Abstract
The phosphatase Rtr1 has been implicated in dephosphorylation of the RNA Polymerase II (RNAPII) C-terminal domain (CTD) during transcription elongation and in regulation of nuclear import of RNAPII. Although it has been shown that Rtr1 interacts with RNAPII in yeast and humans, the specific mechanisms that underlie Rtr1 recruitment to RNAPII have not been elucidated. To address this, we have performed an in-depth proteomic analysis of Rtr1 interacting proteins in yeast. Our studies revealed that hyperphosphorylated RNAPII is the primary interacting partner for Rtr1. To extend these findings, we performed quantitative proteomic analyses of Rtr1 interactions in yeast strains deleted for CTK1, the gene encoding the catalytic subunit of the CTD kinase I (CTDK-I) complex. Interestingly, we found that the interaction between Rtr1 and RNAPII is decreased in ctk1Δ strains. We hypothesize that serine-2 CTD phosphorylation is required for Rtr1 recruitment to RNAPII during transcription elongation.
Introduction
RNA Polymerase II (RNAPII) transcription is precisely regulated by sequential rounds of phosphorylation and dephosphorylation within a specialized C-terminal domain (CTD) in its largest subunit (Rpb1).1–3 The CTD in Saccharomyces cerevisiae consists of 26 repeats of the heptapeptide sequence ‘YSPTSPS’ with increasing numbers of repeats in higher eukaryotes.3–5 Within this sequence, it has been well established that the second and fifth serine in the repeats (hence referred to as Ser2 and Ser5, respectively) are highly phosphorylated and regulate the recruitment of various transcription and chromatin regulatory proteins at specific stages of transcription.3,6 Of note, more recent studies have found that the seventh serine, the first tyrosine, and the fourth threonine in the repeats (hence referred to as Ser7, Tyr1, and Thr4, respectively) are also subjected to dynamic phosphorylation and dephosphorylation during RNAPII transcription.7–11 A number of cyclin-dependent kinases (CDKs) have been shown to phosphorylate Ser2, Ser5, and Ser7 in eukaryotes both in vivo and in vitro.9,12–20 Thr4 phosphorylation has been shown to occur in yeast and human cells and is carried out by CDK9 and Polo-like kinase 3 in humans.21,22 Tyr1 phosphorylation is carried out by Abl kinase in human cells, but the identity of the Tyr1 kinase in yeast remains unknown.11,23 Removal of the CTD phosphorylation marks are carried out by at least four phosphatases in yeast with Rtr1 being responsible for the removal of Ser5-P in early transcription elongation,24 Fcp1 removing Ser2-P in late elongation/termination,25–28 and two components of the cleavage and polyadenylation factor complex, Glc7 and Ssu72, removing Tyr1-P, Ser5-P, and Ser7-P at the transcription termination site.20,29–33
Considering the repetitive nature of the CTD, there are numerous potential combinations of phosphorylated repeats in the CTD that could be specifically recognized to recruit RNAPII regulatory proteins. The precise timing of accessory factor recruitment is essential for the regulation of diverse transcription-coupled processing including: RNAPII elongation, RNA processing, transcription termination, and crosstalk between RNAPII and chromatin. Data from many laboratories suggests that the large number of potential combinations of CTD modification patterns both within and between repeats acts as a ‘CTD code’ to specifically time the recruitment of regulatory factors to RNAPII. This is supported by the identification of proteins that require multiple modifications within single or tandem CTD repeats for targeting to RNAPII.34 This includes: the histone methyltransferase Set2 that requires both Ser2 and Ser5 phosphorylation in up to three tandem repeats for recruitment, as well as the CTD phosphatase Ssu72 and the transcription termination regulator Nrd1 that require Ser5 phosphorylation along with proline 6 in a cis confirmation.31,35–37 The highest degree of CTD modification heterogeneity occurs during transcription elongation and recent results suggest that changes in RNAPII subunit composition might also occur during transcription elongation, adding an additional layer of complexity.5
The CTD phosphatase Rtr1 is recruited to RNAPII specifically during transcription elongation.24 Attempts to characterize the precise role of Rtr1 in the regulation of RNAPII are confounded by a lack of knowledge of the specific proteins and/or CTD modifications that are required for Rtr1 recruitment in vivo. Rtr1 and its human homolog RPAP2 have been implicated in diverse functions including regulation of RNAPII nuclear import, Ser5-P CTD dephosphorylation during transcription elongation, Ser7-P CTD binding, and recruitment of an additional CTD phosphatase.24,38–41 To gain a better understanding of the role of Rtr1 in the regulation of RNAPII function, we have performed an in-depth proteomic analysis of Rtr1 interacting proteins in WT and ctk1Δ yeast. We have discovered that Rtr1 recruitment to RNAPII in vivo requires the activity of the cyclin-dependent kinase complex CTDK-I that phosphorylates Ser2 of the RNAPII CTD. Additionally, we have determined that Rtr1 interacts with a specific hyper-phosphorylated form of RNAPII that is not recognized by the other CTD phosphatases Fcp1 and Ssu72.
Results and discussion
Analysis of the Rtr1 interactome by SAINT
To identify the interacting partners of the CTD phosphatase Rtr1, we employed various affinity purifications followed by multidimensional protein identification technology (MudPIT). For these studies, we generated Rtr1-FLAG and Rtr1-V5 yeast strains in which the epitope tag was integrated into the chromosomal locus for Rtr1. We also utilized Rtr1-TAP from a previous study.42 Each epitope tagged Rtr1 strain was grown to an OD600 ≈ 1–2 prior to lysis by bead beating as previously described.24 Following affinity purification, isolated proteins were digested with trypsin and subjected to three to four technical replicate MudPIT analyses per biological replicate. Technical replicate RAW data files were pooled for FASTA database searching using SEQUEST as previously described.5,43,44 The resulting dataset was filtered to require a false discovery rate of ≤1% for all datasets. The sequence coverage, number of unique peptides, and total number of peptide-spectrum matches (PSMs) for each protein across biological replicates are reported for each affinity and control purification in Table S1 (ESI†). An equal number of mock purifications from the parental strain BY4741 were performed to allow for the application of Significance Analysis of INTeractome (SAINT), a statistical approach that calculates interaction probabilities through the comparison of mock and specific bait affinity purification-mass spectrometry (AP-MS) data.45–52
We performed SAINT analysis using the SAINT-express algorithm through the contaminant repository for affinity purification (CRAPOME, www.crapome.org). This analysis provides three different scoring metrics for each prey identified: an FC-A score (a low stringency fold-change score), FC-B score (a high stringency fold-change score), and a SAINT probability score.45,46,49 The Rtr1 interactome dataset is made up of both single- and double-affinity purifications. A previous global study on kinase and phosphatase interactions found that single-affinity purifications reveal low level or dynamic interactions whereas double-affinity purifications often reveal stable interactions.51 To identify the components of the Rtr1 interactome, we performed SAINT analysis of the single-affinity and double-affinity Rtr1 dataset (Fig. 1 and 2, and Fig. S1, ESI†).
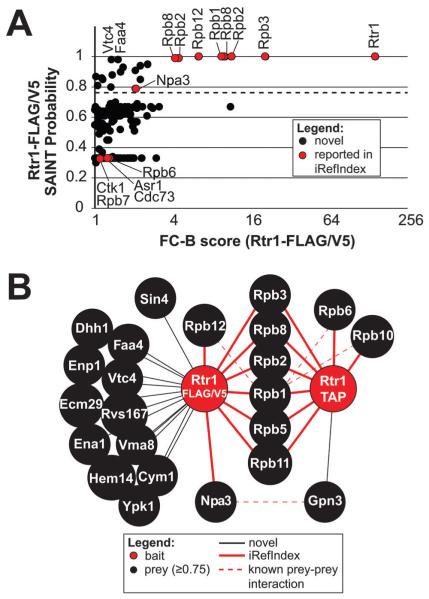
Fig. 1.
Identification of the Rtr1 interactome by AP-MS followed by SAINT. (A) Overview of the interactions identified by SAINT in single-affinity Rtr1 purifications. The graph compares the FC-B score against the SAINT probability scores. The dashed line represents the 0.75 probability cutoff. Proteins of interest are indicated with a label. All identifiers for these data are included in Table S2 (ESI†). (B) A high confidence Rtr1 interaction network is shown illustrating the unique and shared interactions identified in single or double-affinity purifications with a SAINT probability score of ≥0.75. A legend describing all nodes and edges is shown below the network. Previously identified Rtr1 interactions as described in iRef Index are identified with red edges whereas novel interactions are identified with black edges.
