MA, Schulz O, Sehweighoffer E, et al. Syk-dependent cytokine induction by Dectin-1 reveals a novel pattern recognition pathway for C type lectins. Immunity. 2005;22:507–17. doi: 10.1016/j.immuni.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 9.Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: Discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meylan E, Tschopp J, Karin M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442:39–44. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- 11.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, tmaizumi T, Miyagishi M, et al. The RNAhelicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–7. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 12.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–8. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 13.Faustin B, Lartigue L, Bruey JM, Luciano F, Sergienko E, Bailly-Maitre B, et al. Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol Cell. 2007;25:713–24. doi: 10.1016/j.molcel.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 14.Duncan JA, Bergstralh DT, Wang Y, Willingham SB, Ye Z, Zimmermann AG, et al. Cryopyrin/NALP3 binds ATP/dATP, is an ATPase, and requires ATP binding to mediate inflammatory signaling. Proc Natl Acad Sci U S A. 2007;104:8041–6. doi: 10.1073/pnas.0611496104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harton JA, Linhoff MW, Zhang J, Ting JP. Cutting edge: CATERPILLER: A large family of mammalian genes containing CARD, pyrin, nucleotide-binding, and leucine-rich repeat domains. J Immunol. 2002;169:4088–93. doi: 10.4049/jimmunol.169.8.4088. [DOI] [PubMed] [Google Scholar]
- 16.Seong SY, Matzinger P. Hydrophobicity: An ancient damage-associated molecular pattern that initiates innate immune responses. Nat Rev Immunol. 2004;4:469–78. doi: 10.1038/nri1372. [DOI] [PubMed] [Google Scholar]
- 17.Kono H, Rock KL. How dying cells alert the immune system to danger. Nat Rev Immunol. 2008;8:279–89. doi: 10.1038/nri2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matzinger P. Tolerance, danger, and the extended family. Annu Rev Immunol. 1994;12:991–1045. doi: 10.1146/annurev.iy.12.040194.005015. [DOI] [PubMed] [Google Scholar]
- 19.Ferrari D, Pizzirani C, Adinolfi E, Lemoli RM, Curti A, Idzko M, et al. The P2X7 receptor: A key player in IL-1 processing and release. J Immunol. 2006;176:3877–83. doi: 10.4049/jimmunol.176.7.3877. [DOI] [PubMed] [Google Scholar]
- 20.Sitkovsky MV, Ohta A. The ‘danger’ sensors that STOP the immune response: The A2 adenosine receptors? Trends Immunol. 2005;26:299–304. doi: 10.1016/j.it.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 21.Lukashev D, Ohta A, Apasov S, Chen JF, Sitkovsky M. Cutting edge: Physiologic attenuation of proinflammatory transcription by the Gs protein-coupled A2A adenosine receptor in vivo. J Immunol. 2004;173:21–4. doi: 10.4049/jimmunol.173.1.21. [DOI] [PubMed] [Google Scholar]
- 22.Sitkovsky MV, Lukashev D, Apasov S, Kojima H, Koshiba M, Caldwell C, et al. Physiological control of immune response and inflammatory tissue damage by hypoxia-inducible factors and adenosine A2A receptors. Annu Rev Immunol. 2004;22:657–82. doi: 10.1146/annurev.immunol.22.012703.104731. [DOI] [PubMed] [Google Scholar]
- 23.Agresti A, Bianchi ME. HMGB proteins and gene expression. Curr Opin Genet Dev. 2003;13:170–8. doi: 10.1016/s0959-437x(03)00023-6. [DOI] [PubMed] [Google Scholar]
- 24.Wang H, Vishnubhakat JM, Bloom O, Zhang M, Ombrellino M, Sama A, et al. Promflammatory cytokines (tumor necrosis factor and interleukin 1) stimulate release of high mobility group protein-1 by pituieytes. Surgery. 1999;126:389–92. [PubMed] [Google Scholar]
- 25.Rendon-Mitchell B, Ochani M, Li J, Han J, Wang H, Yang H, et al. IFN-gamma induces high mobility group box 1 protein release partly through aTNF-dependentmechanism. J Immunol. 2003;170:3890–7. doi: 10.4049/jimmunol.170.7.3890. [DOI] [PubMed] [Google Scholar]
- 26.Messmer D, Yang H, Telusma G, Knoll F, Li J, Messmer B, et al. High mobility group box protein 1: An endogenous signal for dendritic cell maturation and Thl polarization. J Immunol. 2004;173:307–13. doi: 10.4049/jimmunol.173.1.307. [DOI] [PubMed] [Google Scholar]
- 27.Agnello D, Wang H, Yang H, Tracey KJ, Ghezzi P. HMGB-1, a DNA-binding protein with cytokine activity, induces brain TNF and IL-6 production, and mediates anorexia and taste aversion. Cytokine. 2002;18:231–6. doi: 10.1006/cyto.2002.0890. [DOI] [PubMed] [Google Scholar]
- 28.Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002;418:191–5. doi: 10.1038/nature00858. [DOI] [PubMed] [Google Scholar]
