Abstract
Centrosomes are the principal microtubule organizing centers (MTOCs) of animal cells and comprise a pair of centrioles surrounded by pericentriolar material (PCM). Centriole number must be carefully regulated, notably to ensure bipolar spindle formation and thus faithful chromosome segregation. In the germ line of most metazoan species, centrioles are maintained during spermatogenesis, but eliminated during oogenesis. Such differential behavior ensures that the appropriate number of centrioles is present in the newly fertilized zygote. Despite being a fundamental feature of sexual reproduction in metazoans, the mechanisms governing centriole elimination during oogenesis are poorly understood. Here, we investigate this question in C. elegans. Using antibodies directed against centriolar components and serial-section electron microscopy, we establish that centrioles are eliminated during the diplotene stage of the meiotic cell cycle. Moreover, we show that centriole elimination is delayed upon depletion of the helicase CGH-1. We also find that somatic cells make a minor contribution to this process, and demonstrate that the germ cell karyotype is important for timely centriole elimination. These findings set the stage for a mechanistic dissection of centriole elimination in a metazoan organism.
Keywords: C. elegans, Oogenesis, Centriole, Elimination, CGH-1, Karyotype
INTRODUCTION
The centriole and the related basal body is an evolutionarily ancient organelle characterized by a nine-fold radial arrangement of microtubules (Carvalho-Santos et al., 2010; Hodges et al., 2010; reviewed by Bornens and Azimzadeh, 2007; Strnad and Gönczy, 2008). Centrioles are essential for the formation of flagella and cilia and, thus, for cellular processes that rely on these structures. Furthermore, centrioles are crucial for recruiting the pericentriolar material (PCM) from which microtubules are nucleated in most animal cells. Centriole number is carefully regulated to ensure the proper execution of these fundamental cellular processes. Thus, most proliferating cells have two centrioles early in the cell cycle. At the onset of S phase, a procentriole begins to assemble perpendicular to each centriole. As a result, two pairs of centriole and associated procentriole are present during the remainder of the cell cycle. During mitosis, each spindle pole harbors one such pair. The two centriolar cylinders then disengage from one another and are inherited by each daughter cell.
Fertilization poses a special challenge to this general behavior. Just like fusion of two diploid gametes would result in the generation of tetraploid embryos, the newly fertilized zygote would be endowed with four centrioles if both gametes contributed two each. This could pose a threat to faithful chromosome segregation because the resulting spindle might be tetrapolar. In most metazoan species, including Caenorhabditis elegans and Homo sapiens, this problem is solved by the elimination of centrioles during oogenesis (reviewed by Schatten, 1994; Delattre and Gönczy, 2004; Manandhar et al., 2005; Sathananthan et al., 2006). Electron microscopy (EM) analysis indicates that centrioles are absent in mouse and human oocytes (Hertig and Adams, 1967; Calarco et al., 1972; Szollosi et al., 1972; Szollosi et al., 1986; Sathananthan et al., 1996). By contrast, at least one centriole is maintained during spermatogenesis. The zygote in most vertebrates is endowed with one centriole, which must undergo two cycles of duplication before the first embryonic mitosis (reviewed by Manandhar et al., 2005), whereas two centrioles are contributed by the sperm to the zygote in several other species, including C. elegans. Although differential centriole inheritance in the female versus the male germ line is a fundamental and conserved feature of metazoan sexual reproduction, the underlying mechanisms are poorly understood.
C. elegans is particularly well-suited for an investigation of centriole elimination during oogenesis (hereafter generally referred to as ‘elimination’ for simplicity). First, germ cells (GCs) develop in an assembly-line fashion, such that all stages of oogenesis are present in a single gonad, with their position indicative of their developmental stage (reviewed by Hubbard and Greenstein, 2000). Second, GC development and meiotic progression have been thoroughly investigated in C. elegans, which should facilitate analysis of elimination. Third, proteins that are essential for procentriole formation have been discovered and studied extensively in the early nematode embryo (reviewed by Leidel and Gönczy, 2005). As a result, reagents such as antibodies and GFP fusions are available for probing centriolar components during oogenesis.
In the embryo, molecular epistasis experiments have placed five crucial components in a pathway that leads to procentriole formation (Delattre et al., 2006; Pelletier et al., 2006). SPD-2, which also localizes to the PCM, is present first at the centriole and is needed for recruiting ZYG-1. ZYG-1 is then required for the presence of the interacting components SAS-6 and SAS-5. These two proteins are needed in turn for the recruitment of SAS-4, a stable component that does not exchange with the cytoplasmic protein pool over several cell cycles (Leidel and Gönczy, 2003; Kirkham et al., 2003; Dammermann et al., 2008). By contrast, SAS-5 readily shuttles between centrioles and cytoplasm (Delattre et al., 2004). Although it is known that SAS-4 and SPD-2 foci are absent in mature oocytes (Kirkham et al., 2003; Kemp et al., 2004; Kim and Roy, 2006), a systematic analysis of centriole elimination in C. elegans has not been documented.
Although little is known about the mechanisms governing elimination in any organism, co-suppression of the cyclin-dependent kinase inhibitor CKI-2 has been reported to result in the persistence of maternal centrioles in a fraction of C. elegans embryos (Kim and Roy, 2006). However, more recent findings using a cki-2 deletion allele instead led to the suggestion that CKI-2 is not needed for centriole elimination, raising the possibility that the earlier observation resulted from pleiotropic effects of co-suppression (Buck et al., 2009). Apart from the possible role of CKI-2 in this process, other components and mechanisms that contribute to elimination are not known.
MATERIALS AND METHODS
Nematodes and RNA interference (RNAi)
Nematode culture was carried out according to standard procedures (Brenner, 1974). The following genotypes were used: sas-6(ok2554) (Kitagawa et al., 2011), zyg-1(it25) (Kemp et al., 2007), spd-2(oj29) (O’Connell et al., 1998), sas-5(t2033) (Delattre et al., 2004), sas-5(gk400) (http://www.wormbase.org), sas-4(tm3951) (Kitagawa et al., 2011), cki-2(ok2105) (Buck et al., 2009), fog-1(q253) (Barton and Kimble, 1990) (shifted to the restrictive temperature of 25°C as L1/L2), fog-2(q71) (Schedl and Kimble, 1988), fog-3(q470) (Ellis and Kimble, 1994), tra-1(e1076) (Schedl et al., 1989), her-1(hv1y101) unc-42 and her-1(hv1y101) unc-42 dpy-11;lon-2 (Francis et al., 1995b), analyzed as described (Bean et al., 2004), mog-1(q233) (Graham and Kimble, 1993), fem-3(q96gf) (Barton et al., 1987), cgh-1(ok492) (Audhya et al., 2005), glo-1(zu391) (Hermann et al., 2005), rrf-1(pk1417) (Sijen et al., 2001), GFP-SAS-4 (Kirkham et al., 2003), GFP-SAS-5 (Delattre et al., 2004), GFP-SAS-6 (Leidel et al., 2005), GFP-SPD-2 (Kemp et al., 2004), GFP-TBG-1 (Oegema et al., 2001), GFP –TAC-1 (Bellanger and Gönczy, 2003), VIT-2-GFP (Grant and Hirsh, 1999), NMY-2-GFP (Nance et al., 2003).
RNAi-mediated inactivation using bacterial feeding strains was performed as described (Delattre et al., 2006) using feeding strains from the Ahringer (Kamath et al., 2003) or Vidal (Rual et al., 2004) libraries (denoted ‘A’ and ‘V’, respectively). L4 worms were subjected to sas-5(RNAi) (V) or gld-1(RNAi) (A and V) for 20-22 hours at 20°C and to cgh-1(RNAi) (A) for ∼24 hours or 30-48 hours at 24°C for weak or strong inactivation, respectively. Additional RNAi conditions are reported in supplementary material Table S1.