Fig. 2.
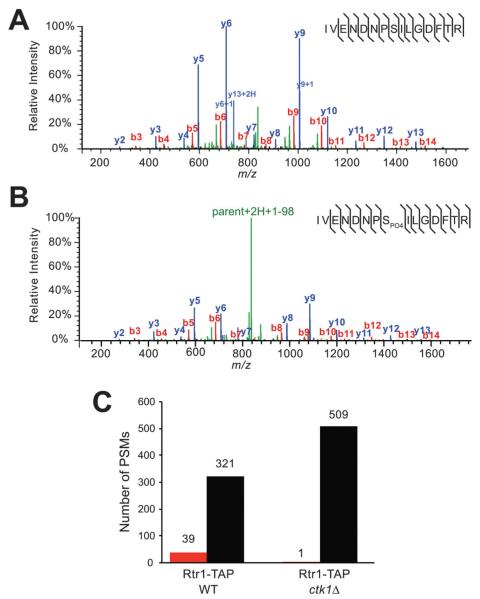
Rtr1 is phosphorylated at serine 217 in vivo. (A) A representative spectrum is shown from Rtr1-TAP for the unmodified IVENDNPSILGDFTR peptide (SEQUEST XCorr = 5.15). The resulting fragment ion series is labelled with b-ions in red and y-ions in blue. Precursor fragments are labelled in green. (B) A representative spectrum is shown from Rtr1-TAP for the phosphorylated IVENDNPSPO4ILGDFTR peptide (SEQUEST XCorr = 5.53). A neutral loss of −98 daltons is observed (green, parent + 2H + 1–98) (C) Number of PSMs detected for either unmodified (black bars) or phosphorylated (red bars) Rtr1 serine 217 in WT or ctk1Δ Rtr1-TAP.
Twenty-two interactions were identified from the single-affinity FLAG/V5 purifications with a SAINT probability of ≥0.75 (Fig. 1). Two of these interaction partners, Idh2 and Rpl33a, were not included in the Rtr1 FLAG/V5 network because they have been reported to be common contaminants of FLAG purifications.51 Although twenty proteins passed the stringency criteria, the high level of enrichment of core RNAPII subunits in Rtr1 single-affinity purifications is easily visualized by comparing many of the “Rpb” FC-B values to those of the other proteins such as Vtc4 and Faa4 (Fig. 1A). To determine the pathways that are enriched in the V5/FLAG set of Rtr1 interacting partners, GO-term enrichment analysis was performed using GOStat.53 The term GO:0005665 for “DNA-directed RNA polymerase II, core complex” was enriched with a p-value of 5.18 × 10−13 along with a number of additional GO terms associated with RNAPII function and transcription (Table S3, ESI†). Sin4 (a subunit of the mediator complex) and Npa3 were both identified as significant interactions and have both been implicated in the regulation of RNAPII function (Fig. 1B).54–57 A number of additional high confidence interactions were significantly enriched in the FLAG/V5 dataset but did not appear to function together in related pathways or complexes as determined by GOStat analysis (Table S3, ESI†). The FLAG/V5 dataset also contained a number of previously identified Rtr1 interacting partners that were not observed in the TAP purification dataset including: Asr1, Cdc73, and Ctk1 (Fig. 1A); however, their SAINT probability scores all fell below the 0.75 cut-off. The highest number of interactions was identified through analysis of the Rtr1-FLAG affinity purifications alone (Table S3, ESI†). A total of 62 interactions were identified with a SAINT probability of ≥0.8. GOStat analysis of the FLAG alone data resulted in similar results as obtained for FLAG/V5 with identification of some additional transcription related proteins including Leo1 (a component of the PAF complex58) (Table S4, ESI†). Recent structural studies have suggested that Rtr1 may not have intrinsic phosphatase activity, but may instead interact with an unknown RNAPII CTD phosphatase.39 No phosphatases were identified as reproducible interacting partners of Rtr1 (Tables S2 and S4 (ESI†), Fig. 1A). Although no protein phosphatases were found to pass the SAINT probability thresholds, the protein phosphatase 2A interacting partner Tap42 was found to be a reproducible interacting partner of Rtr1-FLAG (Table S4, ESI†).59,60
To highlight the overlap between the single- and double-affinity purifications, we generated a network containing the high confidence interactions from both datasets (Fig. 1B). Only 9 interactions were identified with a SAINT probability of ≥0.75 in the double-affinity TAP purification dataset (Fig. 1B and Table S6, ESI†). Of these, Lsm12 was not included in the TAP network since it has been identified as a common contaminant of TAP purifications from yeast.51 Six of the high-confidence Rtr1-TAP interacting proteins overlapped with the proteins identified in the single-affinity datasets and were all subunits of the RNAPII core complex. Two of the remaining three high confidence interactions in Rtr1-TAP were additional RNAPII subunits (Rpb6 and Rpb10). Gpn3, a GPN-loop GTPase, was identified as a novel Rtr1-TAP interaction partner although Gpn3 is known to interact with Npa3.61 Of interest, Npa3 was also identified in Rtr1-TAP purifications although it had a SAINT probability of 0.67, which was below the high confidence interaction cut-off (Fig. S1A and Table S6, ESI†). As expected for a bait protein, Rtr1 showed the highest FC-B score in both the single-affinity and double-affinity purifications and had a SAINT probability of 1 in both datasets. However, the overall FC-B score was significantly higher in the TAP purification dataset than in FLAG/V5, likely due to the second purification step as has been previously shown (compare Fig. 1A and Fig. S1A, ESI†).62 Higher FC-B scores were also observed for the RNAPII subunits in the TAP purification dataset than in FLAG/V5 (Fig. 1A and Fig. S1A, ESI†). For instance, Rpb1 has a FC-B value of 9.66 in the single-affinity purification dataset vs. a FC-B value of 47.26 in the double-affinity dataset (Tables S2 and S3, ESI†). SAINT analysis of the complete Rtr1 interactome dataset (combination of all single and double-affinity purifications) resulted in a network of six RNAPII proteins shown in Fig. S1B (ESI†). Affinity isolated Rtr1 samples often contain low levels of RNAPII interacting proteins and transcription elongation associated factors including: Spt5, Dst1, Rba50, and subunits of the PAF complex (PAF-C). The low abundance of these proteins in Rtr1 purifications suggests that they are likely co-purified as a consequence of their association with RNAPII (Table S1, ESI† (ref. 5)). This idea is supported by data from our previous studies that found Rtr1 as a low-level associated protein in affinity purifications of elongation factors such as Cdc73 (a subunit of PAF-C), Spt4, and Set2.5
Regulation of Rtr1 by the CTDK-I complex
Previous studies in our laboratory have shown that affinity isolated Rtr1 interacts with hyperphosphorylated RNAPII.5,24 Additionally, we observed relative depletion of the Rpb4/7 heterodimer following normalization for protein size. We have previously reported that Rpb4/7 are depleted in Ser2-P RNAPII.5 These data raise a question of whether or not Ser2 CTD phosphorylation stabilizes or is required for Rtr1 recruitment to RNAPII in vivo. Of note, protein microarray studies focused on the identification of protein kinase interacting proteins identified Rtr1 as a direct interacting partner of the CTDK-I subunits Ctk1 and Ctk2 which is primarily responsible for the phosphorylation of serine 2 in the RNAPII CTD in yeast.16,27,63,64 These data could suggest a regulatory role for CTDK-I in the control of Rtr1 function in vivo. To address this, we generated yeast strains with either Rtr1-TAP or Rtr1-FLAG in a CTK1 deletion (ctk1Δ) genetic background. Ctk1 is the catalytic subunit of CTDK-I and deletion of CTK1 in yeast has been shown to abolish the majority of Ser2-P CTD phosphorylation in vivo.17,27 Using these strains, we performed APMS analysis to interrogate the post-translational modification state and interactome of Rtr1 following genetic perturbation of CTDKI (Table S1, ESI†).