- 29.Zong WX, Ditsworth D, Bauer DE, Wang ZQ, Thompson CB. Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes Dev. 2004;18:1272–82. doi: 10.1101/gad.1199904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bierhaus A, Humpert PM, Morcos M, Wendt T, Chavakis T, Arnold B, et al. Understanding RAGE, the receptor for advanced glycation end products. J Mol Med. 2005;83:876–86. doi: 10.1007/s00109-005-0688-7. [DOI] [PubMed] [Google Scholar]
- 31.Urbonaviciute V, Furnrohr BG, Weber C, Haslbeck M, Wilhelm S, Herrmann M, et al. Factors masking HMGB1 in human serum and plasma. J Leukoc Biol. 2007;81:67–74. doi: 10.1189/jlb.0306196. [DOI] [PubMed] [Google Scholar]
- 32.Abeyama K, Stem DM, Ito Y, Kawahara K, Yoshimoto Y, Tanaka M, et al. The N-terminal domain of thrombomodulin sequesters high-mobility group-Bl protein, a novel antiinflammatory mechanism. J Clin Invest. 2005;115:1267–74. doi: 10.1172/JCI22782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamkanfi M, Sarkar A, Vande Walle L, Vitari AC, Amer AO, Wewers MD, et al. Inflammasome-dependent release of the alarmin HMGB1 in endotoxemia. J Immunol. 2010;185:4385–92. doi: 10.4049/jimmunol.1000803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willingham SB, Allen IC, Bergstralh DT, Brickey WJ, Huang MT, Taxman DJ, et al. NLRP3 (NALP3, Cryopyrin) facilitates in vivo caspase-1 activation, necrosis, and HMGB1 release via inflammasome-dependent and -independent pathways. J Immunol. 2009;183:2008–15. doi: 10.4049/jimmunol.0900138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han D, Williams E, Cadenas E. Mitochondrial respiratory chain-dependent generation of superoxide anion and its release into the intermembrane space. Biochem J. 2001;353:411–6. doi: 10.1042/0264-6021:3530411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woo HA, Yim SH, Shin DH, Kang D, Yu DY, Rhee SG. Inactivation of peroxiredoxin I by phosphorylation allows localized H (2)0 (2) accumulation for cell signaling. Cell. 2010;140:517–28. doi: 10.1016/j.cell.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 37.Torres MA, Jones JD, Dangl JL. Reactive oxygen species signaling in response to pathogens. Plant Physiol. 2006;141:373–8. doi: 10.1104/pp.106.079467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dostert C, Petrilli V, Van Bruggen R, Steele C, Mossman BT, Tschopp J. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science. 2008;320:674–7. doi: 10.1126/science.1156995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petrilli V, Papin S, Dostert C, Mayor A, Martinon F, Tschopp J. Activation ofthe NALP3 inflammasomeis triggered by low intracellular potassium concentration. Cell Death Differ. 2007;14:1583–9. doi: 10.1038/sj.cdd.4402195. [DOI] [PubMed] [Google Scholar]
- 40.Zhou R, Tardivel A, Thorens B, Choi I, Tschopp J. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat Immunol. 2010;11:136–40. doi: 10.1038/ni.1831. [DOI] [PubMed] [Google Scholar]
- 41.Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNAmediated by the NALP3 inflammasome. Nat Immunol. 2010 doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar V, Sharma A. Adenosine: An endogenous modulator of innate immune system with therapeutic potential. Eur J Pharmacol. 2009;616:7–15. doi: 10.1016/j.ejphar.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 43.Ralevic V, Bunistock G. Receptors for purines and pyrimidines. Pharmacol Rev. 1998;50:413–92. [PubMed] [Google Scholar]
- 44.Gordon JL. Extracellular ATP: Effects, sources and fate. Biochem J. 1986;233:309–19. doi: 10.1042/bj2330309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robson SC, Kaezmarek E, Siegel JB, Candinas D, Koziak K, Millan M, et al. Loss of ATP diphosphohydxolase activity with endothelial cell activation. J Exp Med. 1997;185:153–63. doi: 10.1084/jem.185.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maxteau F, Communi D, Boeynaems JM, Suarez Gonzalez N. Involvement of multiple P2Y receptors and signaling pathways in the action of adenine nucleotides diphosphates on human monocyte-derived dendritic cells. J Leukoc Biol. 2004;76:796–803. doi: 10.1189/jlb.0104032. [DOI] [PubMed] [Google Scholar]
- 47.Schnurr M, Then F, Galambos P, Scholz C, Siegmund B, Endres S, et al. Extracellular ATP and TNF-alpha synergize in the activation and maturation of human dendritic cells. J Immunol. 2000;165:4704–9. doi: 10.4049/jimmunol.165.8.4704. [DOI] [PubMed] [Google Scholar]