Antibodies
The following primary antibodies raised in rabbits were utilized: 1:600 SAS-4 (Leidel and Gönczy, 2003), 1:20 SAS-5 (Delattre et al., 2004), 1:100 SAS-6 (Leidel et al., 2005), 1:1500 ZYG-1 (Leidel and Gönczy, 2003), 1:1000 TBG-1 (Hannak et al., 2002) (gift from A. Hyman), 1:1000 SPD-2 (Delattre et al., 2006) (gift from M. Glotzer), 1:5000 SPD-5 (Hamill et al., 2002) (gift from B. Bowerman), 1:100 TAC-1 (Bellanger and Gönczy, 2003), 1:1000 ZYG-9 (Gönczy et al., 2001), 1:12,000 PGL-1 (Kawasaki et al., 1998) (gift from S. Strome), 1:200 RME-2 (Grant and Hirsh, 1999) (gift from B. Grant) and 1:1000 phospho-H3-Ser10 (06-570, Millipore). The following primary antibodies raised in mouse were used: 1:100 GFP (3F8.2, Millipore), 1:500 α-tubulin (DM1α, Sigma) and 1:50 IFA-1 (Leung et al., 1999) (ATCC-TIB-131). For 3D structured illumination microscopy analysis, GFP fluorescence was enhanced using GFP-Booster (GBA-488, ChromoTek). Secondary antibodies were goat anti-mouse-Alexa 488, goat anti-rabbit-Alexa 568, donkey anti-rabbit-Alexa 594 and goat anti-mouse-Cy5, all used at 1:500 (Molecular Probes).
Indirect immunofluorescence, microscopy and analysis
Gonads were dissected in 2 μM levamisole in PBS or 0.01% tetramisole in M9, transferred onto polylysine-coated slides, and then fixed and stained as for centrioles in embryos (Delattre et al., 2004). DNA was counterstained with ∼1 μg/ml Hoechst 33258 (Sigma).
For microtubule depolymerization/growth experiments, slides with gonads were transferred to ice-cold lead blocks and left for 20 minutes before being transferred to lead blocks at room temperature for 5 minutes. Gonads were then fixed and stained as above.
Indirect immunofluorescence was imaged on LSM700 Zeiss and SP2 confocal microscopes. Optical sections (0.4-1 μm) were acquired with a 1-2 μm pinhole and 1024×1024 pixel resolution. Planes containing centrioles were maximum-projected using ImageJ or Fiji. Images were processed using Adobe Photoshop, preserving relative image intensities within a series.
SAS-4 centriolar signal was quantified by drawing an 11×11 pixel square around centrioles, followed by Gaussian blur (σ=0.5) and median filtering (radius=0.5). Cytoplasmic and centriolar intensities were determined after automated thresholding using the Fiji Minimum method.
For analysis of centriole persistence, oocytes harboring bivalent chromosomes were scored. Statistics were performed in GraphPad Prism with Fisher’s exact test.
Fluorescence recovery after photobleaching (FRAP)
glo-1(zu391) animals were crossed to GFP-SAS-4 or GFP-SAS-5 worms to reduce gut autofluorescence. Animals were immobilized on 10% agarose pads using 0.1 μm diameter polysterene microspheres (method suggested by C. Fang-Yen, personal communication; Polysciences). FRAP was performed on a LSM700 Zeiss confocal microscope, acquiring 10-20 0.1 μm optical sections at each time point, with bi-directional scanning and 512×512 pixel resolution at 2% laser power. A 5×5 pixel square centered on one or two juxtaposed centrosomes was bleached with 50 iterations and 100% laser power. Stacks were acquired every 30 seconds. Projections were generated in Fiji and the recovery was analyzed qualitatively. Images and movies were then processed by applying Gaussian blur (σ=0.5) and median filtering (radius=0.5).
Transmission electron microscopy (TEM)
Gonads were dissected as for immunofluorescence and prepared as described (Pitt et al., 2000). Briefly, gonads were fixed with 2.2% glutaraldehyde, 0.9% paraformaldehyde in 0.05 M cacodylate buffer (pH 7.4) with 0.09 M sucrose and 0.9 mM MgCl2 and postfixed in 1% osmium tetroxide, 0.8% potassium ferrocyanide in 0.1 M cacodylate buffer (pH 7.2). Specimens were embedded in 1% agarose, stained in 1% uranyl acetate in sodium actetate (pH 5.2) and embedded in eppon resin. Serial sections (70 nm) were contrasted with 0.9% KMnO4, 3% uranyl acetate and lead citrate. Images were taken at 80 kV filament tension using a CM10 Phillips transmission electron microscope and a Morada digital camera (Baker and Masison, 1990). Serial sections were analyzed from top to bottom of the nucleus at 13,500× magnification. At both ends of the nucleus, at least five additional sections were screened. For regions 3 and 4, serial sections of three randomly chosen nuclei were screened. In addition, centriolar pairs were imaged for seven nuclei in regions 2 and 3. In two cases, structures resembling procentrioles were apparent.
Structured illumination microscopy (SIM)
Samples were prepared as for regular immunofluorescence and post-fixed with 4% formaldehyde for 10 minutes. Three-dimensional structured illumination microscopy was performed as described (Schermelleh et al., 2008).
RESULTS
Centriolar proteins required for centrosome duplication in the gonad
Throughout the results section, we refer to four regions of the gonad that correspond to successive stages of GC development (Fig. 1A) (McCarter et al., 1999). Region 1 in the distal end of the gonad contains proliferating syncytial GC nuclei (GCN) that then enter the meiotic cell cycle in the adjacent transition zone. GCN in region 2 have exited premeiotic S phase and are in the pachytene stage of meiosis I. Region 3 is located in the loop of the gonad and contains diplotene GCN. Cellularization of GCs is completed within region 4. Cellularized GCs are in diakinesis and harbor six condensed bivalent chromosomes. Mature oocytes are fertilized when traversing the spermatheca at the proximal end of the gonad.
Fig. 1.
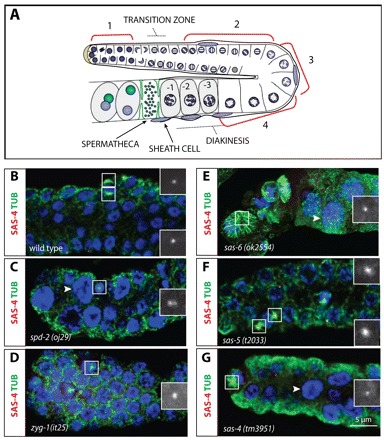
Requirements for centriolar proteins in the hermaphrodite gonad. (A) Schematic representation of the hermaphrodite gonad. For simplicity, the gonad is subdivided into four regions: (1) proliferating GCs; (2) GCs in pachytene; (3) GCs after pachytene; (4) GCs in diplotene after the loop. Note diakinesis oocytes (marked –1, –2, –3) prior to the spermatheca, with six condensed bivalent chromosomes. Sheath cell nuclei are depicted in blue and for simplicity only on one side of the gonad. (B-G) Region 1 from C. elegans gonads at the fourth larval stage (L4) of the indicated genotypes stained for α-tubulin (green), SAS-4 (red in the merged images and shown alone in magnified insets) and DNA (blue). Insets are magnified twofold. Note that the spd-2(oj29) GCN highlighted in C has two SAS-4 foci. Arrowheads point to clearly enlarged GCN.
To evaluate which centriolar proteins to consider in the context of elimination, we tested whether the components essential for centriole formation in early embryos are required in the hermaphroditic germ line. We found occasional monopolar spindles and larger than normal nuclei in region 1 of spd-2(oj29), zyg-1(it25), sas-6(ok2554) and sas-4(tm3951) mutant animals, indicative of defective centrosome duplication (compare Fig. 1B with 1C-E,G). By contrast, no such defects were observed in sas-5(t2033) or sas-5(gk400) homozygous mutant animals, or in sas-5(t2033) animals treated with sas-5(RNAi), even though these conditions all result in a fully penetrant centrosome duplication failure in embryos (Fig. 1F; data not shown) (Dammermann et al., 2004; Delattre et al., 2004). Although we cannot exclude the possibility that residual SAS-5 activity persists in these conditions, these observations suggest that sas-5 is dispensable for centrosome duplication in the hermaphroditic germ line.
Overall, we conclude that SPD-2, ZYG-1, SAS-6 and SAS-4 are required for centrosome duplication in proliferating GCs, as anticipated from the sterility associated with spd-2(oj29) and zyg-1(it25) (O’Connell et al., 1998; Kemp et al., 2007), as well as in sas-6(ok2554) and sas-4(tm3951) (Kitagawa et al., 2011).