Rtr1 is phosphorylated in a Ctk1-dependent manner
Following affinity isolation of Rtr1-TAP, we reproducibly identified both unmodified and phosphorylated spectra for serine 217 within the Rtr1 C-terminal peptide IVENDNPSILGDFTR (compare Fig. 2A to B). In each APMS experiment, the number of PSMs for the unmodified peptide exceeded that of the modified peptide suggesting that only a small percentage of Rtr1 is phosphorylated at serine 217 in vivo (Fig. 2C). Serine 217 is located within the C-terminus of Rtr1, a region that is not strongly conserved between yeast Rtr1 and human RPAP2 (data not shown). However, we hypothesized that serine 217 could be modified as a result of an Rtr1-CTDKI interaction in vivo. Serine 217 does not represent a classical cyclin-CDK consensus motif of serine/threonine followed by a proline. However there is a relevant precedent for cyclin-CDK modification of proline-serine motifs: the modification of Ser7 within the CTD by TFIIH (also a cyclin-dependent kinase).9 To test if serine 217 could be a potential substrate for CTDK-I, we assessed the level of serine 217 phosphorylation in Rtr1-TAP WT and ctk1Δ strains (Fig. 2C). Using this approach, we found that serine 217 phosphorylation decreases in the absence of Ctk1. Although this is not direct evidence that CTDK-I phosphorylates Rtr1, these results show that phosphorylation of Rtr1 at serine 217 is dependent on CTDK-I function in vivo.
To determine if serine 217 regulates the phosphatase activity of Rtr1 in vitro, recombinant WT and S217D (phosphomimetic) Rtr1 were purified from E. coli and used for phosphatase assays with the small molecule substrate DiFMUP. Following dephosphorylation of DiFMUP, the concentration of the fluorescent small molecule DiFMU was monitored over a time course of two hours. As shown in Fig. 3A, Rtr1 S217D did not show any statistically significant difference in phosphatase activity as compared to WT Rtr1. These data show that S217D does not affect the phosphatase activity of Rtr1 in vitro. Although the mutation of serine 217 to aspartic acid did not result in a change in the phosphatase activity of Rtr1 against DiFMUP, it is possible that serine 217 plays a role in the interaction between Rtr1 and RNAPII in vivo. To address this possibility, we introduced WT, S217A, and S217D Rtr1-TAP plasmids into BY4741 (parental) yeast strains and performed immunoprecipitation analysis. As shown in Fig. 3B, Rtr1-TAP was specifically isolated in the Rtr1-TAP containing strains (Fig. 3B, anti-TAP, lanes 6–8). Confirming the interaction of Rtr1 with RNAPII, Rtr1 IP samples also contained Rpb3, the 3rd largest subunit of RNAPII, while no Rpb3 was detected in BY4741 control strain IP samples (Fig. 3B, anti-Rpb3 panel, lanes 5–8). To determine if mutation of serine 217 alters the specific recruitment of Rtr1 to hyperphosphorylated RNAPII, western blot analysis was performed using specific monoclonal antibodies raised against Ser2-, Ser5-, or Ser7-P CTD as indicated. These data show that serine 217 is not required to regulate the interaction between Rtr1 and RNAPII. However, these data clearly show the binding preference of Rtr1 to hyperphosphorylated RNAPII and reveal that Rtr1 can bind to RNAPII when the CTD is modified at Ser2, Ser5, and/or Ser7.
Fig. 3.
Serine 217 phosphorylation does not alter the activity of Rtr1. (A) Mutation of serine 217 to aspartic acid does not alter the phosphatase activity of Rtr1. DiFMUP dephosphorylation assays were performed over a two-hour time course to measure the activity of WT (n = 6) and S217D (n = 9) Rtr1 relative to a GST alone control reaction. The amount of DiFMU produced over time is shown as an average ± standard deviation. (B) Serine 217 is not required for the interaction between RNAPII and Rtr1. TAP-tagged Rtr1 (WT, S217A, or S217D) was immunoprecipitated (IP) using IgG Sepharose resin. Input whole cell extract is shown in the panels to the left. Western blot analysis was performed to assess the interaction of Rtr1 (anti-TAP) with RNAPII. Rpb3 is a core subunit of RNAPII whereas the anti-SerP antibodies are all directed against specific modified forms of the Rpb1 C-terminal domain.
The interaction of Rtr1 with RNAPII is dependent on CTDK-I activity
Since Rtr1 preferentially binds to hyperphosphorylated RNAPII, it is possible that CTDK-I regulates Rtr1 recruitment through modification of the RNAPII CTD. To determine if this could be the case, we performed SAINT analysis of the Rtr1 single and double-affinity purifications to identify changes in the Rtr1 interactome in ctk1Δ. Strikingly, deletion of CTK1 led to a dramatic decrease in the total ion counts of the spectra from co-purifying RNAPII while the overall abundance of Rtr1 remained relatively unchanged (Fig. 4A). SAINT analysis of the Rtr1 WT versus CTK1 deletion dataset revealed decreased SAINT probability values for multiple RNAPII subunits as well as Gpn3 and Npa3 (Fig. 4B and Table S8, ESI†). The same trend was observed for FC-B scores and analysis of raw PSM levels (Fig. S2, ESI†). Although the human homolog of Npa3 has been shown to interact with human Rtr1 directly in vitro, these data suggest that the interaction between Rtr1 and Npa3 is dependent on either their interaction with RNAPII or Ctk1 function in yeast. Interestingly, four proteins were found to have increased SAINT probability scores (≥0.9) in Rtr1 purifications from ctk1Δ. Of these, Hsp26 was not considered as a true Rtr1 interacting protein since it has previously been identified as a common contaminant in FLAG purifications.51 Rtr1 isolated from CTK1 deletion strains was found to have high confidence interactions with: Irr1, a subunit of the cohesin complex; Rrp41, a subunit of the exosome; and Uba2, a subunit of a nuclear SUMO-activating enzyme (Fig. 4B, Fig. S2 and Table S8, ESI†).65–67 Irr1 was a low probability interaction partner of Rtr1 in the WT interactome dataset (SAINT probability = 0.42) but increased to a high probability interaction upon deletion of CTK1 (Fig. 4B). Interestingly, Rrp41 and Uba2 were not detected in Rtr1 WT purifications, but were reproducibly identified in both the FLAG and TAP Rtr1 ctk1Δ datasets (Table S1, ESI†). A previous study found that Rtr1 would interact with Ctk1 and Ctk2 in vitro using protein microarrays. However we were unable to detect a direct interaction between Ctk1 and Rtr1 in vivo or between Ctk1/Rtr1 Ctk2/Rtr1 in vitro (Fig. S3, ESI†). Ctk1 was detected as a low probability Rtr1 interacting protein in the FLAG/V5 dataset (Fig. 1A), suggesting that this interaction may occur transiently in vivo. The dynamic nature of protein interaction with various kinases has been well documented elsewhere.51,64 Since we were unable to confirm the direct interaction between Rtr1 and CTDKI, we propose that Rtr1 recruitment is instead regulated by the phosphorylation status of the RNAPII CTD. Overall, the changes in the Rtr1 interactome in ctk1Δ strains strongly suggest that CTDK-I facilitates the interaction between Rtr1 and RNAPII. We propose that the regulation of Rtr1 recruitment to RNAPII in vivo likely occurs through the phosphorylation of Ser2 in the CTD by CTDK-I.
Fig. 4.
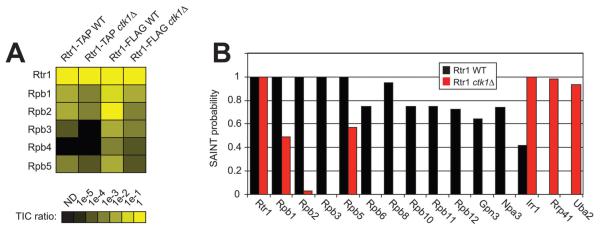
The interaction between Rtr1 and RNAPII is significantly decreased in the absence of Ctk1. (A) The total ion count for each protein of interest was summed from each purification as indicated at the top of the heat map. An intensity scale of the TIC ratio (prey/bait) is shown below the figure. (B) SAINT probability scores for Rtr1 affinity purifications from WT(black bars) and CTK1 deletion (red bars) yeast strains. Multiple subunits of RNAPII are shown (Rpb1, Rpb3, Rpb5, Rpb6, Rpb8, Rpb10, Rpb11, Rpb12) along with the Rtr1 interacting proteins Gpn3 and Npa3 identified in Fig. 1B. Novel Rtr1 interacting partners Irr1, Rrp41, and Uba2 were only identified as significant in ctk1Δ.