- 48.Bours MJ, Swennen EL, Di Virgilio F, Cronstein BN, Dagnelie PC. Adenosine 5′-triphosphate and adenosine as endogenous signaling molecules in immunity and inflammation. Pharmacol Ther. 2006;112:358–404. doi: 10.1016/j.pharmthera.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 49.Kahlenberg JM, Dubyak GR. Mechanisms of caspase-1 activation by P2X7 receptor-mediated K+release. Am J Physiol Cell Physiol. 2004;286:C1100–8. doi: 10.1152/ajpcell.00494.2003. [DOI] [PubMed] [Google Scholar]
- 50.Labasi JM, Petrushova N, Donovan C, McCurdy S, Lira P, Payette MM, et al. Absence of the P2X7 receptor alters leukocyte function and attenuates an inflammatory response. J Immunol. 2002;168:6436–45. doi: 10.4049/jimmunol.168.12.6436. [DOI] [PubMed] [Google Scholar]
- 51.Solle M, Labasi J, Perregaux DG, Stam E, Petrushova N, Koller BH, et al. Altered cytokine production in mice lacking P2X7 receptors. J Biol Chem. 2001;276:125–32. doi: 10.1074/jbc.M006781200. [DOI] [PubMed] [Google Scholar]
- 52.Mariathasan S, Weiss DS, Newton K, McBride J, O’Rourke K, Roose-Girma M, et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–32. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- 53.Kanneganti TD, Lamkanfi M, Kim YG, Chen G, Park JH, Franchi L, et al. Pannexin-1 -mediated recognition of bacterial molecules activates the cryopy’rin inflammasome independent of Toll-like receptor signaling. Immunity. 2007;26:433–43. doi: 10.1016/j.immuni.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 54.Mariathasan S. ASC, Ipaf and Cryopyrin/Nalp3: Bona fide intracellular adapters of the caspase-1 inflammasome. Microbes Infect. 2007;9:664–71. doi: 10.1016/j.micinf.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 55.Martinon F, Mayor A, Tschopp J. The inflammasomes: Guardians of the body. Annu Rev Immunol. 2009;27:229–65. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- 56.Sims JE, Smith DE. The IL-1 family: Regulators of immunity. Nat Rev Immunol. 2010;10:89–102. doi: 10.1038/nri2691. [DOI] [PubMed] [Google Scholar]
- 57.Nakanishi K, Yoshimoto T, Tsutsui H, Okamura H. Interleukin-18 regulates both Thl and Th2 responses. Annu Rev Immunol. 2001;19:423–74. doi: 10.1146/annurev.immunol.19.1.423. [DOI] [PubMed] [Google Scholar]
- 58.Dinarello CA. Immunological and inflammatory functions of the interleukin-1 family. Annu Rev Immunol. 2009;27:519–50. doi: 10.1146/annurev.immunol.021908.132612. [DOI] [PubMed] [Google Scholar]
- 59.Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, et al. Interleukin 17-produeing CD4+effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6:1123–32. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 60.Inohara N, Koseki T, del Peso L, Hu Y, Yee C, Chen S, et al. Nod1, an Apaf-l-like activator of caspase-9 and nuclear factor-kappaB. J Biol Chem. 1999;274:14560–7. doi: 10.1074/jbc.274.21.14560. [DOI] [PubMed] [Google Scholar]
- 61.Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J, et al. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479:117–21. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- 62.Povet JL, Srinivasula SM, Tnani M, Razmara M, Femandes-Alnemri T, Alnemri ES. Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. J Biol Chem. 2001;276:28309–13. doi: 10.1074/jbc.C100250200. [DOI] [PubMed] [Google Scholar]
- 63.Hsu LC, Ah SR, McGillivray S, Tseng PH, Mariathasan S, Humke EW, et al. A NOD2-NALP1 complex mediates caspase-1-dependent IL-lbeta secretion in response to Bacillus anthracis infection and muramyl dipeptide. Proc Natl Acad Sci U S A. 2008;105:7803–8. doi: 10.1073/pnas.0802726105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mariathasan S, Newton K, Monack DM, Vucic D, French DM, Lee WP, et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430:213–8. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- 65.Sutterwala FS, Mijares LA, Li L, Ogura Y, Kazmierczak BI, Flavell RA. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J Exp Med. 2007;204:3235–45. doi: 10.1084/jem.20071239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Suzuki T, Franchi L, Toma C, Ashida H, Ogawa M, Yoshikawa Y, et al. Differential regulation of caspase-1 activation, pyroptosis, and autophagy via Ipaf and ASC in Shigella-infected macrophages. PLoS Pathog. 2007;3:e111. doi: 10.1371/journal.ppat.0030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Suzuki T, Nunez G. A role for Nod-like receptors in autophagy induced by Shigella infection. Autophagy. 2008;4:73–5. doi: 10.4161/auto.5101. [DOI] [PubMed] [Google Scholar]