Progressive loss of focused PCM and microtubule organizing center (MTOC) activity during oogenesis
We set out to determine when centrosomes become undetectable in the hermaphroditic germ line. In the embryo, the PCM assembles in a step-wise fashion, with SPD-5 and SPD-2 recruiting the γ-tubulin protein TBG-1 (Hamill et al., 2002; Kemp et al., 2004; Pelletier et al., 2004), which is needed for the loading of TAC-1 and ZYG-9, which are present primarily in mitosis (reviewed by Oegema and Hyman, 2006).
In the gonad, we found that SPD-5, SPD-2 and TBG-1 all localize to foci in the vicinity of GCN in region 1, and to a lesser extent in regions 2 and 3 (Fig. 2A-C; supplementary material Fig. S1A,B) (Kemp et al., 2004). No focus is detected in region 4 (Fig. 2A-C; supplementary material Fig. S1A,B), although the proteins remain present in the cytoplasm. Moreover, membranes around GCN in regions 2 and 3 are enriched with TBG-1 (Fig. 2C) (Zhou et al., 2009). TAC-1 and ZYG-9 exhibit a more restricted distribution, being present solely in some GCN in region 1 (Fig. 2D; supplementary material Fig. S1C; data not shown). A focus of TAC-1 or of ZYG-9 is not detectable in regions 2-4, in line with the fact that GCN are not proliferating in these regions (Fig. 2D; supplementary material Fig. S1C; data not shown).
Fig. 2.
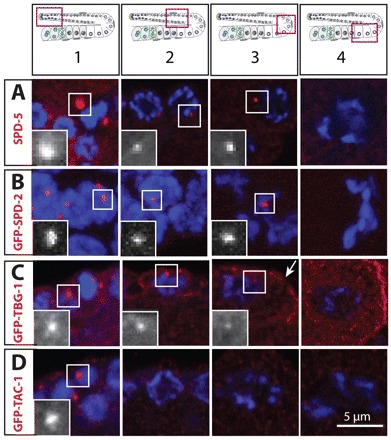
Distribution of PCM proteins. Young adult hermaphrodite gonads stained for the indicated PCM components (red in the merged images and shown alone in magnified insets) and DNA (blue). Insets are magnified twofold. Schematic representations above the panels indicate positions of regions 1-4 in the gonad. The four panels of each row do not necessarily come from the same gonad.
To determine the point during oogenesis at which centrosomes cease to act as MTOCs, we conducted microtubule depolymerization/re-growth experiments. We found that centrosomes can nucleate asters within 5 minutes in region 1, but not in regions 2 or 3 (supplementary material Fig. S2), in accordance with what has been reported previously (Zhou et al., 2009). Therefore, although the PCM is present until the loop region of the gonad, centrosomes lose their capacity to act as MTOCs by the time GCN transition into the meiotic cell cycle.
Loss of focused centriolar proteins during oogenesis
Next, we analyzed the distribution of centriolar proteins. To avoid being mislead by spurious dotty signals in the cytoplasm, we conducted these experiments by simultaneously monitoring the IFA-1 antigen, which marks centrioles in C. elegans embryos (supplementary material Fig. S3) (Leung et al., 1999).
As mentioned above, SPD-2 is localized in a focus in the vicinity of all GCN in region 1 and to a lesser extent in regions 2-3 (Fig. 2A). ZYG-1 has a more restricted distribution, with the protein being detected in a focus only in the vicinity of some GCN in region 1 (Fig. 3A). Such transient distribution is reminiscent of that in early embryos (O’Connell et al., 2001). We found an analogous distribution to that of SPD-2 for SAS-6, SAS-5 and SAS-4. These proteins are present in a tight focus next to nuclei in regions 1-3, but not in region 4, although they remain distributed throughout the cytoplasm (Fig. 3B-D). We then addressed whether SAS-4 and SAS-6 disappear simultaneously during oogenesis. We found that GFP-SAS-4 colocalizes with SAS-6 at all centriolar foci and that the two proteins cease to be detected in a focus in a concomitant manner (supplementary material Fig. S4). Overall, we conclude that centriolar proteins are no longer detected in a focus as GCs move from region 3 to region 4 of the gonad.
Fig. 3.
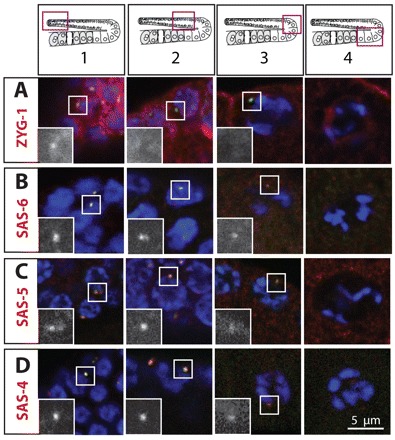
Distribution of centriolar proteins. Young adult hermaphrodite gonads stained for the indicated centriolar proteins (red in the merged images and shown alone in magnified insets), IFA-1 (green) and DNA (blue). Insets are magnified twofold. Schematic representations above the panels indicate positions of regions 1-4 in the gonad. The four panels of each row do not necessarily come from the same gonad. Note that two foci are visible next to the GCN highlighted in C, region 1 and B, region 2.
Next, we investigated whether SAS-4 is a stable component of centrioles during oogenesis, as it is in embryos (Leidel and Gönczy, 2003; Dammermann et al., 2008). This question is of particular interest because centriolar components could be less stable during oogenesis, which might help explain their eventual elimination. We conducted FRAP experiments of GFP-SAS-4 in regions 2 and 3 (Fig. 4A,C). We usually observed no recovery of the signal during the 19 minutes of the experiment (Fig. 4A; supplementary material Movie 1; n=14/16, with the remaining two exhibiting weak recovery). By contrast, centriolar GFP-SAS-5 usually recovered in this time span (Fig. 4B; supplementary material Movie 2; n=15/17), compatible with the observation that SAS-5 shuttles between cytoplasm and centrioles in embryos (Delattre et al., 2004). Overall, these experiments suggest that SAS-4 is a stable component of centrioles during oogenesis.
Fig. 4.
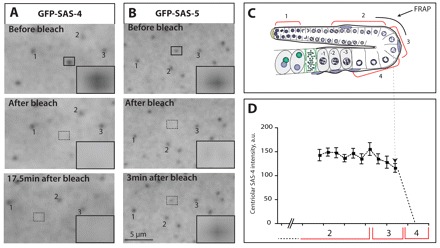
Dynamics of centriolar proteins and kinetics of centriole elimination. (A,B) Images before (top), just after (middle) or at the indicated time after (bottom) photobleaching in region 2/3 of gonads expressing GFP-SAS-4 (A) or GFP-SAS-5 (B). 1, 2 and 3 mark reference centrioles in the field of view that were not bleached. Insets are magnified 3.5-fold. See also supplementary material Movies 1 and 2. (C) Schematic representation indicating the location of the FRAP experiments. (D) SAS-4 centriolar signal intensity in cells located in regions 2, 3 or 4, as indicated, expressed as percentage increase over the surrounding cytoplasmic signal (arbitrary units, a.u.). Dashed line between C and D indicates the approximate location of abrupt SAS-4 signal loss. Shown are mean and s.e.m., n=34.
We then set out to estimate the approximate duration of the elimination process. Quantification of SAS-4 foci in regions 2-4 indicates that the signal intensity is constant in regions 2 and 3. Upon transition into region 4, this intensity rapidly decreases to the levels observed in the cytoplasm from one GCN to the next (Fig. 4D). Given that oocytes are fertilized every ∼23 minutes on average (McCarter et al., 1999), we conclude that elimination occurs within <30 minutes.
Ultrastructural analysis of centrioles during oogenesis
The findings described above establish that centriolar proteins become undetectable in a focus after the loop of the gonad, but do not demonstrate that centrioles are truly absent. It could be that centriolar proteins are no longer detected in a focus, but that centrioles remain present. Alternatively, centrioles could vanish earlier than suggested by the disappearance of focused centriolar proteins. We conducted EM analysis to address these possibilities.