During transcription, some regulatory factors interact with the globular core of RNAPII including the DSIF complex (Spt4/5) and TFIIF.68,69 However, many other proteins are recruited during a specific stage(s) of transcription through interactions with specific phosphorylated forms of the RNAPII CTD. Previous studies using chromatin immunoprecipitation studies (ChIP) have determined that Rtr1 and its human homolog RPAP2 are recruited throughout the transcribed region of active genes and interact with hyper-phosphorylated RNAPII.5,24,40 The phospho-CTD binding preference of Rtr1 is distinct compared to that of the other CTD phosphatases, Ssu72 and Fcp1, which localize to the 3′ end of active genes. To build on our findings, we sought to further characterize the dependence of each CTD phosphatase on specific RNAPII phosphorylation patterns for recruitment to determine if this mechanism was unique to Rtr1. For these experiments, CTD phosphatases were affinity isolated and resulting samples were subjected to western blotting using monoclonal antibodies directed against specific phosphorylated forms of RNAPII (Fig. 5, upper four panels). Western blots for the core RNAPII subunit Rpb3 were also performed to serve as a loading control for RNAPII levels in each immunoprecipitation (IP) (Fig. 5, bottom panel). Additionally, we included Spt4-TAP samples that were isolated with a single IgG sepharose affinity step as a RNAPII interaction control since the Spt4/5 heterodimer is known to interact with RNAPII in a CTD-independent fashion throughout transcription elongation (Fig. 5, lane 1).70,71
Fig. 5.
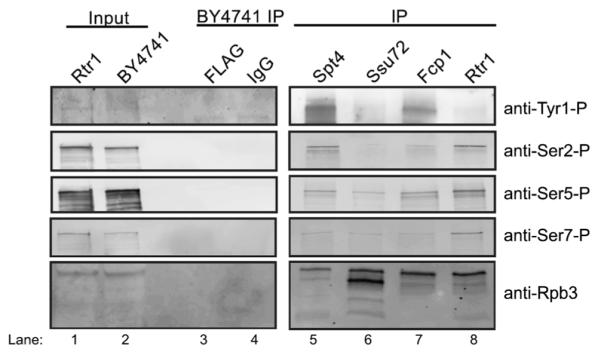
Western blot analysis of CTD phosphatase affinity purifications for identification of the patterns of CTD phosphorylation. The panels to the left show representative input samples for Rtr1-FLAG and BY4741. Control IPs were also performed with both IgG Sepharose and FLAG agarose resin (BY4741 IP). The bait proteins used for affinity purification are indicated at the top of the figure and the antibody used for western blotting is labelled to the right. Spt4-TAP was isolated with IgG sepharose whereas all CTD phosphatases were affinity captured on anti-FLAG agarose. Anti-Rpb3 western blots are included as a loading control for RNAPII levels in each IP. Lane numbers are also given at the bottom of the figure.
Interestingly, we found that Rtr1 IP samples were enriched for Ser2-, Ser5-, and Ser7-P RNAPII while Tyr1-P was relatively low compared to the levels observed in the Spt4-TAP IP (Fig. 5, compare lane 1 to lane 4 relative to Rpb3 levels). These data are consistent with the recruitment of Rtr1/RPAP2 during early transcription elongation prior to the recruitment of Ssu72 and Fcp1. This would also coordinate with the well-described patterns of Ser2 phosphorylation added by CTDK-I.16,20,27,72 These data are also consistent with findings that the recruitment of RPAP2 in human cells requires Ser7-P at some genomic loci.40 Ssu72-FLAG IP samples contained lower levels of Tyr1-, Ser2-, Ser5-, and Ser7-P RNAPII as compared with Spt4 (Fig. 2A, compare lane 1 to lane 2 relative to Rpb3 levels). This pattern was surprising given recent observations that conditional inactivation of Ssu72 (through a temperature sensitive degron) resulted in accumulation of Ser2-P at the 3′ end of transcription units suggesting that Ssu72 dephosphosphorylation could act as a prerequisite for/ or be coupled to Fcp1 removal of Ser2-P.20,32 Additionally, since Ssu72 is known to localize to the polyadenylation site, a region that should be enriched for Ser2-P but depleted for Tyr1-P and Ser5-P (at protein coding genes), we expected to observe Ser2-P in Ssu72 IP samples. Since the recently described Tyr1 phosphatase Glc7 is a subunit of the cleavage and polyadenylation factor (CPF) along with Ssu72, we expect that its CTD binding pattern would mirror that of Ssu72 since they function together in a stable CPF complex.19,33 Fcp1-FLAG IP samples contained Tyr1-P although it was observed at lower levels than observed for Spt4-TAP (Fig. 2A, compare lane 1 to lane 3). This is consistent with Fcp1 ChIP data that shows that Fcp1 recruitment increases within the 3′ end of the transcription unit earlier than the recruitment of Ssu72. The presence of Ser5-P in Fcp1-FLAG samples could also suggest Fcp1 recruitment prior to recruitment of Ssu72 (Fig. 2A, lane 3).
In combination, these data along with data from previous studies, suggest that Rtr1 is recruited during early transcription elongation when all three serine residues are phosphorylated (it is unknown if this is within repeat, between tandem repeats, etc.). The association of Rtr1 with RNAPII appears to decrease as Tyr1 becomes phosphorylated; resulting in a lack of Tyr1-P detection in Rtr1 IP samples (Fig. 5, lane 4). Fcp1 recruitment occurs prior to the 3′ end transcribed genes but after Rtr1 dissociation from RNAPII, consistent with gene-specific and global ChIP studies on Fcp1 localization. Ssu72 is recruited to the termination site as part of the CPF complex, which associates with RNAPII that has been dephosphorylated by Rtr1, Fcp1, and Glc7. The low relative levels of Ser2-P, Ser7-P, and Tyr1-P detected in Ssu72-FLAG samples suggests that coordinated dephosphorylation of the residual CTD-P residues occurs rapidly upon CPF-RNAPII interaction.
Experimental
Yeast strains
The yeast strains used in this study are described in Table S9 (ESI†).24,42,73 All strains were isogenic to BY4741.
TAP purification
This protocol has been previously described.5,24,74,75 In brief, TAP tagged yeast strains were grown to an OD600 ~ 1–2. After cell lysis, heparin and DNase I treatment, and lysate clarification by centrifugation; incubation with IgG Sepharose (GE) was performed for at least 3 hours. The IgG resin was then incubated overnight with AcTEV protease (Invitrogen) to cleave the Protein A ZZ domain and generate an accessible Calmodulin (CAM) Binding Peptide (CBP).74,76 After cleavage, the remaining tagged protein and interacting proteins were incubated with CAM resin for at least 3 hours. The proteins of interest were then eluted from the CAM resin (GE) using elution buffer containing 2 mM EGTA. Spt4-TAP42 used for western blot analysis were eluted from IgG Sepharose resin by AcTEV protease followed by boiling in 1× SDS-PAGE buffer prior to SDS-PAGE separation.
FLAG purification
Approximately 6–12 liters of yeast strains containing Rtr1-FLAG were grown to OD600 ~ 1–2. These cells were centrifuged, washed, and lysed as referenced in the TAP Purification section. After the lysate was clarified, 200 μL of washed anti-FLAG resin (Sigma) was incubated with the lysate for at least 3 hours at 4 °C on a magnetic stir plate. After incubation, the lysate–resin mixture was applied to a 30 mL BioRad Econoprep column and allowed to drain via gravity flow. The FLAG resin was then washed with 30 mL of TAP lysis buffer. After the wash, 1 mL of 50 mM ammonium bicarbonate (pH 8.5) was applied to the column to equilibrate the FLAG resin in the trypsin digestion buffer. The FLAG resin was then resuspended in 500 μL of trypsin digestion buffer (50 mM ammonium bicarbonate, pH 8.5 and 10 μL of 0.1 μg μL−1 Trypsin Gold (Promega)). The buffer–resin mixture was then transferred to a fresh microcentrifuge tube and incubated at 37 °C with shaking overnight. After incubation, the sample was centrifuged to pellet the FLAG resin and the supernatant was removed. The digestion was then quenched with addition of 0.1% formic acid. The same procedure was followed for western blot analysis of anti-FLAG captured CTD phosphatases however proteins were eluted by boiling the agarose-bound complexes in 1× SDS-PAGE buffer.