- 68.Koo IC, Wang C, Raghavan S, Morisaki JH, Cox JS, Brown EJ. ESX-1 -dependent cytolysis in lysosome secretion and inflammasome activation during mycobacterial infection. Cell Microbiol. 2008;10:1866–78. doi: 10.1111/j.1462-5822.2008.01177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Duncan JA, Gao X, Huang MT, O’Connor BP, Thomas CE, Willingham SB, et al. Neisseria gonorrhoeae activates the proteinase cathepsin B to mediate the signaling activities of the NLRP3 and ASC-containing inflammasome. J Immunol. 2009;182:6460–9. doi: 10.4049/jimmunol.0802696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yilmaz O, Sater AA, Yao L, Koutouzis T, Pettengill M, Ojcius DM. ATP-dependent activation of an inflammasome in primary gingival epithelial cells infected by Porphyromonas gingivalis. Cell Microbiol. 2010;12:188–98. doi: 10.1111/j.1462-5822.2009.01390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Master SS, Rampini SK, Davis AS, Keller C, Ehlers S, Springer B, et al. Mycobacterium tuberculosis prevents inflammasome activation. Cell Host Microbe. 2008;3:224–32. doi: 10.1016/j.chom.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Abdul-Sater AA, Koo E, Hacker G, Ojcius DM. Inflammasome-dependent caspase-1 activation in cervical epithelial cells stimulates growth of the intracellular pathogen Chlamydia trachomatis. J Biol Chem. 2009;284:26789–96. doi: 10.1074/jbc.M109.026823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.He X, Mekasha S, Mavrogiorgos N, Fitzgerald KA, Lien E, Ingalls RR. Inflammation and fibrosis during Chlamydia pneumoniae infection is regulated by IL-1 and the NLRP3/ASC inflammasome. J Immunol. 2010;184:5743–54. doi: 10.4049/jimmunol.0903937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shimada K, Crother TR, Karlin J, Chen S, Chiba N, Ramanujan VK, et al. Caspase-1 dependent IL-1 beta secretion is critical for host defense in a mouse model of Chlamydia pneumoniae lung infection. PLoS One. 2011;6:e21477. doi: 10.1371/journal.pone.0021477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Abdul-Sater AA, Said-Sadier N, Padilla EV, Ojcius DM. Chlamydial infection of monocytes stimulates IL-lbeta secretion through activation of the NLRP3 inflammasome. Microbes Infect. 2010;12:652–61. doi: 10.1016/j.micinf.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kanneganti TD, Body-Malapel M, Amer A, Park JH, Whitfield J, Franchi L, et al. Critical role for Cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J Biol Chem. 2006;281:36560–8. doi: 10.1074/jbc.M607594200. [DOI] [PubMed] [Google Scholar]
- 77.Ichinohe T, Lee HK, Ogura Y, Flavell R, Iwasaki A. Inflammasome recognition of influenza virus is essential for adaptive immune responses. J Exp Med. 2009;206:79–87. doi: 10.1084/jem.20081667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Delaloye J, Roger T, Steiner-Tardivel QG, Le Roy D, Knaup Reymond M, Akira S, et al. Innate immune sensing of modified vaccinia virus Ankara (MVA) is mediated by TLR2-TLR6, MDA-5 and the NALP3 inflammasome. PLoS Pathol. 2009;5:e1000480. doi: 10.1371/journal.ppat.1000480. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 79.Gross O, Poeck H, Bscheider M, Dostert C, Hannesschlager N, Endres S, et al. Syk kinase signalling couples to the Nlrp3 inflammasome for anti-fungal host defence. Nature. 2009;459:433–6. doi: 10.1038/nature07965. [DOI] [PubMed] [Google Scholar]
- 80.Hise AG, Tomalka J, Ganesan S, Patel K, Hall BA, Brown GD, et al. An essential role for the NLRP3 inflammasome in host defense against the human fungal pathogen Candida albicans. Cell Host Microbe. 2009;5:487–97. doi: 10.1016/j.chom.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Joly S, Ma N, Sadler JJ, Soli DR, Cassel SL, Sutterwala FS. Cutting edge: Candida albicans hyphae formation triggers activation of the Nlrp3 inflammasome. J Immunol. 2009;183:3578–81. doi: 10.4049/jimmunol.0901323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lamkanfi M, Malireddi RK, Kanneganti TD. Fungal zymosan and mamian activate the cryopynn inflammasome. J Biol Chem. 2009;284:20574–81. doi: 10.1074/jbc.M109.023689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Said-Sadier N, Padilla E, Langsley G, Ojcius DM. Aspergillus fumigatus stimulates the NLRP3 inflammasome through apathway requiring ROS production and the Syk tyrosine kinase. PLoS One. 2010;5:e10008. doi: 10.1371/journal.pone.0010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Franchi L, Eigenbrod T, Nunez G. Cutting edge: TNF-alphamediates sensitization to ATP and silica via the NLRP3 inflammasome in the absence of microbial stimulation. J Immunol. 