We first examined GCN in region 2 that should harbor centrioles. Accordingly, serial sections of three GCN located in region 2 revealed the presence of a pair of centrioles in the vicinity of each GC nucleus (Fig. 5A-F). The orientation of these two centrioles is not perpendicular, indicating that they correspond to disengaged parental centrioles. Super-resolution structured illumination microscopy (SIM) (Schermelleh et al., 2008), which overcomes the resolution limit of conventional microscopy, confirmed the presence of two neighboring SAS-4 foci in most region 2 nuclei (Fig. 5G; 15/20 nuclei).
Fig. 5.
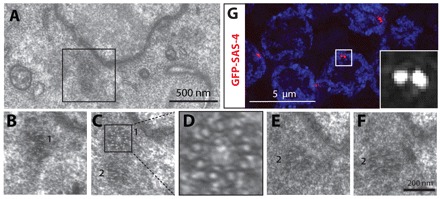
Ultrastructural analysis and super-resolution microscopy of centrioles in pachytene. (A-F) TEM serial sections of centrioles in region 2; B,C,E and F are consecutive sections of the inset in A and show two disengaged mature centrioles (marked 1 and 2). D highlights nine microtubules in one centriole from the inset in C. (G) SIM of centrioles in region 2. Gonad from animal expressing GFP-SAS-4 stained for GFP (red in the merged images and shown alone in magnified inset) and DNA (blue).
Additional EM analysis in favorable specimens showed that procentrioles can be detected in region 2 in the vicinity of the two centrioles (supplementary material Fig. S5A-D). Accordingly, SAS-6, which is recruited to assembling procentrioles in embryos (Dammermann et al., 2004; Leidel et al., 2005; Dammermann et al., 2008), localizes adjacent to SAS-4 foci in region 2 GCN (supplementary material Fig. S5E). Moreover, super-resolution microscopy revealed that some GCN in this region harbor four foci of SAS-4 (supplementary material Fig. S5F-J). Furthermore, we used a complementary approach to address whether procentriole formation is initiated during the meiotic cell cycle. In the absence of gld-1 function, meiotic GCN in region 2 revert to a mitotic fate (Francis et al., 1995a). If procentriole formation does not occur during the meiotic cell cycle, then cells lacking gld-1 function and reverting to a mitotic fate should assemble bipolar spindles with a single centriole in each spindle pole. Contrary to this prediction, two foci of SAS-4 can be detected at spindle poles of gld-1(RNAi) GCs reverted to a mitotic fate (supplementary material Fig. S5K). Taken together, these observations suggest that procentriole formation is initiated during the meiotic cell cycle.
We addressed next whether centrioles and procentrioles are truly absent in region 4, as suggested by the disappearance of the focus of centriolar proteins. First, we observed that the three GCN that were analyzed by serial-section EM in region 3 were each associated with a centriolar pair. By contrast, serial section analysis of three GCN in region 4 established that no recognizable centrioles are present at that stage, although a structure that could be interpreted as a disassembling centriole was detected in one section (data not shown). These observations demonstrate that elimination occurs in diplotene GCN in the loop between regions 3 and 4 during C. elegans oogenesis.
Centriole elimination is delayed in germ cells depleted of the CGH-1 helicase
We set out to investigate the mechanisms contributing to elimination. First, we considered whether the spermatheca or the sperm cells it contains are implicated. We determined that the last oocyte in which a focus of SAS-4 and IFA-1 is detectable in young wild-type animals is oocyte –8 and in older animals oocyte –16 (supplementary material Fig. S6). This suggests that the distance from the spermatheca is not critical for elimination during oogenesis. Furthermore, we found that centrioles disappear normally in fog-1(q253) and fog-2(q71) mutant hermaphrodites that lack sperm cells (data not shown) (Schedl and Kimble, 1988; Barton and Kimble, 1990), demonstrating that sperm cells are dispensable for elimination.
Comparing young and older animals also allowed us to assess whether there is a temporal correlation between elimination and GC cellularization. We found that cellularization is completed on average in oocyte –3 in young animals and –13 in older animals (supplementary material Fig. S6). Thus, in both settings, elimination and cellularization are separated by three to five GCN, indicating that the timing of the two processes is correlated. However, because cellularization is completed after elimination, the former cannot be required for the latter. Accordingly, centrioles do not disappear prematurely in GC of ani-2(RNAi) animals (data not shown), which undergo premature cellularization (Maddox et al., 2005).
We took a candidate approach to identify components that contribute to elimination. Using mutants or RNAi, we tested genes known or suspected to regulate aspects of GC development in regions 3/4 (supplementary material Table S1), scoring the presence of SAS-4 and/or IFA-1. These included genes important for physiological cell death (ced-3, as well as car-1 and cgh-1, which have also been proposed to be required for this process) (Navarro et al., 2001), autophagy (lgg-1, bec-1, vps-34, unc-51, atg-9), translational repression (gld-1, mex-3), meiotic progression through pachytene (daz-1), synaptonemal complex formation (syp-1), and MAP kinase signaling (mpk-1, let-60). In addition, we addressed whether elimination requires protein degradation by inactivating components and regulators of the 26S proteasome (pas-1, pas-2, pas-6, pbs-1, rpn-2, rpn-3, rpt-3).
Because inactivation of these genes, in particular with RNAi, might not have been complete, we cannot rule out the possibility that they might contribute to elimination. With this limitation in mind, we found that whereas all other experimental conditions have no impact (supplementary material Table S1), compromising the RNA helicase CGH-1 alters elimination. Indeed, we found that ∼59% of GCs in diakinesis in the predicted null allele cgh-1(ok492) harbor a SAS-4 focus, whereas this is never the case in the wild type (Fig. 6A,B,D). Given that fertilized embryos are not present in cgh-1(ok492) animals, whether such SAS-4-positive foci would all eventually be eliminated cannot be ascertained. Because gonad morphology is severely disorganized in cgh-1(ok492), we analyzed the milder cgh-1(RNAi) inactivation condition. We found in this case that ∼21% of GCs in diakinesis harbor a SAS-4 focus (Fig. 6C,D). No supernumerary SAS-4-positive foci are observed in embryos derived from cgh-1(RNAi) animals (data not shown), indicating that centrioles are eventually eliminated. We found also that SAS-4 foci are rarely present in diakinesis GCs carrying the null allele cki-2(ok2105), and always disappear thereafter, suggesting that cki-2 contributes only in a minor fashion to timely elimination (Fig. 6D) (Buck et al., 2009).
Fig. 6.
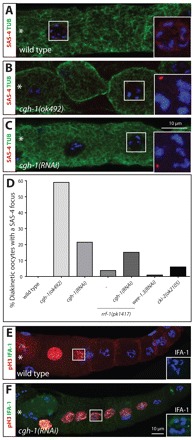
CGH-1 promotes timely centriole elimination. (A-C) Region 4 from young adult hermaphrodites of the indicated genotypes stained for α-tubulin (green), SAS-4 (red in the merged images and shown with DNA in the insets) and DNA (blue). Note that the cgh-1(ok492) diakinesis GC highlighted in B is clearly cellularized. Note also that the inset for the DNA in these panels is a projection to visualize the DNA, whereas the low magnification image represents only one of the z-sections. The most mature GC/oocyte is to the left and marked by an asterisk. (D) Percentage of diakinesis GCs with SAS-4 foci. n and P-values (compared with wild type; Fisher’s exact test): wild type, n=79; cgh-1(ok492), n=22, P<0.0001; cgh-1(RNAi), n=93, P<0.0001; rrf-1(pk1417), n=27, P=0.2547; rrf-1(pk1417) cgh-1(RNAi), n=33, P=0.0018 and P=0.6120 compared with cgh-1(RNAi); wee-1.3(RNAi), n=100, P=1; cki-2(ok2105), n=67, P=0.042. (E,F) Wild-type (E) and cgh-1(RNAi) (F) gonads of young adult hermaphrodites stained for phospho-H3 (red), IFA-1 (green in the merged images and shown with DNA in insets) and DNA (blue). Note that in ∼16% of cgh-1(RNAi) phospho-H3-positive nuclei an IFA-1 focus was observed (n=67) whereas no such cases were observed in the wild type (n=35). Insets are magnified twofold.