Multidimensional protein identification technology
Protein purifications that did not involve on-bead trypsin digestion were TCA precipitated prior to trypsin digestion at 37 °C. TCA precipitated proteins were resuspended in 8 M Urea in 100 mM Tris pH 8.5. Reduction and alkylation were performed with tris(2-carboxyethyl)phosphine (TCEP) and chloroacetamide (CAM) as previously described.5,24,75 The concentration of urea was then reduced to 2 M, followed by trypsin digestion. After the digestions were quenched by addition of 0.1% formic acid, peptides were pressure loaded onto a microcapillary column. The microcapillary column was packed with both strong cation exchange resin and reverse phase resin as previously described.5,43,77 A 10-step MudPIT run was performed for each technical replicate sample on an LTQ Velos or LTQ Velos Pro that was in-line with a Proxeon Easy nLC. The mass spectrometer was set-up in data dependent acquisition mode with the ten most intense ions identified in MS1 selected for MS/MS fragmentation using collision induced dissociation. Dynamic exclusion was set to 90 seconds with a repeat count of 1.
SEQUEST protein database search
RAW data files were subjected to protein database matching using SEQUEST within Proteome Discoverer 1.4 (Thermo). Technical replicate RAW files were pooled together for searches for each biological replicate.75 Database matching was performed using the 08-10-2013 Saccharomyces cerevisiae database which contained 5910 non-redundant annotated yeast protein sequences obtained from the National Center for Biotechnology Information (NCBI). The database also contained 67 common contaminant proteins including human keratins, IgGs, and proteolytic enzymes. Database search parameters were as follows: precursor mass tolerance = 1.4 dalton, fragment mass tolerance = 0.8 dalton, up to 2 missed cleavages were allowed, enzyme specificity was set to fully tryptic, minimum peptide length = 6 amino acids, static modifications: +57 daltons on cysteine (for all digestions that included TCEP and CAM treatment), dynamic modifications = +16 daltons on methionine and +80 daltons on serine, threonine, and tyrosine. All spectra were required to have a false discovery rate of ≤1% to be reported as a PSM in Table S1 (ESI†). FDR calculation was performed using Percolator, which was launched within Proteome Discoverer 1.4 using a beta version of 1.4 SP1.78 Two empirical fold-change scores (FC-A (average mean of replicates) and FC-B (geometric mean of replicates)) and SAINT probability scores were calculated as outlined in detail the Contaminant Repository for Affinity Purification (CRAPome) website and various publications.46,49–52 GOStat analysis was performed as previously described.43 Additional details about the SAINT analysis are provided in the supplementary Methods section (ESI†). Throughout this report, high confidence interactions have a SAINT value of ≥0.75, moderate confidence interactions have a SAINT value of 0.45–0.74, and low confidence interactions have a SAINT value of 0.01–0.44. Proteins with a SAINT probability of 0 were not defined as Rtr1 interaction partners and were not included in the supplementary tables (ESI†). The RAW data files and pepXML files for the experiments presented in this study are deposited in the Peptide Atlas data repository (http://www.peptideatlas.org) under the identifier PASS00445. RAW files for control purifications were also submitted to the CRAPome.
Rtr1 mutagenesis
Mutations were introduced using Agilent QuikChange Lightning Site Directed Mutagenesis kit according to the manufacturers instructions using primers to mutate serine 217 to alanine or aspartic acid as indicated. The primers used for mutagenesis are as follows: RTR1 S217A-5′AAATTGAGAATGATAATCCCGCCATTTTGGGTGATTTCACAAGGG-3′ and RTR1 S217A antisense-5′-CCCTTGTGAAATCACCCAAAATGGCGGGATTATCATTCTCAACAATTT-3′; RTR1 S217D-5′-CAAAATTGTTGAGAATGATAATCCCGACATTTTGGGTGATTTCACAAGG GAA-3′ and RTR1 S217D antisense-5′-TTCCCTTGTGAAATCACCCAAAATGTCGGGATTATCATTCTCAACAATTTTG-3′. Gateway cloning was used to move the RTR1 gene into pAG413GPD-ccdB-TAP (Addgene plasmid 14262) or pDEST24 (Invitrogen) were used as the dsDNA templates for mutagenesis and expression.
Recombinant Rtr1 expression and purification
pDEST24 (Invitrogen) containing the Saccharomyces cerevisiae RTR1 wild-type, S217A, and S217D point mutations were transformed into E. coli Rosetta DE3 pLysS competent cells (Invitrogen). Cells were grown at 37 °C in LB media supplemented with 100 μg mL−1 ampicillin and chloramphenicol until OD600 reached 0.8–1.0. The cells were induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside and grown overnight at 18 °C. The cells were harvested by centrifugation and stored at −80 °C. All protein variants were purified as follows: the cells were resuspended in lysis buffer (50 mM HEPES, pH 7.5; 150 mM NaCl; 10% glycerol; 1 mM β-mercaptoethanol; 10 mM EDTA, 1 mM PMSF). The cells were lysed by passing twice through a French press at 800 psi followed by treatment with DNase I. The slurry was clarified by centrifugation and the supernatant was loaded onto GST beads using the Bio-Rad LP chromatography system. The beads were washed with an increasing gradient of wash buffer (lysis buffer with 300 mM NaCl), and the protein was eluted using with an increasing gradient of elution buffer (lysis buffer with no glycerol and 100 mM reduced glutathione). Peak fractions were pooled and dialyzed against 50 mM HEPES, pH 7.5; 150 mM NaCl; and 1 mM DTT. The dialyzed protein was concentrated with an Amicon Ultra 10K filter column (Millipore). The protein purity and concentration was determined by a BSA curve on an SDS-PAGE gel.
Phosphatase assays
The initial rates of Rtr1 constructs were determined by the hydrolysis of DiFMUP (Molecular Probes). 200 μL reactions of 1 μM Rtr1 constructs versus 10 μM of DiFMUP were carried out at 30 °C in black 96-well plates in a reaction buffer of 50 mM succinic acid, pH 6.0; 140 mM NaCl; 10 mM MgCl2. Continuous assays were performed by measuring fluorescence produced every 20 min for 2 h using SpectraMax M5 Microplate reader. The relative fluorescence (RFU) of each protein construct was normalized for protein concentration and compared against a standard curve of DiFMU to determine the amount of product produced. Background fluorescence was corrected for by subtraction of RFU values obtained from assays performed with GST alone.
Conclusions
Rtr1 is an atypical protein phosphatase that is involved in the regulation of multiple aspects of RNAPII biology. Analysis of the Rtr1 interactome revealed that the most reproducible interacting partner of Rtr1 in WT log phase yeast cell extracts is RNAPII in all affinity capture approaches used (Fig. 1 and Fig. S1, ESI†). Rtr1 also interacts with the GPN-loop GTPase family members Npa3 and Gpn3 (Fig. 1 and Fig. S1, ESI†). We have previously shown that Rtr1 interacts primarily with a 10-subunit form of RNAPII that is hyper-phosphorylated.5 These findings were extended with analysis of Ser7 and Tyr1 phosphorylation analysis that showed that Rtr1 interacts with RNAPII phosphorylated at an unknown percentage of Ser2, Ser5, and Ser7 residues within the CTD (Fig. 5). The interaction of Rtr1 with the 10-subunit form of RNAPII was also reproduced in the affinity experiments included in this study. Neither Rpb4 nor Rpb7 were identified as significant interacting partners in Rtr1-TAP, FLAG, or V5 purifications (Fig. 1 and Fig. S1, Supplemental tables, ESI†). These findings are summarized in the Rtr1 interactome model shown as Fig. 6.
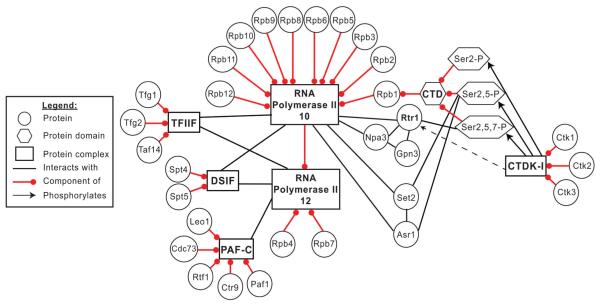
Fig. 6.