2009;183:792–6. doi: 10.4049/jimmunol.0900173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008;9:847–56. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Halle A, Hornung V, Petzold GC, Stewart CR, Monks BG, Reinheckel T, et al. The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat Immunol. 2008;9:857–65. doi: 10.1038/ni.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dostert C, Guarda G, Romero JF, Menu P, Gross O, Tardivel A, et al. Malarial hemozoinis aNalp3 inflammasome activating danger signal. PLoS One. 2009;4:e6510. doi: 10.1371/journal.pone.0006510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Feldmeyer L, Keller M, Niklaus G, Hohl D, Werner S, Beer HD. The inflammasome mediates UVB-induced activation and secretion of interleukin-1beta by keratmocytes. Curr Biol. 2007;17:1140–5. doi: 10.1016/j.cub.2007.05.074. [DOI] [PubMed] [Google Scholar]
- 89.Watanabe H, Gaide O, Petrilli V, Martinon F, Contassot E, Roques S, et al. Activation of the IL-1 beta-processing inflamm asome is involved in contact hypersensitivity. J Invest Dermatol. 2007;127:1956–63. doi: 10.1038/sj.jid.5700819. [DOI] [PubMed] [Google Scholar]
- 90.Schroder K, Zhou R, Tschopp J. The NLRP3 inflammasome: A sensor for metabolic danger? Science. 2010;327:296–300. doi: 10.1126/science.1184003. [DOI] [PubMed] [Google Scholar]
- 91.Yamasaki K, Muto J, Taylor KR, Cogen AL, Audish D, Bertin J, et al. NLRP3/cryopymi is necessary for interleukin-1 beta (IL-lbeta) release in response to hyaluronan, an endogenous trigger of inflammation in response to injury. J Biol Chem. 2009;284:12762–71. doi: 10.1074/jbc.M806084200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cruz CM, Rinna A, Forman HJ, Ventura AL, Perseehini PM, Ojcius DM. ATP activates a reactive oxygen species-dependent oxidative stress response and secretion of proinflammatory cytokines in macrophages. J Biol Chem. 2007;282:2871–9. doi: 10.1074/jbc.M608083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cassel SL, Eisenbarth SC, Iyer SS, Sadler JJ, Colegio OR, Tephly LA, et al. The Nalp3 inflammasome is essential for the development of silicosis. Proc Natl Acad Sci U S A. 2008;105:9035–40. doi: 10.1073/pnas.0803933105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–5. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 95.Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–30. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shimada K, Crother TR, Karlin J, Dagvadoq J, Chiba N, Chen S, et al. Activation of the NLRP3 inflammasome by oxidized mitochondrial DNA during apoptosis. Immunity. 2012;36:401–14. doi: 10.1016/j.immuni.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Martinon F, Burns K, Tschopp J. The inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–26. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 98.Moayeri M, Sastalla I, Leppla SH. Anthrax and the inflammasome. Microbes Infect. 2012;14:392–400. doi: 10.1016/j.micinf.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liao KC, Mogridge J. Expression of Nlrplb inflammasome components in human fibroblasts confers susceptibility to anthrax lethal toxin. Infect Immun. 2009;77:4455–62. doi: 10.1128/IAI.00276-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Miao EA, Ernst RK, Dors M, Mao DP, Aderem A. Pseudomonas aeruginosa activates caspase 1 through Ipaf. Proc Natl Acad Sci U S A. 2008;105:2562–7. doi: 10.1073/pnas.0712183105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Franchi L, Stoolman J, Kanneganti TD, Verma A, Ramphal R, Nunez G. Critical role for Ipaf in Pseudomonas aeruginosa-induced caspase-1 activation. Eur J Immunol. 2007;37:3030–9. doi: 10.1002/eji.200737532. [DOI] [PubMed] [Google Scholar]
- 102.Zamboni DS, Kobayashi KS, Kohlsdorf T, Ogura Y, Long EM, Vance RE, et al. The Bircle cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat Immunol. 2006;7:318–25. doi: 10.1038/ni1305. [DOI] [PubMed] [Google Scholar]
- 103.Miao EA, Alpuche-Aranda CM, Dors M, Clark AE, Bader MW, Miller SI, et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin lbeta via Ipaf. Nat Immunol. 2006;7:569–75. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- 104.Franchi L, Amer A, Body-Malapel M, Kanneganti TD, Ozoren N, Jagirdar R, et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin lbeta in salmonella-infected macrophages. Nat Immunol. 2006;7:576–82. doi: 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- 105.Lightfield KL, Persson J, Brubaker SW, Witte CE, von Moltke J, Dunipace EA, et al. Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nat Immunol. 2008;9:1171–8. doi: 10.1038/ni.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sun YH, Rolan HG, Tsolis RM. Injection of flagellin into the host cell cytosol by Salmonella enterica serotype Typhimurium. J Biol Chem. 2007;282:33897–901. doi: 10.1074/jbc.C700181200. [DOI] [PubMed] [Google Scholar]