Apart from its involvement in mRNA localization and stabilization in the germ line (Boag et al., 2008), CGH-1 also modulates microRNA-target interactions in somatic tissues (Hammell et al., 2009), raising the possibility that it acts in the soma to promote elimination. However, we found that depletion of CGH-1 in rrf-1(pk1417) animals, in which somatic tissues are insensitive to RNAi (Sijen et al., 2001), results in a similar phenotype (Fig. 6D), indicating that CGH-1 acts in the germ line to promote elimination.
Because CGH-1 regulates various aspects of oogenesis (Navarro et al., 2001), we considered whether the presence of SAS-4 in a fraction of diakinesis cgh-1(RNAi) GCs reflects altered specification of the germ line or delayed oocyte development, both of which could indirectly delay elimination. To monitor germline specification, we stained cgh-1(RNAi) gonads with antibodies against the germline-specific P-granules, which we found to be present as in the wild type (supplementary material Fig. S7A,B) (Kawasaki et al., 1998). Additionally, we found that diakinesis oocytes harboring centrioles in cgh-1(ok492) are positive for the yolk receptor RME-2, which is usually found in diakinesis oocytes in the wild type (supplementary material Fig. S7C,D) (Grant and Hirsh, 1999). Therefore, germline specification and development take place, at least in part, upon CGH-1 depletion. To test whether oocyte maturation is delayed, we monitored phospho-histone H3 (phospho-H3) (Hsu et al., 2000), which in the wild type marks the two to three most mature oocytes (Fig. 6E; n=14 gonads, mean=2.5, s.e.m.=0.3). Upon cgh-1(RNAi), we found that six oocytes are marked by phospho-H3 (Fig. 6F; n=11 gonads, mean=6.0, s.e.m.=0.7), indicative of premature, not delayed, oocyte maturation. An analogous conclusion was reached using VIT-2-GFP, which normally marks, on average, the two most mature oocytes (data not shown) (Grant and Hirsh, 1999). To determine whether centrioles generally persist upon premature oocyte maturation, we analyzed wee-1.3(RNAi) animals (Burrows et al., 2006). Although these animals harbor significantly more diakinetic oocytes as a result of premature oocyte maturation (n=15 gonads, mean=5.9, s.e.m.=0.9) than the wild type (n=30 gonads, mean=2.6, s.e.m.=0.1; P<0.0001), only 1/100 diakinetic wee-1.3(RNAi) oocytes exhibits a SAS-4 focus (Fig. 6D). Thus, premature oocyte maturation does not necessarily result in diakinetic oocytes harboring SAS-4 foci. Overall, we conclude that CGH-1 directly or indirectly contributes to timely elimination during oogenesis.
The karyotype of hermaphrodites influences centriole elimination
Next, we investigated whether elimination requires input from the surrounding somatic tissue, for instance the sheath cells that encase GCs in hermaphrodite animals (reviewed by Korta and Hubbard, 2010). First, we tested whether somatic cells are sufficient to induce elimination in GC. We analyzed masculinized mog-1(q233)XX animals, in which hermaphroditic somatic gonads contain male GCs (Graham and Kimble, 1993). As shown in Fig. 7A-D, we found that centrioles are not eliminated in such mutants. Analogous observations were made using masculinized fem-3(q96)XX animals (data not shown) (Barton et al., 1987). Therefore, somatic cells are not sufficient to induce elimination in the germ line. We then addressed whether somatic cells are necessary for this process. We analyzed tra-1(e1076)XX animals, which develop as somatic males but have a hermaphroditic germ line (Schedl et al., 1989). In this case, ∼6% of female diakinetic GCs harbor SAS-4 foci (Fig. 7M; n=47), a result on the borderline of statistical significance (see legend of Fig. 7). Overall, we conclude that the contribution of the soma to centriole elimination is minor.
Fig. 7.
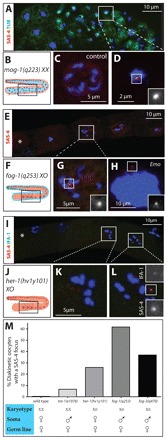
The karyotype contributes to centriole elimination. (A,C,D) Region 3/4 from a mog-1(q233)XX animal (A,D) stained for α-tubulin (green), SAS-4 (red) and DNA (blue). D shows a high magnification view of the area denoted in A, with SAS-4 signal highlighted. A heterozygous diakinetic hermaphrodite oocyte is shown in panel C for comparison. (B) Schematic representation of a mog-1(q233)XX gonad in which hermaphroditic somatic cells (red) encase male GCs (blue). (E,G,H) Region 4 from a fog-1(q253)X0 animal (E,G,H) stained for SAS-4 (red) and DNA (blue). G shows a high magnification view of the area denoted in E, with SAS-4 signal highlighted. The most mature GC/oocyte is to the left and marked by an asterisk. (F) Schematic representation of a fog-1(q253)X0 gonad, in which male somatic cells (blue) encase female GCs (red). (H) Emo oocyte. The number of SAS-4 foci in fog-1(q253)X0 animals ranged from zero to eight, both in diakinesis and Emo oocytes, with the majority having no (32% for diakinesis, 72% for Emo) or one SAS-4 focus (31% for diakinesis, 9% for Emo). We interpret the occurrence of oocytes with more than two SAS-4 foci as centrioles breaking apart or as supernumerary centriole formation during endoreduplicating cycles. (I,K,L) Region 4 from a her-1(hv1y101)X0 animal stained for SAS-4 (red), IFA-1 (green) and DNA (blue). K and L show high magnification views of the areas denoted in I. The most mature GC/oocyte is to the left and marked by an asterisk. (J) Schematic representation of a her-1(hv1y101)X0 gonad in which female X0 GCs (red) are contained in a female gonad (red). (M) Karyotype, sex of the soma and the germ line, as well as percentage of diakinesis GCs with SAS-4 foci. n and P-values (compared with wild type; Fisher’s exact test): wild type, n=79; tra-1(e1076), n=47, P=0.050; fog-1(q253), n=73, P<0.0001; fog-3(q470), n=38, P<0.0001; her-1(hv1y101), n=163, P<0.0001. Insets are magnified twofold.
Intriguingly, we discovered that feminized fog-1(q253)X0 animals, in which female GCs are contained within somatically male gonads (Barton and Kimble, 1990), exhibit a pronounced elimination phenotype. Indeed, we found that ∼62% diakinesis GCs harbor SAS-4 foci in such animals (Fig. 7E-H,M; n=73). Only ∼30% also display a clear focus of IFA-1, raising the possibility that centriole integrity is compromised in the remaining SAS-4-positive oocytes. Further support for defective elimination comes from analysis of Emo GCs in fog-1(q253)X0 animals. In the absence of ovulation, oocytes retained in the gonad undergo successive rounds of S phase and M phase without cell division (‘Emo’ phenotype) (Iwasaki et al., 1996; McCarter et al., 1997). Interestingly, ∼28% of fog-1(q253)X0 Emo GCs harbor SAS-4 foci (Fig. 7H; n=47), suggesting that elimination did not occur in the diakinesis oocytes that gave rise to them. Analogous conclusions were drawn from homozygous fog-3(q470)X0 mutant animals (Fig. 7M), in which female GCs are also contained within a somatically male gonad (Ellis and Kimble, 1994).
Why is the elimination phenotype of fog-1(q253)X0 or fog-3(q470)X0 animals more pronounced than that of tra-1(e1076)XX animals, even though they all harbor female GCs surrounded by a male soma? One difference between the former two and the latter is the karyotype. To address whether the XX karyotype is important for elimination, we analyzed her-1(hv1y101)X0 animals, which develop as somatic and germ line females despite a male karyotype (Francis et al., 1995a). Strikingly, we found that ∼26% diakinesis oocytes in such animals harbor SAS-4 foci (Fig. 7I-M; n=163).
Overall, these results lead us to conclude that the oocyte karyotype contributes in a significant manner to timely elimination of centrioles during C. elegans oogenesis.