The yeast Rtr1 interactome in WT cells. A legend defining all nodes and edges is shown to the left of the network. RNA Polymerase II exists as two complexes in yeast, a predominant (>95%) 12-subunit containing enzyme shown above as “RNA Polymerase II 12” and a 10-subunit containing enzyme shown above as “RNA Polymerase II 10”. The general transcription elongation factors TFIIF and DSIF are able to interact with both forms of RNAPII whereas other factors such as PAF-C have only been found in association with the 12-subunit RNAPII. A number of CTD interacting proteins (Rtr1, Set2, and Asr1) have been found to interact with multiply modified RNAPII CTD. The recruitment of Rtr1 and Set2 to RNAPII requires the activity of CTDK-I. Please note that only a small selection of CTD modification patterns are shown for clarity. In vivo, recruitment of Rtr1 or Set2 might require modification of single or multiple CTD repeats.
We previously reported that Ser2-P modification of the CTD regulates the dissociation of the Rpb4/7 heterodimer and this study suggests that Ser2-P regulates Rtr1 recruitment to RNAPII.5 However, since previous reports have indicated that Rtr1 can directly interact with Ctk1 and Ctk2 on protein microarrays, it is possible that the CTDK-I complex might also play a more direct role in Rtr1 recruitment.64 We conclusively showed that Rtr1 requires Ctk1 activity for maximal recruitment to RNAPII in vivo and for interaction with Npa3 and Gpn3 (Fig. 4 and Fig. S2, ESI†). However, we were not able to observe a direct interaction between Rtr1 and Ctk1 (Fig. S3, ESI†). These data, in combination with our previous studies, support a model in which free nuclear-localized Rtr1 is recruited to RNAPII during early transcription elongation following CTDK-I phosphorylation of Ser2. Npa3 and Gpn3 may interact with Rtr1 through RNAPII or directly. These data suggest that Ser2-P is a prerequisite for Rtr1 recruitment and subsequent dephosphorylation of Ser5-P during early RNAPII transcription elongation (Fig. 6). The requirement for Ser2 phosphorylation would allow for precise timing of a percentage of Ser5-P removal likely following the completion of mRNA capping. These results would explain previous observations that deletion of CTK1 causes increased accumulation of Ser5-P in cell extracts.79 The CTD phosphorylation pattern found in Rtr1-FLAG IPs was unique as compared to the other known CTD phosphatases in yeast Fcp1 and Ssu72. These data suggest that the CTD phosphatases may act as both specific readers and erasers of the CTD code highlighting their important role in the regulation of the RNAPII transcription cycle.
Supplementary Material
Acknowledgements
The authors would like to thank the members of the Mosley lab for critical feedback during this project and during the preparation of this manuscript. We also thank Dattatreya Mellacheruvu for assistance with data analysis through the CRAPome. The biography photograph for A.L.M. was provided by Kristy Vest Photography. This project was supported by funding from the National Institutes of Health grant GM099714 to A.L.M. and GM064440 to G.V. and a Research Support Funds Grant from IUPUI to A.L.M.
Footnotes
Electronic supplementary information (ESI) available: Three figures and nine supplemental tables are available for this work. Table S1 contains all sequence coverage, unique peptide, and PSM information for each AP-MS replicate and is included as a separate file. Tables S2, S4 and S6–S8 contain the FC-A, FC-B, and SAINT probability scores for the Rtr1 affinity purifications. Tables S3 and S5 contain the output obtained from GOStat analysis of significant interaction lists. Table S9 contains all the genotypes and references for the yeast strains used in this study. See DOI: 10.1039/c4mb00109e
References
- 1.Eick D, Geyer M. The RNA Polymerase II Carboxy-Terminal Domain (CTD) Code. Chem. Rev. 2013;113(11):8456–8490. doi: 10.1021/cr400071f. [DOI] [PubMed] [Google Scholar]
- 2.Heidemann M, Hintermair C, Voss K, Eick D. Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochim. Biophys. Acta. 2013;1829(1):55–62. doi: 10.1016/j.bbagrm.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 3.Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol. Cell. 2009;36(4):541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahearn JM, Jr., Bartolomei MS, West ML, Cisek LJ, Corden JL. Cloning and sequence analysis of the mouse genomic locus encoding the largest subunit of RNA polymerase II. J. Biol. Chem. 1987;262(22):10695–10705. [PubMed] [Google Scholar]
- 5.Mosley AL, Hunter GO, Sardiu ME, Smolle M, Workman JL, Florens L, Washburn MP. Quantitative proteomics demonstrates that the RNA polymerase II subunits Rpb4 and Rpb7 dissociate during transcriptional elongation. Mol. Cell. Proteomics. 2013;12(6):1530–1538. doi: 10.1074/mcp.M112.024034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trigon S, Serizawa H, Conaway JW, Conaway RC, Jackson SP, Morange M. Characterization of the residues phosphorylated in vitro by different C-terminal domain kinases. J. Biol. Chem. 1998;273(12):6769–6775. doi: 10.1074/jbc.273.12.6769. [DOI] [PubMed] [Google Scholar]
- 7.Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M, Kremmer E, Eick D. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science. 2007;318(5857):1780–1782. doi: 10.1126/science.1145977. [DOI] [PubMed] [Google Scholar]
- 8.Egloff S, O’Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, Eick D, Murphy S. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science. 2007;318(5857):1777–1779. doi: 10.1126/science.1145989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akhtar MS, Heidemann M, Tietjen JR, Zhang DW, Chapman RD, Eick D, Ansari AZ. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol. Cell. 2009;34(3):387–393. doi: 10.1016/j.molcel.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heidemann M, Eick D. Tyrosine-1 and threonine-4 phosphorylation marks complete the RNA polymerase II CTD phospho-code. RNA Biol. 2012;9(9):55–62. doi: 10.4161/rna.21726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayer A, Heidemann M, Lidschreiber M, Schreieck A, Sun M, Hintermair C, Kremmer E, Eick D, Cramer P. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science. 2012;336(6089):1723–1725. doi: 10.1126/science.1219651. [DOI] [PubMed] [Google Scholar]
- 12.Cho EJ, Takagi T, Moore CR, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 1997;11(24):3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14(19):2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez CR, Cho EJ, Keogh MC, Moore CL, Greenleaf AL, Buratowski S. Kin28, the TFIIH-associated carboxy-terminal domain kinase, facilitates the recruitment of mRNA processing machinery to RNA polymerase II. Mol. Cell. Biol. 2000;20(1):104–112. doi: 10.1128/mcb.20.1.104-112.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keogh MC, Cho EJ, Podolny V, Buratowski S. Kin28 is found within TFIIH and a Kin28-Ccl1-Tfb3 trimer complex with differential sensitivities to T-loop phosphorylation. Mol. Cell. Biol. 2002;22(5):1288–1297. doi: 10.1128/mcb.22.5.1288-1297.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahn SH, Kim M, Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell. 2004;13(1):67–76. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- 17.Kim M, Ahn SH, Krogan NJ, Greenblatt JF, Buratowski S. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 2004;23(2):354–364. doi: 10.1038/sj.emboj.7600053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tietjen JR, Zhang DW, Rodriguez-Molina JB, White BE, Akhtar MS, Heidemann M, Li X, Chapman RD, Shokat K, Keles S, Eick D, Ansari AZ. Chemical-genomic dissection of the CTD code. Nat. Struct. Mol. Biol. 2010;17(9):1154–1161. doi: 10.1038/nsmb.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang DW, Mosley AL, Ramisetty SR, Rodriguez-Molina JB, Washburn MP, Ansari AZ. Ssu72 phosphatase-dependent erasure of phospho-Ser7 marks on the RNA polymerase II C-terminal domain is essential for viability and transcription termination. J. Biol. Chem. 2012;287(11):8541–8551. doi: 10.1074/jbc.M111.335687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bataille AR, Jeronimo C, Jacques PE, Laramee L, Fortin ME, Forest A, Bergeron M, Hanes SD, Robert F. A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase, phosphatase and isomerase enzymes along genes. Mol. Cell. 2012;45(2):158–170. doi: 10.1016/j.molcel.2011.11.024. [DOI] [PubMed] [Google Scholar]