- 107.Zhao Y, Yang J, Shi J, Gong YN, Lu Q, Xu H, et al. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 2011;477:596–600. doi: 10.1038/nature10510. [DOI] [PubMed] [Google Scholar]
- 108.Kofoed EM, Vance RE. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature. 2011;477:592–5. doi: 10.1038/nature10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Case CL, Shin S, Roy CR. Asc and Ipaf Inflammasomes direct distinct pathways for caspase-1 activation in response to Legionella pneumophila. Infect Immun. 2009;77:1981–91. doi: 10.1128/IAI.01382-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–8. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Burckstummer T, Baumann C, Bluml S, Dixit E, Durnberger G, Jahn H, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–72. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 112.Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, et al. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–60. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 113.Muruve DA, Petrilli V, Zaiss AK, White LR, Clark SA, Ross PJ, et al. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452:103–7. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- 114.Sun KH, Yu CL, Tang SJ, Sun GH. Monoclonal anti-double-stranded DNA autoantibody stimulates the expression and release of IL-1 beta, IL-6, IL-8, IL-10 and TNF-alpha from normal human mononuclear cells involving in the lupus pathogenesis. Immunology. 2000;99:352–60. doi: 10.1046/j.1365-2567.2000.00970.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Grenier JM, Wang L, Manji GA, Huang WJ, Al-Garawi A, Kelly R, et al. Functional screening offive PYPAF family members identifies PYPAF5 as a novel regulator of NF-kappaB and caspase-1. FEBS Lett. 2002;530:73–8. doi: 10.1016/s0014-5793(02)03416-6. [DOI] [PubMed] [Google Scholar]
- 116.Wang L, Manji GA, Grenier JM, Al-Garawi A, Merriam S, Lora JM, et al. PYPAF7, a novel PYRIN-containing Apafl-like protein that regulates activation ofNF-kappa B and caspase-1 -dependent cytokine processing. J Biol Chem. 2002;277:29874–80. doi: 10.1074/jbc.M203915200. [DOI] [PubMed] [Google Scholar]
- 117.Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–57. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Normand S, Delanoye-Crespin A, Bressenot A, Huot L, Grandjean T, Peyrin-Biroulet L, et al. Nod-like receptor pyrin domain-containing protein 6 (NLRP6) controls epithelial self-renewal and colorectal carcinogenesis upon injury. Proc Natl Acad Sci U S A. 2011;108:9601–6. doi: 10.1073/pnas.1100981108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.McDonald C, Inohara N, Nunez G. Peptidoglycan signaling in innate immunity and inflammatory disease. J Biol Chem. 2005;280:20177–80. doi: 10.1074/jbc.R500001200. [DOI] [PubMed] [Google Scholar]
- 120.Girardin SE, Philpott DJ. Mini-review: The role of peptidoglycan recognition in innate immunity. Eur J Immunol. 2004;34:1777–82. doi: 10.1002/eji.200425095. [DOI] [PubMed] [Google Scholar]
- 121.Bertin J, Nir WJ, Fischer CM, Tayber OV, Errada PR, Grant JR, et al. Human CARD4 protein is a novel CED-4/Apaf-l cell death family member that activates NF-kappaB. J Biol Chem. 1999;274:12955–8. doi: 10.1074/jbc.274.19.12955. [DOI] [PubMed] [Google Scholar]
- 122.Inohara N, del Peso L, Koseki T, Chen S, Nunez G. RICK, a novel protein kinase containing a caspase recruitment domain, interacts with CLARP and regulates CD95-mediated apoptosis. J Biol Chem. 1998;273:12296–300. doi: 10.1074/jbc.273.20.12296. [DOI] [PubMed] [Google Scholar]
- 123.Hsu YM, Zhang Y, You Y, Wang D, Li H, Duramad O, et al. The adaptor protein CARD9 is required for innate immune responses to intracellular pathogens. Nat Immunol. 2007;8:198–205. doi: 10.1038/ni1426. [DOI] [PubMed] [Google Scholar]