DISCUSSION
Sexual reproduction in metazoan organisms depends on meiosis halving the DNA content in each gamete, but also on a conceptually analogous process ensuring that the proper number of centrioles is preserved across generations (reviewed by Schatten, 1994; Delattre and Gönczy, 2004; Manandhar et al., 2005; Sathananthan et al., 2006). In almost all metazoans analyzed to date, centrioles present in the newly fertilized zygote are of paternal origin or are formed de novo in the embryo. The bias to keep centrioles in sperm can be rationalized by the requirement of centrioles as basal bodies for the sperm flagellum. By contrast, centrioles are usually eliminated during oogenesis. Although elimination is fundamental for successful reproduction, the underlying mechanisms are poorly understood. In this work, we analyzed centriole elimination in C. elegans. In addition to describing the process from a molecular and ultrastructural point of view in the wild type, we establish that the helicase CGH-1 and the karyotype of the germ line contribute to this process. Our work thus sets the stage for a mechanistic dissection of elimination in a metazoan organism.
The timing of centriole elimination seems to be evolutionarily conserved
Although it was clear prior to our work that centrioles are present in proliferating GCs of the C. elegans gonad but not in mature oocytes (Kirkham et al., 2003; Kemp et al., 2004; Kim and Roy, 2006), when centrioles disappear between these two time points had not been systematically analyzed. Here, using antibodies, GFP fusions, as well as serial-section EM, we establish that centrioles are present in pachytene, but disappear during diplotene, prior to diakinesis and oocyte cellularization. In Drosophila, a focus of the centriolar marker GFP-cter-D-PLP is present next to the oocyte nucleus until stage 10, when chromosomes are already in the karyosome state that follows pachytene (Januschke et al., 2006). In mammals, ultrastructural analysis also established that centrioles are present in pachytene, but not thereafter, indicating that centriole elimination occurs during the extended diplotene meiotic arrest (Szollosi et al., 1972).
How are centrioles eliminated in C. elegans? One possibility would be that centrioles disassemble through intermediate structures. We used serial section TEM with conditions optimized for the identification of centrioles, but, in contrast to regions 2 and 3, did not find centrioles in region 4, echoing the immunofluorescence analysis. Only in one instance could we detect a structure in region 4 that might have been a centriolar remnant, although this was far from unambiguous. Intermediates of centriole breakdown have not been observed in mammalian oocytes (Szollosi et al., 1972), suggesting relatively rapid elimination in that case as well.
Together, these findings illustrate that elimination occurs relatively rapidly during the diplotene stage in various species, potentially suggesting commonalities in the underlying mechanisms.
CGH-1 and the germ cell karyotype are important for timely centriole elimination
We found that CGH-1 functions in the germ line to contribute to centriole elimination, as ∼59% diakinesis GCs in the predicted null allele cgh-1(ok492) harbor a focus of SAS-4. CGH-1 associates with a specific set of maternal mRNAs, preventing their degradation (Boag et al., 2008). Therefore, it is possible that one of the CGH-1 targets is an mRNA that encodes a protein promoting elimination. When CGH-1 is compromised, this mRNA would be destabilized, and less of the corresponding protein could promote elimination. It will be interesting to address whether such a model holds true, and, if so, to identify the postulated elimination-promoting factor.
Our experiments indicate that, besides CGH-1, the germ cell karyotype is important for timely elimination, as ∼26% diakinesis GCs in her-1(hv1y101)X0 animals harbor a focus of SAS-4. Why do more diakinesis GCs harbor SAS-4 foci in fog-1(q253)X0 animals (∼62%) or to a lesser extent in fog-3(q470)X0 animals (∼37%) than in her-1(hv1y101)X0 animals? We postulate that this might be due to the former two also having a male soma, which presumably compounds the impact of the X0 karyotype.
The GC karyotype might be decisive because it influences the expression of X-linked genes at the stages of oogenesis during which elimination occurs. Thus, the X chromosome undergoes a burst of transcriptional activation in late-pachytene and early-diplotene, along with Histone 3 Lysine 4 (H3K4) methylation (Kelly et al., 2002; Fong et al., 2002; Bean et al., 2004). Such activation depends on the presence of a pairing partner, as revealed by the analysis of her-1(hv1y101)X0 animals, in which H3K4 marks are loaded with a delay and at low levels onto the unpaired X (Bean et al., 2004). Interestingly, such activating chromatin marks are also absent during XX and X0 spermatogenesis. Therefore, we speculate that timely elimination in XX oocytes requires transcription of an X-linked factor, which promotes elimination. When the karyotype is X0, activation of this factor would occur inefficiently, hence delaying elimination.
Redundant mechanisms for centriole elimination
Currently, we do not know whether CGH-1 acts through the same pathway as the karyotype. However, the existence of partially redundant mechanisms, in part CGH-1-dependent and in part karyotype-dependent, could explain why single genes essential for centriole elimination have not been identified in a genome-wide RNAi-based screen (Sonnichsen et al., 2005).
Interestingly, even though centriole elimination during oogenesis is the rule in most metazoan species, there are exceptions, indicating that redundant mechanisms exist elsewhere. Thus, centrioles persist in Asteria pectinifera starfish oocytes, such that each spindle pole harbors two centrioles at the onset of meiosis I. Three centrioles are extruded into the polar bodies during the two meiotic divisions, whereas the remaining one is eliminated shortly after meiosis II (Uetake et al., 2002; Zhang et al., 2004; Shirato et al., 2006). In the clam Spisula solidissima, maternally derived centrioles are not eliminated but cannot duplicate further in the zygote (Wu and Palazzo, 1999). Thus, there might be more than one way to ensure that maternal centrioles do not contribute to the next generation. In C. elegans, the loss of MTOC function early during oogenesis, followed by centriole elimination at a later time, can also be viewed as two redundant mechanisms ensuring that no microtubule organizing function is contributed to the zygote.
In summary, together with the description of the sequence of events in the wild type, uncovering the contributions of CGH-1 and of the XX karyotype provide molecular entry points for a mechanistic dissection of centriole elimination during oogenesis.
Supplementary Material
Acknowledgments
We thank R. Ciosk, B. Conradt, C. Fang-Yen, D. Hall, E. Lambie, A. Khodjakov, D. Hall and T. Schedl for useful discussions, as well as M. Croisier and S. Rosset (BioEM facility, EPFL) for help with the ultrastructural analysis. We are grateful to B. Conradt, V. Hachet, E. Lambie and T. Schedl for comments on the manuscript. Some strains were obtained from the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources (NCRR).
Footnotes
Funding
This work was supported by post-doctoral fellowships from European Molecular Biology Organization [ALTF 237-2007 to T.M.-D.] and the Human Frontiers Science Program [LT01017/2008-L to T.M.-D.]; a doctoral fellowship from the Boehringer Ingelheim Fonds [to L.v.T.]; funds from the BioImaging Network and the Nanosystems Initiative Munich [to L.S. and H.L.]; and a European Research Council grant [AdG 233335 to P.G.].