- 21.Hintermair C, Heidemann M, Koch F, Descostes N, Gut M, Gut I, Fenouil R, Ferrier P, Flatley A, Kremmer E, Chapman RD, Andrau JC, Eick D. Threonine-4 of mammalian RNA polymerase II CTD is targeted by Polo-like kinase 3 and required for transcriptional elongation. EMBO J. 2012;31(12):2784–2797. doi: 10.1038/emboj.2012.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsin JP, Sheth A, Manley JL. RNAP II CTD phosphorylated on threonine-4 is required for histone mRNA 3′ end processing. Science. 2011;334(6056):683–686. doi: 10.1126/science.1206034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baskaran R, Chiang GG, Mysliwiec T, Kruh GD, Wang JY. Tyrosine phosphorylation of RNA polymerase II carboxyl-terminal domain by the Abl-related gene product. J. Biol. Chem. 1997;272(30):18905–18909. doi: 10.1074/jbc.272.30.18905. [DOI] [PubMed] [Google Scholar]
- 24.Mosley AL, Pattenden SG, Carey M, Venkatesh S, Gilmore JM, Florens L, Workman JL, Washburn MP. Rtr1 is a CTD phosphatase that regulates RNA polymerase II during the transition from serine 5 to serine 2 phosphorylation. Mol. Cell. 2009;34(2):168–178. doi: 10.1016/j.molcel.2009.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Archambault J, Chambers RS, Kobor MS, Ho Y, Cartier M, Bolotin D, Andrews B, Kane CM, Greenblatt J. An essential component of a C-terminal domain phosphatase that interacts with transcription factor IIF in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 1997;94(26):14300–14305. doi: 10.1073/pnas.94.26.14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobor MS, Archambault J, Lester W, Holstege FC, Gileadi O, Jansma DB, Jennings EG, Kouyoumdjian F, Davidson AR, Young RA, Greenblatt J. An unusual eukaryotic protein phosphatase required for transcription by RNA polymerase II and CTD dephosphorylation in S. cerevisiae. Mol. Cell. 1999;4(1):55–62. doi: 10.1016/s1097-2765(00)80187-2. [DOI] [PubMed] [Google Scholar]
- 27.Cho EJ, Kobor MS, Kim M, Greenblatt J, Buratowski S. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 2001;15(24):3319–3329. doi: 10.1101/gad.935901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghosh A, Shuman S, Lima CD. The structure of Fcp1, an essential RNA polymerase II CTD phosphatase. Mol. Cell. 2008;32(4):478–490. doi: 10.1016/j.molcel.2008.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krishnamurthy S, He X, Reyes-Reyes M, Moore C, Hampsey M. Ssu72 Is an RNA polymerase II CTD phosphatase. Mol. Cell. 2004;14(3):387–394. doi: 10.1016/s1097-2765(04)00235-7. [DOI] [PubMed] [Google Scholar]
- 30.Reyes-Reyes M, Hampsey M. Role for the Ssu72 C-terminal domain phosphatase in RNA polymerase II transcription elongation. Mol. Cell. Biol. 2007;27(3):926–936. doi: 10.1128/MCB.01361-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiang K, Nagaike T, Xiang S, Kilic T, Beh MM, Manley JL, Tong L. Crystal structure of the human symplekin-Ssu72-CTD phosphopeptide complex. Nature. 2010;467(7316):729–733. doi: 10.1038/nature09391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang DW, Mosley AL, Ramisetty SR, Rodriguez-Molina JB, Washburn MP, Ansari AZ. Ssu72 phosphatase dependent erasure of phospho-Ser7 marks on the RNA Polymerase II C-terminal domain is essential for viability and transcription termination. J. Biol. Chem. 2012;287(11):8541–8551. doi: 10.1074/jbc.M111.335687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schreieck A, Easter AD, Etzold S, Wiederhold K, Lidschreiber M, Cramer P, Passmore LA. RNA polymerase II termination involves C-terminal-domain tyrosine dephosphorylation by CPF subunit Glc7. Nat. Struct. Mol. Biol. 2014;21(2):175–179. doi: 10.1038/nsmb.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krogan NJ, Kim M, Tong A, Golshani A, Cagney G, Canadien V, Richards DP, Beattie BK, Emili A, Boone C, Shilatifard A, Buratowski S, Greenblatt J. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 2003;23(12):4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kubicek K, Cerna H, Holub P, Pasulka J, Hrossova D, Loehr F, Hofr C, Vanacova S, Stefl R. Serine phosphorylation and proline isomerization in RNAP II CTD control recruitment of Nrd1. Genes Dev. 2012;26(17):1891–1896. doi: 10.1101/gad.192781.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Zhang M. Crystal structure of Ssu72, an essential eukaryotic phosphatase specific for the C-terminal domain of RNA polymerase II, in complex with a transition state analogue. Biochem. J. 2011;434(3):435–444. doi: 10.1042/BJ20101471. [DOI] [PubMed] [Google Scholar]
- 37.Werner-Allen JW, Lee CJ, Liu P, Nicely NI, Wang S, Greenleaf AL, Zhou P. cis-Proline-mediated Ser(P)5 dephosphorylation by the RNA polymerase II C-terminal domain phosphatase Ssu72. J. Biol. Chem. 2011;286(7):5717–5726. doi: 10.1074/jbc.M110.197129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Forget D, Lacombe AA, Cloutier P, Lavallee-Adam M, Blanchette M, Coulombe B. Nuclear import of RNA polymerase II is coupled with nucleocytoplasmic shuttling of the RNA polymerase II-associated protein 2. Nucleic Acids Res. 2013;41(14):6881–6891. doi: 10.1093/nar/gkt455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiang K, Manley JL, Tong L. The yeast regulator of transcription protein Rtr1 lacks an active site and phosphatase activity. Nat. Commun. 2012;3:946. doi: 10.1038/ncomms1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Egloff S, Zaborowska J, Laitem C, Kiss T, Murphy S. Ser7 phosphorylation of the CTD recruits the RPAP2 Ser5 phosphatase to snRNA genes. Mol. Cell. 2012;45(1):111–122. doi: 10.1016/j.molcel.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ni Z, Olsen JB, Guo X, Zhong G, Ruan ED, Marcon E, Young P, Guo H, Li J, Moffat J, Emili A, Greenblatt JF. Control of the RNA polymerase II phosphorylation state in promoter regions by CTD interaction domain-containing proteins RPRD1A and RPRD1B. Transcription. 2011;2(5):237–242. doi: 10.4161/trns.2.5.17803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425(6959):737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 43.Mosley AL, Sardiu ME, Pattenden SG, Workman JL, Florens L, Washburn MP. Highly reproducible label free quantitative proteomic analysis of RNA polymerase complexes. Mol. Cell. Proteomics. 2010;10(2):M110000687. doi: 10.1074/mcp.M110.000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khanna M, Imasaki T, Chikwana VM, Perez-Miller S, Hunter GO, Mosley A, Takagi Y, Hurley TD. Expression and purification of functional human glycogen synthase-1 (hGYS1) in insect cells. Protein Expression Purif. 2013;90(2):78–83. doi: 10.1016/j.pep.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teo G, Liu G, Zhang J, Nesvizhskii AI, Gingras AC, Choi H. SAINTexpress: Improvements and Additional Features in Significance Analysis of Interactome Software. J. Proteomics. 2013 doi: 10.1016/j.jprot.2013.10.023. DOI: 10.1016/j.jprot.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi H, Liu G, Mellacheruvu D, Tyers M, Gingras AC, Nesvizhskii AI. Analyzing protein-protein interactions from affinity purification-mass spectrometry data with SAINT. Curr. Protoc. Bioinf. 2012 doi: 10.1002/0471250953.bi0815s39. ch. 8, p. Unit8 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choi H, Glatter T, Gstaiger M, Nesvizhskii AI. SAINT-MS1: protein-protein interaction scoring using label-free intensity data in affinity purification-mass spectrometry experiments. J. Proteome Res. 2012;11(4):2619–2624. doi: 10.1021/pr201185r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skarra DV, Goudreault M, Choi H, Mullin M, Nesvizhskii AI, Gingras AC, Honkanen RE. Label-free quantitative proteomics and SAINT analysis enable interactome mapping for the human Ser/Thr protein phosphatase 5. Proteomics. 2011;11(8):1508–1516. doi: 10.1002/pmic.201000770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choi H, Larsen B, Lin ZY, Breitkreutz A, Mellacheruvu D, Fermin D, Qin ZS, Tyers M, Gingras AC, Nesvizhskii AI. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat. Methods. 2011;8(1):70–73. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mellacheruvu D, Wright Z, Couzens AL, Lambert JP, St-Denis NA, Li T, Miteva YV, Hauri S, Sardiu ME, Low TY, Halim VA, Bagshaw RD, Hubner NC, Al-Hakim A, Bouchard A, Faubert D, Fermin D, Dunham WH, Goudreault M, Lin ZY, Badillo BG, Pawson T, Durocher D, Coulombe B, Aebersold R, Superti-Furga G, Colinge J, Heck AJ, Choi H, Gstaiger M, Mohammed S, Cristea IM, Bennett KL, Washburn MP, Raught B, Ewing RM, Gingras AC, Nesvizhskii AI. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods. 2013;10(8):730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Breitkreutz A, Choi H, Sharom JR, Boucher L, Neduva V, Larsen B, Lin ZY, Breitkreutz BJ, Stark C, Liu G, Ahn J, Dewar-Darch D, Reguly T, Tang X, Almeida R, Qin ZS, Pawson T, Gingras AC, Nesvizhskii AI, Tyers M. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328(5981):1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kwon Y, Vinayagam A, Sun X, Dephoure N, Gygi SP, Hong P, Perrimon N. The Hippo signaling pathway interactome. Science. 2013;342(6159):737–740. doi: 10.1126/science.1243971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beissbarth T, Speed TP. GOstat: find statistically overrepresented gene ontologies within a group of genes. Bioinformatics. 2004;20(9):1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- 54.Li Y, Bjorklund S, Jiang YW, Kim YJ, Lane WS, Stillman DJ, Kornberg RD. Yeast global transcriptional regulators Sin4 and Rgr1 are components of mediator complex/RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. U. S. A. 1995;92(24):10864–10868. doi: 10.1073/pnas.92.24.10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Staresincic L, Walker J, Dirac-Svejstrup AB, Mitter R, Svejstrup JQ. GTP-dependent binding and nuclear transport of RNA polymerase II by Npa3 protein. J. Biol. Chem. 2011;286(41):35553–35561. doi: 10.1074/jbc.M111.286161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cloutier P, Al-Khoury R, Lavallee-Adam M, Faubert D, Jiang H, Poitras C, Bouchard A, Forget D, Blanchette M, Coulombe B. High-resolution mapping of the protein interaction network for the human transcription machinery and affinity purification of RNA polymerase II-associated complexes. Methods. 2009;48(4):381–386. doi: 10.1016/j.ymeth.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Forget D, Lacombe AA, Cloutier P, Al-Khoury R, Bouchard A, Lavallee-Adam M, Faubert D, Jeronimo C, Blanchette M, Coulombe B. The protein interaction network of the human transcription machinery reveals a role for the GTPase RPAP4/GPN1 and microtubule assembly in nuclear import and biogenesis of RNA polymerase II. Mol. Cell. Proteomics. 2010;9(12):2827–2839. doi: 10.1074/mcp.M110.003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mueller CL, Jaehning JA. Ctr9, Rtf1 and Leo1 are components of the Paf1/RNA polymerase II complex. Mol. Cell. Biol. 2002;22(7):1971–1980. doi: 10.1128/MCB.22.7.1971-1980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Duvel K, Santhanam A, Garrett S, Schneper L, Broach JR. Multiple roles of Tap42 in mediating rapamycin-induced transcriptional changes in yeast. Mol. Cell. 2003;11(6):1467–1478. doi: 10.1016/s1097-2765(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 60.Di Como CJ, Arndt KT. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 1996;10(15):1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- 61.Alonso B, Beraud C, Meguellati S, Chen SW, Pellequer JL, Armengaud J, Godon C. Eukaryotic GPN-loop GTPases paralogs use a dimeric assembly reminiscent of archeal GPN. Cell Cycle. 2013;12(3):463–472. doi: 10.4161/cc.23367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Glatter T, Wepf A, Aebersold R, Gstaiger M. An integrated workflow for charting the human interaction proteome: insights into the PP2A system. Mol. Syst. Biol. 2009;5:237. doi: 10.1038/msb.2008.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mok J, Im H, Snyder M. Global identification of protein kinase substrates by protein microarray analysis. Nat. Protoc. 2009;4(12):1820–1827. doi: 10.1038/nprot.2009.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fasolo J, Sboner A, Sun MG, Yu H, Chen R, Sharon D, Kim PM, Gerstein M, Snyder M. Diverse protein kinase interactions identified by protein microarrays reveal novel connections between cellular processes. Genes Dev. 2011;25(7):767–778. doi: 10.1101/gad.1998811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91(1):35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 66.Benard L, Carroll K, Valle RC, Wickner RB. Ski6p is a homolog of RNA-processing enzymes that affects translation of non-poly(A) mRNAs and 60S ribosomal subunit biogenesis. Mol. Cell. Biol. 1998;18(5):2688–2696. doi: 10.1128/mcb.18.5.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dohmen RJ, Stappen R, McGrath JP, Forrova H, Kolarov J, Goffeau A, Varshavsky A. An essential yeast gene encoding a homolog of ubiquitin-activating enzyme. J. Biol. Chem. 1995;270(30):18099–18109. doi: 10.1074/jbc.270.30.18099. [DOI] [PubMed] [Google Scholar]
- 68.Martinez-Rucobo FW, Sainsbury S, Cheung AC, Cramer P. Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity. EMBO J. 2011;30(7):1302–1310. doi: 10.1038/emboj.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen ZA, Jawhari A, Fischer L, Buchen C, Tahir S, Kamenski T, Rasmussen M, Lariviere L, Bukowski-Wills JC, Nilges M, Cramer P, Rappsilber J. Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. EMBO J. 2010;29(4):717–726. doi: 10.1038/emboj.2009.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hirtreiter A, Damsma GE, Cheung AC, Klose D, Grohmann D, Vojnic E, Martin AC, Cramer P, Werner F. Spt4/5 stimulates transcription elongation through the RNA polymerase clamp coiled-coil motif. Nucleic Acids Res. 2010;38(2):585–596. doi: 10.1093/nar/gkq135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mayer A, Lidschreiber M, Siebert M, Leike K, Soding J, Cramer P. Uniform transitions of the general RNA polymerase II transcription complex. Nat. Struct. Mol. Biol. 2010;17(10):1272–1278. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- 72.Mayer A, Lidschreiber M, Siebert M, Leike K, Soding J, Cramer P. Uniform transitions of the general RNA polymerase II transcription complex. Nat. Struct. Mol. Biol. 2010;17(10):1272–1278. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- 73.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Liebundguth N, Lockhart DJ, Lucau-Danila A, Lussier M, M’Rabet N, Menard P, Mittmann M, Pai C, Rebischung C, Revuelta JL, Riles L, Roberts CJ, Ross-MacDonald P, Scherens B, Snyder M, Sookhai-Mahadeo S, Storms RK, Veronneau S, Voet M, Volckaert G, Ward TR, Wysocki R, Yen GS, Yu K, Zimmermann K, Philippsen P, Johnston M, Davis RW. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285(5429):901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 74.Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24(3):218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 75.Mosley AL, Sardiu ME, Pattenden SG, Workman JL, Florens L, Washburn MP. Highly reproducible label free quantitative proteomic analysis of RNA polymerase complexes. Mol. Cell. Proteomics. 2011;10(2) doi: 10.1074/mcp.M110.000687. M110 000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17(10):1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 77.Florens L, Washburn MP. Proteomic analysis by multidimensional protein identification technology. Methods Mol. Biol. 2006;328:159–175. doi: 10.1385/1-59745-026-X:159. [DOI] [PubMed] [Google Scholar]
- 78.Kall L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods. 2007;4(11):923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 79.Patturajan M, Conrad NK, Bregman DB, Corden JL. Yeast carboxyl-terminal domain kinase I positively and negatively regulates RNA polymerase II carboxyl-terminal domain phosphorylation. J. Biol. Chem. 1999;274(39):27823–27828. doi: 10.1074/jbc.274.39.27823. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.