- 124.Vignal C, Singer E, Peyrin-Biroulet L, Desreumaux P, Chamaillard M. How NOD2 mutations predispose to Crohn’s disease? Microbes Infect. 2007;9:658–63. doi: 10.1016/j.micinf.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 125.Tanabe T, Chamaillard M, Ogura Y, Zhu L, Qiu S, Masumoto J, et al. Regulatory regions and critical residues of NOD2 involved in muramyl dipeptide recognition. EMBO J. 2004;23:1587–97. doi: 10.1038/sj.emboj.7600175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Watanabe T, Asano N, Fiehtner-Feigl S, Gorelick PL, Tsuji Y, Matsumoto Y, et al. NOD 1 contributes to mouse host defense against Helicobacter pylori via induction of type I IFN and activation of the ISGF3 signaling pathway. J Clin Invest. 2010;120:1645–62. doi: 10.1172/JCI39481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shaw MH, Reimer T, Sanchez-Valdepenas C, Warner N, Kim YG, Fresno M, et al. T cell-intrinsic role of Nod2 in promoting type 1 immunity to Toxoplasma gondii. Nat Immunol. 2009;10:1267–74. doi: 10.1038/ni.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Viala J, Chaput C, Boneca IG, Cardona A, Girardin SE, Moran AP, et al. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat Immunol. 2004;5:1166–74. doi: 10.1038/ni1131. [DOI] [PubMed] [Google Scholar]
- 129.Opitz B, Forster S, Hocke AC, Maass M, Schmeck B, Hippenstiel S, et al. Nod1-mediated endothelial cell activation by Chlamydophila pneumoniae. Circ Res. 2005;96:319–26. doi: 10.1161/01.RES.0000155721.83594.2c. [DOI] [PubMed] [Google Scholar]
- 130.Travassos LH, Carneiro LA, Girardin SE, Boneca IG, Lemos R, Bozza MT, et al. Nod1 participates in the innate immune response to Pseudomonas aeruginosa. J Biol Chem. 2005;280:36714–8. doi: 10.1074/jbc.M501649200. [DOI] [PubMed] [Google Scholar]
- 131.Welter-Stahl L, Ojcius DM, Viala J, Girardin S, Liu W, Delarbre C, et al. Stimulation of the cytosolic receptor for peptidoglycan, Nod1, by infection with Chlamydia trachomatis or Chlamydia muridarum. Cell Microbiol. 2006;8:1047–57. doi: 10.1111/j.1462-5822.2006.00686.x. [DOI] [PubMed] [Google Scholar]
- 132.Lysenko ES, Clarke TB, Shchepetov M, Ratner AJ, Roper DI, Dowson CG, et al. Nod1 signaling overcomes resistance of S. pneumoniae to opsonophagocytic killing. PLoS Pathog. 2007;3:ell8. doi: 10.1371/journal.ppat.0030118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Berrington WR, Iyer R, Wells RD, Smith KD, Skerrett SJ, Hawn TR. NOD1 and NOD2 regulation of pulmonary innate immunity to Legionella pneumophila. Eur J Immunol. 2010;40:3519–27. doi: 10.1002/eji.201040518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Frutuoso MS, Hori JI, Pereira MS, Junior DS, Sonego F, Kobayashi KS, et al. The pattern recognition receptors Nod1 and Nod2 account for neutrophil recruitment to the lungs of mice infected with Legionella pneumophila. Microbes Infect. 2010;12:819–27. doi: 10.1016/j.micinf.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 135.Sabbah A, Chang TH, Hamack R, Frohhch V, Tominaga K, Dube PH, et al. Activation of innate immune antiviral responses by Nod2. Nat Immunol. 2009;10:1073–80. doi: 10.1038/ni.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Harton JA, Ting JP. Class II trans activator: Mastering the art of major histocompatibility complex expression. Mol Cell Biol. 2000;20:6185–94. doi: 10.1128/mcb.20.17.6185-6194.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cui J, Zhu L, Xia X, Wang HY, Legras X, Hong J, Ji J, et al. NLRC5 negatively regulates the NF-kappaB and type I interferon signaling pathways. Cell. 2010;141:483–96. doi: 10.1016/j.cell.2010.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Meissner TB, Li A, Kobayashi KS. NLRC5: A newly discovered MHC class I transactivator (CITA) Microbes Infect. 2012;14:477–84. doi: 10.1016/j.micinf.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Benko S, Magalhaes JG, Philpott DJ, Girardin SE. NLRC5 limits the activation of inflammatory pathways. J Immunol. 2010;185:1681–91. doi: 10.4049/jimmunol.0903900. [DOI] [PubMed] [Google Scholar]
- 140.Neerincx A, Lautz K, Menning M, Kremmer E, Zigrino P, Hosel M, et al. A role for the human nucleotide-binding domain, leucine-rich repeat-containing family member NLRC5 in antiviral responses. J Biol Chem. 2010;285:26223–32. doi: 10.1074/jbc.M110.109736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kuenzel S, Till A, Winkler M, Hasler R, Lipinski S, Jung S, et al. The nucleotide-binding oligomerization domain-like receptor NLRC5 is involved in IFN-dependent ‘antiviral immune responses. J Immunol. 2010;184:1990–2000. doi: 10.4049/jimmunol.0900557. [DOI] [PubMed] [Google Scholar]
- 142.Meissner TB, Li A, Biswas A, Lee KH, Liu YJ, Bayir E, et al. NLR family member NLRC5 is a transcriptional regulator of MHC class I genes. Proc Natl Acad Sci U S A. 2010;107:13794–9. doi: 10.1073/pnas.1008684107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Davis BK, Roberts RA, Huang MT, Willingham SB, Conti BJ, Briekey WJ, et al. Cutting Edge: NLRC5-Dependent Activation of the Inflammasome. J Immunol. 2011;186:1333–7. doi: 10.4049/jimmunol.1003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kumar H, Pandey S, Zou J, Kumagai Y, Takahashi K, Akira S, et al. NLRC5 deficiency does not influence cytokine induction by virus and bacteria infections. J Immunol. 2011;186:994–1000. doi: 10.4049/jimmunol.1002094. [DOI] [PubMed] [Google Scholar]