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.075440/-/DC1
References
- Audhya A., Hyndman F., McLeod I. X., Maddox A. S., Yates J. R., 3rd, Desai A., Oegema K. (2005). A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in C. elegans. J. Cell Biol. 171, 267–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker R. E., Masison D. C. (1990). Isolation of the gene encoding the Saccharomyces cerevisiae centromere-binding protein CP1. Mol. Cell. Biol. 10, 2458–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton M. K., Kimble J. (1990). fog-1, a regulatory gene required for specification of spermatogenesis in the germ line of C. elegans. Genetics 125, 29–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton M. K., Schedl T. B., Kimble J. (1987). Gain-of-function mutations of fem-3, a sex-determination gene in C. elegans. Genetics 115, 107–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean C. J., Schaner C. E., Kelly W. G. (2004). Meiotic pairing and imprinted X chromatin assembly in C. elegans. Nat. Genet. 36, 100–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellanger J. M., Gönczy P. (2003). TAC-1 and ZYG-9 form a complex that promotes microtubule assembly in C. elegans embryos. Curr. Biol. 13, 1488–1498 [DOI] [PubMed] [Google Scholar]
- Boag P. R., Atalay A., Robida S., Reinke V., Blackwell T. K. (2008). Protection of specific maternal messenger RNAs by the P body protein CGH-1 (Dhh1/RCK) during C. elegans oogenesis. J. Cell Biol. 182, 543–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornens M., Azimzadeh J. (2007). Origin and evolution of the centrosome. Adv. Exp. Med. Biol. 607, 119–129 [DOI] [PubMed] [Google Scholar]
- Brenner S. (1974). The genetics of C. elegans. Genetics 77, 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck S. H., Chiu D., Saito R. M. (2009). The cyclin-dependent kinase inhibitors, cki-1 and cki-2, act in overlapping but distinct pathways to control cell cycle quiescence during C. elegans development. Cell Cycle 8, 2613–2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows A. E., Sceurman B. K., Kosinski M. E., Richie C. T., Sadler P. L., Schumacher J. M., Golden A. (2006). The C. elegans Myt1 ortholog is required for the proper timing of oocyte maturation. Development 133, 697–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco P. G., Donahue R. P., Szollosi D. (1972). Germinal vesicle breakdown in the mouse oocyte. J. Cell Sci. 10, 369–385 [DOI] [PubMed] [Google Scholar]
- Carvalho-Santos Z., Machado P., Branco P., Tavares-Cadete F., Rodrigues-Martins A., Pereira-Leal J. B., Bettencourt-Dias M. (2010). Stepwise evolution of the centriole-assembly pathway. J. Cell Sci. 123, 1414–1426 [DOI] [PubMed] [Google Scholar]
- Church D. L., Guan K. L., Lambie E. J. (1995). Three genes of the MAP kinase cascade, mek-2, mpk-1/sur-1 and let-60 ras, are required for meiotic cell cycle progression in C. elegans. Development 121, 2525–2535 [DOI] [PubMed] [Google Scholar]
- Dammermann A., Müller-Reichert T., Pelletier L., Habermann B., Desai A., Oegema K. (2004). Centriole assembly requires both centriolar and pericentriolar material proteins. Dev. Cell 7, 815–829 [DOI] [PubMed] [Google Scholar]
- Dammermann A., Maddox P. S., Desai A., Oegema K. (2008). SAS-4 is recruited to a dynamic structure in newly forming centrioles that is stabilized by the gamma-tubulin-mediated addition of centriolar microtubules. J. Cell Biol. 180, 771–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delattre M., Gönczy P. (2004). The arithmetic of centrosome biogenesis. J. Cell Sci. 117, 1619–1630 [DOI] [PubMed] [Google Scholar]
- Delattre M., Leidel S., Wani K., Baumer K., Bamat J., Schnabel H., Feichtinger R., Schnabel R., Gönczy P. (2004). Centriolar SAS-5 is required for centrosome duplication in C. elegans. Nat. Cell Biol. 6, 656–664 [DOI] [PubMed] [Google Scholar]
- Delattre M., Canard C., Gönczy P. (2006). Sequential protein recruitment in C. elegans centriole formation. Curr. Biol. 16, 1844–1849 [DOI] [PubMed] [Google Scholar]
- Eisenmann D. M., Kim S. K. (1997). Mechanism of activation of the C. elegans ras homologue let-60 by a novel, temperature-sensitive, gain-of-function mutation. Genetics 146, 553–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis H. M., Horvitz H. R. (1986). Genetic control of programmed cell death in the nematode C. elegans. Cell 44, 817–829 [DOI] [PubMed] [Google Scholar]
- Ellis R. E., Kimble J. (1994). Control of germ cell differentiation in C. elegans. Ciba Found. Symp. 182, 179–192 [DOI] [PubMed] [Google Scholar]
- Francis R., Barton M. K., Kimble J., Schedl T. (1995a). gld-1, a tumor suppressor gene required for oocyte development in C. elegans. Genetics 139, 579–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R., Maine E., Schedl T. (1995b). Analysis of the multiple roles of gld-1 in germline development: interactions with the sex determination cascade and the glp-1 signaling pathway. Genetics 139, 607–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P., Bellanger J. M., Kirkham M., Pozniakowski A., Baumer K., Phillips J. B., Hyman A. A. (2001). zyg-8, a gene required for spindle positioning in C. elegans, encodes a doublecortin-related kinase that promotes microtubule assembly. Dev. Cell 1, 363–375 [DOI] [PubMed] [Google Scholar]
- Graham P. L., Kimble J. (1993). The mog-1 gene is required for the switch from spermatogenesis to oogenesis in C. elegans. Genetics 133, 919–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B., Hirsh D. (1999). Receptor-mediated endocytosis in the C. elegans oocyte. Mol. Biol. Cell 10, 4311–4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamill D. R., Severson A. F., Carter J. C., Bowerman B. (2002). Centrosome maturation and mitotic spindle assembly in C. elegans require SPD-5, a protein with multiple coiled-coil domains. Dev. Cell 3, 673–684 [DOI] [PubMed] [Google Scholar]
- Hammell C. M., Lubin I., Boag P. R., Blackwell T. K., Ambros V. (2009). nhl-2 Modulates microRNA activity in C. elegans. Cell 136, 926–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannak E., Oegema K., Kirkham M., Gönczy P., Habermann B., Hyman A. A. (2002). The kinetically dominant assembly pathway for centrosomal asters in C. elegans is gamma-tubulin dependent. J. Cell Biol. 157, 591–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann G. J., Schroeder L. K., Hieb C. A., Kershner A. M., Rabbitts B. M., Fonarev P., Grant B. D., Priess J. R. (2005). Genetic analysis of lysosomal trafficking in C. elegans. Mol. Biol. Cell 16, 3273–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertig A. T., Adams E. C. (1967). Studies on the human oocyte and its follicle. I. Ultrastructural and histochemical observations on the primordial follicle stage. J. Cell Biol. 34, 647–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges M. E., Scheumann N., Wickstead B., Langdale J. A., Gull K. (2010). Reconstructing the evolutionary history of the centriole from protein components. J. Cell Sci. 123, 1407–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J. Y., Sun Z. W., Li X., Reuben M., Tatchell K., Bishop D. K., Grushcow J. M., Brame C. J., Caldwell J. A., Hunt D. F., et al. (2000). Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102, 279–291 [DOI] [PubMed] [Google Scholar]
- Hubbard E. J., Greenstein D. (2000). The C. elegans gonad: a test tube for cell and developmental biology. Dev. Dyn. 218, 2–22 [DOI] [PubMed] [Google Scholar]
- Iwasaki K., McCarter J., Francis R., Schedl T. (1996). emo-1, a C. elegans Sec61p gamma homologue, is required for oocyte development and ovulation. J. Cell Biol. 134, 699–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januschke J., Gervais L., Gillet L., Keryer G., Bornens M., Guichet A. (2006). The centrosome-nucleus complex and microtubule organization in the Drosophila oocyte. Development 133, 129–139 [DOI] [PubMed] [Google Scholar]
- Kamath R. S., Fraser A. G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., et al. (2003). Systematic functional analysis of the C. elegans genome using RNAi. Nature 421, 231–237 [DOI] [PubMed] [Google Scholar]
- Karashima T., Sugimoto A., Yamamoto M. (2000). C. elegans homologue of the human azoospermia factor DAZ is required for oogenesis but not for spermatogenesis. Development 127, 1069–1079 [DOI] [PubMed] [Google Scholar]
- Kawasaki I., Shim Y. H., Kirchner J., Kaminker J., Wood W. B., Strome S. (1998). PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell 94, 635–645 [DOI] [PubMed] [Google Scholar]