- 145.Moore CB, Bergstralh DT, Duncan JA, Lei Y, Morrison TE, Zimmermann AG, et al. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature. 2008;451:573–7. doi: 10.1038/nature06501. [DOI] [PubMed] [Google Scholar]
- 146.Tattoli I, Carneiro LA, Jehanno M, Magalhaes JG, Shu Y, Philpott DJ, et al. NLRX1 is a mitochondrial NOD-like receptor that amplifies NF-kappaB and JNK pathways by inducing reactive oxygen species production. EMBO Rep. 2008;9:293–300. doi: 10.1038/sj.embor.7401161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Amoult D, Soares F, Tattoli I, Castanier C, Philpott DJ, Girardin SE. An N-terminal addressing sequence targets NLRX1 to the mitochondrial matrix. J Cell Sci. 2009;122:3161–8. doi: 10.1242/jcs.051193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Allen IC, Moore CB, Schneider M, Lei Y, Davis BK, Scull MA, et al. NLRX1 protein attenuates inflammatory responses to infection by interfering with the RIG-I-MAVS and TRAF6-NF-kappaB signaling pathways. Immunity. 2011;34:854–65. doi: 10.1016/j.immuni.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Xia X, Cui J, Wang HY, Zhu L, Matsueda S, Wang Q, et al. NLRX1 negatively regulates TLR-induced NF-kappaB signaling by targeting TRAF6 and IKK. Immunity. 2011;34:843–53. doi: 10.1016/j.immuni.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Parvatiyar K, Cheng G. NOD so fast: NLRX1 puts the brake on inflammation. Immunity. 2011;34:821–2. doi: 10.1016/j.immuni.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Rebsamen M, Vazquez J, Tardivel A, Guarda G, Curran J, Tschopp J. NLRX1/NOD5 deficiency does not affect MAVS signalling. Cell Death Differ. 2011;18:1387. doi: 10.1038/cdd.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Abdul-Sater AA, Said-Sadier N, Lam VM, Singh B, Pettengill MA, Soares F, et al. Enhancement of reactive oxygen species production and chlamydial infection by the mitochondrial Nod-like famify member NLRX1. J Biol Chem. 2010;285:41637–45. doi: 10.1074/jbc.M110.137885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Williams KL, Taxman DJ, Linhoff MW, Reed W, Ting JP. Cutting edge: Monarch-1: A pjiin/nucleotide-binding domain/leucine-rich repeat protein that controls classical and nonclassical MHC class I genes. J Immunol. 2003;170:5354–8. doi: 10.4049/jimmunol.170.11.5354. [DOI] [PubMed] [Google Scholar]
- 154.Lich JD, Williams KL, Moore CB, Arthur JC, Daws BK, Taxman DJ, et al. Monarch-1 suppresses non-canonical NF-kappaB activation and p52-dependent chemokine expression in monocytes. J Immunol. 2007;178:1256–60. doi: 10.4049/jimmunol.178.3.1256. [DOI] [PubMed] [Google Scholar]
- 155.Williams KL, Lich JD, Duncan JA, Reed W, Rallabhandi P, Moore C, et al. The CATERPILLER protein monarch-1 is an antagonist of toll-like receptor-, tumor necrosis factor alpha-, and Mycobacterium tuberculosis-induced pro-inflammatory signals. J Biol Chem. 2005;280:39914–24. doi: 10.1074/jbc.M502820200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jeru I, Duquesnoy P, Fernandes-Alnemri T, Cochet E, Yu JW, Lackmy-Port-Lis M, et al. Mutations in NALP12 cause hereditary periodic fever syndromes. Proc Natl Acad Sci U S A. 2008;105:1614–9. doi: 10.1073/pnas.0708616105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Consortium TF A candidate gene for familial Mediterranean fever. Nat Genet. 1997;17:25–31. doi: 10.1038/ng0997-25. [DOI] [PubMed] [Google Scholar]
- 158.Chae JJ, Komarow HD, Cheng J, Wood G, Raben N, Liu PP, et al. Targeted disruption of pyrin, the FMF protein, causes heightened sensitivity to endotoxin and a defect in macrophage apoptosis. Mol Cell. 2003;11:591–604. doi: 10.1016/s1097-2765(03)00056-x. [DOI] [PubMed] [Google Scholar]
- 159.McDermott MF. A common pathway in periodic fever syndromes. Trends Immunol. 2004;25:457–60. doi: 10.1016/j.it.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 160.Shoham NG, Centola M, Mansfield E, Hull KM, Wood G, Wise CA, et al. Pyrin binds the PSTPIP1/CD2BP1 protein, defining familial Mediterranean fever and PAPA syndrome as disorders in the same pathway. Proc Natl Acad Sci U S A. 2003;100:13501–6. doi: 10.1073/pnas.2135380100. [DOI] [PMC free article] [PubMed] [Google Scholar]