- Kemp C. A., Kopish K. R., Zipperlen P., Ahringer J., O’Connell K. F. (2004). Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev. Cell 6, 511–523 [DOI] [PubMed] [Google Scholar]
- Kemp C. A., Song M. H., Addepalli M. K., Hunter G., O’Connell K. (2007). Suppressors of zyg-1 define regulators of centrosome duplication and nuclear association in C. elegans. Genetics 176, 95–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. Y., Roy R. (2006). Cell cycle regulators control centrosome elimination during oogenesis in C. elegans. J. Cell Biol. 174, 751–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkham M., Müller-Reichert T., Oegema K., Grill S., Hyman A. A. (2003). SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell 112, 575–587 [DOI] [PubMed] [Google Scholar]
- Kitagawa D., Fluckiger I., Polanowska J., Keller D., Reboul J., Gönczy P. (2011). PP2A phosphatase acts upon SAS-5 to ensure centriole formation in C. elegans embryos. Dev. Cell 20, 550–562 [DOI] [PubMed] [Google Scholar]
- Korta D. Z., Hubbard E. J. (2010). Soma-germline interactions that influence germline proliferation in C. elegans. Dev. Dyn. 239, 1449–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner M. R., Kim S. K. (1998). Genetic analysis of the C. elegans MAP kinase gene mpk-1. Genetics 150, 103–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel S., Gönczy P. (2003). SAS-4 is essential for centrosome duplication in C elegans and is recruited to daughter centrioles once per cell cycle. Dev. Cell 4, 431–439 [DOI] [PubMed] [Google Scholar]
- Leidel S., Gönczy P. (2005). Centrosome duplication and nematodes: recent insights from an old relationship. Dev. Cell 9, 317–325 [DOI] [PubMed] [Google Scholar]
- Leidel S., Delattre M., Cerutti L., Baumer K., Gönczy P. (2005). SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat. Cell Biol. 7, 115–125 [DOI] [PubMed] [Google Scholar]
- Leung B., Hermann G. J., Priess J. R. (1999). Organogenesis of the C. elegans intestine. Dev. Biol. 216, 114–134 [DOI] [PubMed] [Google Scholar]
- MacQueen A. J., Colaiacovo M. P., McDonald K., Villeneuve A. M. (2002). Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 16, 2428–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox A. S., Habermann B., Desai A., Oegema K. (2005). Distinct roles for two C. elegans anillins in the gonad and early embryo. Development 132, 2837–2848 [DOI] [PubMed] [Google Scholar]
- Manandhar G., Schatten H., Sutovsky P. (2005). Centrosome reduction during gametogenesis and its significance. Biol. Reprod. 72, 2–13 [DOI] [PubMed] [Google Scholar]
- McCarter J., Bartlett B., Dang T., Schedl T. (1997). Soma-germ cell interactions in C. elegans: multiple events of hermaphrodite germline development require the somatic sheath and spermathecal lineages. Dev. Biol. 181, 121–143 [DOI] [PubMed] [Google Scholar]
- McCarter J., Bartlett B., Dang T., Schedl T. (1999). On the control of oocyte meiotic maturation and ovulation in C. elegans. Dev. Biol. 205, 111–128 [DOI] [PubMed] [Google Scholar]
- Nance J., Munro E. M., Priess J. R. (2003). C. elegans PAR-3 and PAR-6 are required for apicobasal asymmetries associated with cell adhesion and gastrulation. Development 130, 5339–5350 [DOI] [PubMed] [Google Scholar]
- Navarro R. E., Shim E. Y., Kohara Y., Singson A., Blackwell T. K. (2001). cgh-1, a conserved predicted RNA helicase required for gametogenesis and protection from physiological germline apoptosis in C. elegans. Development 128, 3221–3232 [DOI] [PubMed] [Google Scholar]
- O’Connell K. F., Leys C. M., White J. G. (1998). A genetic screen for temperature-sensitive cell-division mutants of C. elegans. Genetics 149, 1303–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell K. F., Caron C., Kopish K. R., Hurd D. D., Kemphues K. J., Li Y., White J. G. (2001). The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell 105, 547–558 [DOI] [PubMed] [Google Scholar]
- Oegema K., Hyman A. A. (2006). Cell division (January 19 2006), WormBook (ed. The C. elegans Research Community). doi/10.1895/wormbook.1.72.1, http://www.wormbook.org [DOI] [PMC free article] [PubMed]
- Oegema K., Desai A., Rybina S., Kirkham M., Hyman A. A. (2001). Functional analysis of kinetochore assembly in C. elegans. J. Cell Biol. 153, 1209–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier L., Ozlu N., Hannak E., Cowan C., Habermann B., Ruer M., Müller-Reichert T., Hyman A. A. (2004). The C. elegans centrosomal protein SPD-2 is required for both pericentriolar material recruitment and centriole duplication. Curr. Biol. 14, 863–873 [DOI] [PubMed] [Google Scholar]
- Pelletier L., O’Toole E., Schwager A., Hyman A. A., Müller-Reichert T. (2006). Centriole assembly in C. elegans. Nature 444, 619–623 [DOI] [PubMed] [Google Scholar]
- Pitt J. N., Schisa J. A., Priess J. R. (2000). P-granules in the germ cells of C. elegans adults are associated with clusters of nuclear pores and contain RNA. Dev. Biol. 219, 315–333 [DOI] [PubMed] [Google Scholar]
- Rual J. F., Ceron J., Koreth J., Hao T., Nicot A. S., Hirozane-Kishikawa T., Vandenhaute J., Orkin S. H., Hill D. E., van den Heuvel S., Vidal M. (2004). Toward improving C. elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14, 2162–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathananthan A. H., Ratnam S. S., Ng S. C., Tarin J. J., Gianaroli L., Trounson A. (1996). The sperm centriole: its inheritance, replication and perpetuation in early human embryos. Hum. Reprod. 11, 345–356 [DOI] [PubMed] [Google Scholar]
- Sathananthan A. H., Selvaraj K., Girijashankar M. L., Ganesh V., Selvaraj P., Trounson A. O. (2006). From oogonia to mature oocytes: inactivation of the maternal centrosome in humans. Microsc. Res. Tech. 69, 396–407 [DOI] [PubMed] [Google Scholar]
- Schatten G. (1994). The centrosome and its mode of inheritance: the reduction of the centrosome during gametogenesis and its restoration during fertilization. Dev. Biol. 165, 299–335 [DOI] [PubMed] [Google Scholar]
- Schedl T., Kimble J. (1988). fog-2, a germ-line-specific sex determination gene required for hermaphrodite spermatogenesis in C. elegans. Genetics 119, 43–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schedl T., Graham P. L., Barton M. K., Kimble J. (1989). Analysis of the role of tra-1 in germline sex determination in the nematode C. elegans. Genetics 123, 755–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L., Carlton P. M., Haase S., Shao L., Winoto L., Kner P., Burke B., Cardoso M. C., Agard D. A., Gustafsson M. G., et al. (2008). Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science 320, 1332–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato Y., Tamura M., Yoneda M., Nemoto S. (2006). Centrosome destined to decay in starfish oocytes. Development 133, 343–350 [DOI] [PubMed] [Google Scholar]
- Sijen T., Fleenor J., Simmer F., Thijssen K. L., Parrish S., Timmons L., Plasterk R. H., Fire A. (2001). On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107, 465–476 [DOI] [PubMed] [Google Scholar]
- Sonnichsen B., Koski L. B., Walsh A., Marschall P., Neumann B., Brehm M., Alleaume A. M., Artelt J., Bettencourt P., Cassin E., et al. (2005). Full-genome RNAi profiling of early embryogenesis in C. elegans. Nature 434, 462–469 [DOI] [PubMed] [Google Scholar]
- Strnad P., Gönczy P. (2008). Mechanisms of procentriole formation. TrendsCell Biol. 18, 389–396 [DOI] [PubMed] [Google Scholar]
- Szollosi D., Calarco P., Donahue R. P. (1972). Absence of centrioles in the first and second meiotic spindles of mouse oocytes. J. Cell Sci. 11, 521–541 [DOI] [PubMed] [Google Scholar]
- Szollosi D., Mandelbaum J., Plachot M., Salat-Baroux J., Cohen J. (1986). Ultrastructure of the human preovulatory oocyte. J. In Vitro Fert. Embryo Transf. 3, 232–242 [DOI] [PubMed] [Google Scholar]
- Uetake Y., Kato K. H., Washitani-Nemoto S., Nemoto, Si S. (2002). Nonequivalence of maternal centrosomes/centrioles in starfish oocytes: selective casting-off of reproductive centrioles into polar bodies. Dev. Biol. 247, 149–164 [DOI] [PubMed] [Google Scholar]
- Wu X., Palazzo R. E. (1999). Differential regulation of maternal vs. paternal centrosomes. Proc. Natl. Acad. Sci. USA 96, 1397–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q. Y., Tamura M., Uetake Y., Washitani-Nemoto S., Nemoto S. (2004). Regulation of the paternal inheritance of centrosomes in starfish zygotes. Dev. Biol. 266, 190–200 [DOI] [PubMed] [Google Scholar]
- Zhou K., Rolls M. M., Hall D. H., Malone C. J., Hanna-Rose W. (2009). A ZYG-12-dynein interaction at the nuclear envelope defines cytoskeletal architecture in the C. elegans gonad. J. Cell Biol. 186, 229–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


